Abstract
Acute myeloid leukemia (AML) is the most common diagnosed leukemia. In older adults, AML confers an adverse outcome1,2. AML originates from a dominant mutation, then acquires collaborative transformative mutations leading to myeloid transformation and clinical/biological heterogeneity. Currently, AML treatment is initiated rapidly, precluding the ability to consider the mutational profile of a patient’s leukemia for treatment decisions. Untreated patients with AML ≥ 60 years were prospectively enrolled on the ongoing Beat AML trial (ClinicalTrials.gov NCT03013998), which aims to provide cytogenetic and mutational data within 7 days (d) from sample receipt and before treatment selection, followed by treatment assignment to a sub-study based on the dominant clone. A total of 487 patients with suspected AML were enrolled; 395 were eligible. Median age was 72 years (range 60-92 years; 38% ≥75 years); 374 patients (94.7%) had genetic and cytogenetic analysis completed within 7 d and were centrally assigned to a Beat AML sub-study; 224 (56.7%) were enrolled on a Beat AML sub-study. The remaining 171 patients elected standard of care (SOC) (103), investigational therapy (28) or palliative care (40); 9 died before treatment assignment. Demographic, laboratory and molecular characteristics were not significantly different between patients on the Beat AML sub-studies and those receiving SOC (induction with cytarabine + daunorubicin (7 + 3 or equivalent) or hypomethylation agent). Thirty-day mortality was less frequent and overall survival was significantly longer for patients enrolled on the Beat AML sub-studies versus those who elected SOC. A precision medicine therapy strategy in AML is feasible within 7 d, allowing patients and physicians to rapidly incorporate genomic data into treatment decisions without increasing early death or adversely impacting overall survival.
Recurrent mutations seen in older adults with AML are also seen in healthy patients with age-associated clonal hematopoiesis and myelodysplastic syndromes (MDS), with the added cumulative number equating to advanced disease evolution3-6. The mutations that contribute to AML pathogenesis have distinct functional roles, including disrupted apoptosis (TP53), production of oncometabolites (IDH1, IDH2) with resultant epigenetic remodeling, oncogenic signaling (FLT3, KIT, NRAS, KRAS, PTPN11), and epigenetic dysregulation (DNMT3A, TET2, WT1, ASXL1). Use of next-generation sequencing (NGS) has informed our understanding of AML pathogenesis and prognosis, allowing for the characterization of the mutational repertoire of each patient with AML and the dominant leukemic clone.
Current intensive AML chemotherapy is associated with poor outcomes in the absence of allogeneic transplant; 15% of patients aged 18–59 years and 2% of those age ≥60 years are disease-free at 10 years7. Older patients treated with hypomethylating agents are not cured and have a median survival of 6.3 months8-11. To date, molecular data in AML have been used predominately for prognostication and treatment after induction therapy and not for initial therapeutic decisions, outside of the use of FLT3 inhibitors12,13. Conventional treatment of AML involves rapid initiation of therapy, within days, to reduce the perceived risk of death due to disease progression.
This expedited treatment initiation and inability to rapidly obtain mutational data has precluded a precision medicine approach for AML. A retrospective analysis demonstrated that a treatment delay for up to 8 d does not influence overall survival in AML14, but to date, no prospective study has ever examined the feasibility of treatment delay. Based on studies that assigned patients with FLT3 + AML with a delay of 2–3 d, we hypothesized that using cytogenetic and full NGS profiling to guide initial therapy in older patients with AML would be feasible. Based on this hypothesis, the Leukemia & Lymphoma Society (LLS) designed and implemented a precision medicine trial in AML, the Beat AML trial, to prospectively assess the feasibility of assigning treatment based on cytogenetic and molecular results for older patients with AML in ≤7 d.
Results
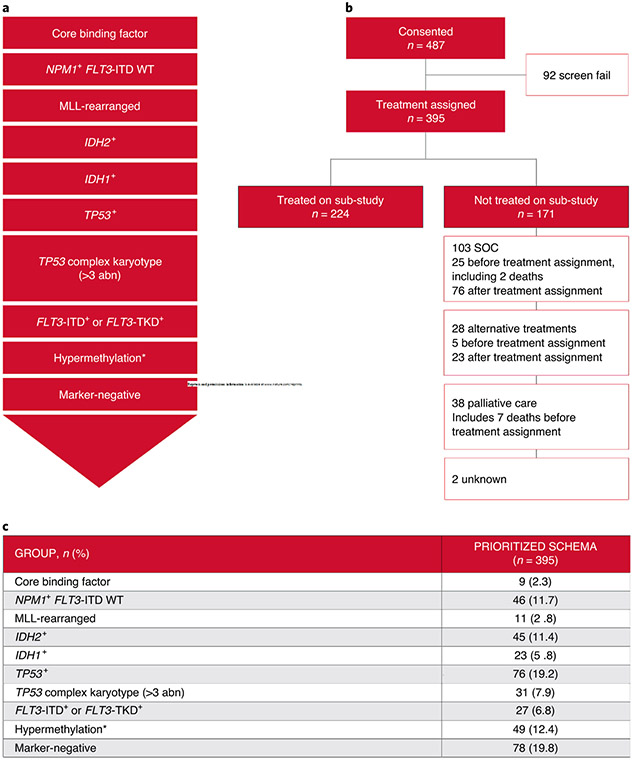
From November 2016 through to January 2019, 487 patients with suspected AML across 14 clinical sites consented to the Beat AML trial. For the top 4 enrolling sites this encompassed 65% (Ohio State University), 84% (Oregon Health & Science University), 40% (Memorial Sloan Kettering) and 80% (University of Maryland) of all eligible patients enrolled on trials at these sites. Of the 487 patients, 395 (81.1%) were eligible and received a treatment assignment; 92 were ineligible (Fig. 1b), most commonly due to an alternative diagnosis (for example, MDS or other) or comorbidities (Supplementary Table 1). The demographics of the 395 eligible patients are shown in Table 1.
Fig. 1 ∣. Overview of the Beat AML trial.
a, AML prioritization by genomic or cytogenetic abnormality into groups. b, Patient distribution following enrollment on the Beat AML trial. c, Genomic assignment of eligible patients with AML by prioritization group. The asterisk denotes the hypermethylation group, which is defined by the TET2/WT1 mutations.
Table 1 ∣.
Baseline characteristics at screening across the different eligible patient groups
| Characteristic | All eligible patients (n = 395) |
Beat AML sub-study (n = 224) |
SOC therapy (n = 103) |
investigational therapy (n = 28) |
Palliative care/unknown (n = 40) |
P1/P2 |
|---|---|---|---|---|---|---|
| Age (years) | 0.009/0.21 | |||||
| Median (range) | 72 (60–92) | 72(60–92) | 71 (60–87) | 71 (61–83) | 77.5 (60–91) | |
| Age ≥75 years, n (%) | 152 (38) | 87 (39) | 36 (35) | 7 (25) | 22 (55) | 0.07/0.54 |
| Sex, n (%) | 0.79/0.90 | |||||
| Female | 175 (44) | 101 (45) | 45 (44) | 10 (36) | 19 (48) | |
| Male | 220 (56) | 123 (55) | 58 (56) | 18 (64) | 21 (53) | |
| Ethnic group, n (%) | 0.94/1.00 | |||||
| White | 347 (90) | 195 (89) | 89 (89) | 26 (93) | 37 (93) | |
| African American | 18 (5) | 10 (5) | 6 (6) | 0 (0) | 2 (5) | |
| Asian | 13 (3) | 8 (4) | 3 (3) | 2 (7) | 0 (0) | |
| Other | 8 (2) | 5 (2) | 2 (2) | 0 (0) | 1 (3) | |
| Unknown | 9 | 6 | 3 | 0 | 0 | |
| Performance status, n (%) | 0.10/0.25 | |||||
| 0 | 83 (23) | 48 (22) | 23 (26) | 10 (36) | 2 (6) | |
| 1 | 183 (50) | 118 (54) | 38 (43) | 11 (39) | 16 (50) | |
| 2 | 90 (25) | 50 (23) | 23 (26) | 6 (21) | 11 (34) | |
| 3 | 9 (2) | 2 (1) | 4 (5) | 1 (4) | 2 (6) | |
| 4 | 1 (<1) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | |
| Not assessed/unknown | 29 | 6 | 15 | 0 | 8 | |
| Hemoglobin (g dl−1) | 0.18/0.61 | |||||
| Median (range) | 8.5 (3.9–15.0) | 8.5 (6.1–13.9) | 8.6 (4.9–15.0) | 8.7 (6.7–12.1) | 8.15 (3.9–12.0) | |
| Not assessed/unknown | 1 | 1 | 0 | 0 | 0 | |
| Platelet (109 l−1) | 0.71/0.45 | |||||
| Median (range) | 60 (5–656) | 59 (7–656) | 65 (5–427) | 60.5 (11–213) | 58.5 (12–320) | |
| Not assessed/unknown | 3 | 3 | 0 | 0 | 0 | |
| WBC (109 l−1) | 0.01/0.16 | |||||
| Median (range) | 4.85 (0.40–231.2) | 4.7 (0.6–194.1) | 6.0 (0.40–231.2) | 1.85 (0.5–49.9) | 8.5 (0.6–168.2) | |
| Not assessed/unknown | 1 | 1 | 0 | 0 | 0 | |
| WBC >50, n (%) | 40 (10) | 20 (9) | 16 (16) | 0 (0) | 4 (10) | 0.07/0.09 |
| Blood blasts, % | 0.21/0.25 | |||||
| Median (range) | 13.9 (0–98.4) (0–98) | 12 (0–96) | 21 (0–98) | 6(0–62) | 18 (0–85) | |
| Not assessed/unknown | 48 | 25 | 14 | 4 | 5 | |
| Bone marrow blasts (%) | 0.10/0.06 | |||||
| Median (range) | 46 (1–99) | 45 (1–98) | 51 (15–99) | 40.5 (3–76) | 42.5 (11–93) | |
| Not assessed/unknown | 26 | 13 | 7 | 2 | 4 | |
| Alanine aminotransferase, U l−1 | 0.76/0.31 | |||||
| Median (range) | 18 (3–203) | 17 (3–180) | 18 (5–203) | 18 (9–106) | 19.5 (5–50) | |
| Not assessed/unknown | 21 | 10 | 6 | 1 | 4 | |
| Aspartate aminotransferase, U l−1 | 0.15/0.93 | |||||
| Median (range) | 21 (6–256) | 21 (8–203) | 21 (6–256) | 18 (11–42) | 24 (9–79) | |
| Not assessed/unknown | 13 | 7 | 4 | 0 | 2 | |
| Bilirubin (mg dl−1) | 0.08/0.62 | |||||
| Median (range) | 0.6 (0.1–10.4) | 0.6 (0.1–2.9) | 0.6 (0.3–10.4) | 0.6 (0.3–3.5) | 0.8 (0.2–3.1) | |
| Not assessed/unknown | 10 | 5 | 3 | 0 | 2 | |
| Creatinine (mg dl−1) | 0.99/0.70 | |||||
| Median (range) | 0.89 (0.34–2.79) | 0.88 (0.34–2.50) | 0.91 (0.39–2.78) | 0.90 (0.44–2.32) | 0.84 (0.47–2.79) | |
| Not assessed/unknown | 7 | 6 | 0 | 0 | 1 | |
| Treatment-related AML, n (%) | 0.57/0.63 | |||||
| No | 331 (84) | 188 (84) | 83 (81) | 26 (93) | 34 (85) | |
| Yes | 63 (16) | 36 (16) | 19 (19) | 2 (7) | 6 (15) | |
| Unknown | 1 | 0 | 1 | 0 | 0 | |
| CBF, n (%) | 0.56/0.46 | |||||
| Absent | 369 (98) | 213 (98) | 91 (96) | 27 (100) | 38 (100) | |
| Present | 9 (2) | 5 (2) | 4 (4) | 0 (0) | 0 (0) | |
| Unknown | 17 | 6 | 8 | 1 | 2 | |
| MLL, n (%) | 0.20/0.14 | |||||
| Absent | 369 (97) | 215 (98) | 91 (95) | 27 (100) | 36 (95) | |
| Present | 11 (3) | 4 (2) | 5 (5) | 0 (0) | 2 (5) | |
| Unknown | 15 | 5 | 7 | 1 | 2 | |
| Complex cytogenetics, n (%) | 0.13/0.78 | |||||
| Absent | 245 (68) | 143 (69) | 63 (71) | 20 (77) | 19 (51) | |
| Present | 115 (32) | 65 (31) | 26 (29) | 6 (23) | 18 (49) | |
| Unknown | 35 | 16 | 14 | 2 | 3 | |
| FLT3-ITD, n (%) | 0.30/0.10 | |||||
| Absent | 357 (90) | 207 (92) | 89 (86) | 26 (93) | 35 (88) | |
| Present | 38 (10) | 17 (8) | 14 (14) | 2 (7) | 5 (13) | |
| NPM1 cytoplasmic, n (%) | 0.61/0.64 | |||||
| WT (VAF < 20%) | 326 (83) | 183 (82) | 87 (84) | 25 (89) | 31 (78) | |
| Mutated (VAF ≥ 20%) | 69 (17) | 41 (18) | 16 (16) | 3 (11) | 9 (23) | |
| IDH2, n (%) | 0.04/0.17 | |||||
| WT (VAF < 20%) | 332 (84) | 179 (80) | 89 (86) | 26 (93) | 38 (95) | |
| Mutated (VAF ≥ 20%) | 63 (16) | 45 (20) | 14 (14) | 2 (7) | 2 (5) | |
| IDH1, n (%) | 0.02/1.00 | |||||
| WT (VAF < 20%) | 357 (90) | 206 (92) | 95 (92) | 20 (71) | 36 (90) | |
| Mutated (VAF ≥ 20%) | 38 (10) | 18 (8) | 8 (8) | 8 (29) | 4 (10) | |
| TP53, n (%) | 0.57/1.00 | |||||
| WT (VAF < 20%) | 311 (79) | 178 (79) | 82 (80) | 23 (82) | 28 (70) | |
| Mutated (VAF ≥ 20%) | 84 (21) | 46 (21) | 21 (20) | 5 (18) | 12 (30) | |
| FLT3-TKD, n (%) | 0.88/0.73 | |||||
| WT (VAF < 20%) | 384 (97) | 220 216 (98) (96) | 101 (98) | 28 (100) | 39 (98) | |
| Mutated (VAF ≥ 20%) | 11 (3) | 4 8 (2) (4) | 2 (2) | 0 (0) | 1 (3) | |
| FLT3 (other), n (%) | 0.80/1.00 | |||||
| WT (VAF < 20%) | 389 (98) | 220 (98) | 102 (99) | 28 (100) | 39 (98) | |
| Mutated (VAF ≥ 20%) | 6 (2) | 4 (2) | 1 (<1) | 0 (0) | 1 (3) | |
| TET2, n (%) | 0.75/0.77 | |||||
| WT (VAF < 20%) | 312 (79) | 179 (80) | 81 (79) | 20 (71) | 32 (80) | |
| Mutated (VAF ≥ 20%) | 83 (21) | 45 (20) | 22 (21) | 8 (29) | 8 (20) | |
| WT1, n (%) | 0.53/0.53 | |||||
| WT (VAF < 20%) | 383 (97) | 217 (97) | 98 (95) | 28 (100) | 40 (100) | |
| Mutated (VAF ≥ 20%) | 12 (3) | 7 (3) | 5 (5) | 0 (0) | 0 (0) |
Fisher’s exact and Kruskal–Wallis tests were used to test for associations between elected treatment groups and categorical or continuous baseline characteristics, respectively. P values from global tests, including the four elected treatment groups, are represented by P1; P values from tests including two elected treatment groups (Beat AML sub-study versus SOC therapy) are represented by P2. All tests were two-sided and P values were not adjusted for multiple testing. CBF, core binding factor; TKD, tyrosine kinase domain; WT, wild type.
During the 7-d period before treatment assignment, patients who did not proceed to the sub-study therapy for the following reasons were considered to have an adverse event of special interest (AESI): (1) death before assignment from AML progression; (2) life-threatening bleeding (central nervous system, acute hemorrhage requiring transfusion); (3) AML progression requiring therapy before assignment; (4) worsening of performance status by two levels or new requirement for intensive care unit care, intubation or surgery related to leukemia (Supplementary Table 2). Treatment assignment was made centrally using a prioritization schema (Fig. 1a) incorporating cytogenetics and somatic mutations present in a dominant clone with a variant allele frequency (VAF) ≥ 0.3 or presence of FLT3-internal tandem duplication (ITD). If no cytogenetic abnormality or mutation with VAF ≥ 0.3 was observed, VAF ≥ 0.2 was used. Patients who did not meet the criteria for assignment based on genetic profiling were assigned to the marker-negative group.
The Beat AML trial was designed to be dynamic, with different arms opening and closing over time. The trial opened with 3 sub-studies and currently has 11 sub-studies (Supplementary Table 3). Patients were centrally assigned to either a genetically defined treatment, marker-negative or alternative therapy. Irrespective of participation in the assigned study, patients were followed for survival. Patients were encouraged to select an alternative therapy (alternative investigational therapy, SOC or palliative care) if the patient with their health-care providers deemed this a better option.
Of the 395 eligible patients, 374 (94.7%, two-sided 95% confidence interval (CI): 92.0–96.7%) were assigned treatment within 7 d of sample receipt. Among the sites participating, this ranged from 90.8% to 100% (Supplementary Fig. 1). Reasons for unsuccessful treatment assignment included suboptimal specimen quality, technical difficulties and instrumentation error. Treatment assignment based on our reported schema and the actual assignment are shown in Fig. 1c and Supplementary Table 4. The majority of patients (71.6%) received a specific treatment assigned by the prioritized schema (Supplementary Table 4); assignments that did not follow the schema were due to either the sub-study not being available, cytogenetic reassignment on central review or choice of an alternative therapy. Study availability affected all arms, but most frequently the marker-negative group where 146 (37%) were classified as marker-negative of whom only 75 truly would have fallen into this group if all treatment arms were open at study onset and throughout (Supplementary Tables 5 and 6). The proportion of patients assigned to the marker-negative group as a result of study availability decreased over time as more studies opened, from 70% in 2016–2017 to 21% in 2018–2019. For each prioritized treatment assignment group, mutational data including co-occurring mutations (at any detectable VAF) are provided in Supplementary Fig. 2a-j. These data show that there were few co-occurring dominant mutations that could have been used for an alternative therapeutic assignment. However, there were exceptions, such as the common co-occurrence of cytoplasmic NPM1 mutations with IDH1 and IDH2 mutations, where chemotherapy15-17 was assigned. In patients without a targetable alteration (‘marker-negative’), we observed a high frequency of ASXL1 (ref. 18), RUNX1 (ref. 19) and spliceosome mutations20,21 that may be amenable to targeted therapies in the future. These data confirm the feasibility of assigning treatment for newly diagnosed patients with AML within 7 d of sample receipt in a multicenter trial with comprehensive molecular profiling.
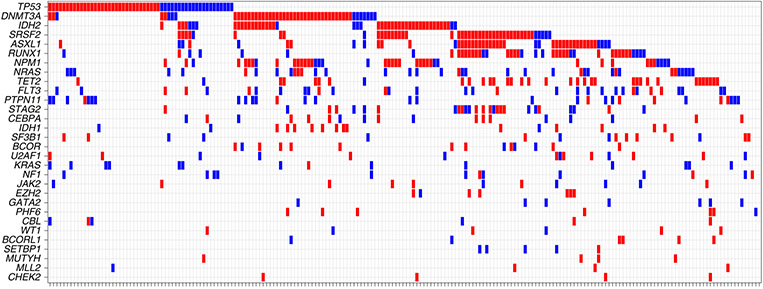
Figure 2 and Supplementary Table 7 show the mutational frequencies of the 30 most commonly mutated genes of eligible patients including DNMT3A (28.0%), TP53 (26.5%), ASXL1 (23.4%) and TET2 (23.2%). The frequency of these mutations and the ten most common mutated genes did not differ between patients screened, eligible for the master trial or treated on assigned sub-study treatment (Supplementary Table 8). When restricting analysis to mutations present at a VAF ≥ 0.3, consistent with their presence in the dominant clone, the most common mutational drivers were DNMT3A (22.7%), TET2 (19.6%), TP53 (19.1%), ASXL1 (19.1%) and SRSF2 (18.4%). Compared to studies in younger adults22-24, a higher frequency of TP53, ASXL1, TET2, RUNX1 and SRSF2 mutations, and a lower frequency of FLT3 and cytoplasmic NPM1 mutations was observed (Supplementary Table 8), highlighting differences in the mutational landscape of AML between older and younger patients.
Fig. 2 ∣. Comutation oncoprint of eligible Beat AML trial patients.
Genes with genomic alterations (short variants and insertions/deletions) are listed in descending order of frequency and each column represents an individual patient. Red indicates that the alteration was present at an allele frequency of ≥30% and blue <30%.
Delaying treatment assignment for up to 7 d to perform molecular profiling is only beneficial if safe and applicable to the vast majority of patients with preserved initial patient outcomes. AESIs identifying a potential decline in patient condition or AML progression requiring more urgent therapy occurred in 26 patients (Supplementary Table 2) in our study. Nine (2.3%) patients died, 32 (8.1%) began therapy before treatment assignment and 38 (9.6%) elected palliative care. The first two patients who died had MLL translocations with a rapidly rising white blood cell (WBC) count indicating disease progression that required urgent therapy within days. These data, coupled with analysis of MLL rearrangements in the Alliance7,15,25, prompted trial modification allowing treatment before full characterization for MLL-rearranged patients. Estimated mortality across all eligible patients at 30 d was 14.1% (95% CI 10.9–18.1%) and mortality was 7.5% (95% CI 5.1–10.9%) when excluding those opting for palliative care. From initial enrollment for screening, 30-d mortality was 3.7% (95% CI 1.9–7.2%) for patients electing to enroll on the Beat AML trial (224), whereas it was 20.4% (95% CI 13.0–31.2%) in (103) patients electing SOC. These prospectively obtained data support that, with notable exceptions, it is safe to delay treatment initiation for up to 7 d in older patients with AML.
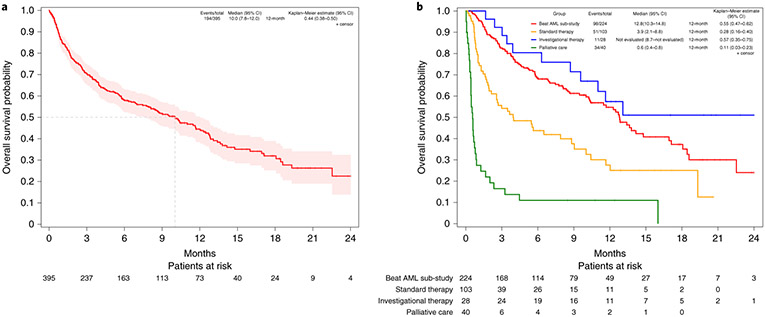
While not a prospective aim in our umbrella study, a common question is how many patients elect to pursue assigned targeted therapy based on genetic profiling. After assignment, 224 (56.7%) patients consented to their assigned Beat AML sub-study and received treatment. Of the 171 patients not enrolling in the assigned sub-study, 103 elected SOC, 28 an alternative protocol with investigational therapy, 38 palliative care and 2 had unknown treatment and were grouped with patients electing palliative care. Patients who opted for palliative care were older and had a higher WBC than other patient groups; those who elected an investigational therapy had lower WBC than those electing a Beat AML-specified therapy or SOC (Table 1); no other clinical characteristics differed based on treatment selection. Other clinical characteristics and most molecular features were not significantly associated with treatment choice (Beat AML or SOC). With respect to IDH2 and IDH1 mutations, IDH2 mutations were more common in patients electing to enroll on the Beat AML trial or SOC, while patients with IDH1 mutations most commonly chose investigational therapy since the IDH1-targeted therapy26 sub-study in Beat AML opened subsequent to the IDH2 sub-study. Importantly, none of the demographic, clinical, laboratory or molecular characteristics were significantly different between the Beat AML and SOC groups. Overall survival is reported for all 395 eligible patients. With a median follow-up of 7.1 months (range: 0–24.8 months), there have been 194 deaths. As shown in Fig. 3a, the estimated median overall survival is 10.0 months (95% CI 7.8–12.0). Overall survival was significantly longer in the Beat AML group (median 12.8, 95% CI 10.3–14.8) compared to either SOC (median 3.9 months, 95% CI 2.1–8.8) or palliative care (median 0.6 months, 95% CI 0.4–0.8) groups, but not significantly different from the investigational therapy group (median not reached) (Fig. 3b). Overall survival estimates at 12 months were 54.7% (95% CI 46.5–62.2), 27.6% (95% CI 16.4–39.9), 11.0% (95% CI 3.5–23.3) and 57.4% (95% CI 35.0–74.6), respectively (Fig. 3b). Multivariable models that controlled for demographic, clinical and molecular variables (Supplementary Table 9) supported these results. Conclusions did not change when patients with AESIs (14 electing SOC, 11 palliative care and 1 investigational therapy) were excluded from the analysis or when the analysis was limited to patients with survival greater than 2 weeks, thus minimizing potential bias due to differential time to elective therapy (Supplementary Tables 10 and 11).
Fig. 3 ∣. Overall survival estimates.
a, All eligible patients on the Beat AML trial. b, By treatment received including assigned Beat AML therapy, SOC (standard therapy), palliative care and alternative investigational therapy. Overall survival estimates were calculated with the Kaplan–Meier method and presented with 95% CIs constructed using the complementary log-log transformation. If a value could not be calculated, not evaluated is indicated. The 2 patients who did not consent to a Beat AML sub-study with unknown treatment were combined with the 38 patients who elected palliative care.
Discussion
Significant progress in understanding the molecular pathogenesis of AML has informed the development of new therapies27. The application of these advances has been impeded by a treatment strategy for AML that mandates rapid treatment often before consideration of targeted therapies can occur. Therefore, we collaboratively implemented a new prospective clinical trial approach aimed at facilitating frontline treatment assignments to specific genomic-defined AML subtypes and demonstrated the feasibility and safety of this approach. The Beat AML trial provided evidence that this new approach to AML therapy is safe for the large majority of individuals and that treatment assignment based on a dominant clone can be applied to virtually all older patients with AML. The recent introduction of multiple targeted therapy toward FLT3, IDH2 and IDH1 mutations along with the identification that European LeukemiaNet high-risk groups may do better with azacitidine versus SOC, provide further evidence for the importance of using this approach in older patients with AML moving forward since high-risk genomic mutations (TP53, RUNX1) are not currently adaptable to rapid PCR-based testing approaches.
This prospective precision medicine trial provided several important insights for future precision medicine trials in AML and other malignancies. First, the trial demonstrated that for the majority of older adults with AML, a delay in therapy to perform detailed molecular profiling was safe. Exceptions to this were patients with rapid proliferative disease or symptoms of leukostasis that are excluded from the study. Second, this approach requires a detailed team-coordinated effort by investigators, patients and caregivers, genomic laboratories, cytogenetic laboratories and a central treatment assignment team. The resources and effort occurred in the context of 14 academic medical centers in the USA with commitment to this treatment approach and ability to monitor patients appropriately during the time of observation. Sites were required to have significant commitment to enrollment and attention to detail of a complicated treatment approach. Third, the majority of patients with AML could be assigned to a specific therapy based on molecular analysis of the dominant AML clone; the ability to direct toward such a therapy increased as more sub-studies were added to the study. Fourth, although not a primary analysis objective of our study, patients who elected to receive the therapy assigned based on the molecular profiling treatment algorithm had a lower early death rate and superior overall survival compared to patients electing to receive SOC. Despite similar demographic and genetic features, the outcome of the SOC group in our trial was poor. At present, the biggest difference in survival between these two groups is within the first three months, which could be reflective of the short follow-up time that may change with additional observations. This could also represent a lower complete remission rate in the SOC arm and the location where they were treated (academic hospital versus community practice). SOC certainly is poor in this patient population, although since initiating our study venetoclax + azacitidine has come forward potentially as a new standard that will result in the adaptation of study assumptions moving forward once phase 3 studies are published. Finally, trials such as this must be rapidly adaptive as was the case with MLL-rearranged patients when early progression was observed in a small number of patients resulting in patients receiving treatment before mutational determination. This study design has the major advantage of allowing efficient testing of new targeted therapies in populations enriched for an enhanced response.
While our study demonstrates the feasibility of precise molecular treatment assignment in older adults with AML, it does not clearly differentiate the benefit of treatment assignment based on a molecular target from better outcome that occurs simply from enrolling on a clinical trial. Such determination will require either randomization of specific large genomic groups to targeted therapy versus SOC or, in less common genomic groups, comparison of treatment with targeted therapy to either real-world data or synthetic controls. While biases clearly exist with the use of real-world data or synthetic controls that can confound the interpretation of outcome, for rare genomic subsets of AML it may not otherwise be possible to demonstrate significant improvement in patient outcomes or a decrease in treatment-related morbidity. Furthermore, during performance of confirmatory phase 3 studies in these genomic groups, it is likely that second-generation molecules with improved pharmaceutical features or new targets will be identified with therapeutic relevance in AML. The treatment arms within the Beat AML trial are designed with these specific considerations in mind and this study will continue to adapt to changes in the AML treatment landscape. Adaptation has required frequent communication with the U.S. Food and Drug Administration (FDA), who were highly responsive and thoughtful in the suggestions made. This study sets the path to establish the safety of precision medicine in AML and sets the stage to extend this same approach to younger patients with this disease and other cancers that are urgently treated as a single disease despite recognition of multiple subtypes.
Methods
Patient eligibility.
Patients aged ≥60 years with suspected AML were eligible (ClinicalTrials.gov NCT03013998). Patients with therapy for antecedent myeloid malignancies were permitted outside of hypomethylating agents, where the biology by both investigators and regulatory authorities was seen to be different. Patients with isolated myeloid sarcoma, acute promyelocytic leukemia, symptomatic AML from central nervous system involvement, leukostasis requiring urgent therapy or symptomatic disseminated intravascular coagulopathy were ineligible. Patients provided written informed consent for the intent to treat.
Study design.
This study is an umbrella protocol designed to assign patients with AML to a specific treatment based on cytogenetic and molecular results returned within 7 d of sample receipt. Once consented, patients underwent bone marrow aspiration and biopsy, local pathology and cytogenetic analysis (centrally reviewed), NGS (Foundation Medicine) and FLT3-ITD ratio assessment (Invivoscribe Technologies). On the basis of cytogenetic and molecular results, each patient was then assigned to a treatment sub-study with its own consent form and end points. Patients were observed either as inpatients or outpatients until treatment assignment. Treatment with hydroxyurea to maintain disease control during this time was encouraged. Outpatients had a mandatory outpatient visit during the 7 d before treatment assignment.
Genomic profiling.
Fresh bone marrow aspirate or peripheral blood specimens were collected in EDTA tubes and extracted the day after procurement. At the time of extraction, smears were prepared and stained with a modified Wright–Giemsa preparation that was reviewed morphologically by an hematopathologist for tumor purity assessment. Genomic profiling employed the FoundationOne Heme platform, which utilizes DNA sequencing to interrogate the entire coding region of 406 genes and select the introns of 31 genes involved in rearrangements as well as RNA sequencing to interrogate 265 genes known to be somatically altered in human hematological malignancies. All four categories of alterations were detected, including single-nucleotide variants (base substitutions and small indels), copy number alterations and rearrangements. The protocols for DNA and RNA extraction, cDNA synthesis, library construction, hybrid capture and variant calling have been published previously28,29 and are described therein (see also Supplementary Information) with associated characterization of platform sensitivity and specificity across alteration types. In brief, genomic DNA and RNA were extracted and fragmented by sonication to approximately 200 base-pair fragment size. A minimum of 22 ng of DNA and 192.5 ng of RNA were required for advancement. Whole-genome shotgun library construction and hybridization-based capture were performed. Selected libraries were sequenced on an Illumina HiSeq 4000, which uses 49 × 49 paired-end reads. A high uniform depth of coverage was achieved with on average >500× coverage by non-PCR duplicate read pairs in DNA with >99% of exons at a coverage >100× and >3 M unique read pairs required for RNA. Samples with a median coverage <150× were considered failed and excluded from analysis. Known germline variants from the 1000 Genomes Project (single nucleotide polymorphism database 135) were removed. Significant nonsynonymous variants were defined as any somatic alteration annotated in the Catalogue Of Somatic Mutations In Cancer database v.62. The mutant allele frequency cutoff used for known somatic variants was 1%, 5% for potential driver somatic variants, 3% for previously described indels and 10% for potential driver indels. Gene amplifications/gains were defined at a copy number ≥6 and gene losses as a copy number of 0. For rearrangements, a minimum of 10 chimera reads was required for known fusions and 50 for potential driver rearrangements.
Trial oversight.
The study was sponsored by Beat AML, LLC, a division of the LLS. Clinical sites were selected by the principals (A.B., R.L., J.C.B. and B.D.) after a detailed application process based on patient volume, institutional commitment, ability to use a central institutional review board and location to enable broad enrollment across the USA. A single scientific review board (Ohio State University) was utilized to meet the National Cancer Institute (NCI)-designated Cancer Center requirement for peer review. This required coordination with the NCI leadership (J. Doroshow) to modify clinical trial oversight rules for non-NCI-sponsored studies applicable to NCI-sponsored Beat AML centers. The institutional review board (Western Institutional Review Board) approved the protocol for all sites. The study was conducted according to the principles of the Declaration of Helsinki (version 2013) and the International Conference on Harmonization Guidelines for Good Clinical Practice. Treatment was assigned by a central team (J.C.B., A.S.M., B.D. and M.S.) after receipt and review of all information. Safety calls with sites, the LLS and clinical research organization staff occurred weekly to review the active status of patients enrolled. These calls were essential to enabling rapid adaptation and feedback to sites, if necessary, about local processes and modification of the trial. An independent data and safety monitoring board periodically reviewed all outcome, treatment and toxicity data. Medical monitors from a clinical research organization reviewed selected data centrally and representatives of the sponsor regularly visited the study sites to monitor study compliance. Clinical data were collected using a combination of data management systems. Central review of serious reported events submitted by pharmaceutical sponsors to determine the relevance to the Beat AML trial was implemented, which greatly diminished (approximately 90%) the requirement of individual event review by study investigators. Contracting was done centrally by the LLS on behalf of all clinical sites to shorten individual agreements between the pharmaceutical companies and institutions.
Statistics.
These were done by a central Beat AML team led by A.B.S. The feasibility of assigning treatment within 7 d of sample receipt was evaluated in the first 109 eligible patients enrolled. This provided a 90% power to rule out a treatment assignment success rate of ≤80% with 95% confidence. Stopping rules are outlined in the Supplementary Information. Unplanned, post-hoc analyses compared demographic, clinical and molecular characteristics among subgroups of patients defined by elected treatment using Fisher’s exact and Kruskal–Wallis tests. Overall survival was defined from the date of consent until the date of death or last contact and estimated using the Kaplan–Meier method. Cox proportional hazards modeled overall survival as a function of the elected treatment group, controlling for important baseline variables. If any variable grossly violated the assumption of proportional hazards, a time-dependent covariate model estimated hazard ratios before and after a particular time point. Multiple imputation estimated missing data and combined results for ten datasets30. No control for multiple comparisons were made for post-hoc analyses. All P values were two-sided; P values <0.05 were considered statistically significant.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
The investigative team thanks the patients and their caregivers who participated in this trial. The study was sponsored by the LLS, which holds the investigational new drug (IND) application for this trial. Funding for the trial was made possible by the Harry T. Mangurian Foundation, many other donors and the sites that enabled resources for rapid turnaround for cytogenetics and other monitoring requirements of patients. The IND and ultimate design and implementation of this trial came forth from integral collaboration of the LLS team and members at the FDA under the leadership of R. Pazdur, D. Przepiorka, A. Deisseroth and A. Farrell, to whom we are greatly indebted. We thank the investigators and coordinators at each of the clinical sites and the patients who participated in this trial and their families who supported them. Sub-studies in this trial were supported by the pharmaceutical sponsor. We thank the pharmaceutical sponsors who paid the cost of performing the specific sub-studies with their investigational drugs. This work was supported by the NCI (grant no. R35 CA198183 to J.C.B.; grant no. R35 CA197594 to R.L.L.) and in part through the NIH/NCI Cancer Center Support grant nos. P30 CA008748, P30CA069533 and P30 CA016058. We acknowledge many references that could not be included based on the limitations of the journal.
Footnotes
Competing interests
A.B., S.M., L.R. and A.Y. are employees of LLS. LLS receives funding from AbbVie, Agios Pharmaceuticals, Alexion Pharmaceuticals, Amgen, Astellas Pharma, AstraZeneca, Boehringer Ingelheim International, Boston Biomedical, Bristol Myers Squibb, Celgene, Genentech, Gilead Sciences, ImmunoGen, Jazz Pharmaceuticals, Johnson & Johnson, Novartis, Pfizer, Pharmacyclics, RTI Health Solutions, Shire and Takeda. R.L.L. has served on the supervisory board of QIAGEN, the scientific advisory boards of Loxo Oncology (until 2019), Imago BioSciences, Mission Bio, Mana Therapeutics, Auron Therapeutics, C4 Therapeutics and Isoplexis (including equity interest). He has received research support from and consulted for Celgene and Roche, was a consultant for Incyte Corporation, Eli Lilly and Company, Janssen Pharmaceutica, Astellas Pharma, MorphoSys and Novartis, and has received research support from Prelude Therapeutics and honoraria from AstraZeneca, Roche, Eli Lilly and Company and Gilead Sciences. A.S.M. has served on the advisory boards of Jazz Pharmaceuticals, AbbVie/Genentech, Astellas Pharma, PTC Therapeutics, Novartis, Agios Pharmaceuticals and Syndax Pharmaceuticals. U.B. has been a consultant for Genetech, Daiichi Sankyo, Takeda, Pfizer, AbbVie/Genetech and Novartis. E.M.S. has served on the advisory boards of Astellas Pharma, AbbVie, Genentech, Daiichi Sankyo, Novartis, Amgen, Seattle Genetics, Syros Pharmaceuticals, Syndax Pharmaceuticals, Agios Pharmaceuticals and Celgene. He is an equity holder in Auron Therapeutics. P.P. has served on the advisory board of Agios Pharmaceuticals. W.S. has served on the advisory boards of Astellas Pharma, Agios Pharmaceuticals, Kite Pharma, Jazz Pharmaceuticals, Servier, MorphoSys and Amgen, and has received speaking honoraria from AbbVie and Pfizer. M.D. has served on the advisory boards of Blueprint Medicines, Takeda, Incyte Corporation and Sangamo Therapeutics. He has been a consultant for Blueprint Medicines, Fusion Pharmaceuticals, Medscape, Novartis, Sangamo Therapeutics and DisperSol Technologies and has received research funding from Blueprint Medicines, Takeda, Novartis, Incyte Corporation, Sun Pharma Advanced Research Company and Pfizer. He has served on the study management committees of Blueprint Medicines and Takeda. W.B. has received research funding from Forma Therapeutics, Xencor and Celyad Oncology and honoraria from AmerisourceBergen. G.S. has served on the advisory boards of Incyte Corporation, ElevateBio, AbbVie, Karyopharm Therapeutics, ONO Pharma UK, Novartis, Evidera, Agios Pharmaceuticals, AstraZeneca, NCI and FDA. He has received research funding from AbbVie, Agios Pharmaceuticals, Actinium Pharmaceuticals, Ambit Biosciences, Amgen, ARIAD Pharmaceuticals, Astellas Pharma, LLS, BioMed Valley Discoveries, Bristol Myers Squibb, Celator Pharmaceuticals, Celgene, Constellation Pharmaceuticals, Forma Therapeutics, Cyclacel Pharmaceuticals, Daiichi Sankyo, Deciphera Pharmaceuticals, California Institute for Regenerative Medicine, Gamida Cell, Gilead Sciences, Incyte Corporation, Janssen Pharmaceutica, Karyopharm Therapeutics, Kite Pharma, Mateon Therapeutics, MedImmune, Millenium, National Marrow Donor Program, NIH, Novartis, Onconova Therapeutics, Onyx Pharmaceuticals, Pfizer, Sangamo Therapeutics, Stemline Therapeutics, Tolero Pharmaceuticals, Trovagene (Cardiff Oncology), University of California, Davis, University of California, San Diego and the University of California Hematologic Malignancies Consortium. He has received stock from Amgen, Bristol Myers Squibb, Pfizer and Johnson & Johnson. He has served on the speakers’ bureaux of Agios Pharmaceuticals, Amgen, Astellas Pharma, Bristol Myers Squibb, Celgene, Sanofi Genzyme, Incyte Corporation, Janssen Pharmaceutica, Jazz Pharmaceuticals, Kite Pharma (Gilead Sciences), Pharmacyclics and Stemline Therapeutics. R.O. has been a consultant for and received honoraria from Jazz Pharmaceuticals, Genentech, Amgen and Revolution Medicines and has received research funding from Daiichi Sankyo, Astellas Pharma, Pfizer, Genentech, Takeda, Novartis, AstraZeneca, MedImmune (AstraZeneca), Spectrum Pharmaceuticals and Mirati Therapeutics. J.F. has been on the advisory boards of Novartis, Servier and Pfizer. He has been a consultant for Revolution Medicine and has received research funding from AbbVie, Boehringer Ingelheim, Actinium Pharmaceuticals, Aprea Therapeutics, Aptose Biosciences, H3 Biomedicine, Kura Oncology, Tolero Pharmaceuticals, Trillium Therapeutics, Xencor and Takeda/Millennium. E.T. has served on the advisory boards of AbbVie, Astellas Pharma, Genentech, Agios Pharmaceuticals and Daiichi Sankyo. He has received clinical trial support from Janssen Pharmaceutica and clinical trial funding and sponsored research from Incyte Corporation. He holds equity in Notable Labs. O.O. has served on the advisory boards of AbbVie, Celgene and Impact Biomedicines. She has received research funding from AbbVie, AstraZeneca, Astex, Agios Pharmaceuticals, Celgene, CTI BioPharma, Incyte Corporation, Janssen Pharmaceutica, Kartos Therapeutics, NS Pharma and OTS. M.A. has served on the advisory boards of Gilead Sciences and Syndax Pharmaceuticals. M.C.F. has been a consultant for Agios Pharmaceuticals, MacroGenics and Daiichi Sankyo and has received research funding from the LLS. J.-A.V. is an employee of Foundation Medicine, a wholly owned subsidiary of Roche Holdings and with equity interest in Roche Holdings. E.S. is an employee of Foundation Medicine with equity interest in Roche Holdings. B.D. has served on the scientific advisory boards of Aileron Therapeutics, Therapy Architects (ALLCRON), Cepheid, Vivid Biosciences, Celgene, RUNX1 Research Program, EnLiven Therapeutics, Gilead Sciences (inactive), Monojul (inactive). He has served on the scientific advisory boards of and received stock from Aptose Biosciences, Blueprint Medicines, Iterion Therapeutics, Third Coast Therapeutics, GRAIL (scientific advisory board, inactive). He is the scientific founder of MolecularMD (inactive, acquired by ICON). He has served on the board of directors of and received stock from Amgen. He has served on the board of directors of Burroughs Wellcome Fund and CureOne, the joint steering committee of Beat AML LLC. He is the founder of VBL Therapeutics, has received clinical trial funding from Novartis, Bristol Myers Squibb and Pfizer and royalties from patent no. 6958335 (Novartis exclusive license), OHSU and the Dana-Farber Cancer Institute (one Merck exclusive license). J.C.B. has received research support from Janssen Pharmaceutica, Genentech, Acerta Pharma and Pharmacyclics and has served on the advisory board of Syndax Pharmaceuticals. He has received funding from the NCI (grant no. R35 CA198183) and in part through an NIH/NCI Cancer Center Support grant no. P30 CA008748. M.R.B., R.C., T.K., B.B., M.B., T.B., V.H.D., N.A.H., M.L., M.M., A.S.R., A.B.S., M.S., T.L.L., C.V. and A.W. declare no conflicting interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-020-1089-8.
Editor recognition statement Javier Carmona was the primary editor on this article, and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-020-1089-8.
Data availability
The data presented represents patient enrollment between 17 November 2016 and 30 January 2018. The trial is active and continues to enroll patients. The LLS is committed to making nonconfidential, nonidentifiable data available to individuals or teams focused on advancing the therapeutic efforts for patients with AML and related blood cancers. These requests will be evaluated by the principals of Beat AML (A.B., R.L.L., B.D. and J.C.B.) for appropriateness.
References
- 1.Karjalainen E & Repasky GA Molecular changes during acute myeloid leukemia (AML) evolution and identification of novel treatment strategies through molecular stratification. Prog. Mol. Biol. Transl. Sci 144, 383–436 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Khwaja A et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers 2, 16010 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Chen J et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat. Med 25, 103–110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai P et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med 24, 1015–1023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 18, 100–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasu S et al. Ten-year outcome of patients with acute myeloid leukemia not treated with allogeneic transplantation in first complete remission. Blood Adv. 2, 1645–1650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone A, Zukerman T, Flaishon L, Yakar RB & Rowe JM Efficacy outcomes in the treatment of older or medically unfit patients with acute myeloid leukaemia: a systematic review and meta-analysis. Leuk. Res 82, 36–42 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Lübbert M et al. Low-dose decitabine versus best supportive care in elderly patients with intermediate- or high-risk myelodysplastic syndrome (MDS) ineligible for intensive chemotherapy: final results of the randomized phase III study of the European Organisation for Research and Treatment of Cancer Leukemia Group and the German MDS Study Group. J. Clin. Oncol 29, 1987–1996 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Fenaux P et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J. Clin. Oncol 28, 562–569 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Kantarjian HM et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J. Clin. Oncol 30, 2670–2677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone RM et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med 377, 454–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uy GL et al. A phase 2 study incorporating sorafenib into the chemotherapy for older adults with FLT3-mutated acute myeloid leukemia: CALGB 11001. Blood Adv. 1, 331–340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoli S et al. Time from diagnosis to intensive chemotherapy initiation does not adversely impact the outcome of patients with acute myeloid leukemia. Blood 121, 2618–2626 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Eisfeld A-K et al. Mutation patterns identify adult patients with de novo acute myeloid leukemia aged 60 years or older who respond favorably to standard chemotherapy: an analysis of Alliance studies. Leukemia 32, 1338–1348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker H et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J. Clin. Oncol 28, 596–604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koszarska M et al. Type and location of isocitrate dehydrogenase mutations influence clinical characteristics and disease outcome of acute myeloid leukemia. Leuk. Lymphoma 54, 1028–1035 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Yang H et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood 131, 328–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mill CP et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood 134, 59–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong JY et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen HD et al. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 78, 5363–5374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardis ER et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med 361, 1058–1066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel JP et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med 366, 1079–1089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaemmanuil E et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisfeld A-K et al. The mutational oncoprint of recurrent cytogenetic abnormalities in adult patients with de novo acute myeloid leukemia. Leukemia 31, 2211–2218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiNardo CD et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med 378, 2386–2398 (2018). [DOI] [PubMed] [Google Scholar]
- 27.DiNardo C & Lachowiez C Acute myeloid leukemia: from mutation profiling to treatment decisions. Curr. Hematol. Malig. Rep 14, 386–394 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Intlekofer AM et al. Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay. Blood Cancer J. 8, 60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frampton GM et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol 31, 1023–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Buuren S Multiple imputation of discrete and continuous data by fully conditional specification. Stat. Methods Med. Res 16, 219–242 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented represents patient enrollment between 17 November 2016 and 30 January 2018. The trial is active and continues to enroll patients. The LLS is committed to making nonconfidential, nonidentifiable data available to individuals or teams focused on advancing the therapeutic efforts for patients with AML and related blood cancers. These requests will be evaluated by the principals of Beat AML (A.B., R.L.L., B.D. and J.C.B.) for appropriateness.