INTRODUCTION
Globally, neurological and mental disorders impact 1 in 3 people across their lifetime.1 Uniquely positioned to improve imaging diagnosis and clinical management for patients with brain diseases, artificial intelligence (AI) is a computational model that parallels human performance on tasks, often without explicit programming. Several classes of AI have been studied extensively, including machine learning (ML) and deep learning (DL), which utilizes artificial neural networks (NN) inspired by neuronal architectures.2,3
Imaging research in AI has grown exponentially. As of 2019, there are ~9,000 PubMed indexed articles that match the search on AI and imaging/radiology and ~5,000 articles on AI and neuroimaging/neuroradiology (Figure 1). Consequently, a thorough discussion on the technology and applications of AI in radiology is beyond the scope of this review and can be found elsewhere.4–8 Discussion of AI for brain tumor, infarct and hemorrhage imaging can be found elsewhere in this issue. Because of the ability of NNs to learn any input-output function,9 whether based on imaging or not, the potential applications of NNs to radiology span not only image-based tasks but also the many non-image-based tasks that radiologists perform.
Figure 1.
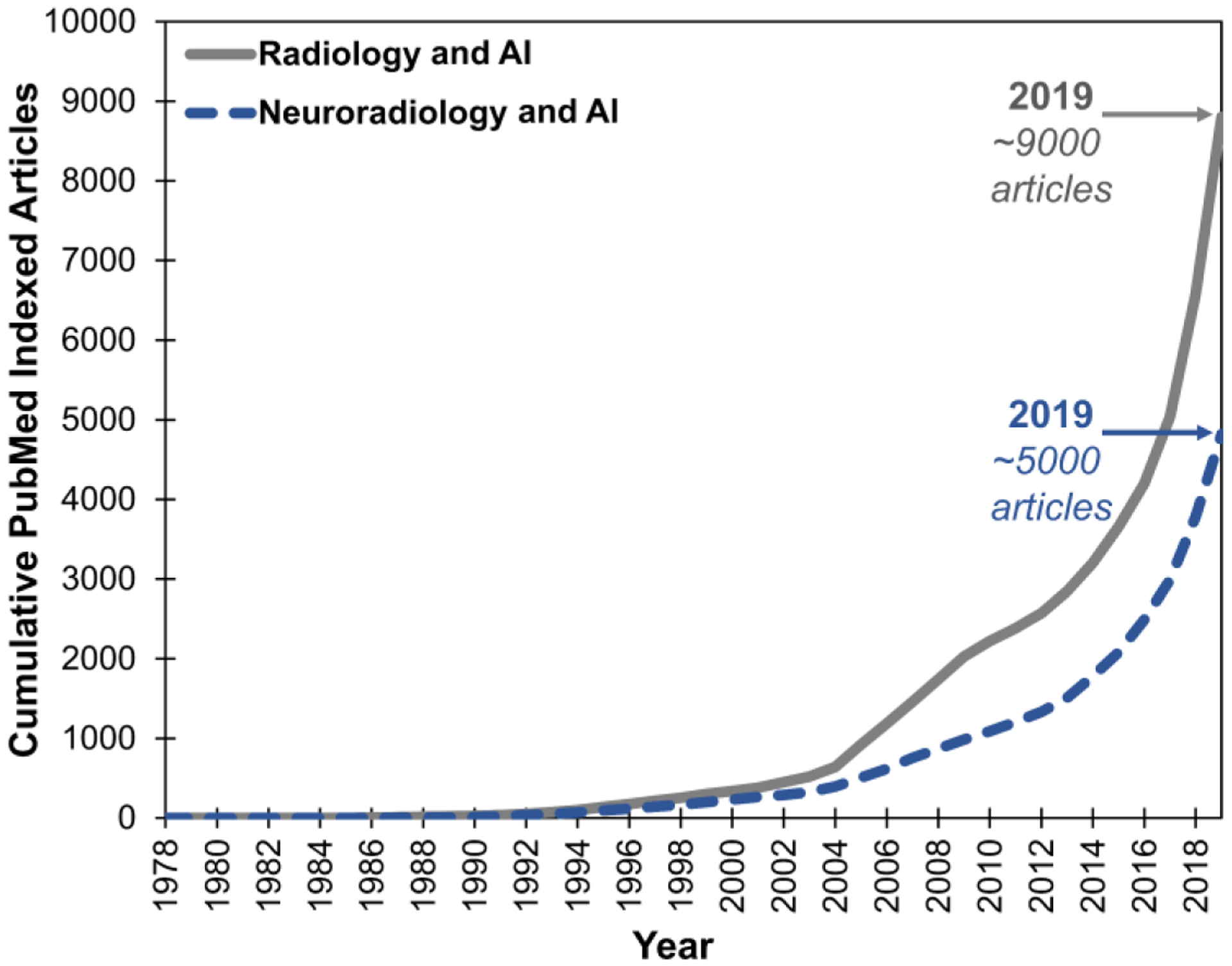
Cumulative frequency of PubMed indexed articles on AI and imaging/radiology (grey line) and AI and neuroimaging/neuroradiology (blue dashed line). Boolean search query for radiology was “AI/ML/DL/NN and clinical/medical and imaging/radiology/body” and for neuroradiology was “AI/ML/DL/NN and clinical/medical and neuroimaging/neuroradiology/brain.” PubMed last accessed November 27, 2019.
Here, we overview a diverse array of current neuroimaging applications of AI, including tasks such as worklist prioritization, lesion detection, anatomical segmentation, safety, quality and precision education, in a diverse set of diseases ranging from neurodegeneration to trauma. Then, we introduce emerging applications, including multimodal integration for neuroradiology. Finally, we discuss challenges and potential solutions for widespread adoption of AI in neuroimaging clinics worldwide.
DIVERSE APPLICATIONS FOR AI IN NEURORADIOLOGY
Throughout the review, we survey a variety of current and emerging applications for AI, ML and DL towards a range of neurological disorders, from trauma to multiple sclerosis, epilepsy to neurodegeneration. Compared to previous reviews of AI in neuroimaging organized by technical developments and AI methods, here our review is organized by clinical tasks relevant to neuroradiology.
1. WORKLIST PRIORITIZATION
Due to the unique structure, function and location of the brain, diagnosis of neurological diseases is largely dependent on imaging. Time from imaging to proper diagnosis and management determines patient outcomes for acute and chronic neurological diseases.10–12
Patients presenting with acute ischemic stroke progress in a sequence from door to imaging to diagnosis and intervention. Shorter door-to-puncture time for intravenous thrombolysis and/or mechanical thrombectomy are associated with better long-term functional outcomes.13,14 One solution to improve door-to-puncture times and outcomes is to reduce imaging-to-diagnosis time. This may be implemented with standardized, automated worklist prioritization. Natural language processing (NLP) algorithms and automated lesion detection networks (described in the next section) may augment efficiency in triaging imaging studies of patients with acute, time-sensitive presentations. NLP and NNs have already been applied to interpretation of chest computed tomography (CT) reports15 and abdominal CT and pathology reports16,17 and such tools may be similarly applied to head CT. Future iterations of such automated detection algorithms may be integrated into radiology workflows to improve efficiency. Indeed, current AI tools on the market can send processed imaging data such as CT perfusion maps via text message to stroke physicians and neurointerventional radiologists in real-time. Further, automated segmentation has been applied to workflow integration for patients presenting with intracranial hemorrhage and such networks could be applied to all patients with head CTs presenting with acute neurologic deficits.18 Despite the high accuracies attained by such networks for detecting findings, challenges of integrating these algorithms into the clinical workflow remain. For example, some intracranial hemorrhages are expected (such as after craniotomy), while others may be unexpected, and therefore much more urgent. Integrating such clinical context into the prioritization score is essential for ensuring that potentially more urgent studies are not de-prioritized.
Worklist prioritization may also improve radiology education. Since radiology trainee performance increases with optimal case exposure and volume,19 AI may assign specific cases to trainees based on their training profile, to promote consistency in individual trainee experiences.20
2. LESION DETECTION
Neurological diagnoses are made through clinical assessments of patient function and detection and interpretation of lesions on neuroimaging. Furthermore, lesion burden is often associated with functional impairment. Hence, accurate detection and characterization of lesions on neuroimaging is vital to expedite diagnosis and management. Across a variety of brain diseases, there are many research applications for AI in segmentation and quantification of image features.
Trauma
Trauma is a common indication for neuroimaging. DL models such as convolutional neural networks (CNNs) have been designed to detect spinal cord contusion injuries. Demonstrating clinical utility, the quantified injury volumes correlate well with motor impairment.21 Many methods utilize CNNs to detect and classify hemorrhages such as epidural and subdural hematomas and subarachnoid and intraparenchymal hemorrhage on head CT scans.22,23 Additionally, ML techniques such as random forest models have been applied to assess white matter hyperintensities in patients with traumatic brain injury.24
Metastatic Disease
Brain metastases occur in about 20% of patients with cancer. Metastatic disease encompasses a clinically and radiographically heterogeneous set of diseases that often has distinct imaging features and warrants management unique from primary tumors. For example, mucinous metastases can have T2w signal hypointensities, metastatic melanoma can display T1w signal hyperintensities and metastatic breast cancer can demonstrate necrotic and cystic lesions.25 Using the Brain Tumor Segmentation (BraTS) challenge datasets, recent DL methods have assessed metastases in multisequence magnetic resonance (MR) studies, including fluid‐attenuated inversion recovery (FLAIR) and pre‐ and post-gadolinium enhanced T1w images.26–28 Such networks are therefore useful for stereotactic surgical planning.
Multiple Sclerosis
Multiple sclerosis (MS) diagnosis and progression depends on neuroimaging, wherein patients present with white matter lesions disseminated over space and time.29 Hence, AI can be extremely beneficial for the quantification and tracking of specific lesions in individual patients. A variety of research AI models (including NNs, k-Nearest Neighbors and support vector machines (SVM)) exist for detecting MS lesions with high specificity and sensitivity.30–32
Diverse Etiologies
Overall, many pathologies demonstrate hyperintense lesions on FLAIR sequence. Methods exist to segment white matter hyperintensities in diseases such as Alzheimer Disease (AD) and small vessel ischemic disease.33,34 However, in the clinical workflow, the diagnosis that a patient presents with is not always known, and methods are required to recognize abnormalities despite diagnostic uncertainty. Recently, our group has shown that a CNN with U-net architecture can perform at near radiologist-level accuracy for detecting lesions across a variety of pathologies, at fractions of the time.35 Such general task-specific, rather than disease-specific, approaches have the potential to be applied universally to neuroimaging. Ultimately, such methods may decrease the rate of perceptual errors in neuroradiology, which are the basis of most diagnostic errors.36,37
3. ANATOMIC SEGMENTATION AND VOLUMETRY
Neuroanatomy is variable across individuals and difficult to segment computationally, particularly when anatomy is distorted by underlying pathology. However, AI has made significant inroads in the segmentation and parcellation of cortical and subcortical structures.38,39 Such anatomical classification AI could be useful for planning stereotactic radiosurgery and surgical resection for pathologies from neoplasia to epileptogenic foci and heterotopias.
Atrophy is essential in diagnosing certain neurodegenerative diseases and AI is well-positioned to enable quantification of anatomical changes. CNNs and SVMs can compare volumetrics of different brain regions including the hippocampus in dementia40–43 and basal ganglia in Huntington disease.44,45 For white matter diseases, current DL networks can detect and measure abnormalities perform volumetrics.34,35 In the future, these tools can be combined with enhanced anatomic linking to calculate changes over time.
4. PATIENT SAFETY AND QUALITY IMPROVEMENT
Protocoling
In addition to diagnosis, patient safety and image quality measures can be improved with AI. The first step of an imaging exam involving a radiologist is often the study protocoling, which can have a major impact on diagnostic accuracy at the time of image interpretation. Methods are being developed for standardizing and improving the protocoling process.46
Contrast Agents
Contrast agents and radiotracers pose non-zero risks to patients. Recent studies suggest as few as two doses of gadolinium-based contrast agents can lead to gadolinium deposition in dentate nuclei, globi pallidi and other deep brain structures.47 Accordingly, CNNs may reduce the gadolinium dose required to interpret enhancing lesions on brain MRI48,49 and even radiation exposure on CT.50,51
For nuclear medicine studies, DL algorithms are being developed to create “synthetic” amyloid positron emission tomography (PET) to reduce radiotracer volume injected. By training CNNs on PET images with lower signal-to-noise ratios, several studies have predicted how amyloid PET images would appear if the patient were given a full radiotracer dose based on PET images representing much lower (<1%) doses of tracer.52–55 Additional studies leverage reconstruction PET with generative adversarial networks (GANs) to promote more rigorous training of AI systems for diverse inputs.56
The clinical impact of such approaches remains to be rigorously studied. While the technical achievements of these dose-reduction algorithms are considerable, the underlying principles of information theory cannot be understated. That is, an AI algorithm cannot create signal if the information is not present in the first place. While much of MR image information is redundant, it is likely that in some unexpected cases, the administration of contrast adds information that cannot be gleaned from the remainder of the sequences. To what extent denoising algorithms will provide the correct predictions in these minority cases remains to be seen. Thus, before their clinical implementation, such algorithms must be diligently evaluated across a diverse spectrum of cases in the real clinical workflow.
Image Reconstruction
Once raw data are acquired, the information contained in sensor space must be reconstructed into an image. Understanding this process as a function that maps an input (sensor space) to an output (the image) reframes this problem as one that could be solved by an AI algorithm, bypassing traditional reconstruction algorithms including the traditional Fourier transform from K-space. Indeed, the AUTOMAP method was recently developed to perform this mapping.57 While promising, such new methods will still need to be evaluated in the context of clinical MRIs to determine that they do not compromise patient safety.
Recall Rates
Proper diagnosis depends on image quality. AI can improve the speed and accuracy of diagnosis by reducing recalls and rescans due to artifacts such as motion. The requisite image quality for different scans depends on the indication and the reading radiologist. Therefore, AI for patient diagnosis should be adapted for each patient and radiologist. Indeed, a recent study demonstrated how DL models can reduce recall and rescan rates for imaging studies of MS and stroke.58
5. PRECISION MEDICAL EDUCATION
“Precision medical education” is the use of tools such as AI to standardize trainees’ medical skills and knowledge yet also individualize the delivery and practice of those skills and knowledge. In line with this paradigm, our institution has developed the Adaptive Radiology Interpretation and Education System (ARIES).59,60 ARIES is a Bayesian network where trainees can select a sub-system network (such as spine), input imaging and clinical features (such as T1 intensity, diffusion restriction and age) and receive a differential diagnosis from expert-derived prior probabilities. Probabilities can be computed from radiographic features alone or with radiographic and clinical data, allowing for a nuanced discussion of diagnosis.
For example, if a patient presents with deficits localized to the cervical spinal cord and imaging reveals a single, homogeneously enhancing and expansile intramedullary lesion that does not have diffusion restriction, the differential diagnosis includes ependymoma and astrocytoma (Figure 2). While imaging features may favor ependymoma, clinical features such as patient age (under 30) and gender (male) may favor astrocytoma.
Figure 2.
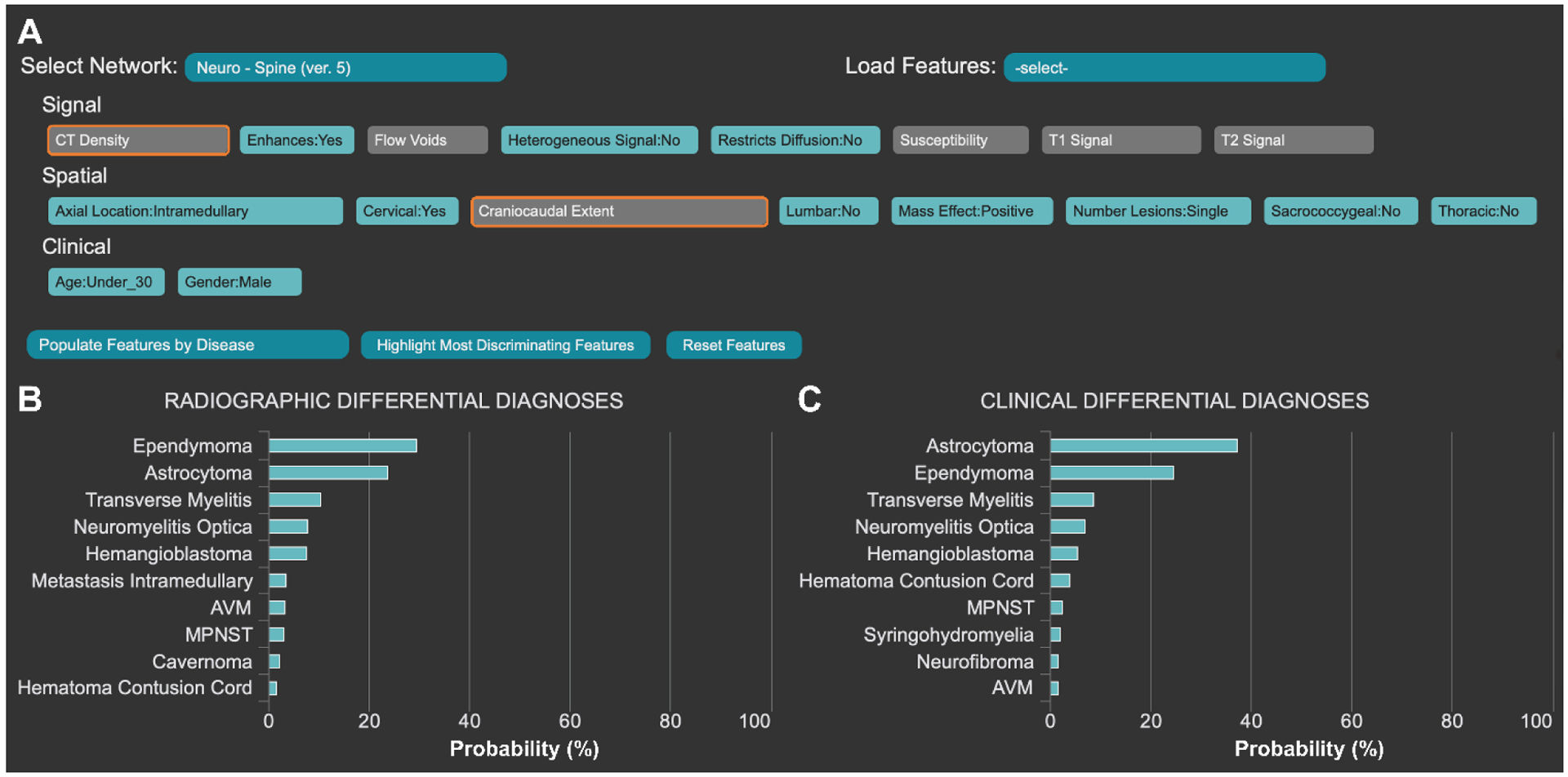
The Adaptive Radiology Interpretation and Education System (ARIES) distinguishes between ependymoma and astrocytoma in the spine network. (A) Features based on signal, spatial and clinical information are selected by the trainee in blue. Unselected features are grey and the most differentiating unanswered features are highlighted in orange. Differential diagnoses by (B) imaging features only vs (C) a combination of clinical and imaging features are derived from manually selected features (A). Probabilities are calculated by a naïve Bayes network.
Bayesian networks can be integrated with lesion and anatomical segmentation methods and teaching files. These AI-augmented teaching databases can create learning profiles for individual trainees, keep track of progress, and recommend relevant cases for trainees to achieve learning milestones.20,61 Studies have shown that AI can assist education through optimally spaced repetition62 and the same may be true for practice case-based learning and high-fidelity simulation training. Because there is little data that AI can improve student outcomes, validation studies must be performed. Notably, AI may hold utility even for people who do not initially benefit from AI (Figure 3). For example, AI may enable adaptive learning for people who do worse or not sufficiently well when using AI and assist in adapting itself until the trainee benefits.
Figure 3.
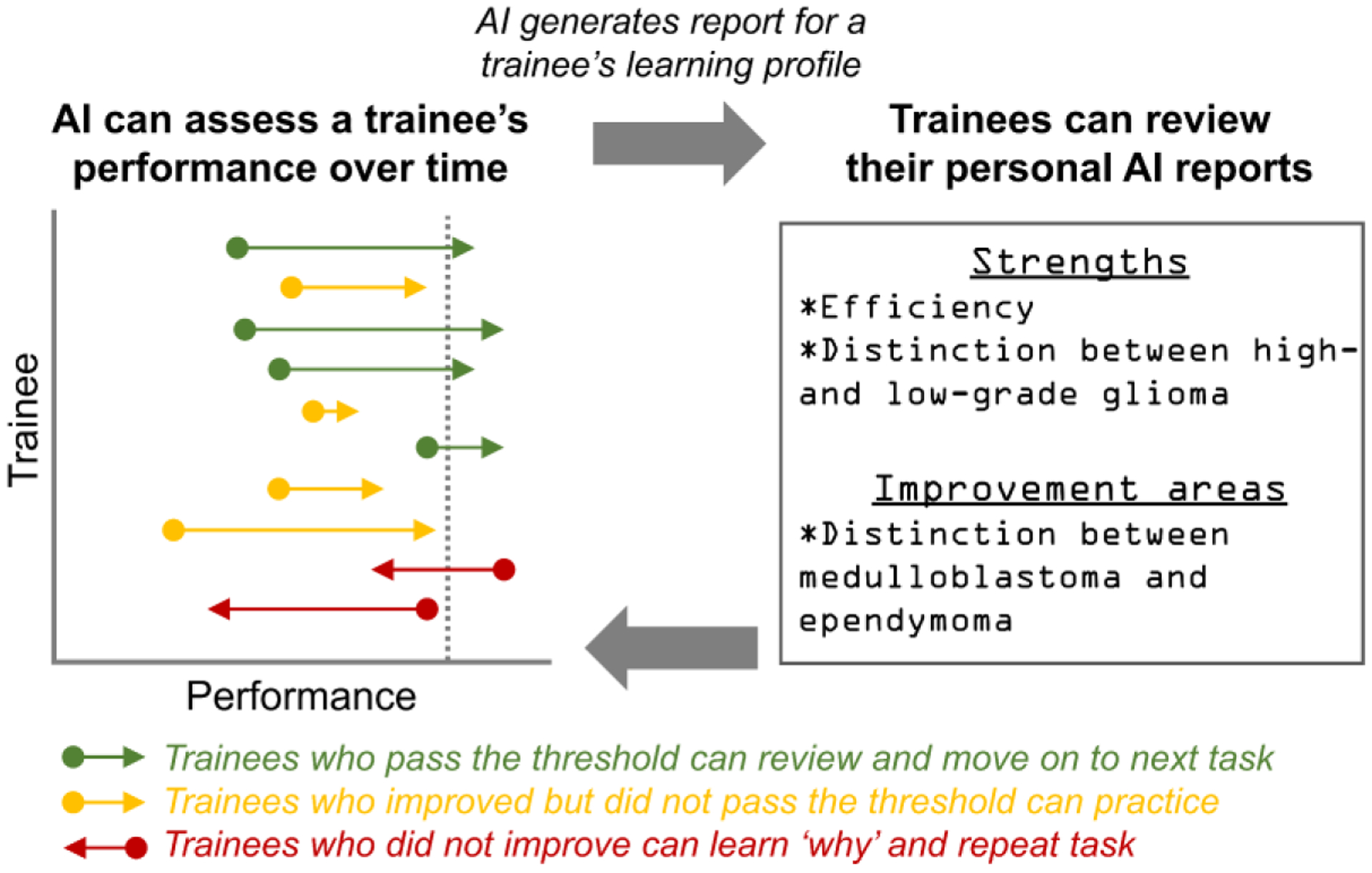
Feedback loop framework for longitudinal AI-augmented precision education.
It is important to emphasize that the usage of AI systems is not to scrutinize trainees or replace educators. Instead, AI should be used as a support to trainees and a supplement to teachers as medicine progresses towards precision education.
6. MULTIMODAL INTREGRATION
Combining multiple imaging modalities improves everyday clinical diagnosis, progression tracking and prognostication. Ongoing studies aim to apply similar models and architectures, trained on multiple forms of data track progress and predict prognosis for many chronic diseases.
Multiple Sclerosis
Diagnosis of MS incorporates detection of lesions from neurologic exams and imaging. Such correlation benefits from conventional to advanced imaging sequences.29 ML tools can integrate magnetization‐prepared rapid gradient‐echo (MPRAGE), magnetization‐prepared two inversion‐contrast rapid gradient‐echo (MP2RAGE), 3D FLAIR and double‐inversion recovery (DIR) for the detection of MS lesions.31 Additionally, joining NNs with graph theory approaches can classify distinct clinical subtypes of MS based on MR and diffusion tensor imaging (DTI).63
Integrated data may not only improve MS diagnosis but also management. Treatment response to disease-modifying biologic therapy such as natalizumab can be predicted with high dimensional ML models.64 In the future, AI used in longitudinal MRIs65 may be integrated with clinical assessment, CSF biomarkers and genetic susceptibility for better diagnosis and management of MS (Figure 4).
Figure 4.
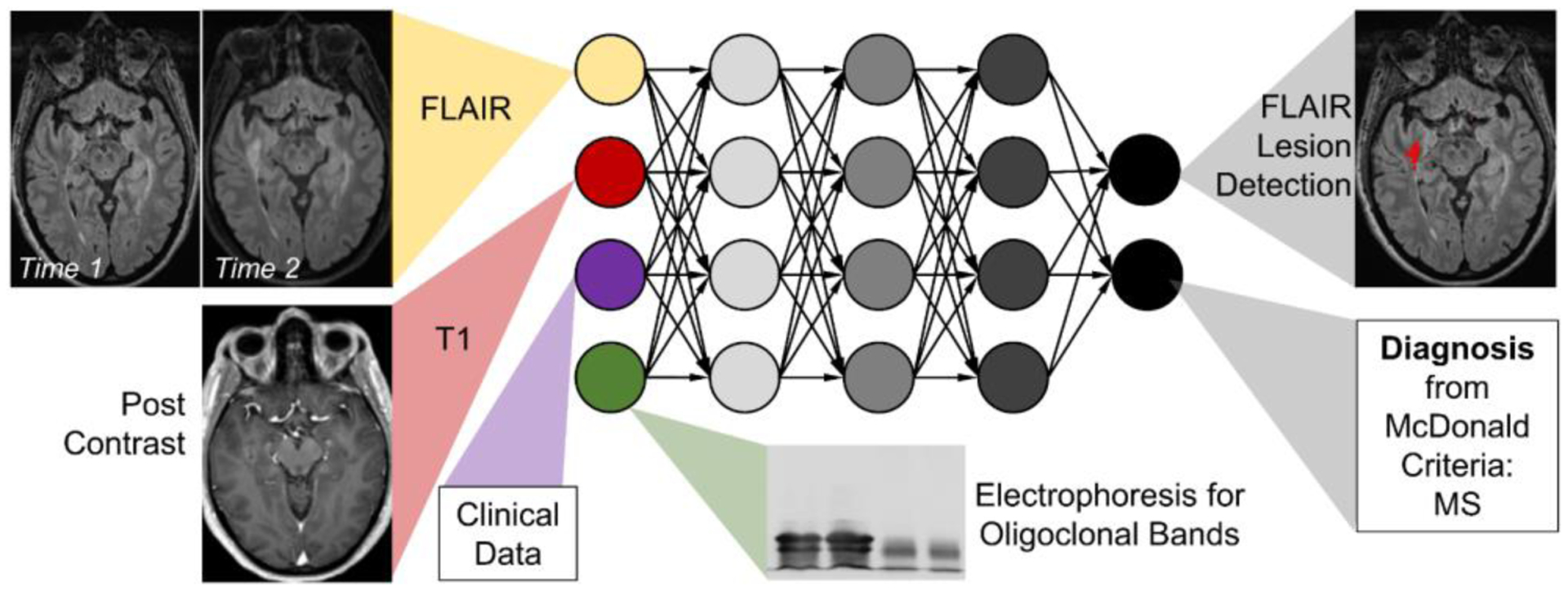
Model of multimodal integration of AI in diagnosis and assessment of disease progression in multiple sclerosis.
Epilepsy
Diagnosis and management in epilepsy may also be augmented with AI. Several SVMs have been shown to detect abnormalities in temporal lobe epilepsy with cortical morphology on T1w MRI and diffusion changes on DTI with kurtosis, mean diffusivity, and fractional anisotropy.66,67 We envision a future AI system to integrate neuroimaging with DL/ML tools for electroencephalography (EEG) event classification68–70 and DL-based wearable devices.71
Since timely anti-convulsant therapy is essential for improved outcomes in epilepsy,72 future AI may be applied to both diagnosis and management. DL models can predict treatment response to surgical resection.73 AI could serve as a resource for detecting ictal events on surface and intracranial EEG, localizing abnormal neurological foci on imaging, and even planning anti-epileptic drug therapy and surgical resection and in post-treatment monitoring (Figure 5).74
Figure 5.
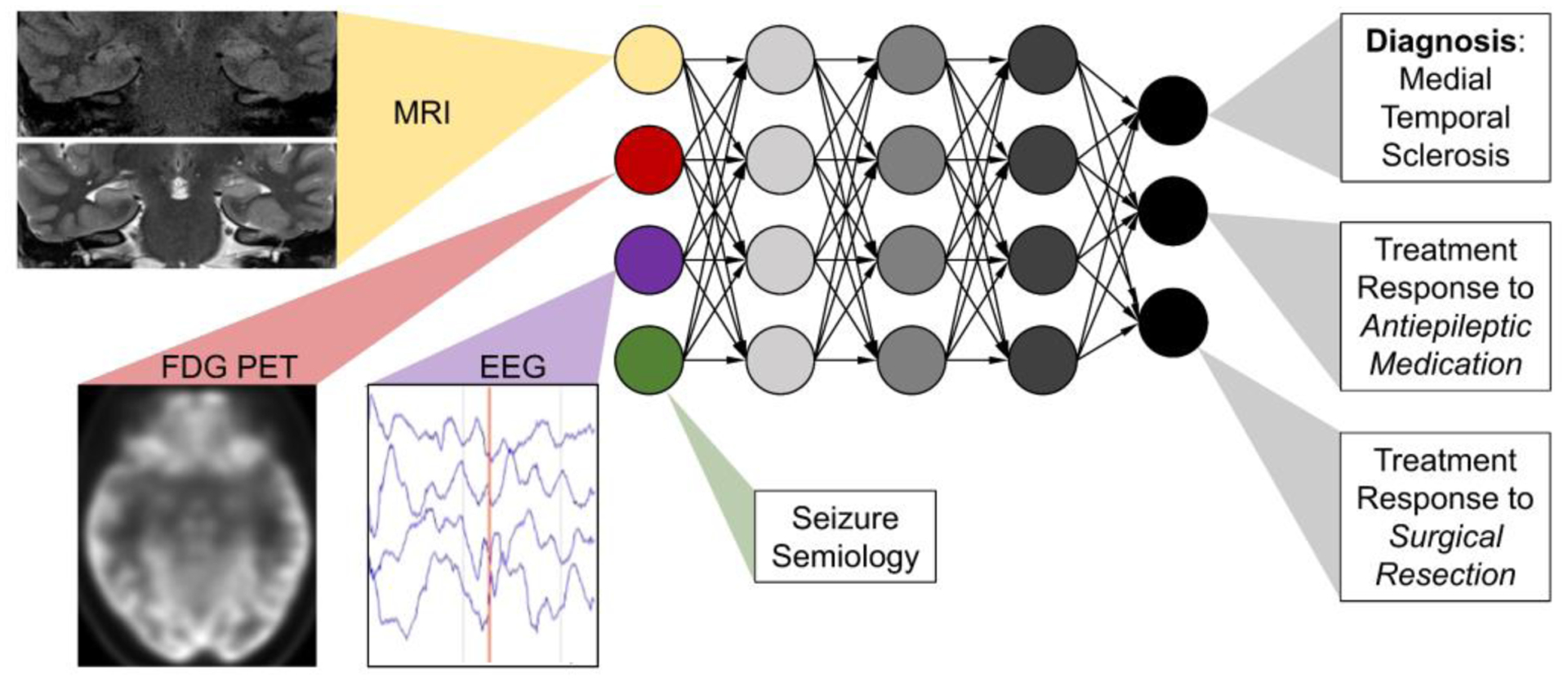
Model of multimodal integration in diagnosis and management of epilepsy. Bilateral mesial temporal sclerosis demonstrates increased signal and volume loss in hippocampal MRI, temporal lobe hypometabolism on 18F-FDG-PET and ictal events on EEG.
Neurodegenerative Disease
About 1 in 3 Americans over the age of 85 die with AD or related dementias.75 However, radiology is poised to improve the diagnosis and management of patients with dementia. Numerous AI networks have trained from large longitudinal datasets such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI), resulting in many diagnostic DL methods for AD, such as models using 18F-fluorodeoxyglucose (FDG) PET76 and structural MRI of the hippocampus77 to predict AD onset from 1 to 6 years in advance. Hence, AI may facilitate diagnosis and progression tracking in dementia.
The diagnosis of dementia types is not always straightforward and AI may assist in differential diagnosis. SVMs can distinguish AD from Lewy body and Parkinson’s dementia.78 Similarly, CNNs can differentiate mild cognitive impairment (MCI) and AD.79 In addition to diagnosis, progression and prognosis, integrative AI can also probe neurobiology. New ML techniques such as Subtype and Stage Inference (SuStaIn) have provided novel neuroimaging and genotype data-driven classifications of diagnostic subtypes and progressive stages for AD and frontotemporal dementia (FTD). SuStaIn has localized distinct regional hotspots for atrophy in different forms of familial FTD caused by mutations in genes including C9orf72, GRN and MAPT.80 Thus, neuroimaging AI tools may encourage novel neurobiological discoveries.
Because of the wide prevalence, long natural history and complex clinical landscape of neurodegenerative disease, the integration of many sources of imaging and clinical information is essential.81 DL has already integrated MRI, neurocognitive and APOE genotype information to predict conversion from MCI to AD.82 Combining several AI systems (including structural MRI and amyloid PET) together may augment diagnosis and management along the complex natural history of AD (Figure 6). We envision that AI tools for imaging will be integrated with AI systems designed to examine serum amyloid markers,83,84 mortality prediction from clinicians’ progress notes and assessments of cognition85 and postmortem immunohistochemistry images.86 This comprehensive suite of AI models may improve many facets of care in neurodegenerative disease.
Figure 6.
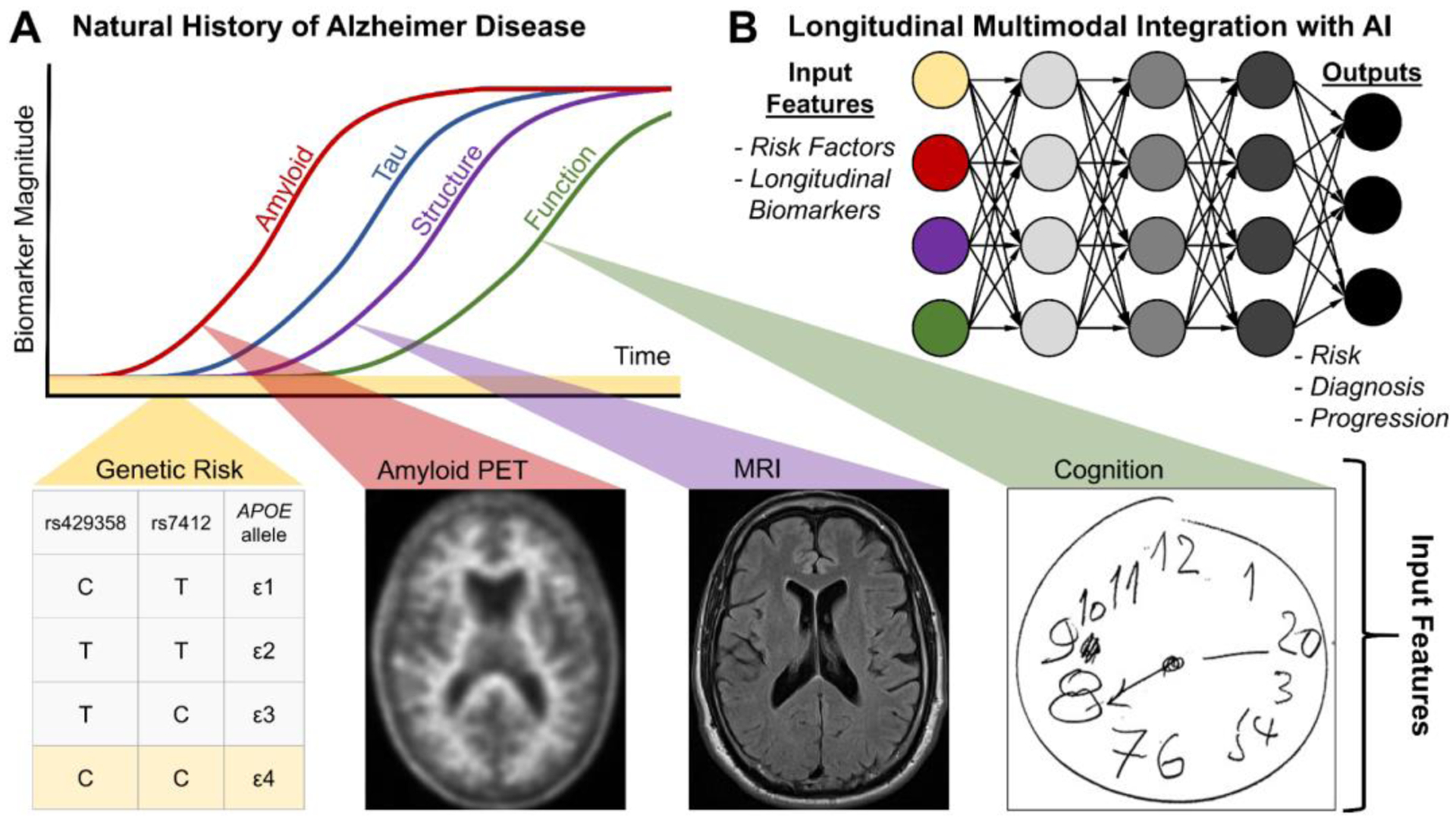
Model of multimodal integration of AI for diagnosis and management in Alzheimer disease. (A) Natural history of Alzheimer disease (adapted from81). (B) AI-enabled multimodal integration of input features for longitudinal (re)assessment of risk, diagnosis and progression.
In Huntington Disease, an autosomal dominant movement disorder, diagnosis and management may be enhanced by incorporating CAG repeat length data with NNs developed for caudate volumetry,45 objective gait assessment87 and EEG.88,89 Such multi-pronged approaches may improve risk stratification, progression monitoring and clinical management in patients and families.
7. TOWARDS A GENERALIZED NEUROIMAGING AI
Currently in its infancy stages, AI has potential to support neuroradiological diagnosis in drafting reports for radiologists to review. Yet, differential diagnosis in neuroradiology is particularly complex. A “generalized” AI capable of end-to-end neuroimaging diagnosis (where inputs are imaging studies and the output is a draft report) will require abilities to: extract imaging features, process clinical notes, parcellate neuroanatomy, construct differential diagnoses, recognize potential sources of artifact, compare with prior studies and more. Future development of more inclusive AI systems may go beyond imaging quantification and incorporate studies from nuclear medicine, electrophysiology, laboratory findings and clinical data. Such generalized AI tools may put these quantified numbers in perspective and eventually generate a comprehensive radiology report, which can be used to make informed clinical decisions in multi-disciplinary discussions. This will require the integration of many different AI systems trained on a vast wealth of neurological conditions.
Even if achievable, an end-to-end AI diagnostic resource for clinicians and radiologists will still require substantial human oversight and supervision. It is essential that radiologists stay in the loop on all aspects of imaging, from ordering the correct study to supervising its quality and interpretation. The significance of the question asked by an imaging order, and its clinical context, cannot be understated; it is currently difficult to imagine an AI algorithm able to account for the unique context in which an imaging study is ordered for a patient. Additionally, as data specialists, radiologists must provide continual, real-time feedback to update the AI system.90 This enables physicians to serve as part of the “checks and balances” between human and machine.
Current research trends (Figure 1) do suggest that early prototypes of such a generalized AI are on the horizon. These proposed integrated and knowledgeable AIs build upon existing multi-organ system AI systems.91,92 An effective generalized AI should also promote precision medical education and produce information that is “interpretable” and “explainable” for trainees and physicians alike.93 While AI is still far from generalized utility, it holds promise to one day improve neuroimaging at all levels, from processing and diagnosis to education and management.
DISCUSSION
AI has myriad neuroimaging applications. Here, we presented several clinical tasks well-suited for AI (including workflow organization, lesion and anatomical segmentation, quality, safety and multimodal integration), for a variety of neurological diseases (trauma, epilepsy, MS, neurodegeneration, etc.)
However, there are many challenges facing AI for such applications. First, effective AI requires immense data for training and validation. Many current neuroimaging databases are disease-specific (such as ADNI or BraTS), so it will be important to integrate many regional and international imaging networks to sufficiently train AI for a variety of diagnoses and patient populations. Second, multimodal AI tools are computationally costly, and significant investment in infrastructure is required. Third, not only are current AI systems all uniquely trained, but they are also uniquely designed. Rigorous best practices are required in designing and validating AI networks. For multimodal integration, it is important to assess the effect of each individual input modality toward output detection and/or diagnosis. Hence, strategies such as “leave-one-out” and accuracy metrics for segmentation and diagnosis should be standardized.94 Fourth, clinical AI must balance accuracy and explainability. While clinical decision support systems rely on known features and algorithms, current DL methods are still “black boxes.” Rigorous methods should be developed to allow for better model introspection, which fall under the recent explainable AI methods; such methods are beyond the scope of this review but will be essential for adoption of AI in healthcare.93,95 Indeed, our group has demonstrated that an AI system combining deep learning and Bayesian networks for brain MRI can generate accurate and interpretable output such as differential diagnoses and lesion characteristics across a variety of neurological disorders96,97. Fifth, adoption will depend on demonstrated improvement in patient outcomes and radiologist efficiency and collective agreement on the benefits of AI. This will require studies designed similar to clinical drug trials for trainees and attending radiologists. Akin to phase 4 studies, AI tools must be monitored after adoption to fine-tune algorithms. Sixth, physicians must be trained in using AI to recommend such algorithm adjustments. It is likely that an emerging subset of physicians will work with the informatics community to continually monitor and update AI tools. Overall, AI has many diverse applications in neuroimaging currently being validated across the new landscape of precision medicine.
KEY POINTS.
Artificial intelligence (AI) has potential to improve efficiency and accuracy in neuroradiology.
AI tools are starting to be validated for tasks such as workflow optimization, segmentation and precision education for a variety of brain diseases.
Future AI may integrate imaging data with additional diagnostic data from the medical record.
SYNOPSIS.
Recent advances in artificial intelligence (AI) and deep learning (DL) hold promise to augment neuroimaging diagnosis for patients with brain tumors and stroke. Here, the authors review the diverse landscape of emerging neuroimaging applications of AI, including workflow optimization, lesion segmentation, and precision education. Given the many modalities used in diagnosing neurologic diseases, AI may be deployed to integrate across modalities (MR imaging, computed tomography, PET, electroencephalography, clinical and laboratory findings), facilitate crosstalk among specialists, and potentially improve diagnosis in patients with trauma, multiple sclerosis, epilepsy, and neurodegeneration. Together, there are myriad applications of AI for neuroradiology.
DISCLOSURE STATEMENT
SM has research grants from Galileo CDS and Novocure. MTD and AMR have nothing to disclose.
REFERENCES
- 1.World Health Organization. Global health estimates 2015: Disease burden by cause, age, sex, by country and by region, 2000–2015. Geneva: 2016. http://www.who.int/healthinfo/global_burden_disease/estimates/en/index1.html [Google Scholar]
- 2.Hassabis D, Kumaran D, Summerfield C, et al. Neuroscience-Inspired Artificial Intelligence. Neuron 2017; 95:245–258. doi: 10.1016/j.neuron.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Kriegeskorte N. Deep Neural Networks: A New Framework for Modeling Biological Vision and Brain Information Processing. Annu Rev Vision Sci 2015;1:417–446. doi: 10.1146/annurev-vision-082114-035447 [DOI] [PubMed] [Google Scholar]
- 4.Erickson BJ, Korfiatis P, Akkus Z, et al. Machine learning for medical imaging. Radiographics 2017; 37: 505–15. doi: 10.1148/rg.2017160130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohli M, Prevedello LM, Filice RW, et al. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 2017;208:754–60. doi: 10.2214/AJR.16.17224 [DOI] [PubMed] [Google Scholar]
- 6.Viera S, Pinaya WHL, Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci Biobehav Rev 2017;74(A):58–75. doi: 10.1016/j.neubiorev.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 7.Zaharchuk G, Gong E, Wintermark M, et al. Deep Learning in Neuroradiology. AJNR Am J Neuroradiol 2018. doi: 10.3174/ajnr.A5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudie JD, Rauschecker AM, Bryan RN, et al. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 2019;290(3):607–618. doi: 10.1148/radiol.2018181928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural Networks 1989;2(5):359–366. doi: 10.1016/0893-6080(89)90020-8 [DOI] [Google Scholar]
- 10.Chowdhury FA, Nashef L, Elwes RD. Misdiagnosis in epilepsy: a review and recognition of diagnostic uncertainty. Eur J Neurol 2008;15:1034–1042. [DOI] [PubMed] [Google Scholar]
- 11.Solomon AJ, Bourdette DN, Cross AH, et al. The contemporary spectrum of multiple sclerosis misdiagnosis: A multicenter study. Neurology 2016;87:1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarnutzer AA, Lee SH, Robinson KA, et al. ED misdiagnosis of cerebrovascular events in the era of modern neuroimaging: A meta-analysis. Neurology 2017; 88: 1468–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lees KR, Bluhmki E, von Kummer R, et al. Time to treatment with intravenous alteplase and outcome in stroke: an updated pooled analysis of ECASS, ATLANTIS, NINDS, and EPITHET trials. Lancet 2010;375:1695–1703 doi: 10.1016/S0140-6736(10)60491-6 [DOI] [PubMed] [Google Scholar]
- 14.Saver JL, Goyal M, van der Lugt A, et al. Time to treatment with endovascular thrombectomy and outcomes from ischemic stroke: a meta-analysis. JAMA 2016;316:1279–1288. doi: 10.1001/jama.2016.13647 [DOI] [PubMed] [Google Scholar]
- 15.Chen MC, Ball RL. Yang L, et al. Deep Learning to Classify Radiology Free-Text Reports. Radiology 2017;286(3):845–852. doi: 10.1148/radiol.2017171115 [DOI] [PubMed] [Google Scholar]
- 16.Winkel DJ, Heye T, Weikert TJ, et al. Evaluation of an AI-Based Detection Software for Acute Findings in Abdominal Computed Tomography Scans: Toward an Automated Work List Prioritization of Routine CT Examinations. Invest Radiol 2019;54(1):55–59. doi: 10.1097/RLI.0000000000000509 [DOI] [PubMed] [Google Scholar]
- 17.Steinkamp JM, Chambers CM, Lalevic D, et al. Automated Organ-Level Classification of FreeText Pathology Reports to Support a Radiology Follow-up Tracking Engine. Radiology: Artificial Intelligence 2019;1(5):e180052. doi: 10.1148/ryai.2019180052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, et al. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. npj Digital Medicine 2018;1:9. doi: 10.1038/s41746-017-0015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bump GM, Heller MT, et al. Resident case volume correlates with clinical performance: finding the sweet spot. Acad Radiol 2019;26:136–140. doi: 10.1016/j.acra.2018.06.023 [DOI] [PubMed] [Google Scholar]
- 20.Duong MT, Rauschecker AM, Rudie JD, et al. Artificial intelligence for precision education in radiology. British J Radiology 2019;92:20190389. doi: 10.1259/bjr.20190389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy DB, Dupont SM, Gros C, et al. Convolutional Neural Network–Based Automated Segmentation of the Spinal Cord and Contusion Injury: Deep Learning Biomarker Correlates of Motor Impairment in Acute Spinal Cord Injury. AJNR Am J Neuroradiol 2019;40(4):37–44. doi: 10.3174/ajnr.A6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma B, Venugopalan K. Classification of hematomas in brain CT images using neural network. 2014 International Conference on Issues and Challenges in Intelligent Computing Techniques (ICICT). doi: 10.1109/ICICICT.2014.6781250 [DOI] [Google Scholar]
- 23.Chang PD, Kuoy E, Grinband J, et al. Hybrid 3D/2D Convolutional Neural Network for Hemorrhage Evaluation on Head CT. AJNR Am J Neuroradiol 2018;39(9):1609–1616. doi: 10.3174/ajnr.A5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone JR, Wilde EA, Taylor BA, et al. Supervised learning technique for the automated identification of white matter hyperintensities in traumatic brain injury. Brain Inj 2016;30:1458–1468. doi: 10.1080/02699052.2016.1222080 [DOI] [PubMed] [Google Scholar]
- 25.Achrol AS, Rennert RC, Anders C, et al. Brain metastases. Nature Reviews Disease Primers 2019:5. doi: 10.1038/s41572-018-0055-y [DOI] [PubMed] [Google Scholar]
- 26.Grøvik E, Yi D, Iv M, et al. Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. J Magnetic Resonance Imaging 2019. doi: 10.1002/jmri.26766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charron O, Lallement A, Jarnet D, et al. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018;95:43–54. doi: 10.1016/j.compbiomed.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Stojadinovic S, Hrycushko B, et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS ONE 12(10): e0185844. doi: 10.1371/journal.pone.0185844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson AJ, Banwell BL. Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 17(2):162–173. doi: 10.1016/S1474-4422(17)30470-2 [DOI] [PubMed] [Google Scholar]
- 30.Yoo Y, Tang LY, Brosch T, et al. Deep learning of joint myelin and T1w MRI features in normal-appearing brain tissue to distinguish between multiple sclerosis patients and healthy controls. Neuroimage Clin 2018;17:169–78. doi: 10.1016/j.nicl.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fartaria MJ, Bonnier G, Roche A, et al. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J Magn Res Imaging 2016;43:1445–1464. doi: 10.1002/jmri.25095 [DOI] [PubMed] [Google Scholar]
- 32.Lao Z, Shen D, Liu D, et al. Computer-assisted segmentation of white matter lesions in 3D MR images using support vector machine. Acad Radiol 2008;15:300–313. doi: 10.1016/j.acra.2007.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilello M, Doshi J, Nabavizadeh SA, et al. Correlating cognitive decline with white matter lesion and brain atrophy: magnetic resonance imaging measurements in Alzheimer’s disease. J Alzheimers Dis 2015;48:987–994. doi: 10.3233/JAD-150400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rachmadi MF, Valdés-Hernández MDC, Agan MLF, et al. Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Comput Med Imaging Graph 2018; 66: 28–43. doi: doi: 10.1016/j.compmedimag.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 35.Duong MT, Rudie JD, Wang J, et al. Convolutional neural network for automated FLAIR lesion segmentation on clinical brain MR imaging. AJNR Am J Neuroradiol 2019;40(8):1282–1290. doi: 10.3174/ajnr.A6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donald JJ, Barnard SA. Common patterns in 558 diagnostic radiology errors. J Med Imaging Radiat Oncol. 2012;56(2):173–178. doi: 10.1111/j.1754-9485.2012.02348.x [DOI] [PubMed] [Google Scholar]
- 37.Waite S, Scott J, Gale B, et al. Interpretive Error in Radiology. AJNR Am J Roentgenol 2017;208(4):739–749. doi: 10.2214/AJR.16.16963 [DOI] [PubMed] [Google Scholar]
- 38.Novosad P, Fonov V, Collins DL, et al. Accurate and robust segmentation of neuroanatomy in T1-weighted MRI by combining spatial priors with deep convolutional neural networks. Hum Brain Mapp 2019. doi: 10.1002/hbm.24803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wachinger C, Reuter M, Klein T. DeepNAT: deep convolutional neural network for segmenting neuroanatomy. Neuroimage 2018;170:434–45. doi: 10.1016/j.neuroimage.2017.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Shi F, Pu F, et al. Hippocampal Shape Analysis of Alzheimer Disease Based on Machine Learning Methods. AJNR Am J Neuroradiol 2007;28(7):1339–1345. doi: 10.3174/ajnr.A0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsao S, Gajawelli N, Zhou J, et al. Feature selective temporal prediction of Alzheimer’s disease progression using hippocampus surface morphometry. Brain Behav 2017;7(7):e00733. doi: 10.1002/brb3.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ataloglou D, Dimou A, Zarpalas D, et al. Fast and Precise Hippocampus Segmentation Through Deep Convolutional Neural Network Ensembles and Transfer Learning. Neuroinformatics 2019. doi: 10.1007/s12021-019-09417-y [DOI] [PubMed] [Google Scholar]
- 43.Xie L, Wisse LEM, Pluta J, et al. Automated segmentation of medial temporal lobe subregions on in vivo T1-weighted MRI in early stages of Alzheimer’s disease. Hum Brain Mapp 2019;40(12):3431–3451. doi: 10.1002/hbm.24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobbs NZ, Henley SM, Wild EJ, et al. Automated quantification of caudate atrophy by local registration of serial MRI: evaluation and application in Huntington’s disease. Neuroimage 2009;47(4):1659–1665. doi: 10.1016/j.neuroimage.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 45.Rizk-Jackson A, Stoffers D, Sheldon S, et al. Evaluating imaging biomarkers for neurodegeneration in pre-symptomatic Huntington’s disease using machine learning techniques. Neuroimage 2011;56(2):788–796. doi: 10.1016/j.neuroimage.2010.04.273 [DOI] [PubMed] [Google Scholar]
- 46.Brown AD, Marotta TR. A Natural Language Processing-based Model to Automate MRI Brain Protocol Selection and Prioritization. Acad Radiol. 2017;24(2):160–166. doi: 10.1016/j.acra.2016.09.013 [DOI] [PubMed] [Google Scholar]
- 47.Kang H, Hii M, Le M, et al. Gadolinium Deposition in Deep Brain Structures: Relationship with Dose and Ionization of Linear Gadolinium-Based Contrast Agents. AJNR Am J Neuroradiol 2018;39:1597–1603. doi: 10.3174/ajnr.A5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong E, Pauly JM, Wintermark M, et al. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reason Imaging 2018;48:330–40. doi: 10.1002/jmri.25970 [DOI] [PubMed] [Google Scholar]
- 49.Kleesiek J, Morshuis JN, Isensee F, et al. Can Virtual Contrast Enhancement in Brain MRI Replace Gadolinium?: A Feasibility Study. Invest Radiol 2019;54(10):653–660. doi: 10.1097/RLI.0000000000000583 [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Zhang Y, Zhang W, et al. Low-dose CT via convolutional neural network. Biomed Opt Express 2017;8:679–94. doi: 10.1364/BOE.8.000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. (2017) 44:1–32. doi: 10.1002/mp.12344 [DOI] [PubMed] [Google Scholar]
- 52.Chen KT, Gong E, Macruz FBdC, Xu J, Boumis A, Khalighi M, Poston KL, et al. Ultra–Low-Dose 18F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs. Radiology 2018;290(3):249–256. doi: 10.1148/radiol.2018180940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen KT, Schürer M, Ouyang J, et al. Quantitative brain imaging using integrated PET/MRI Investigating the optimal method to generalize an ultra-low-dose amyloid PET/MRI deep learning network across scanner models. J Cereb Blood Flow & Metab 2019;39(1S):113–114. doi: 10.1177/0271678X19850985 [DOI] [Google Scholar]
- 54.Kaplan S, Zhu YM. Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging. 2018. doi: 10.1007/s10278-018-0150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang L, Qiao Y, Nie D, et al. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing 2017;267:406–16. doi: 10.1016/j.neucom.2017.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ouyang J, Chen KT, Gong E, et al. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Medical Physics 2019. doi: 10.1002/mp.13626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu B, Liu JZ, Cauley SF, et al. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487–492. doi: 10.1038/nature25988 [DOI] [PubMed] [Google Scholar]
- 58.Sreekumari A, Shanbhag D, Yeo D, et al. A Deep Learning–Based Approach to Reduce Rescan and Recall Rates in Clinical MRI Examinations. AJNR Am J Neuroradiol 2019. doi: 10.3174/ajnr.A5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen P-H, Botzolakis E, Mohan S, et al. Feasibility of streamlining an interactive Bayesian-based diagnostic support tool designed for clinical practice. In: Zhang J, Cook TS, eds. Proceedings of SPIE Medical Imaging 2016. pp.97890C. [Google Scholar]
- 60.Duda JT, Botzolakis E, Chen P-H.et al. Bayesian network interface for assisting radiology interpretation and education. In: Zhang J, Chen P-H, eds. Proceedings of SPIE Medical imaging 2018. pp.26. [Google Scholar]
- 61.Chen P-H, Loehfelm TW, Kamer AP, et al. Toward data-driven radiology education-early experience building multi-institutional academic trainee interpretation log database (MATILDA). J Digit Imaging 2016;29:638–644. doi: 10.1007/s10278-016-9872-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabibian B, Upadhyay U, De A, et al. Enhancing human learning via spaced repetition optimization. Proc Natl Acad Sci USA 2019;116(10):3988–3993. doi: 10.1073/pnas.1815156116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marzullo A, Kocevar G, Stamile C, et al. Classification of Multiple Sclerosis Clinical Profiles via Graph Convolutional Neural Networks. Front Neurosci 2019;12:594. doi: 10.3389/fnins.2019.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanber B, Nachev P, Barkhof F, et al. High-dimensional detection of imaging response to treatment in multiple sclerosis. npj Digital Medicine 2019;2:49. doi: 10.1038/s41746-019-0127-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arani LA, Hosseini A, Asadi F, et al. Intelligent Computer Systems for Multiple Sclerosis Diagnosis: a Systematic Review of Reasoning Techniques and Methods. Acta Inform Med 2018;26(4):258–264. doi: 10.5455/aim.2018.26.258-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rudie JD, Colby JB, Salamon N. Machine learning classification of mesial temporal sclerosis in epilepsy patients. Epilepsy Res. 2015;117:63–69. doi: 10.1016/j.eplepsyres.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 67.Del Gaizo J, Mofrad N, Jensen JH, et al. Using machine learning to classify temporal lobe epilepsy based on diffusion MRI. Brain Behav 2017;7:e00801. doi: 10.1002/brb3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ullah I, Hussain M, Qazi E-u-H, et al. An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Systems Appl 2018;107:61–71. doi: 10.1016/j.eswa.2018.04.021 [DOI] [Google Scholar]
- 69.Zhou M, Tian C, Cao R, et al. Epileptic Seizure Detection Based on EEG Signals and CNN. Front Neuroinformatics 2018;12:95. doi: 10.3389/fninf.2018.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hossain MS, Amin SU, Alsulaiman M, et al. Applying Deep Learning for Epilepsy Seizure Detection and Brain Mapping Visualization. ACM Transactions on Multimedia Computing, Communications, and Applications. 2019;15(1s):10. doi: 10.1145/3241056 [DOI] [Google Scholar]
- 71.Kiral-Kornek I, Roy S, Nurse E, et al. Epileptic Seizure Prediction Using Big Data and Deep Learning: Toward a Mobile System. EBioMedicine 2018;27:103–111. doi: 10.1016/j.ebiom.2017.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krumholz A. Wiebe S, Gronseth GS, et al. Evidence-based guideline: Management of an unprovoked first seizure in adults: Report of the Guideline Development Subcommittee of the American Academy of Neurology and the American Epilepsy Society. Neurology 2015;84(16):1705–1713. doi: 10.1212/WNL.0000000000001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gleichgerrcht E, Munsell B, Bhatia S, et al. Deep learning applied to whole brain connectome to determine seizure control after epilepsy surgery. Epilepsia 2018;59:1643–1654. doi: 10.1111/epi.14528 [DOI] [PubMed] [Google Scholar]
- 74.Nagaraj V, Lee S, Krook-Magnuson E, et al. The Future of Seizure Prediction and Intervention: Closing the loop. J Clin Neurophysiol 2015;32(3):194–206. doi: 10.1097/WNP.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alzheimer’s Association. 2019 Alzheimer’s Disease Facts and Figures. Alzheimers Dement 2019;15(3):321–387. [Google Scholar]
- 76.Ding Y, Sohn JH, Kawczynski MG, et al. A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 2019;290(2):456–464. doi: 10.1148/radiol.2018180958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Habes M, Wolk DA, et al. A deep learning model for early prediction of Alzheimer’s disease dementia based on hippocampal magnetic resonance imaging data. Alzheimer’s & Dementia 2019;1–19. doi: 10.1016/j.jalz.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katako A, Shelton P, Goertzen AL, et al. Machine learning identified an Alzheimer’s disease-related FDG-PET pattern which is also expressed in Lewy body dementia and Parkinson’s disease dementia. Scientific Reports 2018; 8:13236. doi: 10.1038/s41598-018-31653-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basaia S, Agosta F, Wagner L, et al. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage: Clinical;21(2019):101645. doi: 10.1016/j.nicl.2018.101645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young AL, Marinescu RV, Oxtoby NP, et al. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nature Communications 2018;9:4273. doi: 10.1038/s41467-018-05892-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jack CR, Knopman DS, Jagust WJ, et al. Update on hypothetical model of Alzheimer’s disease biomarkers. Lancet Neurol 2013;12(2):207–216. doi: 10.1016/S1474-4422(12)70291-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spasov S, Passamonti L, Duggento A, et al. A parameter-efficient deep learning approach to predict conversion from mild cognitive impairment to Alzheimer’s disease. NeuroImage;189(2019):276–287. doi: 10.1016/j.neuroimage.2019.01.031 [DOI] [PubMed] [Google Scholar]
- 83.Ashton NJ, Nevado-Holgado AJ, Barber IS, et al. A plasma protein classifier for predicting amyloid burden for preclinical Alzheimer’s disease. Science Advances 2019;5(2):eaau7220 doi: 10.1126/sciadv.aau7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goudey B, Fung BJ, Schieber C, et al. A blood-based signature of cerebrospinal fluid Aβ1–42 status. Sci Rep 2019. March 11;9(1):4163. doi: 10.1038/s41598-018-37149-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L, Sha L, Lakin JR, et al. Development and Validation of a Deep Learning Algorithm for Mortality Prediction in Selecting Patients With Dementia for Earlier Palliative Care Interventions. JAMA Network Open 2019;2(7):e196972. doi: 10.1001/jamanetworkopen.2019.6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang Z, Chuang KV, DeCarli C, et al. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nature Communications 2019;10:2173. doi: 10.1038/s41467-019-10212-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ye Q, Xia Y, Yao Z. Classification of Gait Patterns in Patients with Neurodegenerative Disease Using Adaptive Neuro-Fuzzy Inference System. Comput Math Methods Med 2018:9831252. doi: 10.1155/2018/9831252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Odish OFF, Johnsen K, van Someren P, et al. EEG may serve as a biomarker in Huntington’s disease using machine learning automatic classification. Scientific Reports 2018;8:16090. doi: 10.1038/s41598-018-34269-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Tommaso M, De Carlo F, Difruscolo O, et al. Detection of subclinical brain electrical activity changes in Huntington’s disease using artificial neural networks. Clin Neurophysiol 2003;114(7):1237–1245. doi: 10.1016/s1388-2457(03)00074-9 [DOI] [PubMed] [Google Scholar]
- 90.Jha S, Topol EJ. Adapting to Artificial Intelligence: Radiologists and Pathologists as Information Specialists. JAMA 2016;316(22):2353–2354. doi:1001/jama.2016.17438 [DOI] [PubMed] [Google Scholar]
- 91.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 92.Binder T, Tantaoui EM, Pati P, et al. Multi-Organ Gland Segmentation Using Deep Learning. Front Med 2019;6:173. doi: 10.3389/fmed.2019.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holzinger A, Langs G, Denk H, Zatloukal K, Müller H. Causability and explainability of artificial intelligence in medicine. WIREs Data Mining Knowl Discov 2019;9(4):e1312. doi: 10.1002/widm.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walker EA, Petscavage-Thomas JM, Fotos JS, et al. Quality metrics currently used in academic radiology departments: results of the QUALMET survey. Br J Radiol 2017; 90: 20160827. doi: 10.1259/bjr.2016082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tjoa E, Guan C. A Survey on Explainable Artificial Intelligence (XAI): Towards Medical XAI. arXiv 2019:1907.07374v3 [DOI] [PubMed] [Google Scholar]
- 96.Rauschecker AM, Rudie JD, Xie L, et al. Artificial Intelligence System Approaching Neuroradiologist-level Differential Diagnosis Accuracy at Brain MRI. Radiology 2020;295(3):626–637. doi: 10.1148/radiol.2020190283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rudie JD, Rauschecker AM, Xie L, et al. Subspecialty-level deep gray matter differential diagnoses with deep learning and Bayesian networks on clinical brain MRI: a pilot study. Radiology: Artificial Intelligence 2020. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]


