Abstract
Since the bipolar disorder (BD) signals identified by genome-wide association study (GWAS) often reside in the non-coding regions, understanding the biological relevance of these genetic loci has proven to be complicated. Transcriptome-wide association studies (TWAS) providing a powerful approach to identify novel disease risk genes and uncover possible causal genes at loci identified previously by GWAS. However, these methods did not consider the importance of epigenetic regulation in gene expression. Here, we developed a novel epigenetic element-based transcriptome-wide association study (ETWAS) that tested the effects of genetic variants on gene expression levels with the epigenetic features as prior and further mediated the association between predicted expression and BD. We conducted an ETWAS consisting of 20 352 cases and 31 358 controls and identified 44 transcriptome-wide significant hits. We found 14 conditionally independent genes, and 10 genes that did not previously implicate with BD were regarded as novel candidate genes, such as ASB16 in the cerebellar hemisphere (P = 9.29 × 10–8). We demonstrated that several genome-wide significant signals from the BD GWAS driven by genetically regulated expression, and NEK4 explained 90.1% of the GWAS signal. Additionally, ETWAS identified genes could explain heritability beyond that explained by GWAS-associated SNPs (P = 5.60 × 10–66). By querying the SNPs in the final models of identified genes in phenome databases, we identified several phenotypes previously associated with BD, such as schizophrenia and depression. In conclusion, ETWAS is a powerful method, and we identified several novel candidate genes associated with BD.
Keywords: gene expression prediction, epigenetic regulation, bipolar disorder, candidate gene, missing heritability
Introduction
Bipolar disorder (BD) is a severe neuropsychiatric disorder characterized by recurrent episodes of depression and mania that affect thought, perception, emotion, and social behavior. Based on twin studies, the narrow-sense heritability of BD was estimated to be over 70%.1,2 Genome-wide association study (GWAS) has seen great strides and invaluable utilities in revealing initial insights into BD’s genetic architecture. Despite the significant success of GWAS in delineating elements that contribute to the genetic architecture of psychiatric disorders, only a small fraction of this heritability is explained by associated loci,3 leaving a substantial proportion of genetic risk factors uncharacterized.
Most of the identified variants mapped through GWAS reside in non-coding regions of the genome,4 which may be involved in modulating gene regulatory programs.4–8 Recent mechanistic studies have demonstrated that GWAS-identified variants located in the active chromatin regions more frequently and highly enriched with expression quantitative trait loci (eQTL).9,10 Moreover, most common risk variants identified to date are only associated with diseases with modest effect sizes, and many risk variants have not been identified via a typical GWAS, even with a large sample size.11 Transcriptome-wide association study (TWAS) that systematically investigates the association of genetically predicted gene expression with disease risk, providing a powerful approach to identify novel disease risk genes and uncover possible causal genes at loci identified previously by GWAS.12–16
Nevertheless, gene expression is highly regulated in many steps, including transcriptional regulation, splicing, end modification, export, and degradation. Transcriptional regulation of DNA into mRNA can occur on both genetic and epigenetic levels. The epigenetic regulation alters the accessibility of DNA to transcription factors by chemical modification of chromatin. For example, several post-translational modifications that occur on the histones can change chromatin structure and function,17 make it accessible or vice versa to transcription factors. Functional class quantification in 11 diseases from the Wellcome Trust Case Control Consortium, including BD, have shown that 80% of the common variants that contribute to phenotype variability attribute to DNase I hypersensitivity sites.18 They are likely to regulate chromatin accessibility and transcription, further highlighting the importance of transcript regulation at the epigenetic level. Histone modifications are involved in both activation and repression of transcription19 and further linked to diseases.20 Researchers have developed a growing body of computational methods to predict gene expression from histone modification signals of chromatin structure.21–24 Thus, integrating epigenetic features is essential for the prediction of gene expression besides genetic variants.
In this study, we set out to develop a 4-step quantitative pipeline named epigenetic element-based transcriptome-wide association studies (ETWAS), based on the interpretation of epigenetic element, genotype, gene expression, and phenotype. We used ETWAS to investigate the association between gene expression and BD risk using the largest BD cohort currently available (as of 2020); the cohort consisted of 20 352 BD cases and 31 358 controls from Europe. We found that ETWAS outperformed original methods, and we identified 14 conditionally independent genes associated with BD risk in 13 brain tissues. We additionally identified 10 genes that were not previously implicated with BD.
Methods
Data Resources
RNA Sequencing Data Sets
We used transcriptome and high-density genotyping data of European decedent from the Genotype-Tissue Expression (GTEx) study Pilot Project V8 (dbGap accession: phs000424.v8.p2)10 to establish gene expression prediction models (supplementary methods). We also obtained freely available RNA-seq data from 358 European lymphoblastoid cell lines produced by the Genetic European Variation in Health and Disease25 (Geuvadis) as the validation data set to test the prediction models generated in the GTEx whole blood. The tissue abbreviations, sample sizes are listed in supplementary table 1.
Epigenetic Elements
The chromatin states and the DNase I hypersensitive sites (DHS) of relevant tissues were downloaded from the Roadmap Epigenome Project.26 Freely available transcription factor binding sites (TFBS) were obtained from the Encyclopedia of DNA Elements (ENCODE).27 The URLs of all epigenetic data are listed in supplementary table 2.
GWAS Summary Statistics
We used the most recent summary statistics of the Psychiatric Genomics Consortium (PGC) Bipolar Disorder Working Group, comprising 20,352 BD cases and 31,358 controls of European descent (supplementary table 3). Details on participant ascertainment and quality control were previously reported by Eli et al3 (supplementary methods).
ETWAS Framework
Our current study is based on the premise that gene expression is heritable.28 Considering the heritability genes typically enriched for trait associations,14 we estimated gene expression heritability (supplementary methods) and only focused on the significantly heritable genes in further analyses.
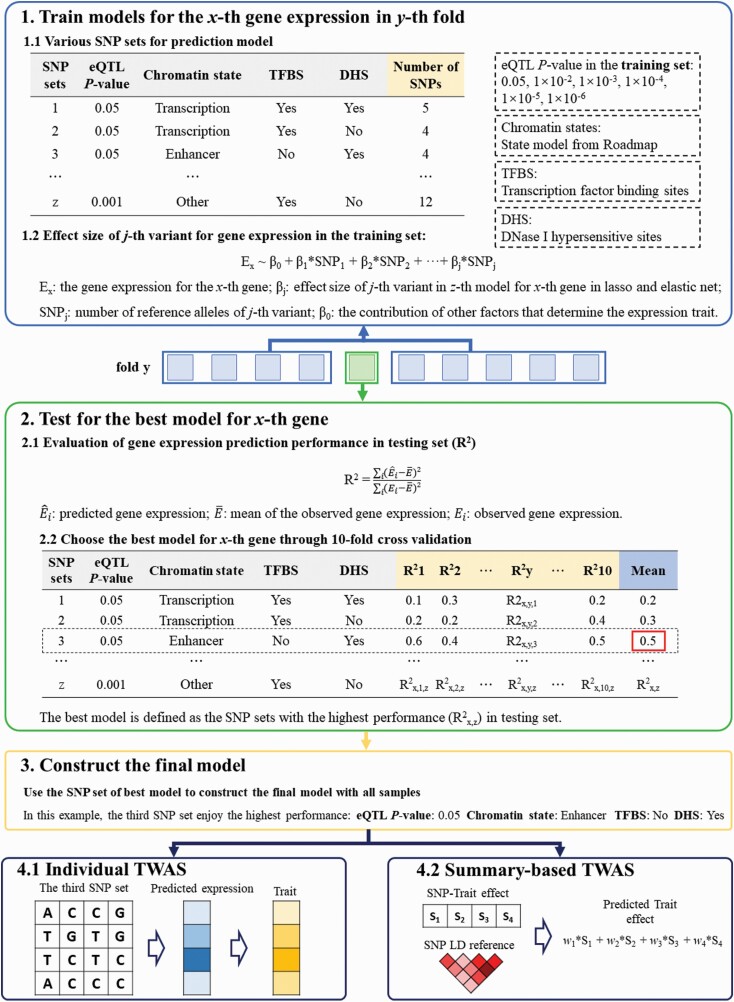
Our prediction framework included 4 main sections that acted sequentially (figure 1). First, for each gene, we divided the SNPs within 1 Mb of the transcription start/end site of the gene into multiple SNP sets according to the eQTL P-value and epigenetic annotations. We constructed multiple elastic net and lasso models with the SNPs in each SNP set using the initial reference data. Second, we evaluated the prediction performance of each SNP set using 10-fold cross-validation R2 between the predicted and observed expression, and the SNP set with the highest mean R2 was selected as the best model. Third, we constructed the final prediction model with the parameters of the best SNP set using all the samples in the reference data. We estimated the associations between predicted expressions and traits with the combination of SNP-trait effect sizes while accounting for linkage disequilibrium among SNPs. The process to evaluate and select the best model is described in greater detail in supplementary methods.
Fig. 1.
Schematic of ETWAS approach. The prediction framework includes 4 sections. First, for each gene, we divide the SNPs within 1 Mb of the transcription start/end site of the gene into multiple SNP sets according to the eQTL P-value and epigenetic annotations. We construct multiple elastic net and lasso models with the SNPs in each SNP set using 10-fold cross-validation R2 in the initial reference data. Second, we evaluate the prediction performance of each SNP set by cross-validation R2 between the predicted and observed expression, and the SNP set with the highest mean R2 is selected as the best model. Third, we construct the final prediction model with the parameters of the best SNP set using all the samples in the reference data. We then estimate the associations between predicted expressions and traits with the combination of SNP-trait effect sizes while accounting for linkage disequilibrium among SNPs.
Model Evaluation
We displayed the analyses for evaluating the ETWAS and the reasons for including them (supplementary figure 1, supplementary methods). After got the gene expression prediction models by ETWAS, we first evaluated the important role of epigenetic annotation in improving the performance of gene expression via 10-fold cross-validation by (1) testing whether the performance of models significantly increases with the number of active annotations increases, (2) calculating the correlation between model performance and SNP number, and (3) comparing the epigenetic annotation distribution of genes’ best models and SNPs. Then, we compared the cross-validation performance between ETWAS and recent work in parallel to ours, which imputed expression used only genetic variants with different models, such as lasso, and elastic net. Finally, we evaluated the prediction models on an independent cohort. Cross-study prediction accuracy was measured by weights derived from the best models with all whole blood samples from GTEx to predict gene expression levels in the Geuvadis dataset.
Model Application
We applied ETWAS to identify genes associated with BD using summary data comprising 20 352 BD cases and 31 358 controls of European descent. Since several of the ETWAS hits overlapped with significant BD loci, we first performed conditional and joint analyses to establish whether these signals were due to multiple-associated features or conditionally independent. We validated the ETWAS identified conditionally independent genes by (1) differential gene expression analyses, (2) calculating partitioned heritability, and (3) phenome-wide association study. We then performed the GWAS catalog enrichment analyses and drug target enrichment analyses to demonstrate ETWAS’s ability to identify BD-related genes. To test for the biological functions of ETWAS identified genes, the FUMA29 prioritized genes with and without ETWAS identified genes were tested against gene sets obtained from MsigDB (GO gene sets and curated gene sets) using hypergeometric tests. At last, we employed ETWAS to identify new expression-trait associations using an early released summary association data for BD in 2012, comprising 7481 BD cases and 9250 controls of European descent, and then looked for genome-wide significant SNPs at these loci in the larger BD GWAS (expanded to 20 352 BD cases and 31 358 controls).
Results
Model Generation and Evaluation
Heritability of Gene Expression
The mean heritability of gene expression was 0.016 for all protein-coding genes in whole blood, and 0.04 in brain tissues ranging from 0.031 to 0.049 in different tissues (supplementary table 1). The low heritability indicated the genotype alone played a less important role in the expression of all genes, which highlights the importance of integrating epigenetic features for the prediction of gene expression besides genetic variants. We identified 2239 significantly heritable genes in whole blood and 19 632 gene-tissue pairs of 9492 genes in 13 brain tissues. Among the significantly heritable genes in the brain, we found that almost half of them (5155/9492) were significant only in one brain tissue (supplementary figure 2). We observed a high correlation of gene expression heritability in different brain tissues, with a correlation coefficient ranging from 0.35 to 0.69 (supplementary figure 3).
Epigenetic Annotation Improved the Performance of Gene Expression Prediction
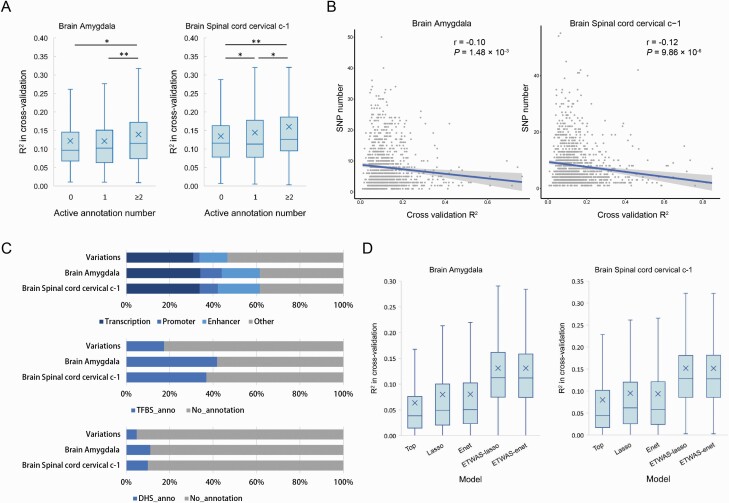
The performance of gene expression prediction was better in higher heritable genes (supplementary figure 4), which could be supplementary to prove the appropriateness of focusing on high heritability genes. We evaluated whether the expression of heritable genes could be accurately imputed in 13 brain tissues from genotype with the epigenetic elements as prior. We noted that the cross-validation performance significantly increased with the number of active annotations increasing in 11 tissues, and the genes with at least 2 active annotations 1.01× to 1.19× outperformed the genes with 0 active annotations (figure 2A, supplementary figure 5). We identified a negative correlation between cross-validation R2 and SNP number in the best models (figure 2B, supplementary table 4), which consistent with the sparsity of the local architecture of gene expression and a handful of genetic variants that contribute to the variability in gene expression.30 The negative correlation between model performance and SNP number indicated that the model performance was not better with the increased SNPs. Thus, the key to improving the performance is to choose the effective SNPs. We think epigenetic annotation plays an important role in selecting effect SNPs. We demonstrated that the best SNP sets for predicting gene expression significantly enriched in active epigenetic elements by comparing the epigenetic annotation frequency between the best SNP sets and all variants used (Pearson’s chi-squared test, P < 0.01) (figure 2C, supplementary figure 6). Specifically, the best SNP sets distributed in the active HMM annotation at a frequency of 1.32–1.45 times that of all variants, while distributed in the TFBS region and DHS region at a frequency of 2.12–2.43 and 1.70–2.40 times that of all variants. We further evaluated ETWAS’s performance and compared them with recent work in parallel to ours. On average, our pipeline attained slightly better performance than using lasso, elastic net, and top SNP (figure 2D, supplementary figure 7). For ETWAS, the average 10-fold cross-validated prediction R2 value ranging from 0.11 to 0.16 in different brain tissues (supplementary table 4).
Fig. 2.
Epigenetic data improves the performance of gene expression prediction in brain tissues. (A) The cross-validation R2 of the best prediction models are sorted according to the active annotation number and group into 3 categories: 0, 1, ≥2. One asterisk (*) indicates P-value smaller than 0.05 (P < 0.05), 2 asterisks (**) indicates P-value smaller than 0.01 (P < 0.01). (B) The correlation between the prediction performance and the SNP number in the best model. The x-axis represents cross-validation R2 and the y-axis represents the SNP number of the best model. (C) The epigenetic annotation distributions of the SNPs used for all genes and the best ETWAS models in 2 brain tissues. (D) Accuracy of individual-level expression imputation models. Accuracy is estimated using cross-validation R2 between predicted and true expression. Box plots indicate the accuracy distribution for 2 brain tissues in 5 methods: top, lasso, elastic net (Enet), ETWAS-lasso, and ETWAS-enet.
ETWAS Performance in a Separate Cohort
We also tested the prediction models on separate cohorts. The average prediction R2 was 0.034. The top 3 genes with the highest performance are illustrated in supplementary figure 8, proving a comparison of the predicted and observed expression. Among these genes, the LDHC trained in GTEx performed best, and the R2 between predicted and observed expression levels in Geuvadis was 0.82. We found the diversity of predicted gene expression depended on the number of SNPs in the final model, and the dispersion of predicted gene expression depended on the SNP weights. A quantile-quantile plot showed expected and observed R2 from ETWAS is given (supplementary figure 9). We identified a substantial departure from the null distribution, indicated that the ETWAS model trained in GTEx whole blood captured a substantial proportion of the transcriptome variability in Geuvadis.
ETWAS Identified New BD Associations
ETWAS Significant Hits
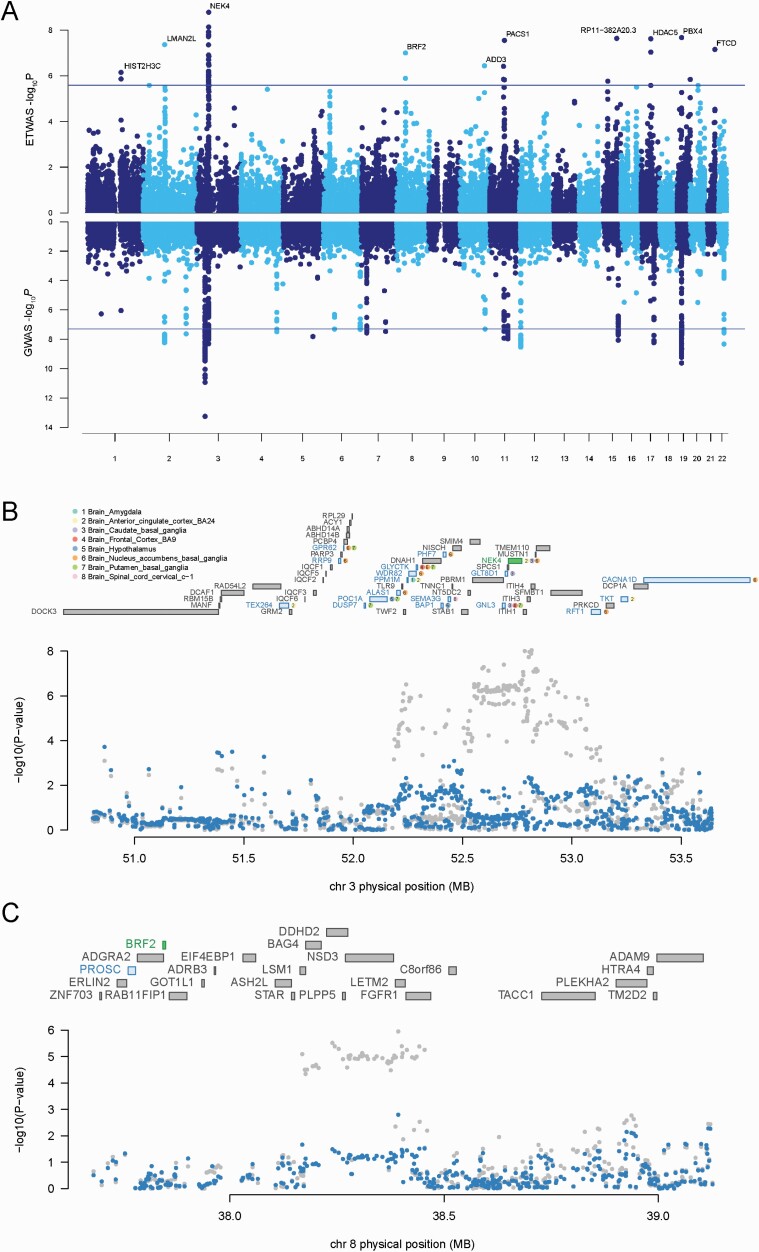
We applied ETWAS to identify genes associated with BD using summary data comprising 20 352 BD cases and 31 358 controls of European descent. A total of 9492 unique genes across 13 brain tissues (resulting in 19 632 distinct tests, supplementary figure 2) were used in this study. We set the significance threshold at P < 2.55 × 10–6 (0.05/19 632) after adjustment for multiple testing corrections by Bonferroni correction. ETWAS identified 34 susceptibility genes associated with BD, comprising 44 total associations (figure 3A, supplementary table 5). For example, the most significant gene, NEK4, associated with BD in 3 tissues (PCBG = 1.66 × 10–9, PACC = 7.38 × 10–9, PNAB = 1.88 × 10–6).
Fig. 3.
ETWAS identifies new BD associations. (A) Manhattan plot of the ETWAS (upper) and GWAS (lower) results for BD (n = 20 352 cases and n = 31 358 controls). The line represents the Bonferroni-corrected significant thresholds, P = 2.55 × 10–6 for ETWAS, and P = 5 × 10–8 for GWAS. (B–C) Regional association of ETWAS hits. Chromosome 3 (B) and chromosome 8 (C) regional association plot. The conditionally significant genes are NEK4 (B) and BRF2 (C). The bottom panel shows a regional Manhattan plot of the GWAS data before (light gray) and after conditioning on the predicted expression of the conditionally significant genes. For color, please see the figure online.
Expression Signals Explained Several BD Loci
We identified 14 conditionally independent genes through conditional analyses (table 1). We observed that NEK4 explained most of the signals at its loci, a region that contained 27 significant ETWAS hits (rs2071044 lead SNPGWASP = 9.10 × 10–9, conditioned on NEK4 lead SNPGWASP = 0.57, explained 0.901 of the variances) (figure 3B). We detailed NEK4’s ETWAS expression models (supplementary figure 10A) as well as genes at the same locus and shared the same variants in the final prediction models.
Table 1.
Conditionally Independent ETWAS Genes for BD
| Gene | Tissue | Best eQTL | aCross-validationR2 | TWAS Z-score | bTWAS P-value | cImplicated in 2019 BD GWAS |
|---|---|---|---|---|---|---|
| NEK4 | CBG | rs2019065 | 0.108 | 6.03 | 1.66 × 10–9 | Yes |
| NEK4 | ACC | rs2255107 | 0.073 | 5.78 | 7.38 × 10–9 | Yes |
| NEK4 | NAB | rs731831 | 0.131 | 4.77 | 1.88 × 10–6 | Yes |
| LMAN2L | SUB | rs11891926 | 0.146 | −5.48 | 4.36 × 10–8 | Yes |
| PBX4 | CER | rs2288865 | 0.072 | 5.60 | 2.13 × 10–8 | Yes |
| ADD3 | CER | rs4918489 | 0.155 | 5.09 | 3.68 × 10–7 | Yes |
| RP11-382A20.3 | CEH | rs8034801 | 0.094 | 5.59 | 2.32 × 10–8 | No |
| HDAC5 | CEH | rs7207464 | 0.093 | 5.58 | 2.41 × 10–8 | No* |
| PACS1 | FRO | rs7114014 | 0.107 | 5.55 | 2.84 × 10–8 | No* |
| FTCD | NAB | rs2839258 | 0.195 | −5.39 | 7.08 ×10–8 | No |
| ASB16 | CEH | rs9910055 | 0.185 | 5.34 | 9.29 × 10–8 | No |
| BRF2 | HYP | rs12549353 | 0.141 | 5.33 | 1.01 × 10–7 | No |
| FADS1 | CEH | rs174568 | 0.197 | −5.07 | 3.90 × 10–7 | No |
| FADS1 | CER | rs174535 | 0.174 | −4.82 | 1.43 × 10–6 | No |
| HIST2H2AA3 | COR | rs2039800 | 0.034 | −4.83 | 1.39 × 10–6 | No |
| ZNF584 | SCC | rs1550813 | 0.168 | 4.82 | 1.45 × 10–6 | No |
| CDAN1 | CER | rs1359003 | 0.073 | 4.78 | 1.73 × 10–6 | No |
Note: CBG, Brain Caudate basal ganglia; ACC, Brain Anterior cingulate cortex BA24; NAB, Brain Nucleus accumbens basal ganglia; CER, Brain Cerebellum; CEH, Brain Cerebellar Hemisphere; FRO, Brain Frontal Cortex BA9; SUB, Brain Substantia nigra; HYP, Brain Hypothalamus; SCC, Brain Spinal cord cervical c-1; COR, Brain Cortex.
aThe cross-validation R2 between predicted and observed gene expression is based on 10-fold cross-validation within training data.
bTo account for multiple testing, we used a significance threshold of 2.55 × 10–6 for the ETWAS analyses.
cWhether there are any genome-wide significant SNPs within 500 kb away from the gene in the discovery dataset. The asterisk (*) indicates the genes implicated in a more massive combined data.
Expression Signals Drove BD ETWAS Loci
Among the conditionally independent genes, we identified 10 genes that were not implicated in the original BD GWAS, which were regarded as novel candidate genes for BD (table 1). BRF2 explained 0.352 of the variances (rs12677998 lead SNPGWASP = 1.10 × 10–6, conditioned on BRF2 lead SNPGWASP = 1.60 × 10–3) (figure 3C). We detailed BRF2’s ETWAS expression models (supplementary figure 10B) as well as genes at the same locus and shared the same variants in the final prediction models.
ETWAS Increased Power to Find BD Associations
Functional Relevance of Identified Genes
Among the 9492 significantly heritable genes, 7730 genes were tested in the PsychENCODE, and 24/34 genes identified by ETWAS after multiple testing adjustments were associated with BD at the expression level or the transcript level. We found 10 of the 14 conditionally independent genes were proved, including 7 novel genes (supplementary table 6). Five of the novel genes can be found annotated phenotypes in the knock-out mice model. Since the phenotypes MGI arranged did not include BD, we listed all the phenotypes of conditionally independent novel genes reported in MGI (supplementary table 7). We found 3 genes (HDAC5, ASB16, and CDAN1) associated with cardiovascular disease (CVD) relevant phenotypes, such as cardiac hypertrophy, abnormal heart morphology, abnormal heart shape, and an enlarged heart. There is an integration of the various factors that putatively underlie the association of BD with CVD,31 which indirectly suggested that the genes we found may be related to BD.
Partitioned Heritability of ETWAS Identified BD Genes
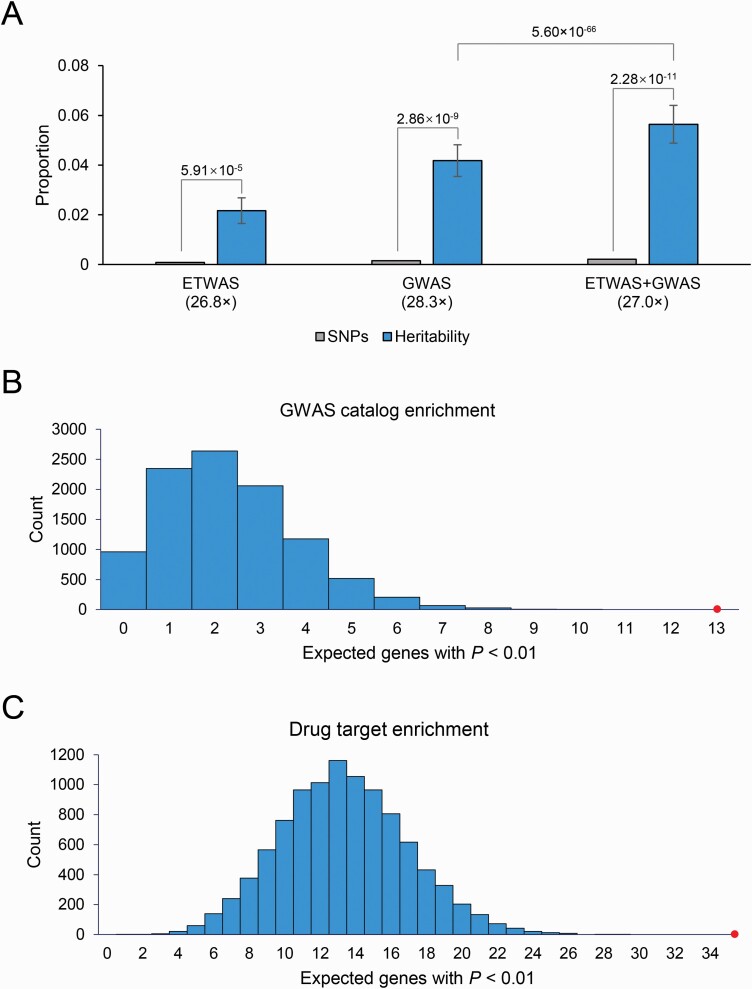
Conditionally independent genes explained 2.17% (SE = 0.52%) of the estimated heritability, a 26.8× enrichment (P = 5.91 × 10–5) compared to the percentage of SNPs. Combined the SNPs from ETWAS and GWAS explained a much larger percentage of heritability (5.64%, SE = 0.76%). We performed a t-test to compare the partitioned heritability of GWAS loci with that of GWAS loci and the ETWAS identified conditionally independent genes. As shown in figure 4A, ETWAS identified genes significantly increased the proportion of explained heritability (P = 5.60 × 10–66). We further identified similar results when compared with the sub-GWAS signals with P-value < 1 × 10–7 and 1 × 10–6 (supplementary figure 11), but not for signals with P-value < 1 × 10–5. These results indicated that most of the genes identified by ETWAS could be covered by signals with P-value less than 1 × 10–5.
Fig. 4.
(A) Partitioned heritability of conditionally independent genes and GWAS loci. The null expectation, equal to the percentage of SNPs in each category (light gray), and P-values report the difference from this expectation. Fold enrichment relative to the null expectation is shown in parentheses below each category. Error bars show standard errors. (B) GWAS catalog enrichment analyses of ETWAS genes. (C) Drug targets enrichment analyses of ETWAS genes. The 2 histograms show the expected number of genes with P < 0.01 based on 10 000 random permutations. The large points show the observed number of previously known BD genes/targets that fall below this threshold. For color, please see the figure online.
Phenome-Wide Association Study
A total of 158 phenotypes, such as cognitive, immunological, metabolic, neurological, psychiatric, were significantly associated with the SNPs in the final model of the BD genes (supplementary figure 12A). We found 38 phenotypes, including 9 psychiatric phenotypes, genetically correlated with BD (P < 0.05) (supplementary figure 12B). After Bonferroni corrections, there were 9 phenotypes such as schizophrenia and depression positive correlated with BD. We also found evidence for the causal effects of 6 phenotypes on BD (supplementary figure 13). For example, schizophrenia significantly increased the prospective risk of BD (OR = 1.55, 95% CI: 1.50–1.61).
GWAS Catalog and Drug Target Enrichment Analyses
We found that BD had a significant enrichment (P < 10–4) of GWAS catalog reported genes (figure 4C) and Open Targets Platform reported drug targets (figure 4D) in the ETWAS results, which suggested that they were likely to be true disease associations even for those failed to meet strict genome-wide significance.
Gene-Set Enrichment Analyses
We prioritized 156 unique genes for BD by FUMA SNP2GENE process (supplementary methods). We noticed that ETWAS identified genes shared biological functions with the GWAS prioritized genes in 18 gene sets, such as “Alzheimer’s disease incipient up” (supplementary table 8). Besides, we identified additional 12 gene sets after adding ETWAS results, such as cardiovascular disease-relevant phenotypes, “cardiac muscle contraction,” “hypertrophic cardiomyopathy (HCM)” and “dilated cardiomyopathy.”
Validation of Novel Loci in Subsequent BD Study
We further employed ETWAS to identify new expression-trait associations using an early released summary association data for BD in 2012, comprising 7481 BD cases and 9250 controls of European descent.32 We identified 4 conditionally independent novel genes (more than 500 kb away from any genome-wide significant SNPs) using the 2012 BD GWAS summary. We then looked for genome-wide significant SNPs at these loci in the larger 2019 BD GWAS3 (expanded to 20,352 BD cases and 31,358 controls). We identified all the 4 novel BD-associated genes contained genome-wide significant SNPs in the 2019 GWAS summary data (P < 5 × 10–8, supplementary table 9). Thus, ETWAS is highly predictive of robust phenotypic associations.
Application of ETWAS to Height
We applied our method in height (~700 000 samples) to confirm the reliability of ETWAS (supplementary results). We identified 700 genes associated with height after Bonferroni correction (supplementary figure 14). Of the 700 identified genes, 9 genes were not proximal to any genome-wide-significant SNP for height, implicated novel genes (supplementary table 10). Information from the MGI and the validation of novel loci in subsequent height study also supported the reliability of ETWAS (supplementary results, supplementary tables 11 and 12).
Discussion
In this study, we developed a pipeline named epigenetic element-based transcriptome-wide association studies (ETWAS). We identified 44 genes with genetically predicted expression levels associated with BD risk. Additionally, through conditional and joint analyses, we identified 14 independent genes associated with BD risk in 13 brain tissues. Ten of these genes were not previously implicated with BD, which were regarded as novel candidate genes.
We applied conditional and joint association methods to identify independent genes. Importantly, the ETWAS expression signals drove the significance for several previously implicated BD loci when conditioned on the ETWAS genes. For example, NEK4 explained 90.1% of the GWAS signal, which suggested that after considering the predicted expression signal of NEK4, there was little residual association signal from the genetic variant in the GWAS locus. We identified 14 conditionally independent genes, 4 (NEK4, LMAN2L, PBX4, and ADD3) implicated in the original BD GWAS, and the rest 10 genes were regarded as novel candidate genes for BD. Zhihui et al33 have reported the associations between psychiatric risk alleles and mRNA expression of NEK4, and the overexpression of NEK4 could reduce mushroom density spines in rat primary cortical neurons, the most mature form of all spines that responsible for long-term memory. For the novel candidate genes, M Ikeda and colleagues performed a GWAS of BD in the Japanese population and highlighted a locus at 11q12.2, a region known to contain regulatory genes for plasma lipid levels, including FADS1.34 Moreover, variations in FADS1 have been related to polyunsaturated fatty acids (PUFA) blood concentrations by candidate gene approaches35 and several pieces of evidence suggested that the omega-3 PUFA was associated with reduced risk for developing BD,36 which indirectly suggested the association between FADS1 and BD. Future studies could interrogate whether expression differences of other candidate genes are consistent with our findings.
Since gene’s heritability provided an upper bound of the predictive accuracy, genes that did not significantly heritable at current sample sizes were not included in this project. Some of the strongly implicated genes in BD risk were not assayed, such as ITIH1, FADS2, and NCAN,3 due to non-significant heritability estimates in any of the brain tissue. We think this could be partly due to the quality of available eQTL data. Although gene expression is amenable to genetic prediction with relatively modest sample sizes because of the sparse genetic architecture of gene expression,30 recent evidence suggested that larger expression reference panels would help increase the total number of significant cis-heritable genes available for prediction.14 We detected the overlap between the ETWAS genes and GWAS reported genes. Among the 19 loci, 9 genes were significantly heritable in at least one brain tissue and qualified for ETWAS analyses. Two genes were identified by ETWAS after multiple testing adjustments, which were LMAN2L (PSUB = 4.36 × 10–8) and ADD3 (PCER = 3.68 × 10–7) (supplementary table 13). Another 2 genes were identified at a nominal significance level, which were RPS6KA2 (PCEH = 0.029) and ZNF592 (PCER = 0.030). For the rest 5 genes, we displayed the final prediction models and found that the identified SNPs did not play a role in the gene expression model. For example, TRANK1 significantly heritable in the cerebellum and hypothalamus, and the GWAS-identified lead SNP, rs9834970, did not contribute to the gene expression prediction model in the cerebellum (PeQTL = 0.61) or the hypothalamus (PeQTL = 0.91) (supplementary figure 15). Since rs9834970 is in the intergenic region and closest to TRANK1, the original GWAS paper assigned the locus to TRANK1. Additionally, our method aims to identify genes associated with the disease. However, GWAS-identified loci are not always associated with diseases by regulating gene expression.37,38 Thus, it is reasonable that some of the GWAS assigned genes were not identified by ETWAS. Therefore, we suggest ETWAS as a complementary method to identify new phenotype-associated genes as well as prioritize candidate genes.
Furthermore, 2 genes (NEK4 and FADS1) were Bonferroni-corrected significant in several brain tissues, while others only significant in the specific tissue. Since expression regulation may be common across tissue types, it was refreshing not to see consistency across panels. For instance, PBX4 had a P-value of 2.13 × 10–8 in the cerebellum but a P-value of 0.069 in the cortex. Similarly, HDAC5 had a P-value of 2.41 × 10–8 in the cerebellar hemisphere but a P-value of 4.90 × 10–3 in the substantia nigra. Although it may be due to tissue-specificity, it is essential to note that it may also be due to specific effects and the quality of the RNA data and panel size of different tissue types from GTEx.
Although we are convinced ETWAS has significant potential to delineate further the biological mechanisms for human complex diseases, our current study’s limitations also need to be addressed. In part due to the historical paucity of eQTL in populations of non-European ancestry, all subjects from the 2 reference panels were limited to be European ancestry, and the results may not apply to other populations. Since the genetic predictors of gene expression are more accurate in populations of similar ancestry,39 further study with a larger sample size of different races with both genotype and gene expression levels is needed. Next, although ultrarare variants have been reported to drive substantial cis heritability of human gene expression,40 it is unrealistic to include singletons in expression prediction models at present. Additionally, BD-associated genes identified by ETWAS does not imply causality, and functional studies are needed to determine underlying mechanisms of risk comprehensively. Larger transcriptome and GWAS datasets for BD are likely to improve statistical power for gene identification in the future. Likewise, transcriptome datasets from specific ancestry could also improve future BD ETWAS approaches.
In conclusion, ETWAS is a powerful method that increases statistical power to identify genes associated with BD. We hope ETWAS could provide novel insights into the identification of additional susceptibility genes and further delineate the biological mechanisms for other human complex diseases.
Supplementary Material
Acknowledgment
The authors have declared that there are no conflicts of interest in relation to the subject of this study.
Funding
This study is supported by the National Natural Science Foundation of China (31970569, 31871264), Innovative Talent Promotion Plan of Shaanxi Province for Young Sci-Tech New Star (2018KJXX-010), and the Fundamental Research Funds for the Central Universities. This study is also supported by the High-Performance Computing Platform of Xi’an Jiaotong University.
References
- 1. Edvardsen J, Torgersen S, Røysamb E, et al. Heritability of bipolar spectrum disorders. Unity or heterogeneity? J Affect Disord. 2008;106(3):229–240. [DOI] [PubMed] [Google Scholar]
- 2. Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C(1):48–58. [DOI] [PubMed] [Google Scholar]
- 3. Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soldner F, Stelzer Y, Shivalila CS, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533(7601):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Claussnitzer M, Dankel SN, Kim KH, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373(10):895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park JH, Wacholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gamazon ER, Wheeler HE, Shah KP, et al. ; GTEx Consortium . A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gusev A, Mancuso N, Won H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50(4):538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barbeira AN, Dickinson SP, Bonazzola R, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manor O, Segal E. Robust prediction of expression differences among human individuals using only genotype information. PLoS Genet. 2013;9(3):e1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 18. Gusev A, Lee SH, Trynka G, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium . Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95(5):535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong XJ, Weng ZP. The correlation between histone modifications and gene expression. Epigenomics-Uk 2013;5(2):113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh R, Lanchantin J, Robins G, Qi YJ. DeepChrome: deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016;32(17):639–648. [DOI] [PubMed] [Google Scholar]
- 22. Dong X, Greven MC, Kundaje A, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13(9):R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frasca M, Bertoni A, Re M, Valentini G. A neural network algorithm for semi-supervised node label learning from unbalanced data. Neural Netw. 2013;43:84–98. [DOI] [PubMed] [Google Scholar]
- 24. Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107(7):2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lappalainen T, Sammeth M, Friedländer MR, et al. ; Geuvadis Consortium . Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kundaje A, Meuleman W, Ernst J, et al. ; Roadmap Epigenomics Consortium . Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monks SA, Leonardson A, Zhu H, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75(6):1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wheeler HE, Shah KP, Brenner J, et al. ; GTEx Consortium . Survey of the heritability and sparse architecture of gene expression traits across human tissues. PLoS Genet. 2016;12(11):e1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldstein BI, Carnethon MR, Matthews KA, et al. ; American Heart Association Atherosclerosis; Hypertension and Obesity in Youth Committee of the Council on Cardiovascular Disease in the Young . Major depressive disorder and bipolar disorder predispose youth to accelerated atherosclerosis and early cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2015;132(10):965–986. [DOI] [PubMed] [Google Scholar]
- 32. Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–U162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang ZH, Zhou DY, Li HJ, et al. The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function, and mushroom dendritic spine. Mol Psychiatr 2020;25(1):48–66. [DOI] [PubMed] [Google Scholar]
- 34. Ikeda M, Takahashi A, Kamatani Y, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23(3):639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Neill CM, Minihane AM. The impact of fatty acid desaturase genotype on fatty acid status and cardiovascular health in adults. Proc Nutr Soc. 2017;76(1):64–75. [DOI] [PubMed] [Google Scholar]
- 36. Messamore E, Almeida DM, Jandacek RJ, McNamara RK. Polyunsaturated fatty acids and recurrent mood disorders: Phenomenology, mechanisms, and clinical application. Prog Lipid Res. 2017;66:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yao C, Chen G, Song C, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9(1):3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He B, Shi J, Wang XW, Jiang H, Zhu HJ. Genome-wide pQTL analysis of protein expression regulatory networks in the human liver. BMC Biol. 2020;18(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mogil LS, Andaleon A, Badalamenti A, et al. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14(8):e1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hernandez RD, Uricchio LH, Hartman K, Ye C, Dahl A, Zaitlen N. Ultrarare variants drive substantial cis heritability of human gene expression. Nat Genet. 2019;51(9):1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.