Abstract
It is becoming more apparent in tissue engineering applications that fine temporal control of multiple therapeutics is desirable to modulate progenitor cell fate and function. Herein, the independent temporal control of the co-delivery of miR-148b and miR-21 mimic plasmonic nanoparticle conjugates to induce osteogenic differentiation of human adipose stem cells (hASCs), in a de novo fashion, is described. By applying a thermally labile retro-Diels–Alder caging and linkage chemistry, these miRNAs can be triggered to de-cage serially with discrete control of activation times. The method relies on illumination of the nanoparticles at their resonant wavelengths to generate sufficient local heating and trigger the untethering of the Diels–Alder cycloadduct. Characterization of the photothermal release using fluorophore-tagged miRNA mimics in vitro is carried out with fluorescence measurements, second harmonic generation, and confocal imaging. Osteogenesis of hASCs from the sequential co-delivery of miR-21 and miR-148b mimics is assessed using xylenol orange and alizarin red staining of deposited minerals, and quantitative polymerase chain reaction for gene expression of osteogenic markers. The results demonstrate that sequential miRNA mimic activation results in upregulation of osteogenic markers and mineralization relative to miR-148b alone, and co-activation of miR-148b and miR-21 at the same time.
Keywords: gold, microRNA, nanoparticles, retro-Diels–Alder, sequential, silver
1. Introduction
Short noncoding RNA molecules participate in post-transcriptional regulation pathways and have gained much attention in regulating gene expressions for a variety of treatments. In particular, microRNAs (miRNA) offer promise in improving repair of tissue damage through well-controlled regenerative processes.[1,2] These short RNA sequences associate with RNA-induced silencing complexes (RISC) and regulate mRNA localization, polyadenylation, and translation.[3] Canonically, they negatively regulate gene expression through degradation or translational inhibition, by binding to the 3′ untranslated region (3′UTR) of mRNA transcripts.[3,4] Modulation of human mesenchymal stem cells (hMSCs), including adipose-derived (hASCs) and bone marrow stem cells (hBMSCs), using miRNAs has widely been studied[5–7] with a keen focus on blueprinting therapies for bone defects and skeletal reconstruction.
MicroRNA-148b was studied[8,9] and identified to upregulate alkaline phosphatase to stimulate osteogenesis activity de novo in hBMSCs and hASCs. Li et al.[10] investigated miR-148b in hASCs and reported noggin (NOG), a well-known bone morphogenetic protein (BMP) gene repressor, as a direct gene target. miR-148b initiated downregulation of NOG and promoted osteogenic differentiation through upregulation of BMP expression.
MicroRNA-21 is another widely studied RNA sequence, particularly in examining cell proliferation of cancer cells and stem cells.[11,12] miR-21 was found to be overexpressed in nonsmall cell lung cancer cells, and played a role in repressing phosphatase and tensin homolog, a tumor suppressor, leading to lung cancer cell growth and invasion.[13] In contrast, overexpression in mouse embryonic stem cells was found to downregulate the “stemness” and pluripotency of cells by directly targeting Sox2.[14]
To circumvent the issue of nonspecificity, a host of drug delivery systems has been proposed,[15–18] from liposomal, and viral vectors, to proteins and nano-vehicles. While their mechanisms are many, their purpose is identical: drug dormancy and inactivation prior to reaching their intended target. From the aforementioned drug vehicles, plasmonic nanoparticles have acquired considerable interest. Silver and gold nanoparticles have been investigated for siRNA delivery, specifically for their release kinetics[19,20] as well as in the delivery of miR-148b[9,21] to induce osteogenesis in hASCs.
While targeted delivery is an important concept for clinical and translational applications of gene therapy systems, the ability to also co-deliver therapies sequentially and temporally opens the door for a wide variety of effective treatments. The presented study demonstrates the novel concept of delivering two different miRNA mimics, miR-21 and miR-148b, on two separate nanoparticle vehicles, photothermally responsive gold (GNPs) and silver nanoparticles (SNPs), in hASCs to demonstrate augmented osteogenesis.
2. Results and Discussion
2.1. Gold and Silver Nanoparticle Photothermal Release
Prior to investigating the osteogenic inductive ability of co-transfecting miR-21 and miR-148b, the photothermal release of the miRNA mimic molecules from the surfaces of SNPs and GNPs was initially characterized. While gold nanoparticles were commercially procured, the hydroxypropyl cellulose (HPC)-stabilized silver nanoparticles were synthesized in house as previously described and characterized.[22,23] The latter yielded particles with an average diameter size of 75.05 ± 0.65 nm, while the gold nanoparticles’ average diameter size was 78.8 ± 2.85 nm. These nanoparticles were used for all experiments while second harmonic generation measurements (SHG) were conducted with a separate batch of each particle. As has been extensively published, metallic particles at the nanoscale can exhibit plasmonic resonances. Noble metals, in particular, demonstrate this phenomenon within visible light frequencies. These sub-100 nm particles have demonstrated high transfection efficiency in mammalian cells,[24] and, in the case of noble metal nanoparticles, the ability to respond to visible light frequencies upon illumination. The latter occurs as a result of a polariton generated between the oscillating conduction band electrons and positively charged lattice.[25] Upon illumination at resonant wavelength of oscillation, these electrons enter an electron–hole pair excited state, which results in electron–electron scattering as well as electron–lattice scattering. The extinction spectra of both nanoparticles used in the present study are reported in Figure 1A,B, along with their transmission electron microscope (TEM) images. The plasmon peak was observed at 405–415 nm for SNPs and 551–557 nm for GNPs. As a result of this plasmonic response, generation of localized heating on the nanoparticle surfaces can be achieved in what is referred to as the photothermal effect. This has been explained by the metal’s ability to absorb far-field radiation, generating enhanced near fields, with plasmonic resonances decaying by re-emission of electrons, and producing hot electron–hole pairs via Landau damping.[26,27] Heat dissipation into the local surroundings arises from electron–electron scattering between “hot” electrons and lower energy electrons, as well as electron–phonon interactions that give rise to lattice energy losses.[25]
Figure 1.
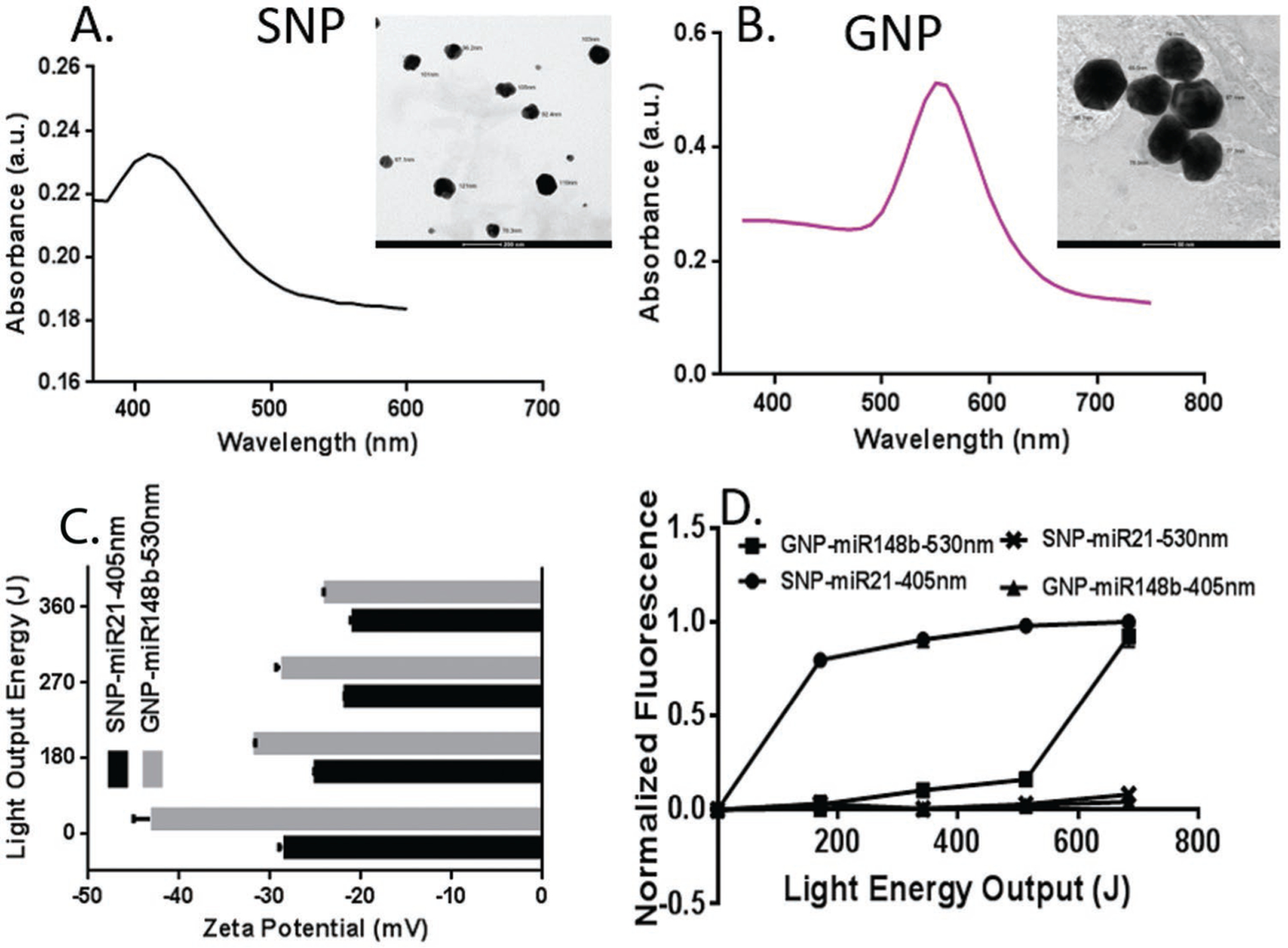
A) TEM image and UV-vis spectra for silver nanoparticles, and B) gold nanoparticles. C) Zeta potential measurements of miRNA-NP conjugates for silver and gold surfaces with increasing doses of incident light. D) Normalized fluorescence measurements of nanoparticle supernatant solutions, irradiated at 405 and 530 nm light wavelengths. Gold nanoparticle surfaces in this experiment were modified with Diels–Alder-linked miRNA mimic-FAM dye molecules, while silver nanoparticles were surface-modified similarly with Cy3-tagged dye molecules.
With any RNA delivery vehicle, a stable conjugation is crucial to protect the structure and function of therapeutics against opsonization and RNase degradation. Characterization of this system and its stability was described in our previous manuscript.[22] By conjugating to the nanoparticle surface, we can enhance the stability of RNA and shield it from RNAse degradation and lysozyme hydrolysis. Additionally, the 2’-O-methyl modified RNA mimics used in the study aid in prolonging the lifetime of RNA in serum containing media.[28] For effective therapeutic activity, however, a controlled and targeted mechanism for the release of molecules is also significant. Early exposure to acidic environments in endo/lysosomal structures within cells will mute nucleic acid molecules’ biological effects, requiring a large deliverable dose to mitigate this loss. While there have been several advances made in the design of nanoparticle systems, their release mechanisms are largely assumed or neglected. To design a nanoparticle vehicle with targeted ability to decage and release miRNA mimics, a thermally labile linker, responsive to the nanoparticles’ photothermal phenomenon was developed that could cleave and release conjugated miRNA mimics at localized temperatures greater than 60 °C.[22,29] The covalently bound linker was based on Diels–Alder chemistry which relies on the cycloadduct formation between a diene molecule and dienophile. The advantage this linker possesses is the ability to selectively release conjugated miRNA upon photo-irradiation of the nanoparticle substrate at their plasmon wavelength, generating adequate localized heat to initiate the retro-Diels–Alder reaction and release the miRNA. This study is the first to our knowledge that demonstrates the proposed delivery vehicle in a biological application.
A type of click chemistry, the Diels–Alder cycloaddition takes place between a diene and dienophile possessing overlapping orbital energies (diene’s lowest unoccupied molecular orbital and dienophile’s highest occupied molecular orbital), generating sigma bonds from π bonds [4 + 2].[30–32] The result is a stable covalently bound linker capable of conjugating molecular cargo to surfaces. In the present study, a cycloadduct between a furan group and maleimide was prepared, with additional modifications on the terminal ends. The thiol terminal group on the furan end allows for the facile linkage between the – SH group and Ag and Au surfaces, as has been described.[33] On the maleimide terminal, a hexanoic acid group was added which enabled the covalent crosslinking between carboxyl ends and the 5’ amine-modified miRNA mimics to yield a robust amide bond. At temperatures 60 °C or higher, it was demonstrated that the retro-Diels–Alder reaction, i.e., the reversal of the cycloadduct, could be triggered.[22,29] Using photo-activation, sufficient localized heat was generated to thermally cleave the linker.
Different miRNA mimics with distinct fluorophores were attached to the SNPs and GNPs using Diels–Alder chemistry and irradiated to quantify the photo-release. For each NP system, 30 uL of 4uM miRNA mimics was added per mL of nanoparticle suspension. Final measurements had shown that miRNA surface quantity on NPs after modification and cleanup was approximately 9271 miRNA molecules per SNP, and 18512 molecules per GNP. A packing rate of almost double that of SNPs was observed for GNPs. This is likely due to the superior GNP surface stability and resistance to surface oxidation that may reduce the favorability of thiol (Au/Ag-SH) linkages in the case of silver. At different light-emitting diode (LED) light energies, at their respective plasmon wavelengths, the zeta potentials of the nanoparticle surfaces were measured and are shown in Figure 1C. To further investigate the extent of release at their plasmon wavelengths, fluorescence measurements of the released miRNA in the supernatant were taken, upon irradiation at 530 and 405 nm with increasing incident light exposure. These results are presented in Figure 1D. In these samples, miR-21 on SNPs were tagged with Cy3 molecules, while miR-148b on GNPs were labeled with fluorescein amidites (FAM). As shown, photo-release was achieved at each particle’s plasmonic wavelength, while almost no release was observed upon reversal of the applied light wavelengths. Furthermore, at equal light energy outputs, the SNPs exhibited much more robust de-caging than the GNPs. To investigate further, the absorbance of the Diels–Alder adduct only was measured and is presented in Figure S1 in the Supporting Information. From this data, high absorbance was observed at ≈400 nm with a discernable local maximum at around 450 nm. This suggests a possibly more complex process involved in the photothermal response of SNPs on the retro-Diels–Alder reaction, with likely a coupled phononic/thermal response and electron-ejection or photon-generating phenomenon triggering the untethering of the cycloadduct. This phenomenon of “hot carriers” has been well described in photocatalytic reactions,[34,35] playing a role in lowering the activation energy barriers in bonds adhered to the surface and has been shown to be distinct from the typical photothermal effect. This relationship between the thermal and nonthermal response was found to be synergistic and co-dependent, increasing super-linearly with increased illumination. Experiments to confirm this are outside the scope of this study but are recommended for future studies. However, the argument that the cycloadduct can sever in response to direct 405 nm LED light alone seems unlikely, as a result of the observed release profile for the GNP-FAM sample at 405 nm. The nonplasmon resonant state of the GNPs at 405 nm led to little or no photo-release. Overall, the release trends in Figure 1D suggest more effective release of the miRNA from SNPs than GNPs at shorter light exposures, offering the ability to co-deliver small molecules at separate and distinct intervals.
2.2. SHG Measurement
SHG is a noninvasive, nondestructive, surface-sensitive, non-linear spectroscopy for characterizing physical and chemical properties at colloidal nanoparticle interfaces.[36–39] In the SHG process, two incident photons of frequency ω add coherently to generate a third photon with a frequency of 2ω. SHG is dipole forbidden in centrosymmetric media, like bulk solution, but can be generated at the surface of nanoparticles where the symmetry is broken. Recently, SHG has been used as an accurate method to determine molecular adsorption and chemical reactions at nanoparticle surfaces,[40,41] ion-transport processes in liposomes,[42–44] and the release of oligonucleotides from the surface of plasmonic nanoparticles.[19,20,22,45]
The release of miRNA from the surface of gold and silver plasmonic nanoparticles was measured in real time using surface-specific SHG spectroscopy. Representative SHG spectra, centered at 400 nm with a full-width half-maximum of approximately 4.5 nm, of SNP-miR-21 complex are shown in Figure 2A. The miRNA-functionalized nanoparticles have higher SHG intensities than the corresponding nanoparticles without miRNA functionalization due to the increased surface charge density through the χ(3) effect, in agreement with our previous results.[19,20,22,45] Upon irradiation at 400 nm, the SNP absorbed energy at the plasmon resonance, leading to the cleaving of the linker and release of miRNA mimic, resulting in a lower SHG intensity, as shown in Figure 2A. Similar results are also obtained for GNP-miR-148b upon irradiation at 530 nm (Figure S2, Supporting Information). These cleaving dynamics were caused by the photothermal response of the plasmonic nanoparticles, by hot electrons generated from the plasmon resonance leading to cleavage of the linker, or likely a combination of these two processes.[46,47]
Figure 2.
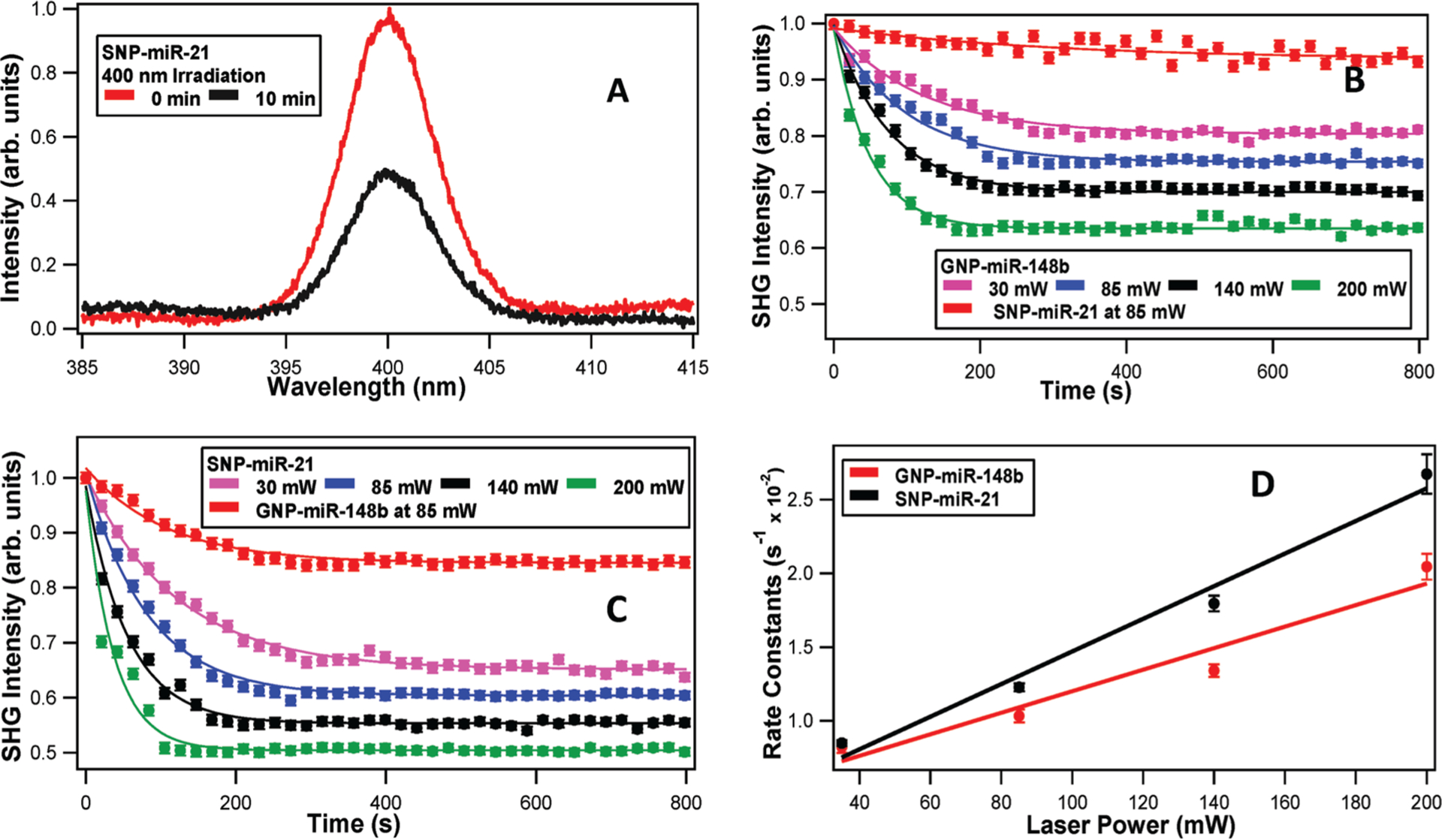
A) Representative SHG spectra of miRNA-functionalized silver nanoparticles at different wavelengths and irradiation times. SHG intensity of the miRNA-functionalized gold and silver nanoparticles as a function of time under varying laser irradiation powers using B) 530 nm and C) 400 nm. D) The obtained photothermal cleaving rate constants as a function of laser power for miRNA-functionalized gold and silver nanoparticles, using 530 and 400 nm irradiation, respectively.
The time-dependent SHG experiments for both sets of nanoparticles were analyzed using power-dependent fits, with higher irradiation powers leading in faster and more complete photothermal cleaving, as shown in Figure 2B,C. The experimentally obtained results were fit to a pseudo first-order exponential function given by ESHG = A + B e−kt , where k is the cleaving rate and A and B are proportionality constants. The values of the fit parameters are summarized in Tables S1 and S2 in the Supporting Information. Figure 2D displays the obtained rate constants plotted as a function of laser power for the gold and silver nanoparticles, with corresponding linear fits. The obtained slopes from the fits are (0.007 ± 0.001) and (0.011 ± 0.001) s−1 mW−1 for the gold and silver nanoparticles, respectively. Similarly, the obtained y-intercepts from the fits are (0.471 ± 0.166) and (0.366 ± 0.147) s−1 for the gold and silver nanoparticles, respectively. The nonzero y-intercepts are a result of small amount of cleaving caused by the 800 nm laser needed for the SHG measurements. These experimental results indicate that the photothermal release of miRNA mimic at the corresponding plasmon resonances was more efficient for silver nanoparticles, as seen earlier in the fluorescence measurements. This result can be attributed to a higher concentration of hot electron formation in silver nanoparticles leading to higher cleaving efficiencies, although more research is needed to study this process in greater detail. Additionally, control experiments show that the SNP-miR-21 complex has only minimal cleaving upon irradiation with 530 nm, as displayed in Figure 2B. In contrast, the GNP-miR-148b complex has a greater amount of cleaving upon irradiation at 400 nm, as shown in Figure 2C. The gold nanoparticle plasmon peak is centered near 530 nm; however, the gold nanoparticles retain substantial absorption near 400 nm. This absorption at 400 nm can raise the local temperature of the gold nanoparticle surface leading to the photothermal release of miRNA and the corresponding decrease in the SHG intensity that was observed.
2.3. In Vitro Photo-Release of GNPs and SNPs
To investigate the photothermal release of the Diels–Alder-linked and FAM-tagged oligonucleotides in vitro between the two sets of nanoparticles, colocalization between the nanoparticles and fluorescent molecules in the transfected cells were examined using confocal microscopy. For all light irradiation experiments, cells were cultured in Opti-MEM reduced serum media. This was due to the possibility of near-UV wavelengths generating toxic photo-products and radical oxygen among amino acid components of Dulbecco’s modified Eagle medium (DMEM).[48] At 405 nm, hASCs transfected with either SNP-FAM or GNP-FAM were illuminated at 50 J of incident light energy, and 565 J at 530 nm. Due to the observed toxic cellular response to the 405 nm LED light source, the 405 nm group was maintained at a much less intense exposure than the GNP samples to mitigate its adverse effects. More importantly, release rates in Figure 1D revealed that 550–600 J was needed for substantial thermal cleavage in GNP samples at 530 nm. For each nanoparticle system, images were taken of nonilluminated samples (NA), samples irradiated at 405 nm (50 J), and at 530 nm (565 J). These images are presented in Figures 3 and 4, for SNP and GNP, respectively. The green fluorescein isothiocynate (FITC) channel was used to view the FAM conjugated to the nanoparticles, while the blue and red channels were selected to view SNPs and GNPs, respectively. The overlay images between the nanoparticles’ and FAM channels are presented as a qualitative assay in determining release of the payload upon photo-activation based on colocalization of the channels. In Figure 3, SNP-FAM 405 nm treated cells showed more intense and punctate spots of green that do not colocalize with the blue pixels contributed by the silver nanoparticles’ plasmon scattering. Due to fluorescence resonance energy transfer (FRET) during conjugation prior to release, the optical scattering intensity of the nanoparticles is likely diminished as a result of the proximally close fluorophore tag on the miRNA molecules. FRET phenomenon occurs as a result of overlapping excitation and emission levels of two optically active molecules, and close distance between the two. This is due to the energy decay from high energy electrons to lower levels transferred to excite the electrons in the neighboring molecule, reducing the effect of photoemission. From this, we anticipate the released sample of SNPs at 405 nm to exhibit more intense blue signals as the scattering phenomenon is no longer weakened by FRET. This is a result of the distance increase between the probes.[49]
Figure 3.
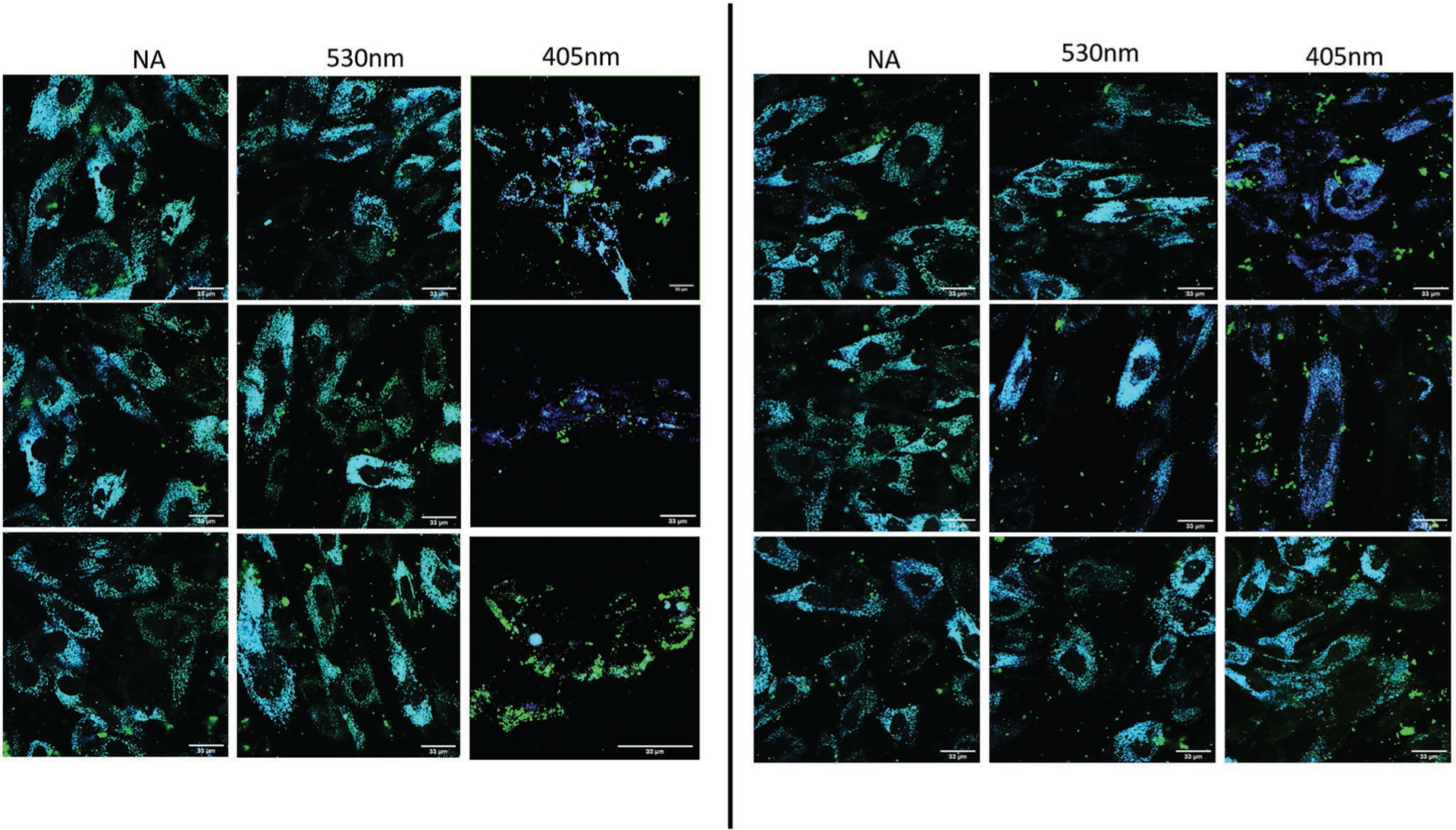
Colocalization of SNPs (blue) and conjugated FAM molecules (green) in hASCs. Irradiation at 400 nm (50 J) and 530 nm (565 nm). 60X magnification used with scale bars annotated at 33 μm.
Figure 4.
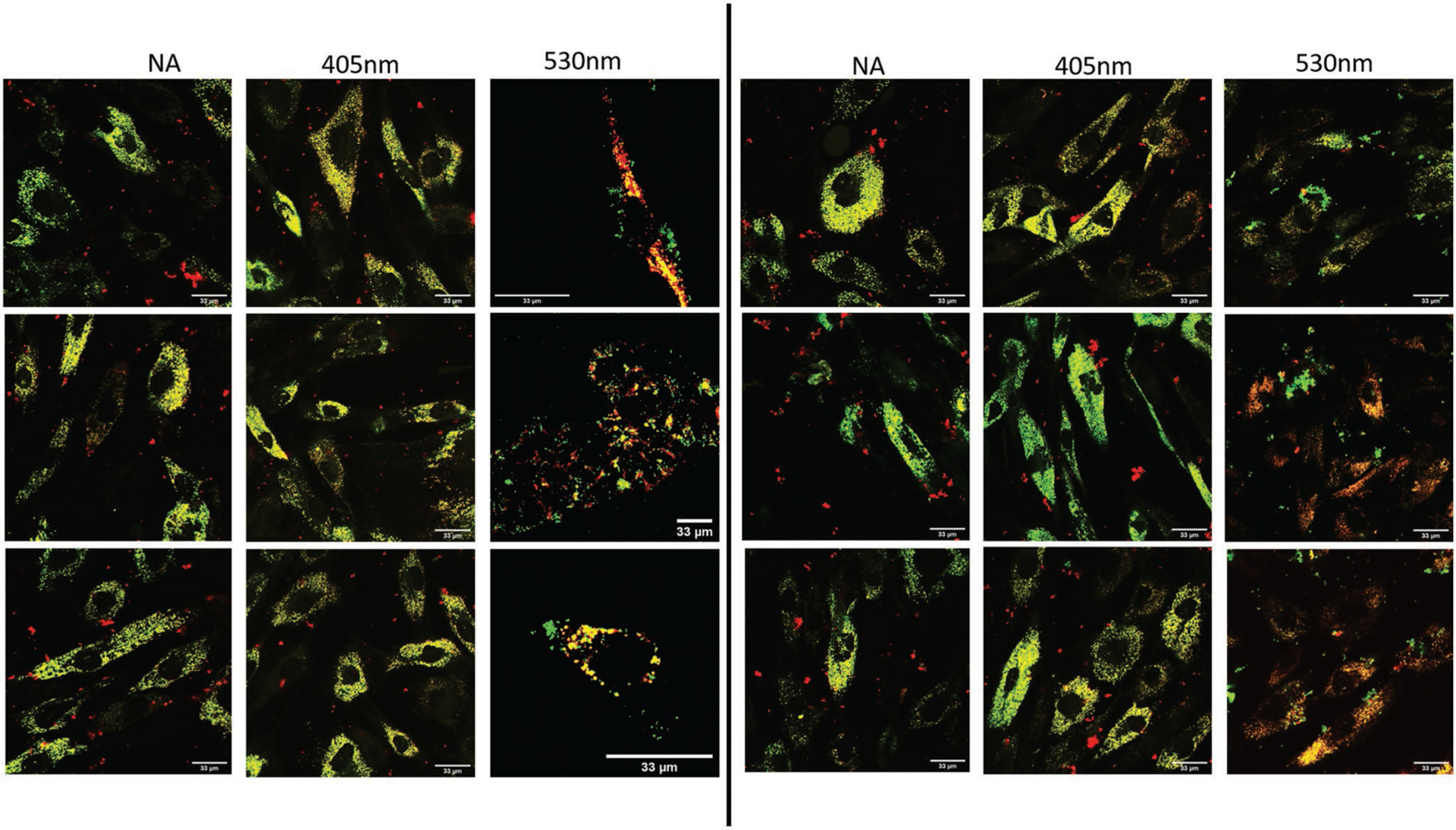
Colocalization of GNPs (red) and conjugated FAM molecules (green) in hASCs. Irradiation at 400 nm (50 J) and 530 nm (565 nm). 60X magnification used with scale bars annotated at 33 μm.
The average calculated Mander’s colocalization coefficients were calculated and plotted in Figure S5 in the Supporting Information but are summarized here: for the nonirradiated SNP samples, 530 nm-irradiated, and 405 nm-irradiated samples were 0.723, 0.653, and 0.436, respectively. Among these groups, the 405 nm was the only statistically significant group. The results indicate thermal release at the particles’ plasmon resonance wavelength, as anticipated.
SNP-treated hASCs were also treated with Texas-Red-tagged transferrin endosomal staining, to investigate the nanoparticle-miRNA conjugates ability to escape from the encapsulations after endocytic uptake.[50,51] Those images along with the z-stack images are presented in Figure S5 in the Supporting Information. Low to moderate amounts of colocalization were found between the red-stained endosomes and green SNP-FAM molecules regardless of illuminated state, suggesting clathrin-mediated endosomal encapsulation to be present but limited. The transfection mechanism of metal nanoparticles has been demonstrated to be nonspecific.[52] Receptor-mediated uptake as well as phagocytosis, macropinocytosis, adhesion, and passive transport, has all been demonstrated to play a role in nanoparticle entry into cells.[53] Whether one mechanism dominates over the others is largely dependent on the particle’s size and shape, surface charge, and chemical moiety.[51,52,54]
In Figure 4, images of hASCs treated with GNP-FAM are represented. Once again, upon inspection of the overlay channels between the FAM and GNPs, a greater degree of de-colocalization is observed in the 530 nm-irradiated sample, GNPs’ plasmonic resonant wavelength. The average calculated Mander’s coefficients, plotted in Figure S5 in the Supporting Information, for the nonirradiated, 405 nm-irradiated, and 530 nm-irradiated samples were 0.800, 0.790, and 0.543, respectively. Only the 530 nm group showed statistical significance.
2.4. Osteogenic Induction
Studies with MSCs suggest miR-21 plays a role in exiting stem cells from their G1-S-G2-M cell cycle and toward nonspecific differentiation. Reports of miR-21 mimics promoting osteogenic differentiation in hBMSCs by targeting the SMAD-7 gene a suppressant of the TGF-β family,[55] and adipogenesis in hASCs via TGF-β/SMAD 3 signaling modulation,[56] have been demonstrated. These results allude to the strong possibility of miR-21 acting to promote differentiation toward the most probable and likely lineage associated with the cell type; adipocytes for hASCs and osteoblasts for hBMSCs. It is, therefore, hypothesized that it is possible to drive cells down a differentiation pathway with one post-transcriptional regulator, miR-148b, and amplify their differentiation effect with miR-21.
The direct targets of miR148b and miR21 were extracted from microRNA.org, and the relevant genes are summarized in Tables S3 and S4 in the Supporting Information. We observe that NOG, an inhibitor of BMP-2, is a direct target of miR148b, and SOX2, a pluripotency marker, is a target for miR21.[10–12]
For effective clinical use of miRNA mimic therapies, there is a need for targeted and precise delivery of these small molecules for treatment to mitigate off-target effects and nonspecific accumulation. There is also the need to protect the vulnerable nucleotide sequences from intra and extra cellular nuclease degradation. In cases of bone defect repair and regeneration, the need for spatial and temporal control was proven necessary as was demonstrated,[57] in which the formation of uncontrolled heterotrophic bone growth from delivered BMPs in other parts of the body became a clinical and health concern.
The miR-21/miR-148b-treated hASC groups listed in Table 1 were maintained at 37 °C incubation for 21 days post-illumination prior to analysis, replenishing the stromal media for all wells every 3 days. To assess mineralization between the different samples, two chemical stain methods were used: xylenol orange (XO) and alizarin red (ARS). While the latter is more common and replete in literature involving osteogenic assays, its nonspecific binding to hydroxyapatite as well as lack of aqueous solubility has led to investigations into alternative assays that can supplement the information provided by ARS, such as XO.[58,59] XO is a calcium-binding stain, highly water soluble, nontoxic at low concentrations, and fluorescent.[58] Accumulations of calcium deposits such as in mineralized nodules produce intense orange fluorescence upon staining of those nodules. All hASC groups were stained overnight with XO, and imaged after rinsing with Dulbecco’s phosphate-buffered saline (DPBS). These results are presented in Figure 5. For the stromal culture medium (SM) control group (A), only a diffusive light stain is observed, reflective of the lack of mineralized nodules of these nondifferentiated hASCs. In the positive control group (C), in which hASCs were left in osteogenic positive control (OM) for 21 days, high mineralization was apparent with intense fluorescence in this sample. In groups E through I, in which miR-21 and miR-148b were delivered to hASCs using SNPs and GNPs, respectively, small mineralized nodule formations were evident. More intense stain was observed in groups H and I, where the second miRNA (148b or 21) was delivered following a 48 h delay after the initial dose. Sporadic staining was evident in Group B, hASCs treated with GNPs-miR-148b and SNPs-miR-21 but without photo-activation. This suggests some premature and random cleaving of the miRNAs during incubation at 37 °C before their removal from media. Remainder control groups and their images can be found in Figure S6 in the Supporting Information.
Table 1.
hASC groups treated with GNP-miR-148b and/or SNP-miR-21, illuminated at different wavelengths.
| Group | Description |
|---|---|
| A | hASCs in stromal medium (SM) only |
| B | hASCs transfected with SNP-miR-21 and GNP-miR-148b, but nonirradiated |
| C | hASCs in osteogenic differentiation medium (OM) |
| D | hASCs transfected with GNP-miR-148b only, and irradiated at 530 nm |
| E | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 405 and 530 nm 16 h post-transfection |
| F | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 405 nm at 16 h and 530 nm at 40 h |
| G | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 530 nm at 16 h and 405 nm at 40 h |
| H | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 405 nm at 16 h and 530 nm at 64 h |
| I | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 530 nm at 16 h and 405 nm at 64 h |
| LA | hASCs transfected with nonmodified SNPs and GNPs, and irradiated at 405 and 530 nm at 16 h |
| miR-21 + 405nm | hASCs transfected with SNP-miR-21 only, and irradiated at 405 nm |
| miR-21 + miR-148b + 405nm | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 405 nm only |
| miR-21 + 148b + 530nm | hASCs transfected with SNP-miR-21 and GNP-miR-148b, and irradiated at 530 nm only |
Figure 5.
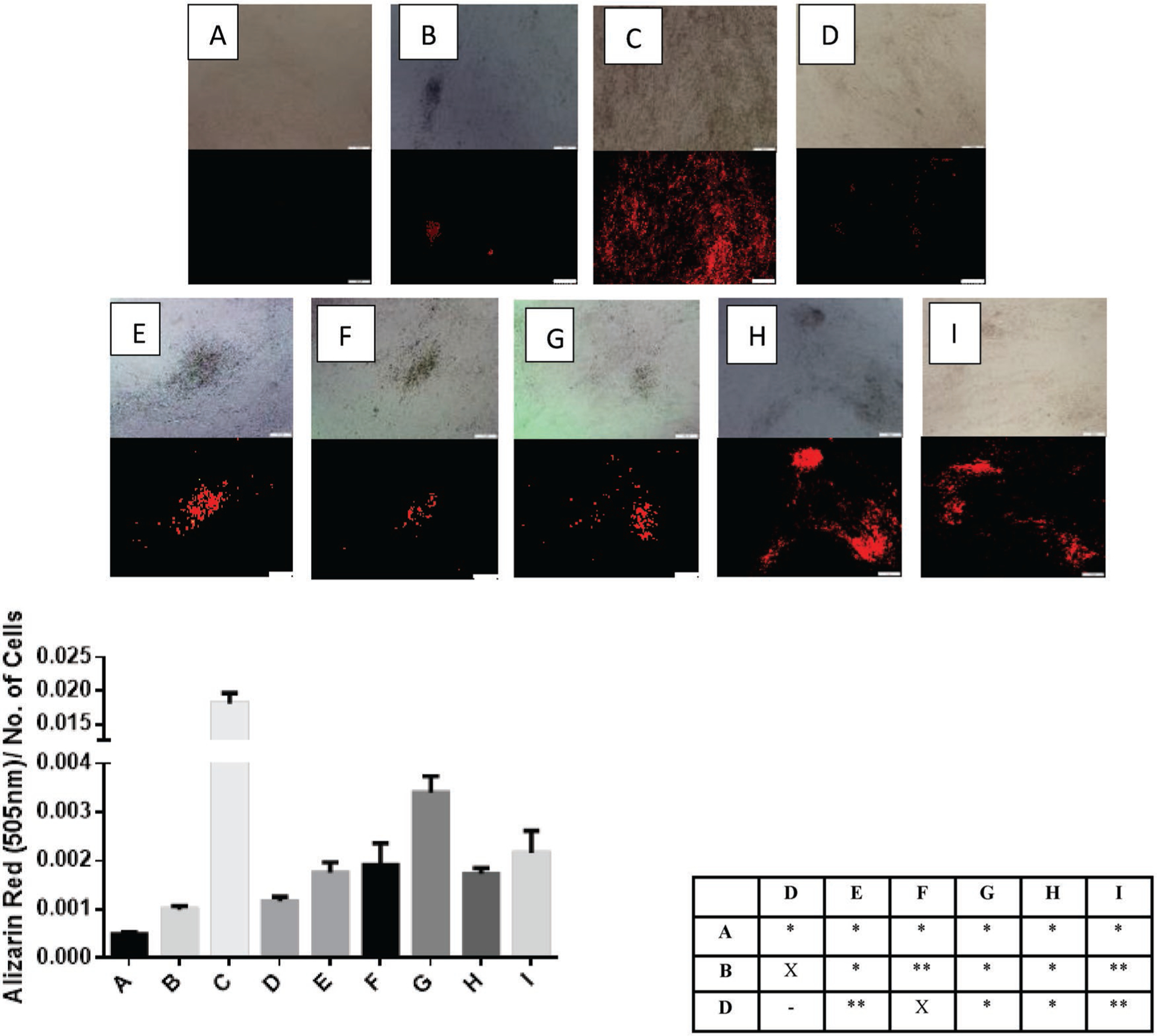
Xylenol orange stained samples, imaged 24 h after applied staining solution (top). All images were collected at constant exposure conditions and magnification as indicated by the scale bar set at 200 μm. (Bottom) Quantification of mineralization using Alizarin Red S staining solution and p-value summary between three negative controls and experimental groups. Values less than 0.05 are denoted as (*), and (**) for those less than 0.1, and X for no statistical significance. The labels indicate the different samples as follows: A = Stromal media control; B = miR-21-SNPs and miR-148b-GNPs without activation; C = Osteogenic differentiation media control; D = miR-21-SNPs and miR-148b-GNPs, light activated at 405 and 530 nm wavelengths of light, simultaneously, 16 h after transfection; E = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 40 h; F = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 40 h; G = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 64 h; H = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 64 h.
To quantify the extent of mineralization of each sample, ARS was applied to paraformaldehyde-fixed hASCs. All ARS images for all groups can be found in Figure S7 in the Supporting Information. In Figure 5, the measured absorbances of ARS after de-staining the samples, normalized to the number of hASCs, are shown. Groups with miR-21 and miR-148b co-delivery regardless of sequence of delivery or duration, showed significantly higher ARS than group A, the SM control. Group D which consisted of hASCs treated with GNP-miR-148b only after 530 nm irradiation, yielded higher ARS than the SM group but lower than the groups with both miRNAs delivered. The statistical differences at a 95% or 90% confidence level between the groups, specifically the three negative controls, A (SM), B (nonirradiated), and D (miR-148b), and the co-delivered groups are summarized in Figure 5. Group C (OM), was statistically significant with respect to all groups with p-values less than 0.001. To determine whether the increased mineralization effect of the combined miRNAs was artifactual or real, osteogenic gene marker expressions were measured and analyzed.
miR-148b has been demonstrated to upregulate alkaline phosphatase (ALP) activity de novo in bone marrow-derived stem cells.[8] To evaluate ALP activity, all experimental groups were measured for ALP gene expression at day 7, summarized in Figure 6. Group A (SM) was used to normalize −ΔΔCt values, with 18 s used as the housekeeping gene. All treated groups exhibited modest upregulation of ALP expression with respect to groups A and B. While miR-148b alone displayed lower ALP upregulation than the other groups except B (nonirradiated), no statistical significance at a 95% confidence level was found between experimental groups at this stage.
Figure 6.
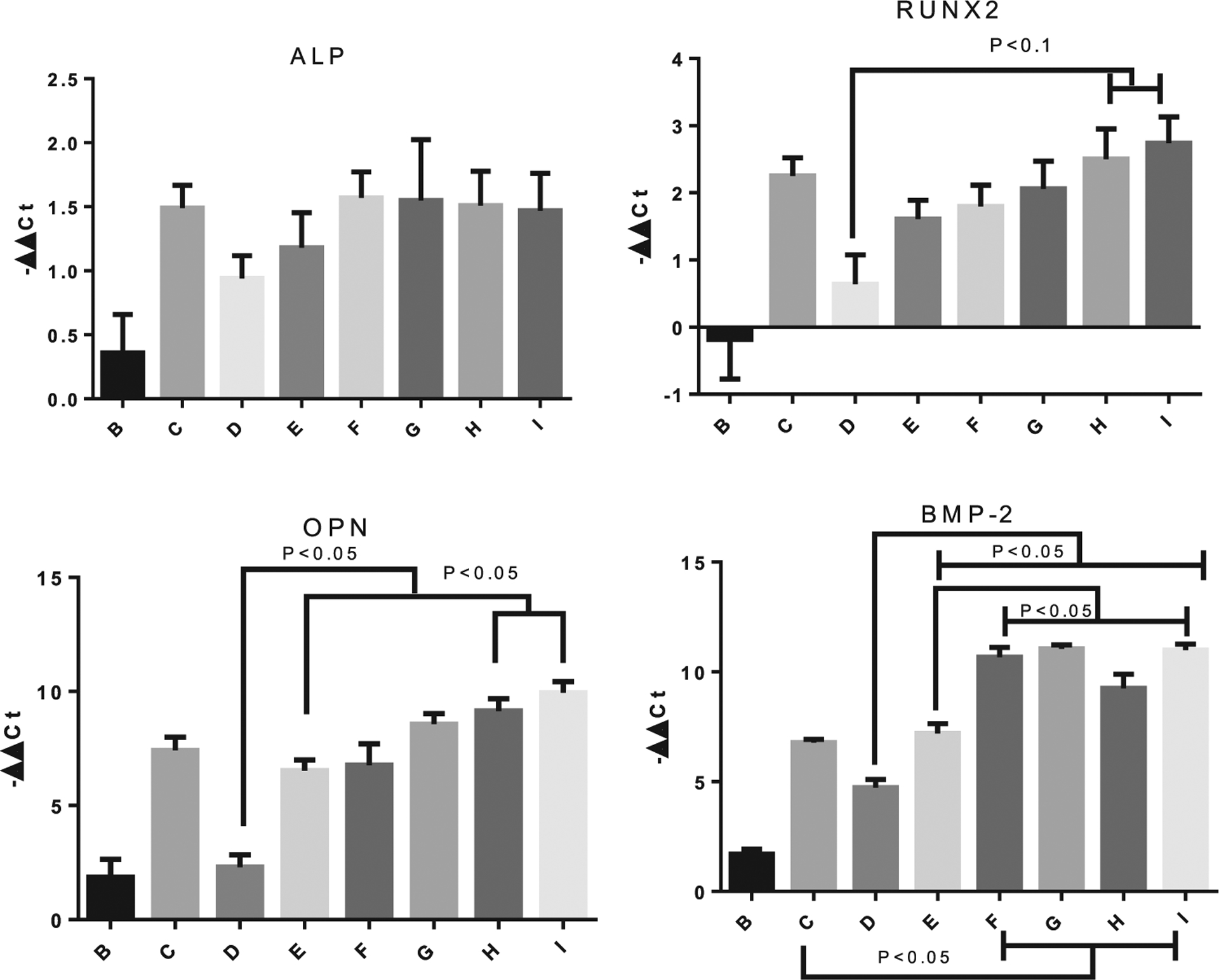
Days 7 and 21 PCR results. ALP expression was measured at day 7, while Runx2, OPN, and BMP-2 were measured at day 21. The legends indicate the different samples as follows: B = miR-21-SNPs and miR-148b-GNPs without activation; C = Osteogenic differentiation media control; D = miR148-GNPs light activated at 530 nm 16 h post-transfection; E = miR-21-SNPs and miR-148b-GNPs, light activated at 405 and 530 nm wavelengths of light, simultaneously, 16 h after transfection; F = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 40 h; G = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 40 h; H = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 64 h; I = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 64 h.
At day 21, gene expression levels of Runx2, BMP-2, and osteopontin (OPN) were measured to elucidate the osteogenic pathways of the different treatments. Runx2 is a mid-stage marker and typically a strong indicator of osteogenesis in stem cells.[60] In Figure 7, group C (OM) showed upregulation at this late time-point with −ΔΔCt mean of 2.25, and similar upregulation for groups E–I. There was no determined statistical differences at a 95% confidence level between the positive control and experimental groups, however, groups H and I exhibited statistical differences at 90% confidence level with respect to group D. Group D, with GNP-miR-148b only, displayed only modest upregulation compared to other groups, with a mean −ΔΔCt value of 0.64.
Figure 7.
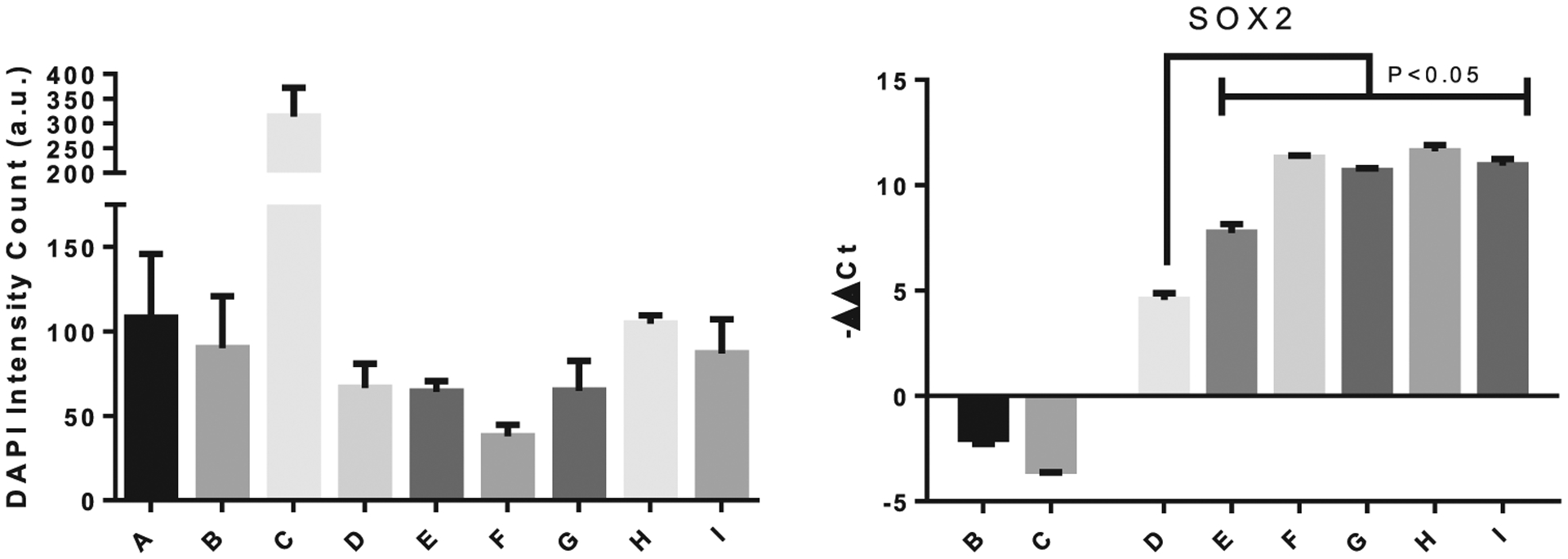
Cell count number, measured by DAPI staining of cells, and Sox2 gene expression at day 21. The legends indicate the different samples as follows: B = miR-21-SNPs and miR-148b-GNPs without activation; C = Osteogenic differentiation media control; D = miR148-GNPs light activated at 530 nm 16 h post-transfection; E = miR-21-SNPs and miR-148b-GNPs, light activated at 405 and 530 nm wavelengths of light, simultaneously, 16 h after transfection; F = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 40 h; G = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 40 h; H = miR-21-SNPs and miR-148b-GNPs, light activated at 405 nm 16 h post-transfection, followed by 530 nm at 64 h; I = miR-21-SNPs and miR-148b-GNPs, light activated at 530 nm 16 h post-transfection, followed by 405 nm at 64 h.
OPN and BMP-2 are mid-to-late stage gene markers, and highly expressed in osteoblast cells.[61] The gene expression results for OPN and BMP-2 displayed the most significant results for the enhanced effect the co-delivery of miR-148b and miR-21 has on osteogenesis (Figure 7). At this time point, groups E through I (miR-21 and 148b co-delivered) all showed nonstatistical differences from group C (OM) for OPN expression, while demonstrating statistical significance from groups D and B (148b only and nonirradiated) and robust upregulation with −ΔΔCt values ranging from 6.63 to 9.95 for the co-delivered groups, compared to 2.31 for miR-148b alone. Among the co-delivered groups, delivery of the miRNAs over a 48 h period (H and I) produced the most substantial and significantly highest OPN upregulation. Interestingly, groups F through I, in which delivery of miRNA mimics was performed sequentially over 24 or 48 h after the initial dose, showed statistically higher expressions of BMP-2, with values ranging from 9.26 to 11.05, than even the positive control group C with a reported value of 6.78. The gene expression results provide compelling evidence for the potent effect of co-delivering miR-21 and miR-148b on inducing an enhanced osteogenic pathway for hASCs.
In examining Sox2 gene expressions at day 7, single release groups of either miR-148b or miR-21 displayed downregulation of the potency marker, as predicted for cells undergoing differentiation (Figure S10, Supporting Information). However, for all miR-21 and miR-148b co-delivered samples (groups E through I), Sox2 is shown to be significantly upregulated. At day 21, this upregulation was amplified even more so, with −ΔΔCt values reaching as high as 11 (Figure 7). Examination of the light-activated only group, presented in Figure S11 in the Supporting Information, also demonstrated a modest increase in Sox2 expression. One explanation is the possibility of induced toxicity post-irradiation that might instigate cells to proliferate and regenerate. There have been reports of far-infrared exposure causing upregulation of Sox2 gene expressions in hBMSCs.[62] The cell count numbers for the different groups are also displayed in Figure 7 with a notable reduction in cell numbers for some of the groups. These results may indicate the effect of miR-21 in arresting the cell cycle pathway and driving them toward a specific lineage. However, the upregulation of Sox2 in groups E through I may also attest to a premature tumorigenic growth response in these stem cells that may not be captured within the timeframe of this study. Sox2 expression was positively correlated with cancer stem cells, specifically in osteosarcomas.[63] Despite the lack of extensive proliferation seen from the cell number trends, the occurrence of osteosarcomas will likely need to be validated in future in vivo trials with extended differentiation profiles.
Overall, histochemical staining and polymerase chain reaction (PCR) results suggest the co-delivery of miR-21 and miR-148b to have an increased effect on the osteogenic differentiation of hASCs. While there exist some discrepancies on the extent of mineralization and gene expressions between simultaneous delivery and sequential delivery at different times and sequences, they were not significant enough to downregulate or nullify miR-148b’s osteogenic ability, at least not within the time points selected for this study. On the contrary, they all demonstrated enhanced faculty in this regard. The implications of this suggest that, under a single dose of miR-21 and miR-148b conjugated to two different nanoparticles, hASC osteogenesis can be upregulated with temporally discrete delivery of the miRNA mimics. Interestingly, sequential delivery of the miRNA versus simultaneous delivery did exhibit greater upregulation of osteogenic markers and mineralization, significantly in some cases. This may be a result of overloading the RISC pathway during simultaneous release, and hence diminishing the efficacy of miRNA targeting, or that the fluctuating expressions of targeted genes and the delivery of miRNA mimics at 64 h coincided with an ideal amount of expression for knockdown or upregulation. To rule out the former hypothesis, miR-148b was delivered with an unrelated, anti-GFP siRNA. The data are presented in Figures S8 and S9 in the Supporting Information. No statistical upregulation from miR-148b alone was observed for this group, but indeed showed identical trends to the osteogenic ability of miR-148b alone. This strongly suggests that the enhancement in osteogenesis was a result of miR-21 and miR-148b co-delivery. To eliminate the influence of photo-induction at 64 h and other time points on osteogenic ability, different control groups comprised of nonmodified nanoparticles light activated at serial time points mirroring the main study are presented in Figure S13 in the Supporting Information, with gene expression and mineralization assays. No statistical differences were observed between illuminations at the distinct time points. However, a modest upregulation in Sox2 expression at day 21 was observed for all groups, which partly supports our hypothesis of the photothermal effect inducing some self-renewal capacity in the hASCs. However, this effect was much larger with the miRNAs delivered, which suggests a more intricate genetic effect on the pluripotency profile of these cells.
The presented model provides a working framework in which the delivery of two nucleic acid therapies could be co-delivered at different time points with a single applied dose. Complete staining assays and PCR results for chemically transfected miR-21 and 148b mimics are presented in Figures S8 and S9 in the Supporting Information. Additionally, the complete set of NP groups, including groups where both sets of NPs are present but illuminated at only a single wavelength, and PCR results used in this study are presented in Figures S10 and S11 in the Supporting Information. Cell counts of NP-transfected and chemically transfected hASCs for each group by measuring DAPI (diamidino-2-phenylindole) intensities are presented in Figure S12 in the Supporting Information.
2.5. Transfection and Uptake Efficiency
Transfection efficiencies measured by flow cytometry, and NP uptake efficiency measured by inductively coupled plasma atomic emission spectroscopy (ICP-AES) are presented in Figure S14 in the Supporting Information. Fluorescence measurements inside the cells indicated SNP transfection at 40%, while GNP at 20% within 4 h after addition. The low numbers are likely due to quenching effects on the fluorophore from the overlapping nanoparticles ‘optical states during conjugation or degradation from exposure of the 5’ fluorophore to Rnase enzymes in subcellular compartments.
Using ICP-AES, SNPs uptake efficiency, calculated by normalizing the particle mass inside cells to the total added, was determined to be 70.71 ± 2.14% at 16 h after transfection. Meanwhile, 40 h after transfection that efficiency decreased to 48.57 ± 14.29% while interestingly increasing again to 102.00 ± 5.00% after 64 h. These values suggest a role of the uptake efficiency in improving osteogenesis, where groups in which the second miRNA was released 64 h post-transfection showed the most dramatic results, possibly due to increased intake of nanoparticles at this time point. In the closed environment of the 2D cell cultures, the nanoparticles are likely undergoing exocytosis and re-entering cells during the incubation cycles prior to media exchange.
2.6. Raman Spectroscopy
To determine the compositional nature of the mineralized nodules formed, Raman spectroscopy was used to identify the unique vibrational modes of hydroxyapatite. In Figure 8, the peaks attributed to hydroxyapatite include the three minor peaks at 400–490, 570–625, and 1020–1095 cm−1, and a single intense peak at around 960 cm−1.[64] Both OM and miR-21/miR-148b samples displayed similar profiles to the hydroxyapatite sample, particularly at the sole signature peak of 960 cm−1. These results give confidence that the mineralization process was forming hydroxyapatite and not some other calcium salt as a result of nonbone-related biomineralization. As a negative control, the SM group was tested and displayed no hydroxyapatite crystal formation.
Figure 8.
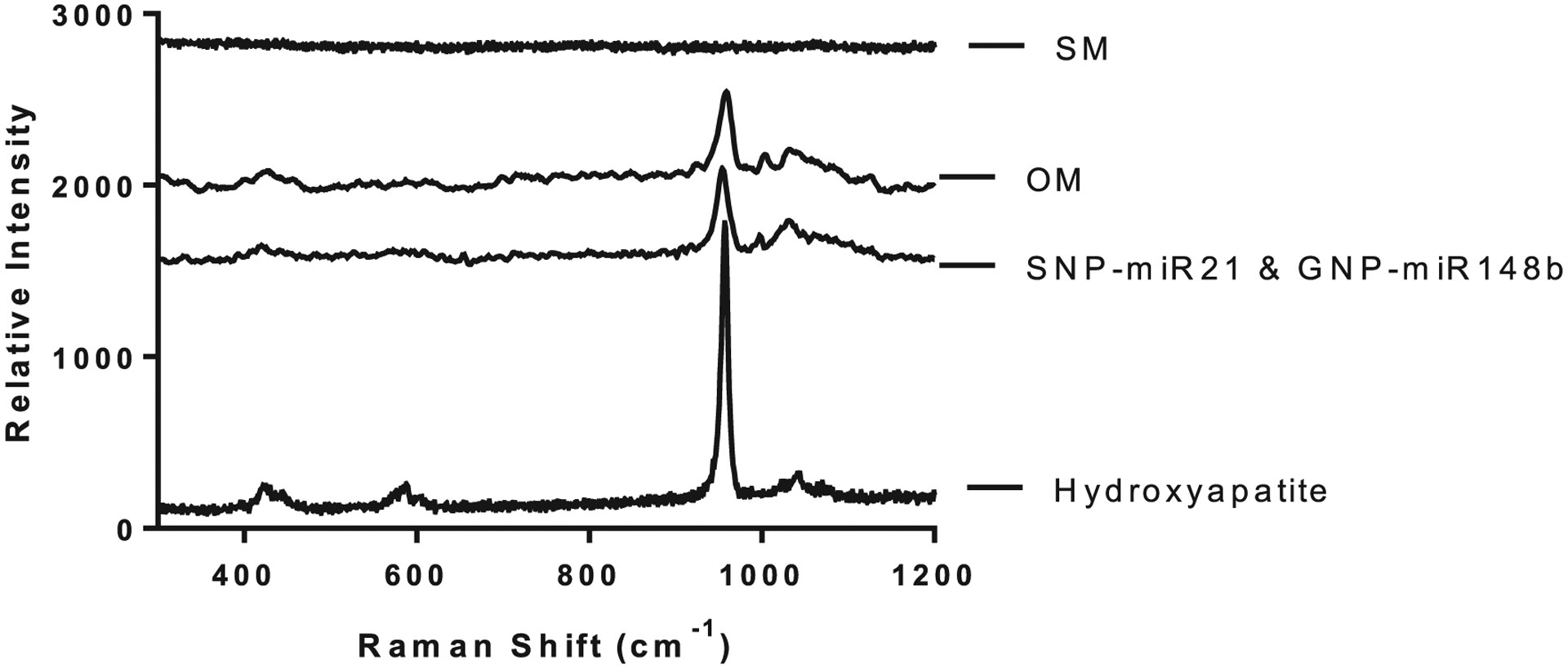
Raman spectroscopy results for Groups A (SM), C (OM), and I (SNP-miR-21 and GNP-148b) with hydroxyapatite powder used as a reference.
3. Conclusion
A co-delivery system consisting of GNPs and SNPs systems was designed with the ability to carry and photothermally release their miRNA payloads at distinct times and wavelengths of light. Mineralization assays demonstrated higher staining with both miR-21 and miR-148b delivery, with increased gene expressions of Runx2, BMP-2, and OPN. Herein, we demonstrated the potent ability of the retro-Diels–Alder chemistry to temporally release genes and drive augmented osteogenesis in a single dose of our developed gene therapy. Systems providing spatiotemporal control have the potential to improve outcomes in surgical reconstruction and regenerative medicine by precise modulation of tissue repair processes. Regulation of gene expression with miRNA mimics is a promising tool for enhancing control of tissue repair processes particularly when combined with endogenous MSC progenitors. Spatiotemporally directing the proliferation and osteogenic differentiation of these progenitors is of particular interest in the design of therapies for segmental bone defects, spinal fusion, and craniofacial reconstruction. The capability to target damaged tissues selectively and modulate potency and osteogenic differentiation provides a clinically relevant tool to stimulate osteogenic progenitors and improve segmental defect repair. While the results presented suggest promise, further studies in vivo are needed to address differences in diffusion, transfection efficiencies, and particle longevity in an active immune system. We hope to investigate this further in future works.
4. Experimental Section
Materials:
6-Maleimidohexanoic acid (90%), 2-furanmethanethiol (98%), methanol (>99.9%), isopropanol (>99.9%), dichloromethane (99.8%), N-hydroxysuccinimide (NHS, 98%), silver nitrate (99%), formaldehyde (36.5–38%), sodium hydroxide (98%, pellets), HPC (Mn = 80 000, 99%), antifoam A (100%), tris(2-carboxyethyl) phosphine hydrochloride solution (TCEP) (0.5 m), ARS staining solution (40 × 10−3 m), XO tetrasodium salt, glacial acetic acid, and GNPs stabilized in 0.1 × 10−3 m PBS were purchased and used as received from Sigma Aldrich (St. Louis, MO). EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride), Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM F-12), DPBS without Ca/Mg, lipofectamine RNAiMAX transfection reagent, DEPC-treated water, DAPI, transferrin from human serum/Texas Red conjugate and Opti-MEM reduced serum medium were purchased from ThermoFisher Scientific (Waltham, MA), FBS from Lonza, and hASCs at passage zero were acquired from LaCell (New Orleans, LA). Custom amine-modified oligonucleotides (miR-21: 3’ C6-NH2 2’OMe CAA CAC CAG UCG AUG GGC UGU Cy3 5’; miR-148b: 5’ C6-NH2 2’OMe UCA GUG CAU CAC AGA ACU UUG U 6-FAM 3’; anti-GFP siRNA: 5’ GCAAGCUGACCCUGAAGUU 3’) were ordered from Trilink Bio Technologies (San Diego, CA). Mounted 405 and 530 nm LED lights from ThorLabs, Inc. (Newton, NJ) were used for photo-activation experiments.
Cell Culture:
Commercially acquired hASCs from three different donors were cryopreserved at passage 0, and later thawed for subsequent use for in vitro experiments at passage 2. The cells were passaged in 25 cm2 flasks (BD Falcon, Franklin Lakes, NJ) in SM, which consisted of DMEM F-12, supplemented with 10% FBS and 1% antibiotic solution (10 000 μg mL−1 with penicillin and streptomycin) at 37 °C and 5% CO2.[9] For the OM, SM was supplemented with dexamethasone (1 × 10−9 m), ascorbic acid (50 × 10−6 m), and β-glycerophosphate (20 × 10−3 m).
GNP and SNP Modification:
HPC-SNPs were synthesized as described,[22,23] while PBS-stabilized, 80 nm diameter GNPs were purchased from Sigma-Aldrich. To conjugate both sets of nanoparticles with the miR-21 and miR-148b mimics, the surfaces were first conjugated with thiol- and carboxyl-terminated Diels–Alder cycloadducts as outlined in Ref. [22]. In the present study, miR-21 was coupled with silver, while miR-148b was coupled with gold. Briefly, 6-maleimidohexanoic acid (2.11 g) and 2-furanmethanethiol (0.5 g) were mixed in 20 mL methanol and DCM (1:1) for 7 days. The linkers were then mixed with the nanoparticles at approximately 8E9 particles per mL, in 1 mL aliquots for 24 h at room temperature to allow for surface attachment through thiol linkage. For oligonucleotide addition, the nanoparticles were first centrifuged (10 000 g, 10 min) and resuspended in isopropanol thrice, followed by mixing with 100 × 10−3 m (100 μL) EDC/NHS and the respective 5’amine miRNA mimics (4 × 10−6 m, 30 μL) for 24 h at room temperature to allow for covalent coupling. Verification of conjugation was tested after centrifugation and resuspension in DEPC water, by displacing the ligands with TCEP treatment (10 μL), and measuring supernatant fluorescence at 525 nm for FAM, and 575 nm for Cy3.
Photothermal Release of Oligonucleotides:
To test and compare the photothermal release rates of SNPs and GNPs, light irradiation studies were set up. Equal concentrations (200 ppm) and volumes (100 μL) of each conjugated nanoparticle were loaded separately into a 35 mm Petri dish, after which they were irradiated at either 405 or 530 nm wavelength light. The measured power outputs for each LED light source were reported as 570 mW at 405 nm, and 317 mW at 530 nm. The irradiation times were selected to normalize the total energy at each wavelength. 25 μL of each sample was used at each time point, and later collected, diluted, and spun down to measure the supernatant fluorescence for either FAM or Cy3. All measurements were normalized to a nonirradiated control as well as a TCEP-treated sample used to represent the total particle loading. Additionally, zeta potential measurements of the nanoparticles were carried out to monitor the change in surface charge as the negatively charged molecules were cleaved and removed.
SHG Measurement:
Nanoparticles for SHG measurements were prepared as follows: GNPs and SNPs were prepared in colloidal aqueous suspension using chemical reduction of chloroauric acid and silver nitrate, respectively.[19,20,65–67] All chemicals were purchased from Alfa Aesar (Haverhill, MA) and used without further purification in ultrapure water. The colloidal gold nanoparticle sample was prepared by a seeded-growth method using hydroquinone and sodium citrate as reducing agents.[19,65,66] Briefly, an aqueous solution of 300 μL of 25 × 10−3 m chloroauric acid in 30 mL of ultrapure water was brought to reflux under vigorous stirring conditions, followed by the addition of 900 μL of 34 × 10−3 m sodium citrate to synthesize gold seeds. The colloidal gold seed solution underwent a color change from pale yellow to deep red after 15 min and was then removed from heating and cooled to room temperature. For the seeded-growth synthesis of gold nanoparticles, 200 μL of prepared gold seeds and 100 μL of 25 × 10−3 m chloroauric acid was added to 10 mL of water, followed by the addition of 30 μL of 34 × 10−3 m sodium citrate, and 100 μL of 30 × 10−3 m hydroquinone at room temperature under vigorous stirring conditions for 60 min. For the silver nanoparticle synthesis, an aqueous solution was first made by mixing 250 μL of 59 × 10−3 m silver nitrate, 1 mL of 34 × 10−3 m sodium citrate, 30 μL of 98 × 10−6 m potassium iodide, and 1.25 mL of ultrapure water. This solution was added to 100 μL of 100 × 10−3 m ascorbic acid solution in 47.5 mL of boiling water and the mixture was refluxed for 60 min under vigorous stirring conditions to prepare the silver nanoparticles.[20,67] The nanoparticles were further modified with the Diels–Alder-linked miRNA mimics as described above.
The experimental setup for the SHG, which was consisted of an ultrafast laser system, an optical setup, and a high-sensitivity spectroscopy detector, was described previously.[19,20,45] A portion of the titanium:sapphire oscillator laser beam, centered at 800 nm with 75 fs pulses at 80 MHz repetition rate and an average power of 2.6 W, was used to seed the amplifier laser which pumped an optical parametric amplifier (OPA) for tunable-wavelength laser light. The beam from the OPA was used for irradiation studies at 400 and 530 nm, corresponding to the plasmon resonances of the SNPs and GNPs, respectively. For SHG measurements, the remaining portion of the 800 nm oscillator laser was attenuated to an average power of 790 mW and was focused to a 1 cm quartz cuvette containing the nanoparticle sample. An integrated data acquisition program controlled a magnetic stir bar and a beam block to open and shut every 20 s for collecting the SHG and background spectra in order to measure the background-subtracted SHG signal as a function of time.
In Vitro Photo-Release of SNPs and GNPs:
To monitor the in vitro release of the fluorescently-labeled payload, hASCs were transfected with each of the nanoparticles. For this experiment, the FAM fluorophore was used on both nanoparticle sets. The hASCs were initially seeded at a density of 3 00 000 cells per dish, on 35 mm glass-bottom Petri dishes in SM and incubated at 37 °C, 5% CO2. After 24 h, 100 uL each of the conjugated GNPs and SNPs were added separately to the cells for transfection and incubated for an additional 4 h. The groups tested included hASCs with GNPs-FAM nonirradiated, SNPs-FAM nonirradiated, GNPs-FAM irradiated at 530 nm, GNPs-FAM irradiated at 405 nm, SNPs-FAM irradiated at 530 nm, and SNPs-FAM irradiated at 405 nm. Three dishes were used for each group (NA/450 nm/530 nm), for each nanoparticle set, gold and silver, with two images taken per dish for a total of 18 images per set. The merged nanoparticle and fluorophore confocal images are presented in the main text. Additional images, along with rotated z-stacks to highlight the de-localization of the probes, are provided in Figure S5 in the Supporting Information.
All cells treated with SNPs were additionally stained with the endosomal tracking agent transferrin, tagged with Texas Red, as described in literature.[24,50] The transfected cells were placed in Opti-MEM medium prior to LED irradiation. Cells were irradiated at the 405 nm LED light (50 J), and at 530 nm (565 J). Post-irradiation, the dishes were incubated at 37 °C for an additional 4 h prior to imaging. The dishes were mounted in an Olympus FV10i-LIV confocal microscope at objective 60X phase contrast water immersion/NA1.2. SNPs-FAM samples were viewed at blue (405 nm/420–520 nm) and FITC (495 nm/519 nm) channels, while GNPs-FAM samples were viewed at POPO-3 (533 nm/574 nm) and FITC (495 nm/519 nm) channels.
The Manders coefficient was calculated using FIJI software. Initially, background subtraction was run by selecting a background region of interest and subtracting it from the image. Furthermore, the threshold was calculated prior to running the Coloc 2 plug-in in FIJI. The threshold-accounted coefficients were used for reporting, specifically for the green channel in which the fluorophore was imaged.
Osteogenic Induction:
To assess the effect of co-delivering miR-21 and miR-148b on the osteogenic induction of hASCs, 2 × 105 cells per well were seeded in 24-well tissue culture plates (BD Falcon, Franklin Lakes, NJ). Prior to treatment, the hASCs were left to incubate in SM at 37 °C for 24 h. A total of 13 groups in duplicates were set up to measure different effects of sequence and dosage time, as well as controls. These are summarized in Table 1. Only results for groups A through I are depicted in the main article, while comprehensive results with the remaining four control groups can be found in the Supporting Information. Prior to transfection, all groups to be irradiated were placed in Opti-MEM solution. For transfection, 5 μL of SNP-miR-21 (≈8E9 particles mL−1, 0.123 × 10−6 m miR-21) and/or 10 μL GNP-miR-148b (≈4E9 particles mL−1, 0.128 × 10−6 m miR-148b) suspended in Opti-MEM were added to the respective groups for 16 h prior to photo-release. Where indicated, the treated cells were exposed to 405 nm LED light (50 J), and/or 530 nm LED light (565 J). The cells were maintained in the same Opti-MEM at 37 °C in between the different photo-release wavelengths, as well as for an additional 24 h after the final irradiation step, prior to replacing media with SM. Thereafter, the culture media was replaced every 3 days with SM. Evaluation of osteogenesis was conducted at day 7 and day 21 using PCR, and histochemical staining at day 21.
For investigating the effect of the miRNAs alone, additional groups in which the miRNA mimics were chemically transfected using RNAiMAX transfection reagent only, per the manufacturer’s protocol, were set up and are presented in the Supporting Information. Included with this set was an unrelated anti-GFP control siRNA co-delivered with miR-148b.
Histochemical Staining:
To evaluate mineralization of the cultured cells after 21 days, ARS and XO staining were conducted independently. In the latter case, the wells were rinsed with DPBS and fixed with 4% paraformaldehyde solution for 15 min at room temperature. They were then rinsed again and stained with 40 × 10−3 m ARS staining solution for 20 min.[9] The dye was removed and wells were washed with DPBS five times prior to imaging on an Olympus light microscope. For quantification, 10% acetic acid was used for de-staining, as outlined in ref. [68]. Briefly, 200 uL of 10% acetic acid was added to each well, and incubated at room temperature for 30 min. The well contents were scraped and collected in microcentrifuge tubes, followed by heating at 85 °C for 10 min. After leaving on ice for 5 min, the solid components were centrifuged out, the supernatant collected and measured at 505 nm absorbance. All values were normalized to the number of hASCs at day 21, measured by DAPI staining.
For XO staining, a 20 × 10−3 m stock solution was prepared, and transferred to the live cells in SM at a final concentration of 20 × 10−6 m. The stained samples were left overnight for 24 h at 37 °C, prior to rinsing with DPBS and imaging.
Quantitative Real Time Polymerase Chain Reaction (qPCR):
To measure gene expressions at day 7 and day 21, RNA was extracted from the hASCs with PureLink RNA Mini Kit (Thermo Fisher), following the manufacturer’s protocol. The extracted RNA was converted to cDNA prior to amplification using the Thermo Scientific Verso cDNA Synthesis Kit, and running it on the QuantStudio 3 Real-Time PCR system.
For PCR amplification of the cDNA, the following primer sequences (Integrated DNA Technologies, Coralville, IA) were used to assess osteogenic and pluripotent gene expressions (forward and reverse): ALP, 5′-AATATGCCCTGGAGCTTCAGAA-3′ and 5′-CCATCCC-ATCTCCCAGGAA-3′; RUNX2, 5′-GCAAGGTTCAACGATCTGAGATT-3′ and 5′-AGACGGTTATGGTCAAGGTGAAA-3′; OPN, 5′-TTCCAAGTAAG-TCCAACGAAAG-3′ and 5′-GTGACCAGTTCATCAGATTCAT-3′; BMP-2, 5′-ACTACCAGAAACGAGTGGGAA-3′ and 5′-GCATCTGTTCTCGGAAAACCT-3′;SOX2, 5′-TACAGCATGTCCTACTCGCAG-3′ and 5′-GAGGAAG-AGGTAACCACAGGG-3′. All genes were normalized to the 18s rRNA housekeeping gene and −ΔΔCt values were calculated using the ΔΔCt method.[69]
Transfection and Uptake Efficiency:
The transfection efficiency by fluorescence measurement was performed using flow cytometry analysis. 4 h after transfection, the cells were trypsinized and fixed with 4%paraformaldehyde, and suspended in 96-well plates for analysis. Cy3-conjugated NPs were used for all measurements. Red fluorescence of the cells was measured using BD SORP LSRFortessa Cytometer (Franklin Lakes, NJ). 50 000 events were counted, with two replicates used for each analyzed group. The percentage of hASCs transfected with either Cy3-GNPs or Cy3-SNPs was determined using the phycoerythrin (PE) channel to view the fluorophore, while FITC was used for NP plasmon scattering. The samples were run at a flow rate of 0.5 μL s−1 and all analyses were performed using FlowJo software.
To measure the uptake of silver nanoparticles at the proposed illumination time points (16 h, 40 h, and 64 h), ICP-AES (Perkin Elmer Optima 5300 DV Optical Emission Spectrometer, Walthon, MA), was used to measure silver quantities inside the cells. After fixation, the cells were dehydrated overnight at 37 °C, and digested with 37% HCl solution. 7% diluted samples were prepared for analysis.
Raman Spectroscopy:
To empirically corroborate the hydroxyapatite identity of the mineralized nodules produced by hASCs after 21 days, the following groups were prepared for Raman analysis: A, C, and I, as well as commercial hydroxyapatite powder as a reference. The adhered hASCs were detached and loaded onto a glass microscope slide in DPBS suspension prior to analysis. A Horiba LabRam system was used for all measurements, using 488 nm laser and 50X objective. The selected scan range used was 300–1200 cm−1.
Statistical Analysis:
Results presented were expressed as mean ± SEM, and analyzed by analysis of variance for comparison of three or more groups, in which a p-value < 0.05, or 0.1 where indicated, was deemed significant.
Supplementary Material
Acknowledgements
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program under Award No. W81XWH-18-1-0115 and the NIH (RDE024790A). The opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. The authors of this study are grateful to Lisa Bernsten and Cong Chen for many insightful discussions about the roles of miR-21 and miR-148b. The authors thank Maxwell Wetherington for assisting with Raman Spectroscopy, Trevor Clarke for his help with transmission electron microscopy, and Laura Jean Liermann for her assistance with ICP-AES.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/smll.201902864.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Mohammad Abu-Laban, The Department of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802, USA.
Prakash Hamal, The Department of Chemistry, Louisiana State University, Baton Rouge, LA 70803, USA.
Julien H. Arrizabalaga, The Department of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802, USA
Anoosha Forghani, The Department of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802, USA.
Asela S. Dikkumbura, The Department of Chemistry, Louisiana State University, Baton Rouge, LA 70803, USA
Raju R. Kumal, John and Willie Leone Family Department of Energy and Mineral Engineering, Pennsylvania State University, University Park, PA 16802, USA
Louis H. Haber, The Department of Chemistry, Louisiana State University, Baton Rouge, LA 70803, USA
Daniel J. Hayes, The Department of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802, USA Materials Research Institute, Millennium Science Complex, Pennsylvania State University, University Park, PA 16802, USA; The Huck Institute of the Life Sciences, Millennium Science Complex, Pennsylvania State University, University Park, PA 16802, USA.
References
- [1].Sen C, Ghatak S, Am. J. Pathol 2015, 185, 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Frith JE, Porrello ER, Cooper-White JJ, Stem Cells Transl. Med 2014, 3, 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Catalanotto C, Cogoni C, Zardo G, Int. J. Mol. Sci 2016, 17, 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kapinas K, Delany AM, Arthritis Res. Ther 2011, 13, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fang S, Deng Y, Gu P, Fan X, Int. J. Mol. Sci 2015, 16, 8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tang Y, Zhang L, Tu T, Li Y, Murray D, Tu Q, Chen JJ, J. Cell. Mol. Med 2018, 22, 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen S, Zheng Y, Zhang S, Jia L, Zhou Y, Stem Cell Rep. 2017, 8, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Aldred SF, Fedorov Y, PLoS One 2009, 4, e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qureshi AT, Monroe WT, Dasa V, Gimble JM, Hayes DJ, Biomaterials 2013, 34, 7799. [DOI] [PubMed] [Google Scholar]
- [10].Li K-C, Lo S-C, Sung L-Y, Liao Y-H, Chang Y-H, Hu Y-C, J. Tissue Eng. Regener. Med 2017, 11, 3068. [DOI] [PubMed] [Google Scholar]
- [11].Xu J, Zhang W, Lv Q, Zhu D, Oncol. Rep 2015, 33, 3108. [DOI] [PubMed] [Google Scholar]
- [12].Sekar D, Saravanan S, Karikalan K, Thirugnanasambantham K, Lalitha P, Islam V, Curr. Pharm. Biotechnol 2015, 16, 43. [DOI] [PubMed] [Google Scholar]
- [13].Zhang J, Wang J, Zhao F, Liu Q, Jiang K, Yang G, Clin. Chim. Acta 2010, 411, 846. [DOI] [PubMed] [Google Scholar]
- [14].Singh SK, Marisetty A, Sathyan P, Kagalwala M, Zhao Z, Majumder S, Stem Cell Res. 2015, 15, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alavi M, Karimi N, Safaei M, Adv. Pharm. Bull 2017, 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lockney D, Franzen S, Lommel S, Methods Mol. Biol 2011, 726, 207. [DOI] [PubMed] [Google Scholar]
- [17].Bruno BJ, Miller GD, Lim CS, Ther. Delivery 2013, 4, 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hubbell JA, Chilkoti A, Science 2012, 337, 303. [DOI] [PubMed] [Google Scholar]
- [19].Kumal RR, Landry CR, Abu-Laban M, Hayes DJ, Haber LH, Langmuir 2015, 31, 9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kumal RR, Abu-Laban M, Landry CR, Kruger B, Zhang Z, Hayes DJ, Haber LH, Langmuir 2016, 32, 10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qureshi AT, Doyle A, Chen C, Coulon D, Dasa V, Del Piero F, Levi B, Monroe WT, Gimble JM, Hayes DJ, Acta Biomater. 2015, 12, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Abu-Laban M, Kumal RR, Casey J, Becca J, LaMaster D, Pacheco CN, Sykes DG, Jensen L, Haber LH, Hayes DJ, J. Colloid Interface Sci 2018, 526, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Qureshi AT, Monroe WT, Lopez MJ, Janes ME, Dasa V, Park S, Amirsadeghi A, Hayes DJ, J. Appl. Polym. Sci 2011, 120, 3042. [Google Scholar]
- [24].Brown P, Qureshi A, Moll A, Hayes D, Monroe W, ACS Nano 2013, 7, 2948. [DOI] [PubMed] [Google Scholar]
- [25].Brongersma ML, Halas NJ, Nordlander P, Nat. Nanotechnol 2015, 10, 25. [DOI] [PubMed] [Google Scholar]
- [26].Manjavacas A, Liu J, Kulkarni V, Nordlander P, ACS Nano 2014, 8, 7630. [DOI] [PubMed] [Google Scholar]
- [27].Li X, Xiao D, Zhang Z, New J. Phys 2013, 15, e023011. [Google Scholar]
- [28].Barnaby SN, Lee A, Mirkin CA, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bakhtiari A, Hsiao D, Jin G, Gates B, Branda N, Angew. Chem., Int. Ed 2009, 48, 4166. [DOI] [PubMed] [Google Scholar]
- [30].Dewar M, Pierini A, J. Am. Chem. Soc, 1984, 106, 203. [Google Scholar]
- [31].Alston P, Ottenbrite R, Cohen T, J. Org. Chem 1979, 44, 3739. [Google Scholar]
- [32].Csende F, Fülöp F, Stájer G, Curr. Org. Synth 2008, 5, 173. [Google Scholar]
- [33].Pakiari A, Jamshidi Z, J. Phys. Chem. A 2010, 114, 9212. [DOI] [PubMed] [Google Scholar]
- [34].Zhou L, Swearer D, Zhang C, Robatjazi H, Zhao H, Henderson L, Dong L, Christopher P, Carter E, Nordlander P, Halas N, Science 2018, 362, 69. [DOI] [PubMed] [Google Scholar]
- [35].Zhang X, Li X, Reish M, Zhang D, Su N, Gutierrez Y, Moreno F, Yang W, Everitt H, Liu J, Nano Lett. 2018, 18, 1714. [DOI] [PubMed] [Google Scholar]
- [36].Eisenthal KB, Chem. Rev 2006, 106, 1462. [DOI] [PubMed] [Google Scholar]
- [37].Gonella G, Dai H-L, Langmuir 2014, 30, 2588. [DOI] [PubMed] [Google Scholar]
- [38].Haber LH, Kwok SJ, Semeraro M, Eisenthal KB, Chem. Phys. Lett 2011, 507, 11. [Google Scholar]
- [39].Hamal P, Nguyenhuu H, Don VS, Kumal RR, Kumar R, McCarley RL, Haber LH, J. Phys. Chem. B 2019, 123, 7722. [DOI] [PubMed] [Google Scholar]
- [40].Khoury RA, Ranasinghe JC, Dikkumbura AS, Hamal P, Kumal RR, Karam TE, Smith HT, Haber LH, J. Phys. Chem. C 2018, 122, 24400. [Google Scholar]
- [41].Jen S-H, Dai H-L, Gonella G, J. Phys. Chem. C 2010, 114, 4302. [Google Scholar]
- [42].Kumal RR, Nguyenhuu H, Winter JE, McCarley RL, Haber LH, J. Phys. Chem. C 2017, 121, 15851. [Google Scholar]
- [43].Srivastava A, Eisenthal KB, Chem. Phys. Lett 1998, 292, 345. [Google Scholar]
- [44].Liu Y, Yan EC, Zhao X, Eisenthal KB, Langmuir 2001, 17, 2063. [Google Scholar]
- [45].Kumal RR, Abu-Laban M, Hamal P, Kruger B, Smith HT, Hayes DJ, Haber LH, J. Phys. Chem. C 2018, 122, 19699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goodman AM, Hogan NJ, Gottheim S, Li C, Clare SE, Halas NJ, ACS Nano 2017, 11, 171. [DOI] [PubMed] [Google Scholar]
- [47].Hatef A, Fortin-Deschênes S, Boulais E, Lesage F, Meunier M, Int. J. Heat Mass Transfer 2015, 89, 866. [Google Scholar]
- [48].Stoein JD, Wang RJ, Proc. Natl. Acad. Sci. U. S. A 1974, 71, 3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Umadevi M, in Surface Plasmon Enhanced, Coupled and Controlled Fluorescence (Ed: Geddes CD), Wiley, New York: 2017, Ch. 11. [Google Scholar]
- [50].Kapur A, Medina SH, Wang W, Palui G, Ji X, Schneider JP, Mattoussi H, ACS Omega 2018, 3, 17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S, Adv. Mater 2009, 21, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li Y, Monteiro-Riviere N, Nanomedicine 2016, 11, 3185. [DOI] [PubMed] [Google Scholar]
- [53].Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC, J. Controlled Release 2014, 190, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang S, Gao H, Bao G, ACS Nano 2015, 9, 8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim Y, Hwang S, Bae Y, Jung J, Stem Cells 2009, 27, 3093. [DOI] [PubMed] [Google Scholar]
- [56].Li X, Guo L, Liu Y, Su Y, Xie Y, Du J, Zhou J, Ding G, Wang H, Bai Y, Liu Y, Biochem. Biophys. Res. Commun 2017, 493, 928. [DOI] [PubMed] [Google Scholar]
- [57].Hustedt JW, Blizzard DJ, Yale J Biol. Med 2014, 87, 549. [PMC free article] [PubMed] [Google Scholar]
- [58].Wang Y-H, Liu Y, Maye P, Rowe DW, Biotechnol. Prog 2006, 22, 1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Moester M, Schoeman M, Oudshoorn I, van Beusekom M, Mol I, Kaijzel E, Löwik C, de Rooij K, Biochem. Biophys. Res. Commun 2014, 443, 80. [DOI] [PubMed] [Google Scholar]
- [60].Xu J, Li Z, Hou Y, Fang W, Am. J. Transl. Res 2015, 7, 2527. [PMC free article] [PubMed] [Google Scholar]
- [61].Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y, Li W, Huang Y, Zhang X, Shao C, Roberts A, Rabson A, Ren G, Zhang Y, Wang Y, Denhardt D, Shi Y, Stem Cells 2014, 32, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jeong Y-M, Cheng XW, Lee S, Lee KH, Cho H, Kang JH, Kim W, Sci. Rep 2017, 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Maurizi G, Verma N, Gadi A, Mansukhani A, Basilico C, Oncogene 2018, 37, 4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cuscó R, Guitián F, de Aza S, Artúsa L, J. Eur. Ceram. Soc 1998, 18, 1301. [Google Scholar]
- [65].Perrault SD, Chan WC, J. Am. Chem. Soc 2009, 131, 17042. [DOI] [PubMed] [Google Scholar]
- [66].Brown KR, Walter DG, Natan MJ, Chem. Mater 2000, 12, 306. [Google Scholar]
- [67].Li H, Xia H, Wang D, Tao X, Langmuir 2013, 29, 5074. [DOI] [PubMed] [Google Scholar]
- [68].Gregory C, Gunn W, Peister A, Prockop D, Anal. Biochem 2004, 329, 77. [DOI] [PubMed] [Google Scholar]
- [69].Livak K, Shmittgen T, Methods 2001, 25, 402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


