Abstract
Nitrogen (N2) is the most important source of mineral N for plant growth, which was mainly transported by nitrate transporters (NRTs). However, little is known about the NRT gene family in potato. In this study, StNRT gene family members were identified in potato. In addition, we performed StNRT subfamily classification, gene structure and distribution analysis, and conserved domain prediction using various bioinformatics tools. Totally, 39 StNRT gene members were identified in potato genome, including 33, 4 and 2 member belong to NRT1, NRT2, and NRT3, respectively. These 39 StNRT genes were randomly distributed on all chromosomes. The collinearity results show that StNRT members in potato are closely related to Solanum lycopersicum and Solanum melongena. For the expression, different members of StNRT play different roles in leaves and roots. Especially under sufficient nitrogen conditions, different members have a clear distribution in different tissues. These results provide valuable information for identifying the members of the StNRT family in potato and could provide functional characterization of StNRT genes in further research.
Introduction
Nitrogen (N) play an essential role that affects plant growth and development, which is an important component of chlorophyll, amino acids, nucleic acids, and secondary metabolites [1]. Nitrate (NO3¯) is the most important source of mineral N for plants growth [2]. NO3¯ can act as a signaling molecule that regulates gene expression in many processes, such as plant growth, root system architecture development [3], leaf growth and development [4,5], seed dormancy [6], and flowering time [7]. Plants can uptake NO3¯ from soil and store it in vacuoles through a series of transport pathways [8–10], but mainly performed by nitrate transporters (NRTs) that are encoded by a multigene family [11]. According to their affinity for the substrate, NRTs are divided into two systems: the low-affinity transport system (LATS) (via nitrate transporter 1 family, NRT1) [12,13] and the high-affinity transport system (HATS) (via nitrate transporter 2 family, NRT2) [14,15]. Therefore, plants had evolved a series of NRT gene family members to make better use of NO3¯. There were three NRT gene subfamilies: NRT1, NRT2, and NRT3 [16]. Till now, several studies have elucidated NRT genes functions and evolutionary history in many plant species such as Arabidopsis thaliana [17,18], rice [19], poplar [20] and pineapple [21]. Our previous study found that NRT gene responded positively to nitrogen deficiency stress [22]. Besides that, Pieczynski et al reported that some NRT family members were not only involved in the nitrogen transportation, but also responded to drought [23]. Phylogenetic studies have revealed that NRT1 families gather a large number of genes and could be divided in 8 to 10 subfamilies [13,24], which had been shown to incorporate transporters not only for NO3¯, but also for peptides, amino acids, nitrite, glucosinolates, abscisic acid and gibberelins [2]. As compared to NRT1 families, NRT2 families analyzed in various species contain a much lower number of genes. In A. thaliana, there are seven members of the NRT2 gene family from NRT2.1 to NRT2.7 [17,25]. Gene structure of the AtNRT family members were reviewed by Okamoto, but the functions of NRT1 and NRT2 transporters are largely unknown [25]. Further physiological analysis is needed to understand the precise role of individual NRT gene, in particular for potato, because there were no systematic reports on the NRT gene family members in potato.
As for potato, large amount of nitrogen is needed in the growth and development. Therefore, it can provide theoretical basis for potato breeding to understand the family members of StNRT and their relationship. In this study, StNRT gene family members were identified in potato. In addition, we performed StNRT subfamily classification, gene structure and distribution analysis, and conserved domain prediction using various bioinformatics tools. This study could be helpful for further functional study of StNRT genes and molecular breeding of potato.
Materials and methods
Genome-wide identification of NRT proteins and genes
A total of 60 AtNRT family members sequences from Arabidopsis thaliana were collected from TAIR (https://www.arabidopsis.org/) and some previous studies [25,26]. Also, according to Tsay’s report, 81 OsNRTs were collected [26]. All these collected NRT members were used as queries to search against sequence homologs in the potato genome from the Ensemblplants (http://plants.ensembl.org/info/website/ftp/index.html). The candidate StNRT members were identified using BLAST method and HMMER 3.0 software (http://hmmer.janelia.org/). Then, the candidate members were further confirmed according to Uniport database (https://www.uniprot.org/) and those without NRT gene annotation were discarded. To identify the domains of the candidate members, online programmes NCBI conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/Structure/cdd/wrpsb.cgi) was used with expect value <0.05 and the results were displayed with TBtools (V0.67, https://github.com/CJ-Chen/TBtools) [27].
Chromosomal localization and gene duplication of potato StNRT genes
All the candidate StNRTs were mapped on potato chromosomes and displayed by TBtools software according to the potato StNRT gene positions in the annotation file from ensemble plant genome database. To identify the duplicated and tandem repeated genes, ClustalW alignment comparison of all StNRT members was carried out with a threshold of similarity >75% and their genomic locations. The tandem duplicated genes were restricted within the range of 100 kb distance [28].
StNRTs structure, conserved domain, motif, and phylogenetic analysis
StNRTs structure were analyzed by aligning the coding sequence (CDS) regions to the genomic DNA sequences. The gene structure and conserved domains obtained from CDD database of all the members were displayed using the TBtools software. The motifs were predicted via the Multiple Expectation Maximisation for Motif elicitation (MEME) online tool (http://meme-suite.org/tools/meme). As for molecular weight (MW) and the theoretical isoelectric point (pI) prediction, the online tool ExPASy (https://www.expasy.org/tools/) were used basing on the proteins sequence of all the StNRT members.
Phylogenetic tree construction
To evaluate the evolution relationship of all the family members of StNRTs, phylogenetic tree was constructed via MEGA (version 7.0.26). Firstly, the ClustalX program was used to perform multiple sequence alignments of the StNRTs of Arabidopsis thaliana and potato. Then, Maximum Likelihood (ML) tree was constructed basing on the optimal model prediction results with 1000 bootstrap tests.
Identification of gene synteny
Gene synteny analysis were performed by BLAST and the Multiple Collinearity Scan toolkit (MCScanX) [29] according to Song’s report [30]. Briefly, the sequence of potato candidate gene family members were searched against itself using BLASTP with an E-value cut-off of 1 × 10−10 and identity >75%. Then, the acquired BLASTP results were next used as the MCScanX input to assess the collinear blocks. For the gene synteny among different genomes, we selected 4 plant genomes for collinear analysis, including Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum and Solanum melongena. The assembly of Arabidopsis thaliana, Oryza sativa and Solanum lycopersicum were obtained from Ensemblplants (http://plants.ensembl.org/info/website/ftp/index.html) and the assembly sequence of Solanum melongena was obtained from China National Genebank (CNGB, https://www.cngb.org/index.html). The analysis process refered to the instruction of MCScanX software.
Transcriptome expression analysis
The Illumina RNA-seq data were downloaded from the SRA database (https://submit.ncbi.nlm.nih.gov/subs/sra/) with the submission number of SRS4186597 (the data was up loaded in our previous study [22]) to study the expression patterns of all the identified StNRT members in response to nitrogen deficiency. Briefly, Potato cultivar cv. Shepody was treated with sufficient-N- and deficient-N-fertilizer. Then, leaf and root transcriptomes were analyzed and differentially expressed genes (DEGs) in response to N deficiency were identified. We compared the expression differences of these StNRT members between the sufficient N fertilizer group and the deficient N group in leaf and root. The sequence data used was obtained from Solanum tuberosum cv. Shepody. The expression of StNRT members were showed in a heatmap via TBtools software.
Cis-element enrichment analysis
The upstream sequences (2 kb) of the StNRT sequences were retrieved and then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify six regulatory elements, abscisic acid (ABA)-responsive elements (ABRE), ACE, CAT-box, estrogen response element (ERE), MYB and MYC. Then GSDS 2.0 (http://gsds.cbi.pku.edu.cn/index.php) was used to plot the location of these elements.
Results
Identification and analysis of StNRT genes
A total of 46 and 47 StNRT peptides sequence obtained in BLAST and HMMER3 analysis results, respectively, of which 46 members were the common genes. According to the annotation information of uniport database, all these 44 genes belong to NRT family. After removing the duplicate sequence, 39 genes were obtained. All these 39 sequences were reserved and submitted to CDD to confirm the conserved domain. The results showed that nine domains were identified and seven of them were MFS-related domains. These 39 sequences were named based on their chromosomal locations (Table 1). The lengths of the StNRT proteins ranged from 203 (StNRT07) to 653 amino acids (StNRT33) with mean length of 559.10. The conserved domain results showed that the StNRT genes in potato contained the same domains with that of Arabidopsis thaliana and Oryza sativa (S1a and S1b Fig) and most of the genes contained complete domains (Fig 1). The molecular weights of StNRT genes were between 22.65 kDa (StNRT07) and 71.9 kDa (StNRT33). Theoretical pI value range from 6.03 (StNRT20) to 9.36 (StNRT34).
Table 1. StNRT genes identified in potato and their sequence characteristics.
| StNRT ID | Protein ID | Gene ID | Chromosomal localization | Gene length (bp) | Amino acid length (aa) | pI* | MW (kD)* | CDS length (bp)* | |
|---|---|---|---|---|---|---|---|---|---|
| StNRT01 | PGSC0003DMP400002994 | PGSC0003DMG400001671 | 67991306 | 67996008 | 4702 | 554 | 6.39 | 66441.93 | 1665 |
| StNRT02 | PGSC0003DMP400047815 | PGSC0003DMG402027501 | 74630384 | 74633515 | 3131 | 593 | 9.12 | 63512.61 | 1782 |
| StNRT03 | PGSC0003DMP400036006 | PGSC0003DMG400020708 | 79794672 | 79799593 | 4921 | 530 | 8.8 | 67005.04 | 1593 |
| StNRT04 | PGSC0003DMP400012236 | PGSC0003DMG400006913 | 26506508 | 26508700 | 2192 | 512 | 9.03 | 45858.84 | 1539 |
| StNRT05 | PGSC0003DMP400002522 | PGSC0003DMG400001393 | 46706587 | 46710386 | 3799 | 596 | 7.99 | 57116.7 | 1791 |
| StNRT06 | PGSC0003DMP400031706 | PGSC0003DMG400018193 | 51862141 | 51866797 | 4656 | 584 | 6.23 | 35792.95 | 1755 |
| StNRT07 | PGSC0003DMP400031592 | PGSC0003DMG400018129 | 51871879 | 51873422 | 1543 | 590 | 9.29 | 22651.25 | 1773 |
| StNRT08 | PGSC0003DMP400043961 | PGSC0003DMG400025339 | 53135911 | 53138548 | 2637 | 552 | 8.63 | 66220.39 | 1659 |
| StNRT09 | PGSC0003DMP400043959 | PGSC0003DMG400025337 | 53160486 | 53162884 | 2398 | 424 | 9.02 | 65517.71 | 1275 |
| StNRT10 | PGSC0003DMP400043958 | PGSC0003DMG400025336 | 53169753 | 53173630 | 3877 | 589 | 9.32 | 66164.32 | 1770 |
| StNRT11 | PGSC0003DMP400005176 | PGSC0003DMG400002865 | 388737 | 394987 | 6250 | 582 | 8.99 | 64393.43 | 1749 |
| StNRT12 | PGSC0003DMP400020732 | PGSC0003DMG400011693 | 68832667 | 68834950 | 2283 | 585 | 9.09 | 65181.01 | 1758 |
| StNRT13 | PGSC0003DMP400020731 | PGSC0003DMG400011692 | 68837821 | 68840580 | 2759 | 590 | 9.15 | 64547.23 | 1773 |
| StNRT14 | PGSC0003DMP400025609 | PGSC0003DMG400014539 | 2690484 | 2695466 | 4982 | 581 | 8.86 | 65222.23 | 1746 |
| StNRT15 | PGSC0003DMP400030769 | PGSC0003DMG400017620 | 6018942 | 6027657 | 8715 | 585 | 8.99 | 68498.12 | 1758 |
| StNRT16 | PGSC0003DMP400030807 | PGSC0003DMG400017637 | 6118146 | 6121903 | 3757 | 553 | 8.4 | 67732.91 | 1662 |
| StNRT17 | PGSC0003DMP400029708 | PGSC0003DMG400016996 | 9714775 | 9717165 | 2390 | 580 | 8.81 | 54353.09 | 1743 |
| StNRT18 | PGSC0003DMP400044048 | PGSC0003DMG400025395 | 8311895 | 8316339 | 4444 | 500 | 8.35 | 68301.19 | 1503 |
| StNRT19 | PGSC0003DMP400064739 | PGSC0003DMG400042635 | 31438515 | 31443669 | 5154 | 608 | 8.72 | 65380.81 | 1827 |
| StNRT20 | PGSC0003DMP400054567 | PGSC0003DMG401031322 | 47048581 | 47054364 | 5783 | 627 | 6.03 | 62394 | 1884 |
| StNRT21 | PGSC0003DMP400011673 | PGSC0003DMG400006606 | 56077004 | 56080118 | 3114 | 606 | 8.55 | 60265.93 | 1821 |
| StNRT22 | PGSC0003DMP400052968 | PGSC0003DMG400030438 | 57021923 | 57025262 | 3339 | 203 | 8.12 | 64066.11 | 612 |
| StNRT23 | PGSC0003DMP400039866 | PGSC0003DMG400022993 | 5775617 | 5778627 | 3010 | 319 | 8.87 | 65275.63 | 960 |
| StNRT24 | PGSC0003DMP400001318 | PGSC0003DMG402000668 | 45033113 | 45036254 | 3141 | 566 | 9.16 | 65119.92 | 1701 |
| StNRT25 | PGSC0003DMP400022078 | PGSC0003DMG400012479 | 479008 | 483551 | 4543 | 605 | 8.85 | 65321.31 | 1818 |
| StNRT26 | PGSC0003DMP400008499 | PGSC0003DMG400004795 | 52567376 | 52571923 | 4547 | 584 | 9.15 | 65171.39 | 1755 |
| StNRT27 | PGSC0003DMP400024739 | PGSC0003DMG400014054 | 395745 | 398087 | 2342 | 597 | 8.37 | 63771.68 | 1794 |
| StNRT28 | PGSC0003DMP400035282 | PGSC0003DMG400020318 | 402706 | 405477 | 2771 | 591 | 8.66 | 63508.52 | 1776 |
| StNRT29 | PGSC0003DMP400030052 | PGSC0003DMG400017204 | 56806392 | 56815251 | 8859 | 585 | 9.31 | 68098.05 | 1758 |
| StNRT30 | PGSC0003DMP400041720 | PGSC0003DMG400024120 | 37692715 | 37696235 | 3520 | 595 | 6.57 | 66177.4 | 1788 |
| StNRT31 | PGSC0003DMP400019579 | PGSC0003DMG400011085 | 54161108 | 54166759 | 5651 | 555 | 9.15 | 66165.74 | 1668 |
| StNRT32 | PGSC0003DMP400064264 | PGSC0003DMG400042160 | 55059494 | 55061848 | 2354 | 611 | 9.04 | 57804.32 | 1836 |
| StNRT33 | PGSC0003DMP400048707 | PGSC0003DMG400028026 | 19452106 | 19456164 | 4058 | 599 | 9.06 | 71898.12 | 1800 |
| StNRT34 | PGSC0003DMP400002117 | PGSC0003DMG400001145 | 41898143 | 41899905 | 1762 | 575 | 9.36 | 57595.05 | 1728 |
| StNRT35 | PGSC0003DMP400044035 | PGSC0003DMG400025389 | 44400838 | 44404224 | 3386 | 653 | 9.03 | 61353.52 | 1962 |
| StNRT36 | PGSC0003DMP400000603 | PGSC0003DMG400000303 | 4387465 | 4391552 | 4087 | 574 | 8.44 | 62005.02 | 1725 |
| StNRT37 | PGSC0003DMP400025448 | PGSC0003DMG400014449 | 48920928 | 48925553 | 4625 | 568 | 7.03 | 64698.13 | 1707 |
| StNRT38 | PGSC0003DMP400025503 | PGSC0003DMG400014473 | 48944132 | 48959296 | 15164 | 534 | 8.08 | 61286.58 | 1605 |
| StNRT39 | PGSC0003DMP400045945 | PGSC0003DMG400026455 | 50643015 | 50647292 | 4277 | 570 | 8.97 | 66547.84 | 1713 |
*pI, Isoelectric point; MW (kD), Molecular weight; CDS length (bp), Coding DNA Sequence length.
For the column chromosomal localization, the number in the left-hand list is the starting position and the right-hand list is the end position.
Fig 1. Conserved domain identification of StNRT genes.
The nine type domains are displayed in different colored boxes. The information for each domain is shown on the right. The length of gene/protein can be estimated using the scale at the bottom.
Phylogenetic analysis of potato StNRT genes
To decipher the evolutionary relationships and functional associations of NRT genes in potato, the multi-species phylogenetic tree was constructed based on the full-length amino acid sequences of NRTs from potato, Arabidopsis thaliana, and rice with the Maximum Likelihood method. In total, 60 sequences from Arabidopsis thaliana, 81 sequences from rice, 39 sequences from potato were assessed in the phylogenetic tree (Fig 2). The phylogenetic analysis revealed that all the NRTs could be divided into four groups: NRT1, NRT2, and NRT3.1 and NRT3.2. There were 33 StNRT genes belong to NRT1. There were 4 StNRT genes belong to NRT2, including StNRT04, StNRT17, StNRT32 and StNRT34. In addition, we identified two StNRT3 gene: StNRT06 (StNRT3.2) and StNRT07 (StNRT3.1). In addition, we found that StNRT genes in potato prefers to cluster with the AtNRT genes of Arabidopsis thaliana, rather than Oryza sativa.
Fig 2. The phylogenetic tree of potato, rice and Arabidopsis thaliana NRT genes.
The 39 potato, 81 rice and 60 Arabidopsis thaliana NRT protein sequences were aligned by Clustal X and the phylogenetic tree was constructed using MGA 7.0 by the Maximum likelihood (ML) method. The Bootstrap value was 1,000 replicates. The colored background indicates the different subfamily. Different geometric makers represent NRTs of different plants.
Chromosome localization and duplication of the StNRT gene family
The location analysis showed that all these 39 StNRT members were randomly distributed on the 12 potato chromosomes (Fig 3). Chromosomes 02, 07 and 08 contain two StNRT genes, while chromosome 03 and 06 contains the most StNRT genes (5 in each) among all potato chromosomes. Gene duplication events have driven the expansion of potato StNRT genes, with 13 genes found in 6 duplicated blocks and 26 StNRT genes located outside of the duplicated blocks. Six pairs of genes, including StNRT06/−07, 08/−09/−10, 12/−13, 15/−16, 27/−28, and 37/−38 were separated by less than a 100-kb region on chromosome 03, 03, 04, 05 and 09 and 12, respectively.
Fig 3. The chromosomal location and gene duplication of StNRT family member in potato.
The ordinate indicates the chromosome length (amino acid length). The tandem duplicated genes are marked by blue rectangles.
Gene structure and motifs in StNRT gene family
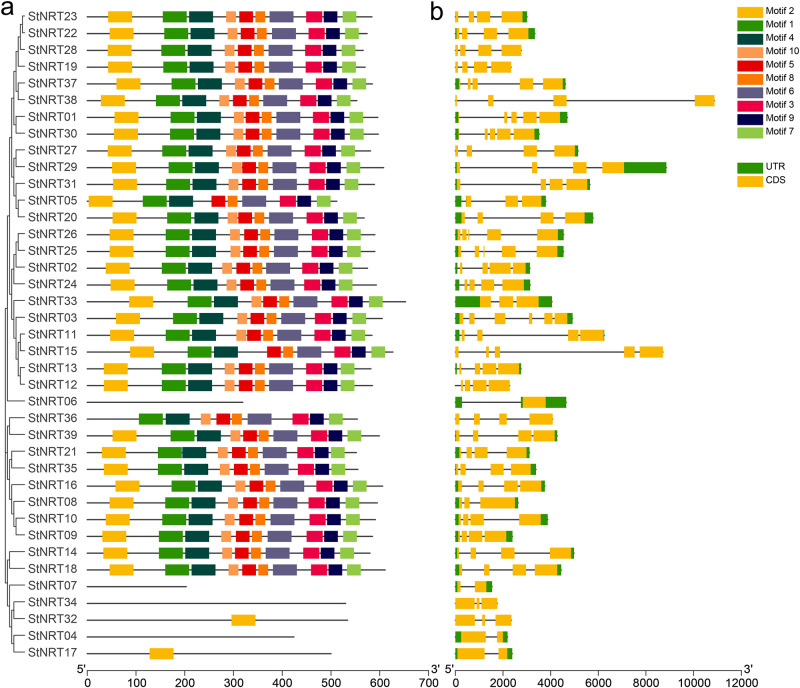
Conserved motifs were analyzed for all the 39 StNRT members using MEME software and 10 motifs were identified (Fig 4a). There were no motifs found on StNRT04, StNRT06, StNRT07 and StNRT34. Only one motif found on StNRT17 (Motif 2) and StNRT32 (Motif 2). Interestingly, these five genes mentioned above contained the PLN00028 domain (the typical characteristics of NRT gene). To identify the motifs that contained PLN00028 domain, we further compared the gene sequences of Arabidopsis thaliana and potato. The results showed that these genes in Arabidopsis thaliana and potato had the consistent motifs (S2 Fig). For genes structure, most genes consist of 4 exons (Fig 4b). But some genes are composed of five or more exons, such as StNRT25, StNRT26, StNRT03, StNRT15, etc. In addition, there was only one exon found in StNRT06.
Fig 4. Conserved motif and gene structure analysis.
(a). Distributions of conserved motifs in StNRT members. Ten putative motifs are indicated in different colored boxes. (b). Exon organization of StNRT members. Yellow boxes represent exons and black lines with same length represent introns.
Collinearity analysis of StNRT members
In order to study the locus relationship between the orthologous of different chromosomes, collinearity analysis was performed. The analysis showed that StNRT25 and StNRT26 were highly conserved in chromosome 8. StNRT08 and StNRT16 were highly conserved between chromosome 3 and 5 (Fig 5a). For StNRT members locus relationship between potato and Arabidopsis thaliana, we found that four StNRT genes had homologous genes in Arabidopsis thaliana (Fig 5b). However, no homologous genes found in Oryza sativa (Fig 5c). When comparing potatoes to their near-source species, we found that all StNRT members of potato had orthologous genes in eggplant and tomato (Fig 5d and 5e). Especially in tomato, the chromosomal position of the orthologous genes of all StNRT members was also highly consistent with that of potato.
Fig 5. Collinearity analysis of StNRT members.
(a). Collinearity analysis between different chromosomes within the potato genome. Different colors indicate different chromosomes. (b), (c), (d) and (e) showed the collinearity analysis between potato and Arabidopsis thaliana, potato and rice, potato and tomato, potato and eggplant, respectively. The red line indicates members of the StNRT gene with collinearity, and the gray line indicates other genes. Stu, Solanum tuberosum; Ath, Arabidopsis thaliana; Osa, Oryza sativa; Sly, Solanum lycopersicum; Sme, Solanum melongena.
Expression patterns of StNRT genes in different tissues
Using the RNA-seq data, we showed the expression (FPKM values) of 39 StNRT genes in a heatmap in different groups and tissues (Fig 6). The expression results showed that most of the StNRT members had a different expression pattern between leaf and root. In addition, the expression of some genes in the nitrogen-deficient group were higher than that in the normal nitrogen fertilizer group in root, and the expression profiles of these genes in the leaves are just the opposite. In leaf, StNRT02 and StNRT23 were up-regulated in nitrogen-deficient group, but StNRT27, StNRT39, StNRT17, StNRT26, StNRT18, StNRT04, StNRT05, StNRT14, StNRT16, StNRT24, and StNRT37 were down-regulated in nitrogen-deficient conditions. In root, genes like StNRT35, StNRT21, StNRT34 were down-regulated by nitrogen-deficient treatment, while StNRT11, StNRT22, StNRT30 and StNRT31 were up-regulated.
Fig 6. Expression of StNRT members in response to Nitrogen-deficiency.
The Illumina RNA-seq data were downloaded from the SRA database (https://submit.ncbi.nlm.nih.gov/subs/sra/) with the submission number of SRS4186597. Gene expression heatmap obtained by unsupervised comparison of genes differentially expressed in leaf and root. The heatmaps indicate high or low expression levels as green or blue colors, respectively. XN and X represented “Shepody” treated with and without N, respectively. a and b represented leaf and root, respectively.
Analysis of Cis-acting element in StNRT genes’ promoters
After identifying the Cis-acting elements in StNRT genes’ promoters, we found that MYB, MYC and ERE were the most three elements in all StNRT members (Fig 7). StNRT13 and StNRT23 had less elements than other members, StNRT13 contained three elements (MYB, MYC and ERE) and StNRT23 contained four elements (three MYC and one ABRE). StNRT31 (18 elements), StNRT26 (17 elements) and StNRT18 (16 elements) were the top three genes that contained the most Cis-acting elements.
Fig 7. Predicted cis-elements in StNRT promoters.
Promoter sequences (upstream 2000 bp) of 39 StNRT20 genes are analyzed by PlantCARE. The upstream length to the translation starts site can be inferred according to the scale at the bottom. Different color boxes represent different elements.
Discussion
Nitrate is necessary for plant growth and development. Understanding the gene function and evolution of NRT family members is important for plant research. Several studies have elucidated the NRT genes functions and evolutionary history in many plant species such as Arabidopsis thaliana [17,18], rice [19], poplar [20] and pineapple [21]. In this present study, 39 StNRT genes were identified including 33 StNRT1, 4 StNRT2, and 2 StNRT3. Acordding to previous studies, there were 24 AtNRTs in Arabidopsis thaliana and 48 candidate NRT genes in pineapple [18,21]. In total, we identified 39 StNRTs in our results, which is within a reasonable range.
As we know, the formation of gene family mainly includes the following ways: 1). whole genome duplication or polyploidization [31]; 2). tandem duplications (of one to a few adjacent genes) [32]; 3). wegmental duplication [33]; 4). transposable elements (TE) [34]; and 5). exon duplication and shuffling [35]. In this study, there were 39 StNRT members randomly distributed on the 12 potato chromosomes. Of which 13 genes found in 6 duplicated blocks. The gene family members that located in the same block might be formed by tandem duplications. These 13 StNRT genes mgiht reveal an early form of gene family member formation. It is speculated that the duplicated genes located in the same block might have closer gene homology, structure and function, which was also confirmed by the evolutionary tree and gene structure analysis in this study. In addition, we found that StNRT25 and StNRT26, StNRT08 and StNRT16 are collinear in the potato genome (Fig 5a), indicating that the formation of these genes may be due to segmental duplication or transposable elements.
Gene structure is related its function. Previous studies have shown that there are five conserved domains in the protein sequences of Arabidopsis thaliana NRT genes [36], which was consistent with our research. Most of the NRT genes are contained in MFS family, which has 12 transmembrane domains [37]. In this study, we found most StNRT genes contained MFS family domains. In plants, NRT proteins transport a wide variety of substrates: nitrate, peptides, amino acids, dicarboxylates, glucosinolates, IAA, and ABA [38]. Due to the long intron of StNRT38 and StNRT15, the squence length was greater than other StNRT members in potato; moreover, it contained a longer MFS family domain, suggesting that the function of these genes might be more complex. In addition, we found that StNRT32, StNRT34, StNRT17 and StNRT04 contained the same domain PLN00028, and these four genes belong to NRT2 subfamily, indicating that NRT2 subfamily might works through PLN00028 domian.
The collinearity analysis showed that these StNRT members in potato are closely related to Solanum lycopersicum and Solanum melongena. Especially for Solanum lycopersicum, the NRT genes also have a good correspondence in the position of the chromosome in both potato and tomato, indicating the close relationship between tomato and potato. These results suggested that StNRT family expanded through segmental duplication events during evolution, and the evolutionary events among potato, Solanum lycopersicum and Solanum melongena might be at an early stage.
Gene expression patterns can provide insights into gene function. Our results showed that most of the StNRT members expressed in leaf and root. Some genes were expressed differently in different organs, such as StNRT09, StNRT10, StNRT13, StNRT21 etc. Our present study identified that several StNRT members were down-regulated by N deficiency (e.g. StNRT30, StNRT17, StNRT39) in leaf, but up-regulated in root. Tiwari et al reported that StNRTs were the most down-regulated in roots under low N conditions [39]. According to our previous study, the NRT transcripts showed different expression profiles in different potato breeds, especially for varieties with different sensitivity to N deficiency [22]. Hence, we inferred that this might be due to the genetic differences in different potato breeds. However, the different expression profiles indicated that the NRTs are crucial for the acquiring N and its conversion to ammonia [40]. NRT2 family is known to control N uptake and transport and is widely distributed in plants [41]. Lezhneva et al [40] reported that the Arabidopsis thaliana AtNRT2.5 was only expressed in the shoot and root of Arabidopsis thaliana in response to N deficiency. Arabidopsis thaliana has 7 NRT2 family members, and NRT2.7 is the only NRT2 member located on the tonoplast membrane in the seeds, and it functions out of interaction with NAR2.1 in transporting nitrate [42,43]. However, the expression profiles of StNRT34, StNRT17 and StNRT04 were decreased in potato leaf by N deficiency, suggesting the increased N metabolism. Different members of StNRT play different roles in leaves and roots. Especially under sufficient nitrogen conditions, different members have a clear distribution in different organizations. However, in the Nitrogen-deficiency conditions, all members of the StNRT family are widely expressed.
The Cis-acting elements in StNRT showed that most of the StNRTs might be regulated by TFs like MYB, MYC and ERE. MYB, MYC and ERE are TFs that known to play roles in abiotic stress [44,45]. The widespread recognition site of MYB, MYC and ERE also indicates that these three TFs might be the regulatory factors for StNRT. Similarly, Bai et al also found the MYB element exists in the promoter region of pepper NRT gene [46], which makes our speculation more credible.
Conclusion
A total of 39 StNRT gene family members were identified in the potato genome, including 33 StNRT1, 4 StNRT2, and 2 StNRT3. The collinearity results show that StNRT members in potato are closely related to Solanum lycopersicum and Solanum melongena. For the expression, Different members of StNRT play different roles in leaves and roots. Especially under sufficient nitrogen conditions, different members have a clear distribution in different organizations. And most of the StNRTs might be regulated by TFs like MYB, MYC and ERE.
Supporting information
(a) and (b) shows the Conserved domain identification of NRT genes in Arabidopsis thaliana and rice, respectively. The abscissa represents the amino acid length. Different colors represent different domains.
(TIF)
Distributions of conserved motifs in StNRT members. Ten putative motifs are indicated in different colored boxes.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
The author thanks Jilin Agricultural University Potato Innovation Team platform.The research was funded Jilin Potato Genetic breeding and Improved seed Breeding Innovation team 20200301025RQ.
References
- 1.Amtmann A, Armengaud P. Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Current Opinion in Plant Biology. 2009;12(3):275–83. doi: 10.1016/j.pbi.2009.04.014 [DOI] [PubMed] [Google Scholar]
- 2.O’Brien José A, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, et al. Nitrate Transport, Sensing, and Responses in Plants. Molecular Plant. 2016;9(6):837–56. doi: 10.1016/j.molp.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 3.Araya T, Kubo T, von Wirén N, Takahashi H. Statistical modeling of nitrogen-dependent modulation of root system architecture in Arabidopsis thaliana. Journal of Integrative Plant Biology. 2016;58(3):254–65. doi: 10.1111/jipb.12433 [DOI] [PubMed] [Google Scholar]
- 4.Urlić B, Jukić Špika M, Becker C, Kläring H-P, Krumbein A, Goreta Ban S, et al. Effect of NO3 and NH4 concentrations in nutrient solution on yield and nitrate concentration in seasonally grown leaf lettuce. Acta Agriculturae Scandinavica, Section B—Soil & Plant Science. 2017;67(8):748–57. doi: 10.1080/09064710.2017.1347704 [DOI] [Google Scholar]
- 5.Qiqige S, Jia L, Qin Y, Chen Y, Fan M. Effects of different nitrogen forms on potato growth and development. Journal of Plant Nutrition. 2017;40(11):1651–9. doi: 10.1080/01904167.2016.1269345 [DOI] [Google Scholar]
- 6.Fawcett RS, Slife FW. Effects of Field Applications of Nitrate on Weed Seed Germination and Dormancy. Weed Science. 1978;26(6):594–6. Epub 2017/06/12. doi: 10.1017/S0043174500064626 [DOI] [Google Scholar]
- 7.Castro Marín I, Loef I, Bartetzko L, Searle I, Coupland G, Stitt M, et al. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta. 2011;233(3):539–52. doi: 10.1007/s00425-010-1316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends in Plant Science. 1998;3(10):389–95. doi: 10.1016/S1360-1385(98)01311-9 [DOI] [Google Scholar]
- 9.Lejay L, Gojon A. Chapter Six—Root Nitrate Uptake. In: Maurel C, editor. Advances in Botanical Research. 87: Academic Press; 2018. p. 139–69. [Google Scholar]
- 10.Shitan N, Yazaki K. Chapter Nine—New Insights into the Transport Mechanisms in Plant Vacuoles. In: Jeon KW, editor. International Review of Cell and Molecular Biology. 305: Academic Press; 2013. p. 383–433. doi: 10.1016/B978-0-12-407695-2.00009-3 [DOI] [PubMed] [Google Scholar]
- 11.Tavares OCH, Santos LA, Ferreira LM, Sperandio MVL, da Rocha JG, García AC, et al. Humic acid differentially improves nitrate kinetics under low- and high-affinity systems and alters the expression of plasma membrane H+-ATPases and nitrate transporters in rice. Annals of Applied Biology. 2017;170(1):89–103. doi: 10.1111/aab.12317 [DOI] [Google Scholar]
- 12.Andrews M, Raven JA, Lea PJ. Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Annals of Applied Biology. 2013;163(2):174–99. doi: 10.1111/aab.12045 [DOI] [Google Scholar]
- 13.Léran S, Varala K, Boyer J-C, Chiurazzi M, Crawford N, Daniel-Vedele F, et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science. 2014;19(1):5–9. doi: 10.1016/j.tplants.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 14.Vidmar J, Zhuo D, Siddiqi M, Glass A. Isolation and Characterization of HvNRT2.3 and HvNRT2.4, cDNAs Encoding High-Affinity Nitrate Transporters from Roots of Barley. Plant physiology. 2000;122:783–92. doi: 10.1104/pp.122.3.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerezo M. Major Alterations of the Regulation of Root NO3- Uptake Are Associated with the Mutation of Nrt2.1 and Nrt2.2 Genes in Arabidopsis. Plant Physiology—PLANT PHYSIOL. 2001;127:262–71. doi: 10.1104/pp.127.1.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dechorgnat J, Chi Tam N, Armengaud P, Jossier M, Diatloff E, Filleur S, et al. From the soil to the seeds: The long journey of nitrate in plants. Journal of experimental botany. 2011;62:1349–59. doi: 10.1093/jxb/erq409 [DOI] [PubMed] [Google Scholar]
- 17.Orsel M, Krapp A, Daniel-Vedele F. Analysis of the NRT2 Nitrate Transporter Family in Arabidopsis. Structure and Gene Expression. Plant Physiology. 2002;129(2):886. doi: 10.1104/pp.005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lezhneva L, Kiba T, Feria-Bourrellier A-B, Lafouge F, Boutet-Mercey S, Zoufan P, et al. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. The Plant Journal. 2014;80(2):230–41. doi: 10.1111/tpj.12626 [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Hu B, Yuan D, Liu Y, Che R, Hu Y, et al. Expression of the Nitrate Transporter Gene OsNRT1.1A/OsNPF6.3 Confers High Yield and Early Maturation in Rice. The Plant Cell. 2018;30(3):638. doi: 10.1105/tpc.17.00809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai H, Euring D, Volmer K, Janz D, Polle A. The Nitrate Transporter (NRT) Gene Family in Poplar. PLOS ONE. 2013;8(8):e72126. doi: 10.1371/journal.pone.0072126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Yan M, Hu B, Priyadarshani SVGN, Hou Z, Ojolo S, et al. Characterization and the Expression Analysis of Nitrate Transporter (NRT) Gene Family in Pineapple. Tropical Plant Biology. 2018. doi: 10.1007/s12042-018-9209-z [DOI] [Google Scholar]
- 22.Zhang J, Wang Y, Zhao Y, Zhang Y, Zhang J, Ma H, et al. Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLOS ONE. 2020;15(10):e0240662. doi: 10.1371/journal.pone.0240662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieczynski M, Wyrzykowska A, Milanowska K, Boguszewska-Mankowska D, Zagdanska B, Karlowski W, et al. Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnology Journal. 2018;16(2):603–14. doi: 10.1111/pbi.12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Wittgenstein NJJB, Le CH, Hawkins BJ, Ehlting J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evolutionary Biology. 2014;14(1):11. doi: 10.1186/1471-2148-14-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto M, Vidmar JJ, Glass ADM. Regulation of NRT1 and NRT2 Gene Families of Arabidopsis thaliana: Responses to Nitrate Provision. Plant and Cell Physiology. 2003;44(3):304–17. doi: 10.1093/pcp/pcg036 [DOI] [PubMed] [Google Scholar]
- 26.Tsay Y-F, Chiu C-C, Tsai C-B, Ho C-H, Hsu P-K. Nitrate transporters and peptide transporters. FEBS Letters. 2007;581(12):2290–300. doi: 10.1016/j.febslet.2007.04.047 [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Chen H, He Y, Xia R. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018:289660. doi: 10.1101/289660 [DOI] [Google Scholar]
- 28.Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics. 2012;287(6):495–513. doi: 10.1007/s00438-012-0696-6 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. Epub 2012/01/06. doi: 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song X, Duan W, Huang Z, Liu G, Wu P, Liu T, et al. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Scientific reports. 2015;5:14631-. doi: 10.1038/srep14631 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends in Genetics. 2006;22(11):597–602. doi: 10.1016/j.tig.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 32.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–41. Epub 2005/03/09. doi: 10.1016/j.pbi.2005.01.001 . [DOI] [PubMed] [Google Scholar]
- 33.Yin G, Xu H, Xiao S, Qin Y, Li Y, Yan Y, et al. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biology. 2013;13(1):148. doi: 10.1186/1471-2229-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology. 2000;42(1):251–69. doi: 10.1023/A:1006344508454 [DOI] [PubMed] [Google Scholar]
- 35.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nature Genetics. 2005;37(9):997–1002. doi: 10.1038/ng1615 [DOI] [PubMed] [Google Scholar]
- 36.Nogia P, Tomar A, Sidhu GK, Mehrotra R, Mehrotra h, editors. Elucidation of an Array of Nitrate Transporter Paralogs in Arabidopsis Thaliana Genome2016. [Google Scholar]
- 37.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annual Review of Biophysics. 2015;44(1):257–83. doi: 10.1146/annurev-biophys-060414-033901 [DOI] [PubMed] [Google Scholar]
- 38.Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014;19(1):5–9. Epub 2013/09/24. doi: 10.1016/j.tplants.2013.08.008 . [DOI] [PubMed] [Google Scholar]
- 39.Tiwari JK, Buckseth T, Zinta R, Saraswati A, Singh RK, Rawat S, et al. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Scientific Reports. 2020;10(1):1152. doi: 10.1038/s41598-020-58167-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lezhneva L, Kiba T, Feria‐Bourrellier AB, Lafouge F, Boutet‐Mercey S, Zoufan P, et al. The Arabidopsis nitrate transporter NRT 2.5 plays a role in nitrate acquisition and remobilization in nitrogen‐starved plants. The Plant Journal. 2014;80(2):230–41. doi: 10.1111/tpj.12626 [DOI] [PubMed] [Google Scholar]
- 41.Kant S. Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Seminars in Cell & Developmental Biology Academic Press. 2017;74:pp. 89–96. doi: 10.1016/j.semcdb.2017.08.034 [DOI] [PubMed] [Google Scholar]
- 42.Kotur Z, Unkles SE, Glass AD. Comparisons of the Arabidopsis thaliana High-affinity Nitrate Transporter Complex AtNRT2. 1/AtNAR2. 1 and the Aspergillus nidulans AnNRTA: structure function considerations. Israel Journal of Plant Sciences. 2017;64(3–4):21–31. [Google Scholar]
- 43.Kotur Z, Mackenzie N, Ramesh S, Tyerman SD, Kaiser BN, Glass AD. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2. 1. New Phytologist. 2012;194(3):724–31. doi: 10.1111/j.1469-8137.2012.04094.x [DOI] [PubMed] [Google Scholar]
- 44.Nakashima K, Yamaguchi‐Shinozaki K. Regulons involved in osmotic stress‐responsive and cold stress‐responsive gene expression in plants. Physiologia plantarum. 2006;126(1):62–71. [Google Scholar]
- 45.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. Epub 2003/01/02. doi: 10.1105/tpc.006130 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai H, Euring D, Volmer K, Janz D, Polle A. The nitrate transporter (NRT) gene family in poplar. PloS one. 2013;8(8):e72126–e. doi: 10.1371/journal.pone.0072126 . [DOI] [PMC free article] [PubMed] [Google Scholar]