Abstract
In lung adenocarcinoma, oncogenic EGFR mutations co-occur with many tumor suppressor gene alterations, however the extent to which these contribute to tumor growth and response to therapy in vivo remains largely unknown. By quantifying the effects of inactivating ten putative tumor suppressor genes in a mouse model of EGFR-driven Trp53-deficient lung adenocarcinoma, we found that Apc, Rb1, or Rbm10 inactivation strongly promoted tumor growth. Unexpectedly, inactivation of Lkb1 or Setd2 – the strongest drivers of growth in a Kras-driven model – reduced EGFR-driven tumor growth. These results are consistent with mutational frequencies in human EGFR- and KRAS-driven lung adenocarcinomas. Furthermore, Keap1 inactivation reduced the sensitivity of EGFR-driven tumors to the EGFR inhibitor osimertinib and mutations in the KEAP1 pathway were associated with decreased time on tyrosine kinase inhibitor treatment in patients. Our study highlights how the impact of genetic alterations differ across oncogenic contexts and that the fitness landscape shifts upon treatment.
Introduction
During tumor evolution, cancer cells accumulate alterations in oncogenes and tumor suppressor genes, which contribute to many of the hallmarks of cancer (1). Despite their extensive genomic complexity, tumors are frequently classified based on the presence of a single oncogenic driver mutation, while the function of co-incident tumor suppressor gene alterations is largely ignored. There is emerging evidence that the interplay between oncogenic drivers and tumor suppressor genes may influence tumor fitness and impact treatment response (2). However, the combinatorially vast landscape of genomic alterations makes it difficult, except in the most extreme cases, to glean information about the epistatic interactions between tumor suppressor genes and oncogenes from human cancer sequencing data alone (3). This complexity makes inferring the relationship between genotype and therapy response even more tenuous.
Recently, high-throughput, tractable systems that combine autochthonous mouse modeling and genome editing have been developed to directly uncover the functional consequences of genetic alterations during tumorigenesis in vivo (4–10). However, very few studies have investigated the biological consequences of inactivating tumor suppressor genes in the context of different oncogenic drivers in vivo, and existing knowledge about the role of specific genetic alterations in tumor suppressor genes has been primarily inferred from correlative human studies (11–13).
In lung adenocarcinoma, EGFR and KRAS are the two most frequently mutated oncogenic driver genes and occur within a background of diverse putative tumor suppressor gene alterations (2, 11, 12, 14). Among these, TP53 is the most commonly mutated tumor suppressor gene in both oncogenic EGFR- and KRAS-driven lung adenocarcinoma, consistent with the importance of disrupting this pathway during lung cancer development (2, 13, 15–18). Interestingly, many other putative tumor suppressor genes are mutated at different frequencies in oncogenic EGFR- and KRAS-driven lung adenocarcinomas (2, 11, 15). Whether these differences are due to different fitness effects that depend on the oncogenic context has never been tested experimentally. Indeed, previous studies on tumor suppressor genes in lung cancer models in vivo have been primarily conducted in the context of oncogenic Kras-driven tumors, while the functional importance of different tumor suppressor genes in EGFR-driven lung tumors remains largely unstudied (Supplementary Fig. S1A).
In addition to driving growth, inactivation of tumor suppressor genes may affect therapeutic responses. In advanced EGFR mutant lung adenocarcinomas, treatment with EGFR tyrosine kinase inhibitors (TKIs) is the first line of therapy (19–22). Response rates to TKIs are high, however, there is large variability in the depth and duration of response between patients, and acquired resistance inevitably occurs (14). Genomic alterations, including those in RB1 and TP53, have been found to correlate with clinical outcomes on TKI treatment (13, 17, 23). However, given the complexity and diversity of genomic alterations in these tumors, the functional contribution of tumor suppressor genotypes to TKI sensitivity and drug resistance remains poorly understood.
To quantify the functional importance of a panel of ten diverse putative tumor suppressor genes on oncogenic EGFR-driven lung tumor growth in vivo, we coupled multiplexed CRISPR–Cas9-mediated somatic genome editing and tumor barcode sequencing (Tuba-seq) with a novel genetically engineered mouse model of EGFRL858R-driven Trp53-deficient lung cancer. Through the comparison of tumor suppressor effects in oncogenic EGFR- and Kras-driven lung cancer models, we uncovered prevalent epistasis between the oncogenic drivers and tumor suppressors, which explains the different mutational spectra of these tumor suppressor genes in oncogenic EGFR- and KRAS-driven human lung adenocarcinomas. Moreover, we established a direct causal link between tumor suppressor genotypes and differential responses to osimertinib treatment in EGFR-driven lung adenocarcinoma that is supported by correlative human mutational datasets.
Results
Development of a lentiviral-Cre based mouse model of oncogenic EGFR-driven lung adenocarcinoma
The use of lentiviral vectors to initiate tumors in genetically engineered mouse models of human cancer enables control of tumor multiplicity, tumor barcoding to map clonality, and the delivery of lentivirus-encoded cDNAs, shRNAs, and sgRNAs to modify neoplastic cells (6, 8, 24, 25). The simplicity of viral-Cre initiated models of oncogenic Kras-driven lung cancer has enabled the analysis of many genes that co-operate to drive tumor growth within these autochthonous mouse models (Supplementary Fig. S1A) (26). To permit the generation of virally-initiated oncogenic EGFR-driven Trp53-deficient lung tumors, we bred mice with an rtTA/tetracycline-inducible transgene encoding the common lung adenocarcinoma-associated EGFRL858R mutant (TetO-EGFRL858R), a Cre-regulated rtTA transgene (Rosa26CAGs-LSL-rtTA3-IRES-mKate, abbreviated R26RIK), and homozygous Trp53 floxed alleles (p53flox/flox) (27–29). In these TetO-EGFRL858R;Rosa26RIK;p53flox/flox (EGFR;p53) mice, lentiviral-Cre transduction of lung epithelial cells leads to the expression of rtTA and mKate as well as the inactivation of Trp53. Co-incident doxycycline treatment induces the expression of oncogenic EGFR (Fig. 1A).
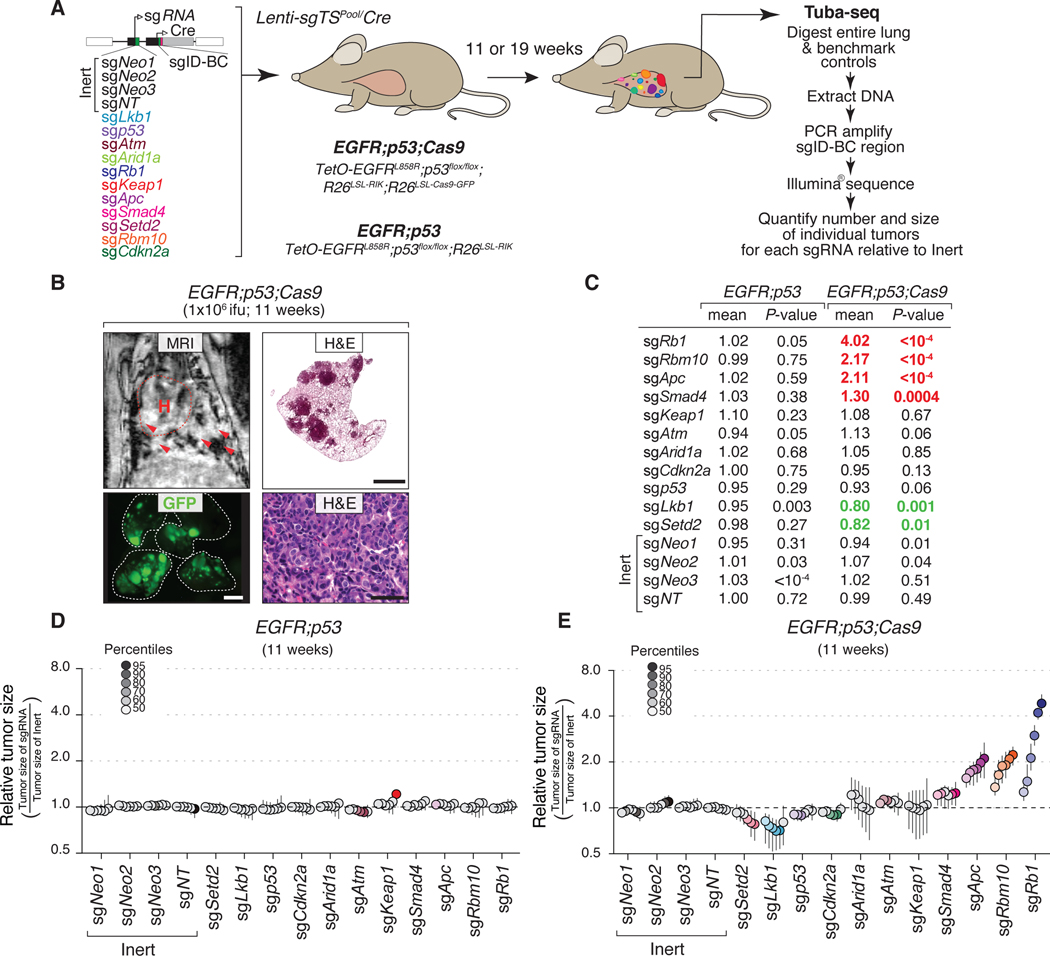
Figure 1. Lentiviral Cre-mediated lung tumor initiation in EGFR;p53 and EGFR;p53;Cas9 mice.
A. Schematic of the TetO-EGFRL858R, R26LSL-RIK, and p53flox alleles in EGFR;p53 mice, prior to and following Cre-mediated recombination. Conditional expression of oncogenic EGFRL858R is under the control of a tetracycline response element (TetO), which is induced by rtTA in the presence of doxycycline (Dox). Lentiviral-Cre inactivates Trp53 and enables the expression of the reverse tetracycline-regulated transactivator (rtTA3) and mKate (from the R26LSL-RIK allele). Cre also allows expression of Cas9 and GFP (from the R26LSL-Cas9-GFP allele) in EGFR;p53;Cas9 mice.
B. MRI showing tumor development 11 weeks after tumor initiation in EGFR;p53 mice (top left panel) with 2×106 ifu Lenti-Cre. The dashed red line surrounds the heart (H) and red arrows indicate areas of tumor. Hematoxylin and eosin (H&E) staining shows lung adenocarcinoma development. Scale bars = 1.2 mm and 100 μm in the top and bottom panels, respectively. Images are from a representative mouse (N = 5).
C. Immunostaining for EGFRL858R, mKate and Cas9 in tumors in EGFR;p53 and EGFR;p53;Cas9 mice. The dashed lines indicate areas of tumor. Scale bars = 200 μm.
D. MRI and H&E showing tumor development 16 weeks after tumor initiation in EGFR;p53;Cas9 mice with 1×105 ifu Lenti-Cre. Tumors are positive for GFP in EGFR;p53;Cas9 mice and lungs are indicated by the white dashed line. H&E image scale bars = 1.2 mm and 100 μm in top and bottom right panels, respectively. GFP image scale bar = 2.5 mm.
We initiated tumors with a lentiviral-PGK-Cre vector (30) in EGFR;p53 mice and used magnetic resonance imaging (MRI) to monitor tumor development (Fig. 1B). Tumors were first detectable in EGFR;p53 mice 8 weeks after tumor initiation. Histological analysis of lungs 11 weeks after tumor initiation confirmed the development of multifocal lung adenocarcinomas (Fig. 1B, bottom panels). Tumors stained positively for EGFRL858R and mKate, as well as surfactant protein C (SP-C) and the lung lineage-defining transcription factor NKX2.1/TTF-1 (Fig. 1C, top panels and Supplementary Fig. S1B) (31–33). Importantly, in this model, tumors were more focal than the diffuse tumors that rapidly develop in the previous CCSP-rtTA;TetO-EGFRL858R model (27), likely due to tumor initiation from fewer cells in the virally initiated model (Fig. 1B, top right panel). Lentivirus-induced tumors in EGFR;p53 mice were more poorly differentiated than those typically observed in the CCSP-rtTA;TetO-EGFRL858R model (Fig. 1B, bottom panels) as shown by increased prevalence of a micropapillary pattern, which is known to be highly aggressive in human adenocarcinoma (27). Thus, this new lentiviral-Cre-initiated model recapitulates the genetic and histopathological features of human oncogenic EGFR-driven TP53-deficient lung tumors.
Multiplexed quantification of tumor suppressor gene function in EGFR-driven lung tumors
To enable somatic genome editing in the EGFR;p53 model, we further incorporated a conditional Cas9 allele (Rosa26LSL-Cas9-GFP) (4) to generate TetO-EGFRL858R;R26RIK/LSL-Cas9-GFP;p53flox/flox (EGFR;p53;Cas9) mice (Fig. 1A). Lentiviral-Cre delivery to EGFR;p53;Cas9 mice initiated multifocal lung adenomas and adenocarcinomas that expressed EGFRL858R, mKate, Cas9 and GFP (Figs. 1C, D). Tumors in EGFR;p53;Cas9 mice were histologically similar to those in EGFR;p53 mice (Figs. 1B, D).
We used an improved version of our Tuba-seq approach to quantify tumor suppressor gene function in oncogenic EGFR-driven lung tumors (Supplementary Methods). Genomic integration of barcoded lentiviral vectors uniquely tags each transduced cell and all of the neoplastic cells within the resulting clonal tumors (8). Each barcode encodes an 8-nucleotide sgID region specific to the sgRNA followed by a random 15-nucleotide barcode (sgID-BC); thus, high-throughput sequencing of this sgID-BC region from bulk tumor-bearing lung can be used to quantify the number of cells in each tumor of each genotype (Supplementary Methods) (8). With this approach, the absolute number of neoplastic cells in each tumor is calculated by normalizing the number of reads of each unique barcode to the number of reads from benchmark control cells added to each sample (Fig. 2A, Supplementary Fig. S2A and Supplementary Methods). Thus, Tuba-Seq enables the parallel analysis of the impact of multiple tumor suppressor gene alterations on tumor growth in vivo.
Figure 2. Multiplexed somatic CRISPR–Cas9-mediated genome editing uncovers tumor suppressor gene effects on EGFR-driven lung tumors.
A. Experimental strategy. Tumors were allowed to develop for either 11 weeks in EGFR;p53;Cas9 and EGFR;p53 mice or 19 weeks in EGFR;p53;Cas9 mice after intra-tracheal administration of Lenti-sgTSPool/Cre. Whole lungs were collected for tumor barcode deep sequencing (Tuba-seq) and histology. The number of neoplastic cells in each tumor (tumor size) was calculated from barcode sequencing of bulk tumor-bearing lungs. Barcode read number was normalized to benchmark control cells that have known barcodes and were added at a known number to each sample (Supplementary Methods).
B. MRI, H&E, and GFP images showing tumor development in EGFR;p53;Cas9 mice 11 weeks after tumor initiation with 1×106 ifu of Lenti-sgTSPool/Cre. H&E image scale bars = 1.2 mm and 100 μm in top and bottom right panels, respectively. Lungs are indicated by the white dashed lines. GFP image scale bar = 2.5 mm. Images are from a representative mouse (N = 10).
C. Relative log-normal (LN) mean size of tumors with each sgRNA in EGFR;p53 (N = 5) and EGFR;p53;Cas9 mice (N = 10) 11 weeks after tumor initiation (normalized to the tumors with inert sgRNAs). P-values were calculated from bootstrapping. P-values < 0.05 and their corresponding means are highlighted in EGFR;p53;Cas9 mice for sgRNAs that positively (red) and negatively (green) affect tumor growth when the effects are equal to or differ >10% compared to the size of tumors with inert sgRNAs.
D. Relative size of tumors at the indicated percentiles of each genotype in EGFR;p53 mice 11 weeks after tumor initiation with the Lenti-sgTSPool/Cre. These mice lack the R26LSL-Cas9-GFP allele; therefore, all sgRNAs are functionally inert. 95% confidence intervals are shown. Percentiles calculated from bootstrapping that are significantly different from the tumors with inert sgRNAs are in color.
E. Relative size of tumors of each genotype in EGFR;p53;Cas9 mice 11 weeks after tumor initiation with the Lenti-sgTSPool/Cre. The relative size of tumors at the indicated percentiles were calculated from the tumor size distribution of all tumors from ten mice. 95% confidence intervals are shown. Percentiles were calculated from bootstrapping and are in color if significantly different from the tumors with inert sgRNAs.
To assess the function of ten diverse putative tumor suppressor genes, which are frequently altered in human lung adenocarcinoma, we initiated tumors in EGFR;p53 and EGFR;p53;Cas9 mice with a pool of barcoded Lenti-sgRNA/Cre vectors (Lenti-sgTSPool/Cre; Fig. 2A). In addition to Lenti-sgRNA/Cre vectors targeting each putative tumor suppressor gene, this pool contains negative control vectors, including four Lenti-sgInert/Cre vectors and a vector with an sgRNA targeting Trp53 (which is already inactivated by Cre-mediated recombination in EGFR;p53;Cas9 mice, Fig. 1A) (8). Lenti-sgTSPool/Cre-initiated tumors were first detectable by MRI 4 weeks after tumor initiation in EGFR;p53;Cas9 mice. At 11 weeks after tumor initiation, when tumors were readily detectable in all mice, tumor-bearing lungs were collected for Tuba-seq analysis and histology (Fig. 2B and Supplementary Fig. S1E).
Putative tumor suppressor genes have distinct effects on EGFR-driven lung tumor growth
We used Tuba-seq to quantify the number of neoplastic cells in clonal tumors of each genotype in the EGFR;p53 model and used two summary statistics to describe the tumor size distribution (percentiles within the tumor size distributions and the log-normal mean (Supplementary Methods; Figs. 2C-E and Supplementary Figs. S3A, B) (8). Effects of the sgRNAs on tumor growth were assessed by examining the significance of the differences in the tumor size distribution compared to controls and also the magnitude of the effects. Tumors with each sgRNA in EGFR;p53 mice, which lack the Cas9 allele, had very similar size distributions (Fig. 2D). Furthermore, tumors initiated with each Lenti-sgInert/Cre vector or Lenti-sgp53/Cre in EGFR;p53;Cas9 mice had very similar tumor size profiles (Figs. 2C, E).
Inactivation of Rb1, Apc, and Rbm10 most dramatically enhanced the growth of oncogenic EGFR-driven Trp53-deficient lung tumors (Figs. 2C, E). While the importance of RB1 inactivation in EGFR-driven lung adenocarcinomas has begun to be investigated (23, 34), very few studies have addressed the functional importance of the APC pathway on EGFR-driven lung adenocarcinomas (35). Interestingly RBM10 is an RNA binding protein and splicing regulator that is poorly studied in cancer in general and has not previously been implicated as a critical regulator of EGFR-driven lung cancer growth (12, 36–38). Rbm10 is an X-linked gene, however, we did not uncover any sex-specific effects of Rbm10 inactivation on tumor size (Supplementary Fig. S2B). More broadly, both the LN mean and the tumor size at the 95th percentile relative to inert tumors were similar between male and female mice across the TSGs studied (Supplementary Fig. S2B).
Surprisingly, inactivation of either Lkb1 or Setd2 – which are strong tumor suppressor genes in analogous oncogenic Kras-driven lung tumor models – dramatically reduced tumor growth relative to sgInert tumors (8, 39, 40). These effects were consistent across multiple percentiles within the tumor size distribution and as assessed by the LN mean of tumor sizes (Figs. 2C, E). Notably, the significant effects observed with sgSetd2, sgLkb1, sgSmad4, sgApc, sgRbm10, and sgRb1 were all recaptured even after we simulated a 50% reduction in cutting efficiency of sgRNAs or when we used other strategies for subsampling underscoring the robustness of our findings (Supplementary Figs. S2C, D and Supplementary Figs. S4A, B). These results suggest that in specific contexts, inactivation of genes presumed to be tumor suppressors can have deleterious effects on cancer growth. Other tumor suppressor genes (Atm, Arid1a, Cdkn2a and Keap1) did not significantly alter tumor growth in the context of this experiment (Figs. 2C, E).
To assess tumor suppressor gene function at a later time point of tumor growth, we initiated tumors in EGFR;p53;Cas9 mice with 10-fold less Lenti-sgTSPool/Cre and performed Tuba-seq after 19 weeks of tumor growth (Supplementary Figs. S1C-H). At this time point, the histology of the tumors was similar to that observed after 11 weeks of tumor initiation (Supplementary Figs. S1C, D). Tuba-seq analysis confirmed the tumor-suppressive function of Rbm10, Apc, and Rb1 (Supplementary Figs. S3A-D). Since we used a 10-fold lower viral titer for this experiment, there were proportionally fewer tumors (Supplementary Fig. S1F), which reduced the resolution of the Tuba-seq analysis. Thus, while inactivation of the other genes did not significantly affect tumor growth at this time-point, we cannot exclude that these genes may influence tumor growth. Interestingly, despite the decreased statistical power, inactivation of Cdkn2a or Arid1a had a positive effect on tumor growth at this 19-week time point (but not at the 11-week time point), suggesting a potential role of these tumor suppressor genes during a later phase of tumorigenesis in this model (Supplementary Figs. S3A, B).
Validation of Apc and Rbm10-mediated tumor suppression
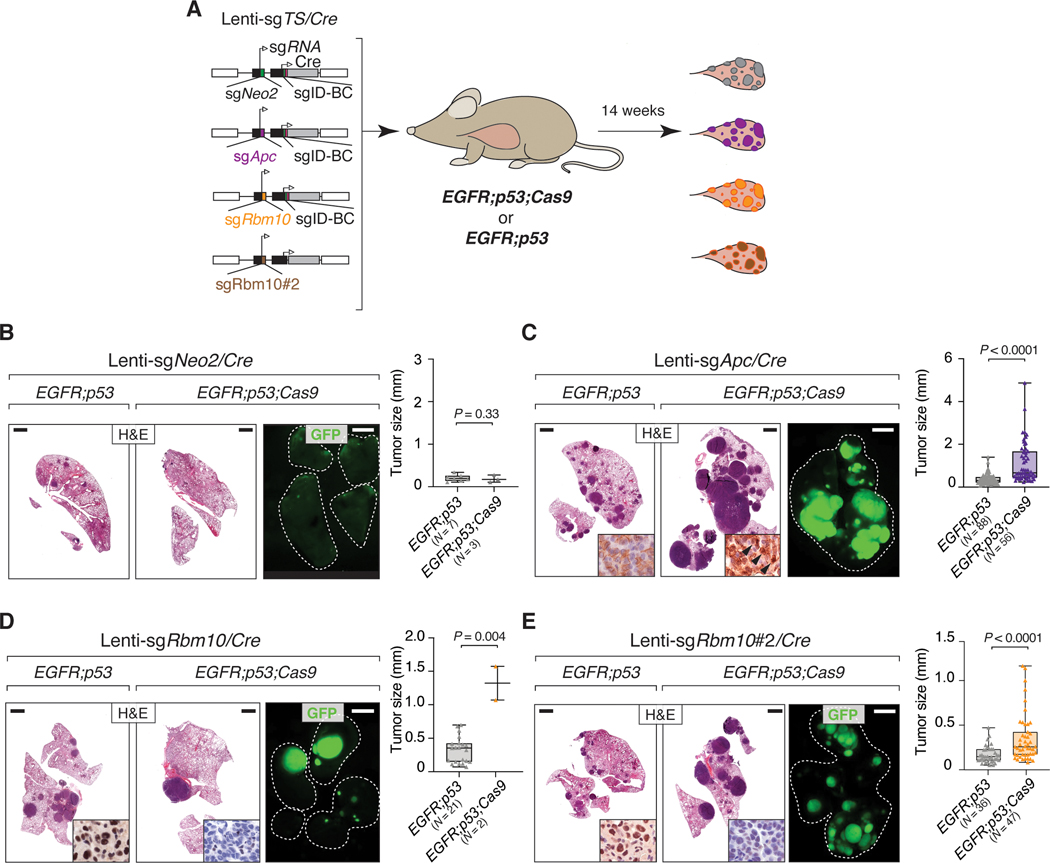
We performed further experiments to confirm the role of two less-well studied tumor suppressors, Apc and Rbm10, on the growth of EGFR-driven tumors. We initiated lung tumors in EGFR;p53 and EGFR;p53;Cas9 mice with Lenti-sgApc/Cre, Lenti-sgNeo2/Cre (sgInert), and two Lenti-sgRbm10/Cre vectors each with a unique sgRNA targeting Rbm10 (N = 3 EGFR;p53 mice/group and N = 5 EGFR;p53;Cas9 mice/group; Fig. 3A). We used two sgRNAs targeting Rbm10 to increase the power of our findings, and because the tumor suppressive role of Rbm10 remains entirely uncharacterized in EGFR-driven lung cancer. Inactivation of either Apc or Rbm10 in EGFR;p53;Cas9 mice gave rise to tumors that were larger than control sgNeo2 tumors in EGFR;p53;Cas9 mice (Supplementary Figs. S5A, B). Moreover, by quantifying the size of individual sgApc- or sgRbm10 tumors in histological sections, we observed that EGFR;p53;Cas9 tumors were larger than tumors initiated in EGFR;p53 mice (Figs. 3B-E). Lenti-sgApc/Cre-initiated tumors in EGFR;p53;Cas9 mice had more cancer cells with stabilization and nuclear localization of β-catenin as well as increased expression of Sox9 (consistent with Apc inactivation) (Supplementary Figs. S5C, D) (41). Furthermore, at least 50% of Lenti-sgRbm10/Cre-initiated tumors in EGFR;p53;Cas9 mice lacked or had lower Rbm10 protein levels (Supplementary Fig. S5E). Tumors with either Apc or Rbm10 inactivation were histologically similar to tumors in EGFR;p53 mice at this time point and had papillary/acinar or micropapillary structures with medium/high nuclear grade (Supplementary Fig. S6A, B). All tumors were positive for adenocarcinoma markers (NKX2.1/TTF-1 and SP-C) and were negative for neuroendocrine markers Synaptophysin (SYP) and UCHL1 and a squamous cell carcinoma marker (p63) (Supplementary Fig. S7). Cancer cells were highly proliferative, while apoptotic cells were rare in tumors in EGFR;p53;Cas9 mice initiated with either Lenti-sgApc/Cre or Lenti-sgRbm10/Cre (Supplementary Fig. S7). These results further confirm the importance of these tumor suppressor genes in constraining EGFR-driven tumor growth in vivo. Collectively, our findings underscore the value of coupling Tuba-seq and CRISPR–Cas9-mediated somatic genome editing with our virally-induced mouse model to dissect gene function in oncogenic EGFR-driven lung cancer.
Figure 3. Apc and Rbm10 inactivation enhance the growth of oncogenic EGFR-driven lung tumors in vivo.
A. Experimental strategy for tumor suppressor gene validation. Tumors were initiated in EGFR;p53 and EGFR;p53;Cas9 mice with Lenti-sgTS/Cre vectors carrying sgApc, sgRbm10, sgRbm10#2, or sgNeo2 as a control (EGFR;p53 N = 3 mice/group, EGFR;p53;Cas9 N = 5 mice/group, 1×105 ifu/mouse).
B-E. H&E and GFP images show that Apc (C) and Rbm10 (D, E) inactivation enhances tumor growth compared to the controls (B). The histology of the left and auxiliary lobes was analyzed. Lungs are indicated by the white dashed lines. Scale bars = 1.2 mm and 2.5 mm for histology and GFP images, respectively. Tumor size was calculated by measuring the longest diameter of each tumor. The number of tumors studied are reported on the X-axis. P-values were calculated using a one-tailed Mann-Whitney U test. Horizontal lines show the median and whiskers indicate the minimum and the maximum for the dataset. The box represents values in the first and the third quartile. The panels at the bottom right of the H&E images show nuclear accumulation of β-catenin (C) and the absence of Rbm10 protein expression (D, E) reflective of Apc and Rbm10 inactivation in tumors in EGFR;p53;Cas9 mice, respectively.
Oncogenic drivers shape the fitness landscape of tumor suppression
The extent to which different oncogenic drivers affect the landscape of tumor suppression is almost entirely unknown. We approached this question by comparing the fitness landscape of tumor suppression within the contexts of oncogenic EGFR- and Kras-driven lung tumors. We repeated an experiment previously performed by our group in which we inactivated the same panel of tumor suppressor genes in KrasLSL-G12D/+;p53flox/flox;R26LSL-Tomato;H11LSL-Cas9 (Kras;p53;Cas9) mice and used library preparation methods and the analytical pipeline identical to those used for the lungs from EGFR;p53 and EGFR;p53;Cas9 mice (Supplementary Fig. S8A) (29, 30, 42). Inactivation of Lkb1, Setd2, and Rb1 were particularly strong drivers of oncogenic Kras-driven Trp53-deficient tumor growth, while inactivation of Rbm10, Apc, Cdkn2a and Arid1a also modestly increased tumor growth (Figs. 4A, B). These results are largely consistent with our previous Tuba-seq analysis of Kras;p53;Cas9 mice, as well as other studies on these genes in oncogenic Kras-driven lung cancer mouse models (Supplementary Fig. S1A) (9, 39–41, 43–47).
Figure 4. Dramatic differences in the effects of tumor suppressor genes in oncogenic EGFR- versus KRAS-driven lung cancer.
A. Relative size of tumors of each genotype in KrasG12D;p53flox/flox;R26LSL-Tomato;H11LSL-Cas9 (Kras;p53;Cas9) mice 14 weeks after tumor initiation with Lenti-sgTSPool/Cre relative to tumors with inert sgRNAs. The relative size of tumors at the indicated percentiles was calculated from the tumor size distribution of all tumors from six mice. 95% confidence intervals are shown. P-values were calculated from bootstrapping. Percentiles that are significantly different from the tumors with inert sgRNAs are in color.
B. LN mean for tumors with each sgRNA in Kras;p53;Cas9 mice 14 weeks after tumor initiation (normalized to the inert tumors). P-values were calculated by bootstrapping and significant values are highlighted in red when P < 0.05 and the effects are >10% compared to the size of tumors with inert sgRNAs.
C. The relative effect of inactivating each tumor suppressor gene on tumor sizes (comparison to tumors with inert sgRNAs) at the indicated percentile in the EGFR;p53;Cas9 (N = 10) and Kras;p53;Cas9 (N = 6) models. Lkb1 and Setd2 inactivation greatly increased tumor size only in the Kras;p53;Cas9 model. Error bars indicate the standard deviation.
D. The frequency of tumor suppressor gene alterations that co-occur with EGFR or KRAS and TP53 mutations in human lung adenocarcinomas (LUADs, data from AACR Project GENIE).
E. LKB1 and SETD2 alterations co-occur significantly more frequently in KRASonc;TP53mut than in EGFRonc;TP53mut human lung adenocarcinomas. The frequency of RBM10 and APC alterations were not significantly different between in KRASonc;TP53mut and EGFRonc;TP53mut tumors. The odds ratios represent the strength of the observed frequencies of tumor suppressor gene alterations that co-occur in KRASonc;TP53mut cases compared to EGFRonc;TP53mut cases. P-values were calculated using a Fisher’s exact test.
Inactivation of several of the tumor suppressor genes (e.g., Rb1) had similar effects on EGFR- and Kras-driven tumors suggesting that these tumor suppressor genes limit lung adenocarcinoma growth regardless of the oncogenic context (Fig. 4C and Supplementary Fig. S8B). However, inactivation of either Lkb1 or Setd2 greatly increased the growth of oncogenic Kras-driven lung tumors but decreased the growth of oncogenic EGFR-driven lung tumors (Figs. 2C, E, Figs. 4A-C and Supplementary Fig. S8B). Thus, the consequences of tumor suppressor gene inactivation in specific contexts are not limited to the magnitude of tumor-suppressive effects but can also be manifested as opposite effects (known as the sign epistasis) even when the oncogenic drivers (in this case, EGFR and KRAS) are traditionally thought to be within a linear pathway.
Profound epistasis between tumor suppressor genes and oncogenic drivers shapes mutational patterns in human lung adenocarcinoma
To compare our functional data with the spectrum of tumor suppressor gene mutations found in human lung adenocarcinomas, we queried data from the AACR Project GENIE database (48). We calculated the frequency of tumor suppressor gene mutations that co-occur with oncogenic EGFR (L858R, Exon 19 deletions, L861Q, G719X) or oncogenic KRAS (at codons 12, 13 or 61) mutations and TP53 mutations. This analysis revealed that RB1, RBM10, and APC are frequently altered in EGFR/TP53 mutant lung adenocarcinomas. Interestingly, RB1 mutations are more frequent in EGFR/TP53 tumors compared to KRAS/TP53 tumors (7.5% versus 3.1%). However, Rb1 inactivation was a major driver of tumor growth in both EGFR and Kras mutant tumors in mice (Figs. 2C, E and Figs. 4A-C). This difference between mouse and human may be related to the higher frequency of alterations in CDKN2A in human KRAS/TP53 mutant tumors (7.3%, Fig. 4D) that would disrupt the same cell cycle pathway as RB1 inactivation. LKB1/STK11 and SETD2, are among the most frequently mutated tumor suppressor genes in KRAS/TP53 mutant lung adenocarcinomas (Fig. 4D) (49, 50). Further supporting a difference in the function of LKB1 and SETD2 in EGFR/TP53 versus KRAS/TP53 mutant lung adenocarcinomas, mutations in these genes occurred at significantly higher frequencies in KRAS/TP53 mutant tumors compared to EGFR/TP53 tumors (Fig. 4E). This asymmetry in the mutation frequency of LKB1 or SETD2 within oncogenic EGFR- or KRAS-driven lung tumors is also significant when we extend our analysis to include all tumors regardless of TP53 mutation status (Supplementary Fig. S8C). Collectively, the mouse and human data indicate that the mutational patterns can reflect the consequences of profound epistasis between tumor suppressor genes and oncogenic drivers.
Keap1 inactivation limits the response of tumors to osimertinib
Genetically engineered mouse models have provided insight into the biology of EGFR-driven lung tumors and have proven valuable for studying mechanisms of resistance to EGFR TKIs, especially on-target mechanisms of resistance (27, 51–53). The TKI osimertinib was recently approved for the first-line treatment of EGFR-driven lung adenocarcinomas. However, pathways involved in modulating the depth of response and mechanisms of resistance to osimertinib are still under investigation (54, 55). To determine how tumor suppressor genes influence the therapeutic response of lung tumors to EGFR inhibition, we treated EGFR;p53;Cas9 mice with Lenti-sgTSPool/Cre-initiated tumors with osimertinib for two weeks starting at 9 weeks after tumor initiation (Fig. 5A). Osimertinib treatment greatly reduced the overall tumor burden relative to vehicle-treated EGFR;p53;Cas9 mice (Supplementary Figs. 9A-D). Residual neoplastic cells were sparse, as determined by staining for EGFRL858R and those cells were not proliferating (Supplementary Figs. S9E-F). The overall tumor response was similar when the 2-week treatment was started 17 weeks after tumor initiation (Supplementary Figs. S9G-J). Residual tumors in EGFR;p53;Cas9 mice retained lung adenocarcinoma features and did not have evidence of phenotypic changes (such as small cell lung cancer (SCLC) transformation) after two weeks of osimertinib treatment (Supplementary Fig. S10A).
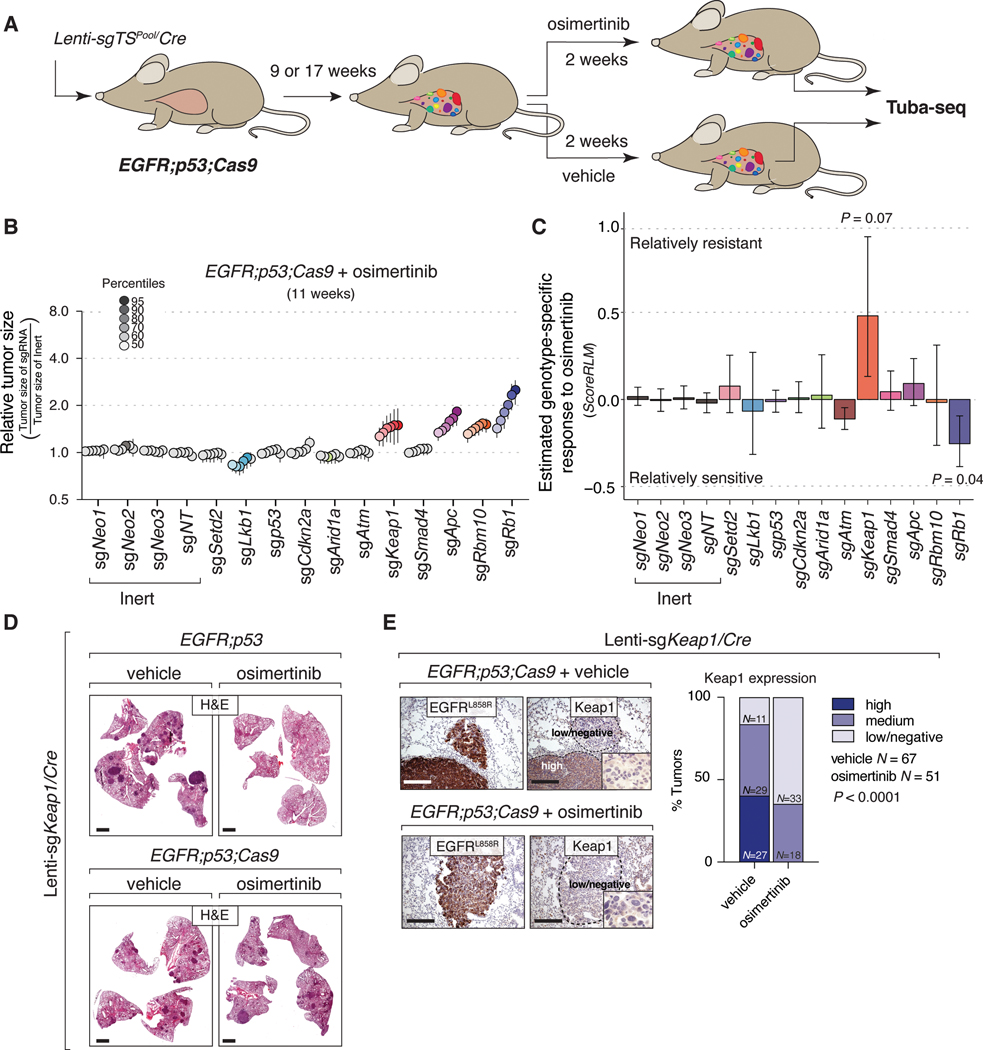
Figure 5. Identification of genotype-specific responses to osimertinib treatment.
A. Experimental strategy. Tumors were initiated with Lenti-sgTSPool/Cre in EGFR;p53;Cas9 mice for nine or 17 weeks and were treated for two weeks with either vehicle or osimertinib (25 mg/kg, 5 days/week).
B. Relative size of the tumors at the indicated percentiles for each genotype in EGFR;p53;Cas9 mice after treatment with osimertinib. 95% confidence intervals are shown. P-values were calculated from bootstrapping. Percentiles that are significantly different from the tumors with inert sgRNAs are in color.
C. Estimate of the genotype-specific treatment response (ScoreRLM) calculated by comparing the LN mean of tumors treated with osimertinib to the LN mean of vehicle-treated tumors in EGFR;p53;Cas9 mice 11 weeks after tumor initiation (Supplementary Methods). Error bars indicate the standard deviation. P-values were calculated from bootstrapping.
D. H&E staining of tumor bearing-lungs in EGFR;p53 and EGFR;p53;Cas9 mice with Lenti-sgKeap1/Cre initiated tumors (N = 8 mice/group). Scale bars = 1.2 mm.
E. Immunostaining of Lenti-sgKeap1/Cre initiated tumors in EGFR;p53;Cas9 mice for EGFRL858R and Keap1 after vehicle or osimertinib treatment (N = 4 mice/treated-group). Tumors are still detectable after two weeks of treatment with osimertinib and mostly express medium or low/negative levels of Keap1 compared to tumors treated with vehicle in EGFR;p53;Cas9 mice (bar graph). The dashed lines indicate areas of tumors and the level of Keap1 is indicated with a label (high, medium, or low/negative). Scale bars = 200 μm. P-values were calculated using a Chi-squared test.
To quantify the impact of inactivating each tumor suppressor gene on the response to osimertinib in vivo, we performed Tuba-seq on the lungs from osimertinib-treated EGFR;p53;Cas9 mice 11 and 19 weeks after tumor initiation and compared the results to Tuba-seq results from vehicle-treated controls (Fig. 5B and Supplementary Figs. S11A, B). Consistent with the imaging data and histological analysis, osimertinib treatment greatly reduced tumor burden as assessed by Tuba-seq (Compare Figs. 2C, E, Fig. 5B and Supplementary Figs. S3A, B and S11A, B; Supplementary Methods).
After two weeks of osimertinib treatment, inactivation of Apc, Rb1 or Rbm10 was still associated with larger tumors while the size distribution of tumors with inactivation of other TSGs remained similar to that of tumors with inert sgRNAs (compare Fig. 5B and Fig. 2E, Supplementary Fig. S11A and Fig. 2C, Supplementary Fig. S11B and Supplementary Figs. S3A, B). The striking exception was tumors with sgKeap1. In vehicle-treated mice, the size distribution of sgKeap1 tumors was almost identical to that of tumors with inert sgRNAs; however, in osimertinib-treated mice, sgKeap1 tumors were significantly larger than the tumors with inert sgRNAs. This suggests that inactivation of Keap1 limits responses to osimertinib (Fig. 5B). Osimertinib resistance conferred by Keap1 inactivation was also observed at 19 weeks after tumor initiation (Supplementary Figs. 11A, B).
We applied an analytical approach that we previously developed and validated to quantify the genotype-specific responses (Fig. 5C and Supplementary Figs. S11C-G; Supplementary Methods) (56). By comparing the LN mean of the observed tumor size distributions in osimertinib-treated mice with the expected tumor size distribution based on the overall drug effects, we can estimate genotype specific drug responses (ScoreRLM; Fig. 5C and Supplementary Fig. S11C). At 11 weeks after tumor initiation, following two weeks of osimertinib treatment sgRb1 tumors were 25% smaller than expected (P = 0.04). Conversely, tumors with Keap1 inactivation were 48% larger than expected (P = 0.07; Fig. 5C). The effect of Keap1 inactivation was even greater at 19 weeks after tumor initiation, where tumors were 274% larger than expected (P = 0.13; Supplementary Fig. S11C). Given the magnitude of the ScoreRLM for sgKeap1 at both time points (ScoreRLM = 0.57 and 1.90 after two weeks of treatment at 11 and 19 weeks after tumor initiation, respectively), we combined the two independent P-values and confirmed that Keap1 inactivation significantly reduced the therapeutic response to osimertinib (Fisher’s method, P-value = 0.05). Other statistical measures of genotype-specific responses, including relative tumor number (ScoreRTN) and relative geometric mean (ScoreRGM) did not significantly differ between the treated and untreated groups (Supplementary Fig. S11D; Supplementary Methods). Our analytical methods allow us to uncover when effects are more pronounced on larger tumors (ScoreRTN and ScoreRGM have much lower sensitivity when the effects are greater on larger tumors; Supplementary Figs. S11E-G; Supplementary Methods). Thus, our data are consistent with the resistance conferred by Keap1 inactivation being more pronounced in larger tumors.
Keap1-deficient tumors have reduced sensitivity to osimertinib which correlates with clinical outcomes
To further investigate these findings, we initiated tumors with Lenti-sgKeap1/Cre in EGFR;p53 and EGFR;p53;Cas9 mice followed by treatment with osimertinib or vehicle (Figs. 5D, E). Osimertinib-treatment reduced the size and number of tumors in EGFR;p53 mice compared to vehicle-treated EGFR;p53 mice (Fig. 5D and Supplementary Figs. S12A, B). Conversely, osimertinib treatment of Lenti-sgKeap1/Cre-initiated tumors in EGFR;p53;Cas9 mice did not decrease tumor size or number compared to vehicle-treated EGFR;p53;Cas9 mice (Figs. 5D, E and Supplementary Figs. S12A, B). Consistent with the inefficiency of CRISPR–Cas9-mediated genome editing in somatic cells, some tumors initiated with Lenti-sgKeap1/Cre in EGFR;p53;Cas9 mice retained Keap1 protein (Fig. 5E, top panels). However, the tumors that remained in osimertinib-treated EGFR;p53;Cas9 mice all had medium to low/negative Keap1 levels (Fig. 5E). Together, these data indicate that while Keap1 inactivation is not positively selected for during oncogenic EGFR-driven tumor growth, osimertinib treatment selects for cancer cells with low or absent Keap1, thus reducing the therapeutic response to the drug.
To correlate our findings with clinical data, we analyzed the effects of KEAP1 pathway alterations on patient outcomes to EGFR inhibition in EGFR/TP53 mutant lung adenocarcinomas. Oncogenic EGFR-driven tumors with KEAP1 pathway inactivation have been suggested to be less responsive to TKIs, and we confirmed this association in EGFR/TP53 tumors (Figs. 6A, B) (57). Mutations in the KEAP1/NFE2L2/CUL3 pathway were associated with a significantly shorter time on EGFR TKI therapy compared with matched patients with KEAP1/NFE2L2/CUL3 wildtype tumors (5.8 versus 14.3 months, P = 0.01; Log-rank test, Fig. 6A; Supplementary Table S1 and Supplementary Table S2). This remained significant even after adjustment for potential confounders such as age, sex, race and smoking status (Supplementary Table S3). Among several other tumor suppressor genotypes, KEAP1 pathway alterations were the most significant driver of limited sensitivity after correction for multiple hypothesis testing (Fig. 6B).
Figure 6. KEAP1 inactivation correlates with reduced therapeutic response to TKIs in human EGFR-driven lung adenocarcinomas.
A. Kaplan-Meier curve showing time to TKI treatment failure for patients with EGFR/TP53 lung adenocarcinomas that do or not have mutations in the KEAP1/NFE2L2/CUL3 axis. Time to treatment failure is the time from the initiation of TKI treatment to the date of discontinuation of TKI due to progression, toxicity or death. The P-value was calculated using the Log rank test.
B. Forest plot of the time on treatment hazard ratios for tumors with KEAP1, RB1, APC, ARID1A or CDKN2A alterations in EGFR/TP53 mutant lung adenocarcinomas. Hazard ratios were calculated from multivariate regression analysis, 95% confidence intervals are shown.
C. KEAP1, NFE2L2, and CUL3 alteration frequency in oncogenic EGFR and TP53 mutant lung tumors in the Yale Lung Rebiopsy Program dataset (Methods). OncoPrint of oncogenic EGFR samples prior to first-line TKI (erlotinib, gefitinib, afatinib) treatment (PRE-TKI, left panel) (N = 22), and at acquired resistance to TKIs (TKI-R, right panel) (N = 27). In each row, the percentage and the type of alterations in each gene are indicated for each patient. For the TKI-R samples, known resistance mechanisms such as acquisition of T790M in EGFR, are reported. Cases with paired PRE-TKI and TKI-R samples are indicated by the patient number highlighted in red.
We also analyzed a dataset of oncogenic EGFR lung adenocarcinoma patient samples collected through the Yale Lung Rebiopsy Program (YLR) prior to first-line TKI treatment and after the development of resistance to TKIs (Fig. 6C; Supplementary Table S4). Among 18 patients with EGFR and TP53 mutant tumors, two had KEAP1 alterations at relapse (TKI-R samples). One case had an acquired missense mutation (Y206N) that lies in a domain of KEAP1 involved in forming the complex with Cullin3 to mediate ubiquitination and degradation of NRF2 (encoded by NFE2L2) (58). The other tumor, which was analyzed only at resistance, had heterozygous loss of KEAP1 (Fig. 6C). Moreover, we also observed two cases of NFE2L2 copy number gain (one detected prior to treatment and maintained at resistance, the other detected only at resistance), and one case of CUL3 heterozygous loss at resistance. Taken together, our in vivo functional results are consistent with human data and support a role for alterations in the KEAP1 pathway in reducing EGFR TKI sensitivity.
Discussion
The genomic landscape of lung adenocarcinoma is combinatorially complex, with a large number of genomic alterations in oncogenic drivers and tumor suppressor genes occurring in various combinations (2, 3). While critical for precision medicine approaches, dissecting the functional contribution of these co-occurring genomic alterations to tumor growth and response to therapy is extremely challenging. Using somatic CRISPR–Cas9-mediated gene editing and Tuba-seq (8, 9), we evaluated the fitness effects of inactivating ten tumor suppressor genes that are commonly altered in lung adenocarcinoma. We uncovered distinct roles for specific tumor suppressor genes during tumor growth in the different oncogenic contexts and on sensitivity to the TKI osimertinib in vivo.
EGFR and KRAS are the most frequently mutated oncogenic drivers in lung adenocarcinoma, collectively occurring in ~40–50% of all patients (2). Oncogenic alterations in these two genes are mutually exclusive, consistent with them being in the same canonical receptor tyrosine kinase (EGFR-RAS-MAPK) signaling pathway (59, 60). For this reason, one might anticipate that inactivating tumor suppressor pathways in EGFR-driven and KRAS-driven lung cancers would have similar biological consequences. It is tempting to make a simple assumption that tumor suppressor genes have a constant marginal effect independent of other genomic alterations, or in other words, that the fitness landscape of tumor suppression is smooth and that epistatic interactions are infrequent and/or small in magnitude. Here, we tested this assumption by comparing the fitness landscape of tumor suppression in EGFR-driven Trp53-deficient and in Kras-driven Trp53-deficient lung adenocarcinoma and found distinct patterns of epistasis between tumor suppressor genes and oncogenes. Inactivation of Rb1, Rbm10 and Apc had a similar effect on EGFR and KRAS-driven lung tumors (Figs. 2C, E, Figs. 4A-C and Supplementary Fig. S8B). In contrast, while Lkb1 and Setd2 were amongst the most potent tumor suppressor genes in Kras/p53 tumors, inactivation of Lkb1 or Setd2 led to reduced tumor growth of EGFR/p53 tumors (Figs. 2C, E and Figs. 4A, B). This observation is consistent with human data, where alterations in either LKB1 or SETD2, are much less common in lung tumors with oncogenic EGFR than in tumors with oncogenic KRAS (Figs. 4D, E and Supplementary Fig. S8C). Thus, we reveal a highly context-dependent fitness landscape of tumor suppression that depends on the nature of the oncogenic driver. Investigation of the mechanisms that underlie the sign epistasis of LKB1 and SETD2 may uncover new biological insights and vulnerabilities of EGFR mutant tumors. More broadly, sign epistasis within seemingly similar cancer contexts could help identify genetic interactions for further functional investigation and should be considered when interpreting cancer genomic data.
Concurrent TP53 and RB1 mutations have been linked to the transformation of lung adenocarcinomas to SCLC that is observed in some EGFR mutant tumors upon the emergence of TKI resistance (23, 61, 62). Additional histological changes have also been described in TKI-resistant tumors, including squamous cell carcinoma differentiation (63). In our screen and validation studies, we found that the tumors had lung adenocarcinoma features and did not express neuroendocrine or squamous cell carcinoma markers (Supplementary Figs. S7, S10A). We also examined a small cohort of EGFR;p53;Cas9 mice with tumors initiated using Lenti-sgRb1/Cre, and these tumors were positive for adenocarcinoma markers and negative for neuroendocrine and squamous cell carcinoma markers at baseline (Supplementary Fig. S10B). Long-term TKI treatment studies in these new models will provide us with the opportunity to determine how tumors on different tumor suppressor gene backgrounds evolve through TKI treatment and whether specific tumor suppressor genes contribute to altering the histological properties of TKI resistant tumors.
A major advantage of autochthonous genetically engineered mouse models of human cancer is that they can be used to study the impact of inactivation of putative tumor suppressor genes on therapy responses in vivo (10, 56, 64). We found that inactivation of Keap1 decreases sensitivity to osimertinib in vivo (Fig. 5). This finding was supported in a clinical cohort of EGFR/TP53 mutated lung adenocarcinomas, in which patients with tumors harboring mutations in KEAP1/NFE2L2/CUL3 had a significantly shorter time to treatment discontinuation with EGFR TKIs compared with matched controls (Figs. 6A, B and Supplementary Figs. S13A-D) (17, 57). These results are consistent with inactivation of this tumor suppressor gene limiting the response to therapy and explains the occasional presence of alterations in KEAP1 pathway genes in TKI-resistant human tumors.
The reduced sensitivity of tumors initiated with Lenti-sgKeap1 may be associated with the accumulation and transactivation of genes regulated by NRF2 (65–67). Indeed, we found that Keap1-deficient tumors had increased nuclear Nrf2 and Nqo1 expression relative to Keap1-proficient tumors, suggesting the presence of enhanced Nrf2 transcriptional activity (Supplementary Fig. S12C) (65–67). Activation of both the MAPK/ERK and AKT/mTOR pathways downstream of EGFR were strongly decreased after two weeks of treatment with osimertinib, thus excluding that the reduced sensitivity could be due to sustained activation of these pathways (Supplementary Fig. S12D). NRF2 is a pleiotropic transcription factor that can lead to increased production of anti-oxidants, regulate ferroptosis and alter cellular metabolism (68). Which of the functional outputs of activation of the NRF2 pathway are responsible for the decreased sensitivity to osimertinib in EGFR mutant tumors remains to be determined. Experiments in lung adenocarcinoma cell lines treated with targeted therapies against the EGFR-MAPK pathway suggest that KEAP1 loss leads to decreased levels of drug-induced ROS and increased glutathione synthesis (66). A systematic analysis of the Keap1 pathway in EGFR mutant genetically engineered mouse models and patient-derived tumors will be necessary to fully elucidate how NRF2 mediates TKI resistance and reveal potential therapeutic vulnerabilities. Indeed, there are potential therapeutic avenues for treating KEAP1-deficient tumors that should be explored in EGFR mutant tumors. In Kras-driven lung tumors, KEAP1/NRF2-dependent metabolic reprogramming leads to a dependency on glutaminolysis, which can be targeted via glutaminase inhibition (69–73). Future studies will be important to establish whether similar metabolic rewiring occurs in Keap1-deficient EGFR mutant tumors that can be leveraged therapeutically.
Genetic alterations in KEAP1 occur in less than 2% of EGFR mutant tumors at baseline and are found in less than 10% of TKI-resistant EGFR mutant lung adenocarcinomas (Fig. 6C and Supplementary Fig. S13E) (17, 74). It is possible that non-genetic alterations that increase NRF2-dependent gene expression also occur in EGFR mutant tumors and that examination of the mutational status of the pathway underestimates the frequency with which it is disrupted (17, 75–78). Collectively, these results raise the possibility that NRF2 should be investigated as a potential target to reduce or delay the onset of drug resistance in EGFR-driven lung adenocarcinomas. More broadly, our study demonstrates that our approach can be used to identify clinically relevant pathways that modulate response to therapy in vivo. By uncovering the driving forces of the heterogeneity of responses to therapy observed in patients, these types of studies could help define high-risk versus low-risk patient populations and guide therapeutic interventions.
This study provides insight into the complex interplay between tumor suppressor genes and other co-occurring mutations in EGFR-driven lung adenocarcinoma tumorigenesis and thus has significant clinical implications. Across many cancers, including lung adenocarcinoma, most tumor suppressors are mutated in a low percentage of tumors. Several of the tumor suppressor genes that we investigated were amongst the most frequently mutated genes in EGFR-driven TP53 mutant lung cancer (e.g., RBM10 and RB1) (Fig. 4D and Supplementary Fig. S14A) (79, 80). However, even the tumor suppressor genes that we found to have an effect on in vivo tumor growth are not as frequently altered in human tumors as might be expected from the experimental findings. This could be related to the overall higher mutation burden in human tumors versus mouse models (81) with human tumors harboring alterations in genetically redundant pathways (cell cycle regulation, WNT pathway) or inactivated by epigenetic mechanisms (11, 35, 82, 83). Finally, the type of mutational process that gives rise to the mutations could also affect the frequency with which certain TSGs are mutated (84, 85). Tuba-seq allows the analysis of genetic effects without being confounded by the frequency with which alterations in that gene are found.
The lack of genomic complexity in the genetically engineered mouse models (81) allows us to evaluate interactions between co-occurring alterations by avoiding confounding factors pervasive in human genomic data (i.e., tumor mutation load, mutation frequency, additional mutations) and environmental factors such as smoking. Our data provide clear quantitative data on mutual exclusivity and synergistic biological effects of genetic alterations that mirror observations in patient specimens, highlighting the strength of these interactions. Additional studies in models with a higher tumor mutation burden, more reflective of that present in human tumors, will be important to uncover the extent to which additional genetic alterations further modulate tumor growth and drug sensitivity in specific oncogenic driver/tumor suppressor gene contexts. Notably, tumors with other oncogenic drivers (e.g., ALK rearrangements) also have unique spectra of co-occurring tumor suppressor gene alterations, further suggesting wide-spread interactions between tumor suppressor gene pathways and oncogenic drivers (Supplementary Fig. S14B). Future in vivo Tuba-seq studies should investigate tumor models driven by other oncogenes to uncover a broader understanding of the genetic interactions between diverse oncogenes and large panels of tumor suppressor genes. Precise mapping of the fitness consequences of combinations of genetic alterations during tumor evolution will help uncover the biological and clinical relevance of specific alterations during carcinogenesis and identify pathways that can be exploited as therapeutic targets to prevent or overcome resistance to targeted therapies.
Methods
Mice and tumor initiation
TetO-EGFRL858R; p53flox/flox; Rosa26CAGs-LSL-rtTA3-IRES-mKate, Rosa26CAGs-LSL-Cas9-GFP, KrasLSL-G12D, Rosa26LSL-tdTomato, and H11LSL-Cas9 mice have been described (4, 25, 27–30, 42, 86, 87). EGFR;p53 and EGFR;p53;Cas9 were on a mixed BL6/129/FVB background and Kras;p53;Cas9 mice were on a mixed BL6/129 background. Approximately equal numbers of males and females were used for each experiment and the number of mice used for each experiment is listed in each figure legend. Lung tumors were initiated by intratracheal administration of Lentiviral-Cre vectors as previously described (26). Tumor burden was assessed by magnetic resonance imaging, fluorescence microscopy, lung weight, and histology, as indicated. Doxycycline was administered by feeding mice with doxycycline-impregnated food pellets (625 ppm; Harlan-Teklad). Osimertinib (AstraZeneca, Cambridge, UK) was resuspended in 0.5% (w/v) methylcellulose (vehicle) and was administered orally (per os, 25 mg/kg 5 days a week). All animals were kept in pathogen-free housing under guidelines approved by either the Yale University Institutional Animal Care or the Stanford University Institutional Animal Care and Use Committee guidelines.
Production, purification, and titering of lentivirus
The barcoded vectors in the Lenti-sgTSPool/Cre have been previously described (Supplementary Table S5) (8). The second Lenti-sgRbm10/Cre vector used in the validation experiments was generated by site-directed mutagenesis (Supplementary Table S6). Briefly, Lentiviral-U6-sgRNA/Cre vectors contain an 8-nucleotide defined sequence (sgID) that identifies the sgRNA followed by a 15-nucleotide random barcode (BC) to uniquely tag each tumor (8). To avoid barcode-sgRNA uncoupling driven by lentiviral template switching during reverse transcription of the pseudo-diploid viral genome, each barcoded Lenti-sgRNA/Cre vector was generated separately (88, 89). We cultured HEK293T cells in Dulbecco’s Modified Eagle Medium with 10% Fetal Bovine Serum and transfected them with individual barcoded Lenti-sgRNA/Cre plasmids (sgLkb1, sgp53, sgApc, sgAtm, sgArid1a, sgCdkn2a, sgKeap1, sgNeo1, sgNeo2, sgNeo3, sgNT1, sgRb1, sgRbm10, sgRbm10#2 unbarcoded, sgSetd2, or sgSmad4) along with pCMV-VSV-G (Addgene #8454) envelope plasmid and pCMV-dR8.2 dvpr (Addgene #8455) packaging plasmid using polyethylenimine. We treated the cells with 20 mM sodium butyrate 8 hours after transfection, changed the culture medium 24 hours after transfection, and collected supernatants 36 and 48 hours after transfection. Subsequently, we removed the cell debris with a 0.45 μm syringe filter unit (Millipore SLHP033RB), concentrated each lentiviral vector by ultracentrifugation (25,000 g for 1.5 hours at 4°C), resuspended the virus in PBS, and stored the virus at −80°C. To determine the titer of each vector, we transduced Rosa26LSL-YFP mouse embryonic fibroblasts (a gift from Dr. Alejandro Sweet-Cordero/UCSF), determined the percentage of YFP-positive cells by flow cytometry, and normalized the titer to a lentiviral preparation of known titer. The cell line was authenticated at UCSF and tested negative for mycoplasma contamination. Lentiviral vectors were thawed and pooled immediately prior to delivery to mice. All these plasmids are available at https://www.addgene.org/Monte_Winslow/.
Lentiviral titers and time of analysis
Anticipated growth rates were determined by monitoring tumor development through magnetic resonance imaging in pilot experiments and the analysis time points were selected to ensure that tumors were detectable by MRI such that their response to treatment could be evaluated. Viral titers used in the experiments were chosen to balance the total tumor burden across mice at the time of analysis after tumor initiation. For analysis of tumor growth 11 weeks after tumor initiation, the Lenti-sgTSPool/Cre titer administered to EGFR;p53 mice was 2×106 infectious units (ifu)/mouse, while for EGFR;p53;Cas9 mice we used 1×106 ifu/mouse. We reasoned that using a higher viral titer in the control EGFR;p53 mice would increase our confidence that any differences observed between EGFR;p53 and EGFR;p53;Cas9 mice were due to inactivation of tumor suppressor genes in the latter model. For the 19-week time point in EGFR;p53;Cas9 mice we initiated tumors with 1×105 ifu/mouse. Two weeks before collection, mice were treated with either vehicle or osimertinib. For the validation experiments in which we used a single vector to initiate tumors (Lenti-sgApc/Cre, Lenti-sgRbm10/Cre, Lenti-sgRbm10#2/Cre or Lenti-sgNeo2/Cre (sgInert)) we used 1×105 ifu/mouse and analyzed the mice after 14 weeks of tumor growth (Supplementary Table S6). For the validation with Lenti-sgKeap1/Cre virus (1×105 ifu/mouse), both EGFR;p53 and EGFR;p53;Cas9 mice were treated 15 weeks after tumor initiation and lungs were collected after two weeks of treatment with either vehicle or osimertinib. Kras;p53;Cas9 were analyzed 14 weeks after tumor initiation with 2.2×104 ifu/mouse.
Magnetic resonance imaging
All procedures were performed in accordance with protocols approved by the Yale University IACUC and in agreement with the NIH Guide for the Care and Use of Laboratory Animals. Respiratory gated, gradient-echo MR images of mice were collected with a 4T (31-cm bore) small-animal Bruker horizontal-bore spectrometer (Bruker AVANCE). All data were collected as previously described (51). Tumor volume was quantified by calculating the area of visible lung opacities present in each image sequence per mouse using BioImage Suite 3.01 (90). For the drug treatment studies, once tumors were detected using MRI, mice were assigned to different groups to ensure a comparable tumor volume distribution prior to treatment between the vehicle-treated and the osimertinib-treated group.
Isolation of genomic DNA from mouse lungs and preparation of sgID-BC libraries
Genomic DNA was isolated from bulk tumor-bearing lung tissue from each mouse as previously described (8). Briefly, three benchmark control cell lines (~5×105 cells per cell line) carrying unique sgID-BCs, were added (“spiked-in”) to each sample prior to lysis to enable the calculation of the absolute number of neoplastic cells in each tumor from the number of sgID-BC reads. Following homogenization and overnight protease K digestion, genomic DNA was extracted from the lung lysates using standard phenol-chloroform and ethanol precipitation.
sgID-BC sequencing libraries were prepared by PCR amplifying the sgID-BC region from total genomic DNA. To enable the identification and subsequent computational elimination of index hopped reads after high-throughput sequencing, the sgID-BC region of the integrated Lenti-sgRNA-BC/Cre vectors was PCR amplified using unique dual indexing primer pairs. To increase the sequence diversity, we added 6–9 Ns before the sequence-specific primers (56). For each mouse, we performed eight 100 μl PCR reactions per sample (4 μg DNA per reaction, 32 μg per mouse) using Q5 High-Fidelity 2x Master Mix (New England Biolabs, M0494X). The PCR products were purified with Agencourt AMPure XP beads (Beckman Coulter, A63881) using a double size selection protocol. The concentration and quality of the purified libraries were determined using the Agilent High Sensitivity DNA kit (Agilent Technologies, 5067–4626) on the Agilent 2100 Bioanalyzer (Agilent Technologies, G2939BA). The libraries were pooled based on lung weight (to have sequencing depth more evenly distributed across samples), cleaned up and size-selected using AMPure XP beads, and sequenced on the Illumina®HiSeq 2500 platform to generate paired-end 150 bp reads (Admera Health). To increase the diversity at the beginning of sequencing reads and improve sequencing quality, 5%−15% PhiX control DNA was added to the library.
Histology and immunohistochemistry
The auxiliary lobe of the right lung was collected for each experimental mouse from the mice transduced with the Lenti-sgTSPool/Cre pool whereas, both the auxiliary and left lobes were collected for the validation experiments with individual sgRNAs (sgApc, sgRbm10, sgRbm10#2 and sgNeo2). Right and auxiliary lobes were collected for the experiment with Lenti-sgKeap1/Cre virus. Lung lobes were fixed in 4% paraformaldehyde overnight at room temperature, placed in 70% ethanol, and paraffin-embedded and sectioned (Histology @ Yale). Four micrometer sections were used for hematoxylin and eosin (H&E) staining and immunohistochemistry. Tumor sizes were determined by measuring the longest diameter for each tumor in H&E stained sections. Tumor size and tumor area were quantified using ImageJ. The limited tissue collected for histological analysis reduced the overall number of tumors that could be measured. P-values were calculated from the Mann-Whitney U test. The following antibodies were used for immunohistochemistry: anti-EGFRL858R (1:200, CST, 3197), anti-SP-C (1:200, abcam, AB40876), anti-TTF-1/Nkx2–1 (1:200, abcam, AB76013), anti-phospho-histone H3 (1:200, CST, 9701), anti-cleaved Caspase 3 (1:300, CST, 9664), anti-mKate (1:2500, Evrogen, AB233), anti-Cas9 (1:500, Novus Biologicals, 7A9–3A3, NBP2–36440), anti-β-catenin (1:500, CST, 8814), anti-Rbm10 (1:200, abcam, AB224149), and anti-Keap1 (1:500, abcam, AB227828), anti-Sox9 (1:500, Merck Millipore, AB5535), anti-Nrf2 (1:500, abcam, AB137550), anti-Nqo1 (1:500, abcam, AB28947), anti-phospho-Erk (1:100, CST, 4370), anti-phospho-Akt (1:100, CST, 4060), anti-UCHL1 (1:500, Sigma Millipore, HPA005993), anti-Synaptophysin (1:400, Abcam, AB32127) and anti-p63 (1:2500, Abcam, AB124762). Nuclear protein expression in tumors was binned as high (over 75% of positive nuclei in a tumor area), medium (between 25 and 75% of positive nuclei) or low/negative (positive nuclear staining below 25%) whereas, cytosolic protein levels were binned as high (over 75% of positive cells), medium (between 25 and 75% of positive cells) or low/negative (positive staining below 25%). Statistical analyses were performed with GraphPad Prism 8 and QuPath.
Analysis of human lung tumor data using GENIE data
The AACR Project GENIE is a registry that contains CLIA-/ISO-certified genomic data collected from the records of more than 9,000 patients who were treated at each of the consortium’s participating institutions (48). Data from GENIE version 4.1-public were accessed through the Synapse Platform. We generated a comprehensive list of all missense, nonsense, and frameshift mutations for all screened genes across all participating centers in the GENIE project, documenting these mutations at the gene and amino acid levels. Based on the sets of genes included in the different screening panels that contribute to Project GENIE, we annotated all lung adenocarcinomas within the database as being wild-type, mutated, or not screened for each gene. From this complete catalog of mutations in each tumor, we determined the rates of co-occurrence of known oncogenic KRAS mutations (G12X, G13X, Q61X) or EGFR mutations (L858R, Exon 19 deletions, L861Q, G719X) with missense, nonsense, or frameshift mutations in our set of ten tumor suppressor genes. Furthermore, we determined the frequency of co-incident tumor suppressor gene mutations in tumors with oncogenic KRAS and inactivating TP53 mutations (KRAS/TP53) and in tumors with oncogenic EGFR and inactivating TP53 mutations (EGFR/TP53) using GENIE version 8.1 (Supplementary Fig. S14A). A similar analysis was also performed on tumors with ALK-rearrangements and TP53 mutations using GENIE version 8.1 (Supplementary Fig. S14B). The pipelines for these analyses are publicly available at www.github.com/dgmaghini/GENIE and https://github.com/lichuan199010/Tuba-seq-analysis-and-summary-statistics which allow users to input OncoTree codes of interest, generate mutational profiles for the corresponding GENIE tumors, and identify the co-occurrence of up to two background mutations (for example, EGFR and TP53) with mutations in other genes of interest. Analyses were performed using Phyton 3.6 and R3.6.
Stanford cohort of EGFR mutant lung adenocarcinomas
Patients with lung cancer who were evaluated at the Stanford Cancer Center and had their tumors analyzed using the Stanford Solid Tumor Actionable Mutation Panel (STAMP)(6) were included in the analysis after obtaining written informed consent. This retrospective study was conducted under a molecular analysis protocol approved by the Stanford University Institutional Review Board. All STAMP cases performed between 2015 and 2019 were included; during this time, there were two different assays used, with one covering 198 genes (302 kb) and the other covering 130 genes (232 kb). STAMP was done as the standard of care and thus at the discretion of the treating physician and could have occurred at the time of diagnosis or at the time of progression.
From the available STAMP cases, patients were selected if they had stage IV non-small cell lung cancer (NSCLC) and had a pathogenic EGFR and p53 mutation. Patients were excluded if they had incomplete data or were lost to follow up prior to analysis of the primary endpoint, if they elected not to receive treatment or if they received adjuvant tyrosine kinase inhibitor therapy for early-stage disease. Within this cohort, patients were further selected for the presence of at least one of the tumor suppressor genes investigated in the preclinical setting (KEAP1, LKB1/STK11, SETD2, SMAD4, RB1, APC, ARID1A and/or CDKN2A). To ensure that there was enough power to conduct an analysis, genes were analyzed individually if there were at least 8 patients with tumors with a mutation of that gene who met inclusion criteria. Due to small numbers, patients with tumors with LKB1 (N = 1), SETD2 (N = 0) and SMAD4 (N = 1) mutations were not analyzed separately. However, as two tumors harboring LKB1 and SMAD4 mutations were also EGFR/P53 mutant, they were included as part of the control arm. Univariate analysis of time to treatment failure was completed on cases with stage IV, EGFR/P53 mutant NSCLC who were treated with a tyrosine kinase inhibitor (13) and had mutations in either KEAP1-pathway component (KEAP1, NFE2L2, CUL3), RB1, APC, ARID1A or CDKN2A. Mutations that were significant on univariate analysis were identified and multivariate analyses accounting for 1) co-founding variables and 2) co-mutations were run. Demographic data including sex, age at diagnosis, smoking history, and ethnicity were extracted (Supplementary Tables S1-S3). For each tumor suppressor, patients were matched 1:3 with a wildtype cohort on the basis of sex, smoking history, ethnicity, age and treatment type. Time to Treatment Failure (TTF) was determined by subtracting the date of discontinuation of TKI due to progression, toxicity or death, from the date of initiation of TKI and reported in months. Statistical analysis was performed using GraphPad Prism 8 and R studio. The Kaplan-Meier method was used to estimate TTF. Comparison of survival curves was made using the Log-rank test. Significance was defined as P < 0.05. Hazard ratios (HR) were generated from multivariate regression analysis performed in R and reported with a 95% confidence interval (CI).
Yale cohort of EGFR mutant lung adenocarcinomas
Patients with EGFR-mutant lung adenocarcinoma were enrolled to a Yale University IRB approved protocol after obtaining written informed consent allowing the collection and analysis of clinical data, archival and fresh tissue, blood and the generation of patient-derived xenografts. This study was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule). Patients who received targeted therapy (erlotinib, gefitinib, or afatinib) as first line therapy, either alone or in combination with other therapies such as chemotherapy or cetuximab, were included (Supplementary Table S4). For genomic studies, formalin-fixed paraffin embedded tissue was macro-dissected to enrich for tumor material. All of the tumor samples (N = 29) analyzed had matched normal tissue and cancer cell purity >20% as assessed by the LOH (Loss of Heterozygosity) events from the whole exome sequencing data.
Whole Exome Sequencing
DNA was extracted from and analyzed as previously described (91). Briefly, genomic DNA was captured on the NimbleGen 2.1M human exome array and subjected to 74-bp paired-end reads on the Illumina HiSeq2000 instrument. The mean coverage for normal was 109.1x and the mean coverage for tumor was 189x with 92.78% and 95.56% for the bases covered at least 20 independent sequence reads, respectively. Sequence reads were mapped to the human reference genome (GRCh37) using the Burrow-Wheeler Aligner-MEM (BWA-MEM) program. Sequence reads outside the targeted sequences were discarded and the statistics on coverage were collected from the remaining reads using in-house per scripts.
Somatic Mutation Calling
For all matched tumor-normal pairs, somatic point mutations and indels were called by MuTect2 using Bayesian classifiers. For all somatic mutations called, we extracted base coverage information in all samples and considered the mutations that were supported by at least two independent sequence reads covering non-reference alleles and present in more than 5% of all sequencing reads. Identified variants were further filtered based on their presence in repositories of common variations (1000 Genomes, NHLBI exome variant server and 2,577 non-cancer exomes sequenced at Yale) and annotated using ANNOVAR program (92). All somatic indels were visually inspected to remove the false positive calls.
Somatic Copy Number Variation Analysis
Copy number analysis was performed as previously described (91). Briefly, copy number variants were identified from the whole exome sequencing data using EXCAVATOR software that normalizes the non-uniform whole exome sequencing data taking GC-content, mappability, and exon-size into account (93). The Hidden Markov Model was utilized to classify each copy number variant segment into five copy number states (homozygous deletion, heterozygous deletion, normal copy number, homozygous copy gain or multiple copy gain). Tumor purity was estimated from LOH using in-house per scripts.
Data and code availability
The datasets generated during and/or analyzed in the current study are available in the NCBI Gene Expression Omnibus database (GSE146550) and dbGAP (phs002334.v1.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002334.v1.p1). GENIE genomic data analysis is publicly available at www.github.com/dgmaghini/GENIE. More GENIE analyses and other data supporting the findings are available at https://github.com/lichuan199010/Tuba-seq-analysis-and-summary-statistics or upon request.
Supplementary Material
Statement of significance.
By modeling complex genotypes in vivo, this study reveals key tumor suppressors that constrain the growth of EGFR mutant tumors. Furthermore, the study shows that KEAP1 inactivation reduces the sensitivity of these tumors to tyrosine kinase inhibitors. Thus, our approach identifies genotypes of biological and therapeutic importance in this disease.
Acknowledgements
G.F was supported by an American Italian Cancer Foundation (AICF) postdoctoral fellowship. C.L. is the Connie and Bob Lurie Fellow of the Damon Runyon Cancer Research Foundation (DRG-2331). H.C. was supported by a Tobacco-Related Disease Research Program Postdoctoral Fellowship (28FT-0019). D.A. was supported by NIH F31 CA203488. K.H. was supported by NIH F32 CA210516. This work was supported by NIH R01 CA231253 (to M.M.W and D.A.P.), NIH R01 CA234349 (to M.M.W and D.A.P.), NIH R01 CA120247 (to K.P.), NIH P50 CA196530 (to K.P.), the Ginny and Kenneth Grunley Fund for Lung Cancer Research (KP) and the Yale Cancer Center Class of ‘61 Research Award (KP). Gilead Sciences, Inc. supported the sequencing of a subset of the Yale Cancer Center specimens. The authors would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of study authors. We thank members of the Politi, Winslow and Petrov Laboratories for helpful feedback and support in particular, Jacqueline Starrett and Emily Forrest for editing, Fernando de Miguel for creating resources to help with the WES analysis. We also thank Henning Stehr for providing the sequencing data from the STAMP cohort and the Bosenberg Laboratory for sharing their Leica M205 FA stereomicroscope.
Conflict of interest disclosure statement
K. Politi is co-inventor on a patent licensed to Molecular MD for EGFR T790M mutation testing (through MSKCC). K. Politi has received Honoraria/Consulting fees from Takeda, NCCN, Novartis, Merck, AstraZeneca, Tocagen, Maverick Therapeutics, Dynamo Therapeutics, Halda and research support from AstraZeneca, Kolltan, Roche and Symphogen. M. M. Winslow has received honoraria from the speakers bureaus of Genentech, and Amgen. M. M. Winslow and D. A. Petrov have ownership and are consultants/advisory board members for D2G Oncology. M. Diehn reports research funding from Varian Medical Systems and Illumina, ownership interest in CiberMed, patent filings related to cancer biomarkers, paid consultancy from Roche, AstraZeneca, and BioNTech, and travel/honoraria from RefleXion and Illumina.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 2.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19(9):495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Haar J, Canisius S, Yu MK, Voest EE, Wessels LFA, Ideker T. Identifying Epistasis in Cancer Genomes: A Delicate Affair. Cell. 2019;177(6):1375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang CH, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK, et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat Med. 2017;23(3):291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winters IP, Chiou SH, Paulk NK, McFarland CD, Lalgudi PV, Ma RK, et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun. 2017;8(1):2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers ZN, McFarland CD, Winters IP, Naranjo S, Chuang CH, Petrov D, et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat Methods. 2017;14(7):737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers ZN, McFarland CD, Winters IP, Seoane JA, Brady JJ, Yoon S, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat Genet. 2018;50(4):483–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winters IP, Murray CW, Winslow MM. Towards quantitative and multiplexed in vivo functional cancer genomics. Nat Rev Genet. 2018;19(12):741–55. [DOI] [PubMed] [Google Scholar]
- 11.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21(10):2213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017;7(6):596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376(22):2109–21. [DOI] [PubMed] [Google Scholar]
- 17.Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res. 2018;24(13):3108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J Thorac Oncol. 2019;14(4):606–16. [DOI] [PubMed] [Google Scholar]
- 19.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. [DOI] [PubMed] [Google Scholar]
- 20.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378(2):113–25. [DOI] [PubMed] [Google Scholar]
- 23.Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol. 2019; 14(10):1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19(1):72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caswell DR, Chuang CH, Yang D, Chiou SH, Cheemalavagu S, Kim-Kiselak C, et al. Obligate progression precedes lung adenocarcinoma dissemination. Cancer Discov. 2014;4(7):781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4(7):1064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20(11):1496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dow LE, Nasr Z, Saborowski M, Ebbesen SH, Manchado E, Tasdemir N, et al. Conditional reverse tet-transactivator mouse strains for the efficient induction of TRE-regulated transgenes in mice. PLoS One. 2014;9(4):e95236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–25. [DOI] [PubMed] [Google Scholar]
- 30.Chiou SH, Kim-Kiselak C, Risca VI, Heimann MK, Chuang CH, Burds AA, et al. A conditional system to specifically link disruption of protein-coding function with reporter expression in mice. Cell Rep. 2014;7(6):2078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puglisi F, Barbone F, Damante G, Bruckbauer M, Di Lauro V, Beltrami CA, et al. Prognostic value of thyroid transcription factor-1 in primary, resected, non-small cell lung carcinoma. Mod Pathol. 1999;12(3):318–24. [PubMed] [Google Scholar]
- 32.Tan D, Li Q, Deeb G, Ramnath N, Slocum HK, Brooks J, et al. Thyroid transcription factor-1 expression prevalence and its clinical implications in non-small cell lung cancer: a high-throughput tissue microarray and immunohistochemistry study. Hum Pathol. 2003;34(6):597–604. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67(13):6007–11. [DOI] [PubMed] [Google Scholar]
- 34.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama S, Sng N, Carretero J, Welner R, Hayashi Y, Yamamoto M, et al. beta-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 2014;74(20):5891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52(5):720–33. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13(4):466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Sun Y, Huang Y, Song F, Huang Z, Bao Y, et al. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci Rep. 2017;7:40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, et al. Systematic In Vivo Inactivation of Chromatin-Regulating Enzymes Identifies Setd2 as a Potent Tumor Suppressor in Lung Adenocarcinoma. Cancer Res. 2017;77(7):1719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster K, Venkateswaran N, Rabellino A, Girard L, Pena-Llopis S, Scaglioni PP. Nullifying the CDKN2AB locus promotes mutant K-ras lung tumorigenesis. Mol Cancer Res. 2014;12(6):912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt A, Knittel G, Welcker D, Yang TP, George J, Nowak M, et al. ATM Deficiency Is Associated with Sensitivity to PARP1- and ATR Inhibitors in Lung Adenocarcinoma. Cancer Res. 2017;77(11):3040–56. [DOI] [PubMed] [Google Scholar]
- 45.Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, et al. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One. 2009;4(5):e5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho VM, Schaffer BE, Karnezis AN, Park KS, Sage J. The retinoblastoma gene Rb and its family member p130 suppress lung adenocarcinoma induced by oncogenic K-Ras. Oncogene. 2009;28(10):1393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter DM, Yates TJ, Ruiz-Torres M, Kim-Kiselak C, Gudiel AA, Deshpande C, et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature. 2019;569(7756):423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res. 2018;24(2):334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pirazzoli V, Ayeni D, Meador CB, Sanganahalli BG, Hyder F, de Stanchina E, et al. Afatinib plus Cetuximab Delays Resistance Compared to Single-Agent Erlotinib or Afatinib in Mouse Models of TKI-Naive EGFR L858R-Induced Lung Adenocarcinoma. Clin Cancer Res. 2016;22(2):426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pirazzoli V, Nebhan C, Song X, Wurtz A, Walther Z, Cai G, et al. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Rep. 2014;7(4):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Starrett JH, Guernet AA, Cuomo ME, Poels KE, van Alderwerelt van Rosenburgh IK, Nagelberg A, et al. Drug Sensitivity and Allele Specificity of First-Line Osimertinib Resistance EGFR Mutations. Cancer Res. 2020;80(10):2017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramalingam SS, Yang JC, Lee CK, Kurata T, Kim DW, John T, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2018;36(9):841–9. [DOI] [PubMed] [Google Scholar]
- 55.Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2019; 382:41–50. [DOI] [PubMed] [Google Scholar]
- 56.Li C, Lin W, Rizvi H, Cai H, McFarland CD, Rogers ZN, et al. Quantitative in vivo analyses reveal a complex pharmacogenomic landscape in lung adenocarcinoma. BioRxiv 2020. [Preprint] doi: 10.1101/2020.01.28.923912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hellyer JA, Stehr H, Das M, Padda SK, Ramchandran K, Neal JW, et al. Impact of KEAP1/NFE2L2/CUL3 mutations on duration of response to EGFR tyrosine kinase inhibitors in EGFR mutated non-small cell lung cancer. Lung Cancer. 2019;134:42–5. [DOI] [PubMed] [Google Scholar]
- 58.Cleasby A, Yon J, Day PJ, Richardson C, Tickle IJ, Williams PA, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19(15):4273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Unni AM, Lockwood WW, Zejnullahu K, Lee-Lin SQ, Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife. 2015;4:e06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zakowski MF, Ladanyi M, Kris MG, Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome G. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355(2):213–5. [DOI] [PubMed] [Google Scholar]
- 62.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res. 2020;26(11):2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9(2):137–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamadori T, Ishii Y, Homma S, Morishima Y, Kurishima K, Itoh K, et al. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31(45):4768–77. [DOI] [PubMed] [Google Scholar]
- 66.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park SH, Kim JH, Ko E, Kim JY, Park MJ, Kim MJ, et al. Resistance to gefitinib and cross-resistance to irreversible EGFR-TKIs mediated by disruption of the Keap1-Nrf2 pathway in human lung cancer cells. FASEB J. 2018:fj201800011R. [DOI] [PubMed] [Google Scholar]
- 68.Wu WL, Papagiannakopoulos T. The Pleiotropic Role of the KEAP1/NRF2 Pathway in Cancer. Annual Review of Cancer Biology. 2020;4(1):413–35. [Google Scholar]
- 69.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47(12):1475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23(11):1362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, et al. LKB1 and KEAP1/NRF2 Pathways Cooperatively Promote Metabolic Reprogramming with Enhanced Glutamine Dependence in KRAS-Mutant Lung Adenocarcinoma. Cancer Res. 2019;79(13):3251–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romero R, Sánchez-Rivera FJ, Westcott PMK, Mercer KL, Bhutkar A, Muir A, et al. Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nature Cancer. 2020;1(6):589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Binkley MS, Jeon YJ, Nesselbush M, Moding EJ, Nabet BY, Almanza D, et al. KEAP1/NFE2L2 Mutations Predict Lung Cancer Radiation Resistance That Can Be Targeted by Glutaminase Inhibition. Cancer Discov. 2020;10(12):1826–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fabrizio FP, Mazza T, Castellana S, Sparaneo A, Muscarella LA. Epigenetic Scanning of KEAP1 CpG Sites Uncovers New Molecular-Driven Patterns in Lung Adeno and Squamous Cell Carcinomas. Antioxidants (Basel). 2020;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, Lu Q, Cai J, Wang Y, Lai X, Qiu Y, et al. Nestin regulates cellular redox homeostasis in lung cancer through the Keap1-Nrf2 feedback loop. Nat Commun. 2019;10(1):5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q, Ma J, Lu Y, Zhang S, Huang J, Chen J, et al. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36(37):5321–30. [DOI] [PubMed] [Google Scholar]
- 78.Marcar L, Bardhan K, Gheorghiu L, Dinkelborg P, Pfaffle H, Liu Q, et al. Acquired Resistance of EGFR-Mutated Lung Cancer to Tyrosine Kinase Inhibitor Treatment Promotes PARP Inhibitor Sensitivity. Cell Rep. 2019;27(12):3422–32 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–37 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173(2):371–85 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McFadden DG, Politi K, Bhutkar A, Chen FK, Song X, Pirun M, et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci U S A. 2016;113(42):E6409–E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20(27):3528–32. [DOI] [PubMed] [Google Scholar]
- 83.Usadel H, Brabender J, Danenberg KD, Jeronimo C, Harden S, Engles J, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62(2):371–5. [PubMed] [Google Scholar]
- 84.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15(9):585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill AJ, McFaline-Figueroa JL, Starita LM, Gasperini MJ, Matreyek KA, Packer J, et al. On the design of CRISPR-based single-cell molecular screens. Nat Methods. 2018;15(4):271–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Annunziato S, Kas SM, Nethe M, Yucel H, Del Bravo J, Pritchard C, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016;30(12):1470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Papademetris X, Jackowski MP, Rajeevan N, DiStasio M, Okuda H, Constable RT, et al. BioImage Suite: An integrated medical image analysis suite: An update. Insight J. 2006;2006:209. [PMC free article] [PubMed] [Google Scholar]
- 91.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu D, Olman V, Wang L, Xu Y. EXCAVATOR: a computer program for efficiently mining gene expression data. Nucleic Acids Res. 2003;31(19):5582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed in the current study are available in the NCBI Gene Expression Omnibus database (GSE146550) and dbGAP (phs002334.v1.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002334.v1.p1). GENIE genomic data analysis is publicly available at www.github.com/dgmaghini/GENIE. More GENIE analyses and other data supporting the findings are available at https://github.com/lichuan199010/Tuba-seq-analysis-and-summary-statistics or upon request.