Abstract
Arbitrarily primed PCR with three primers and pulsed-field gel electrophoresis were used to characterize a set of 75 clinical Legionella pneumophila serogroup 1 isolates, with no apparent epidemiological link, obtained from 24 hospitals in Paris, France, from 1987 to 1997. Unexpectedly, 25 clinical isolates from 15 hospitals had an identical profile (termed type A) by both methods. The same profile was subsequently found in 16 of 64 randomly selected environmental L. pneumophila serogroup 1 isolates from 15 different sites in the Paris area. There was no evidence of geographic clustering or a peak incidence of type A isolation. Type A has not been found in France outside the Paris area, suggesting that a particular type of L. pneumophila serogroup 1 is specifically present in the Paris water distribution network.
Legionella pneumophila is a common cause of nosocomial and community-acquired pneumonia, being transmitted by inhalation of aqueous aerosols. Most cases of Legionnaires’ disease are sporadic, but outbreaks can also occur (1, 2, 3, 7, 8, 11, 12). L. pneumophila is ubiquitous in the aqueous environment, and most outbreaks are linked to contaminated hot water systems in well-defined areas (e.g., hotels, hospitals, and whirlpool spas) and cooling towers (1, 2, 7, 11, 12, 23). Most clinical infections are due to L. pneumophila serogroup 1 (17). Strain subtyping methods are used to confirm the environmental source of infections and include monoclonal antibody typing, plasmid analysis, and multilocus enzyme electrophoresis (6, 15, 16, 19). More recently, techniques based on genomic DNA polymorphisms have been described, such as restriction fragment length polymorphism analysis with or without the use of probes (ribotyping), pulsed-field gel electrophoresis (PFGE), and arbitrarily primed PCR (AP-PCR) (3, 9, 10, 13, 14, 16, 18, 21, 22, 25–27). PFGE has a high discriminatory power, readily distinguishing epidemic from sporadic isolates. PFGE is expensive and time-consuming in comparison with AP-PCR, which is suitable for large sets of isolates (13, 21). The discriminatory power of AP-PCR as an epidemiological tool for investigation of Legionnaires’ disease outbreaks has been previously evaluated with three different primers and found to be similar to that of PFGE (13). The DNA amplification profiles generated by AP-PCR and the DNA restriction profiles given by PFGE were identical for epidemiologically related L. pneumophila isolates, whereas unrelated cases generally gave different profiles except for one cluster of four isolates from the Paris area (13).
Molecular typing techniques have never been applied to a large set of L. pneumophila clinical isolates collected in a large city. In the present study we determined the AP-PCR and PFGE profiles of 75 L. pneumophila isolates from patients admitted to various Paris hospitals between 1987 and 1997. Although these patients had no identified epidemiological link, we found that 33% of them were infected by the same subtype.
MATERIALS AND METHODS
Strains.
A total of 75 clinical L. pneumophila serogroup 1 isolates were studied. They were isolated from bronchoalveolar fluid or bronchial aspirates of patients admitted to 24 hospitals in the Paris area with community-acquired (63 cases) or hospital-acquired (12 cases) legionellosis. Legionellosis was classified as nosocomial when it occurred more than 10 days after hospital admission. The clinical isolates were obtained between 1987 and 1997. Culture and identification of L. pneumophila were carried out in the microbiology laboratory of R. Poincaré Hospital (Garches, France).
Furthermore, 64 environmental isolates were randomly chosen from a collection of strains isolated between 1986 and 1997 by the Laboratoire d’Hygiène de la Ville de Paris from water supplies and cooling towers in 43 sites in the Paris area (25-km radius) including hospitals, other buildings, and swimming pools. One to 17 isolates per year were included (median, 4).
Clinical specimens were plated on buffered charcoal-yeast extract agar supplemented with α-ketoglutaric acid. Environmental strains were isolated as recommended by the French norm for isolation of Legionella in water (AFNOR T90-431) (5). Serogrouping was done by direct immunofluorescence with antisera from the Centers for Disease Control and Prevention (Atlanta, Ga.). Strains were stored at −80°C until use.
AP-PCR assay.
The AP-PCR assay was performed at R. Poincaré Hospital as previously described (13). Primer B1245 was used to screen the strains. Primers IR6110 and Leg2 were used for strains yielding the same pattern with primer B1245. Band patterns were compared visually. Two isolates were considered to have the same AP-PCR type when the patterns obtained with the three primers were indistinguishable. Very weak bands (not apparent on the photographs and/or not detected reproducibly) were not taken into account. In doubtful cases, the amplifications were repeated after a new extraction, and the patterns were compared after comigration on the same agarose gel.
Chromosomal PFGE analysis.
PFGE analysis was performed by the French National Reference Center for Legionellosis. Genomic DNA was prepared as previously described with some modifications (13). Briefly, legionellae were treated with proteinase K (50 μg/ml) in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8) for 24 h at 55°C, and DNA was digested with 20 IU of SfiI restriction enzyme (Boehringer Mannheim, Meylan, France) for 16 h at 50°C. Fragments of DNA were separated in a 0.8% agarose gel prepared and run in 0.5× Tris-borate-EDTA buffer (pH 8.3) in a contour-clamped homogeneous field apparatus (CHEF DRII system; Bio-Rad, Ivry sur Seine, France) with a constant voltage of 150 V. Runs were carried out with constant pulse times (25 s) at 10°C for 11 h and increasing pulse times (35 to 60 s) at 10°C for 11 h. Isolates with patterns which differed by no more than three restriction fragments were considered to have the same pulsotype, while organisms differing by more than three restriction fragments were considered sufficiently divergent to warrant a separate pulsotype designation (24).
Statistical analysis.
Statistical analysis was done with Epi-Info 5 software (Centers for Disease Control and Prevention). Results for patients infected by type A strains were compared with the results for patients infected by other types by using the χ2 test with Yates’ correction. Fisher’s exact test was used to calculate P values when expected numbers were below five.
RESULTS
Clinical isolates.
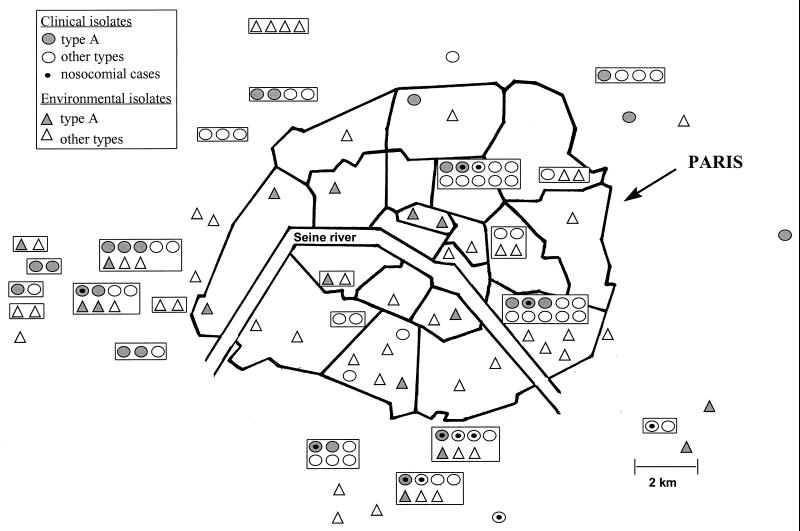
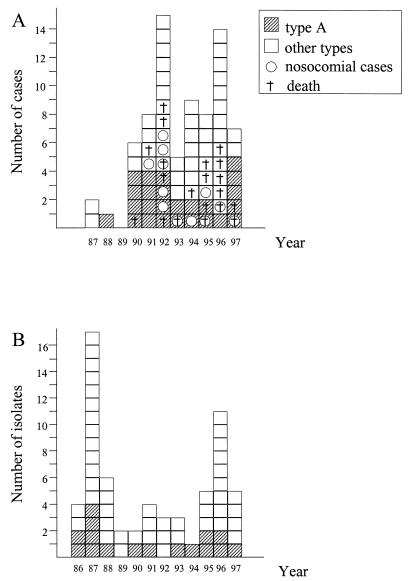
Primer B1245 was chosen to screen the 75 clinical isolates as it discriminates unrelated strains of L. pneumophila serogroup 1 (13). Unexpectedly, an identical pattern was obtained for 28 isolates, while different patterns were obtained for the other 47 isolates. To confirm the molecular identity of these 28 isolates, they were compared by the AP-PCR method, using the other two primers (IR6110 and Leg2), and by PFGE after SfiI digestion. All these techniques gave identical profiles for 25 of the 28 isolates, while the other isolates were considered different from one another. This suggested that a particular L. pneumophila type (designated type A) infected 25 (33%) of the 75 patients. The type A L. pneumophila isolates were isolated from patients admitted to 15 hospitals between 1988 and 1997 (Fig. 1). There was no obvious peak incidence (Fig. 2A). Patients infected by type A did not differ significantly from patients infected by other types in terms of age, sex, risk factors, need for mechanical ventilation, and mortality (Table 1). Among the 25 type A infections, 19 were acquired in the community and 6 were acquired in hospitals. Type A was found in the water supply of three of the six hospitals concerned. In the first case, a type A strain was found in eight hot water samples, including samples from the shower and the hot water tap in the patient’s bedroom. In the second case, a type A strain was isolated from the main hot water flow-back drain but not from two peripheral sites where nonserogroup 1 L. pneumophila was isolated. In the last case, a type A strain was isolated from a hot water sample obtained 4 years earlier, during an epidemiological investigation conducted following the occurrence of three nosocomial cases identified on the basis of serological criteria.
FIG. 1.
Geographic distribution of hospitals where L. pneumophila serogroup 1 infections were detected between 1987 and 1997 and sites where environmental L. pneumophila serogroup 1 strains were isolated. Isolates from the same hospital are boxed.
FIG. 2.
Annual incidence of clinical (A) and environmental (B) L. pneumophila serogroup 1 isolates.
TABLE 1.
Comparison between patients infected by type A strains and those infected by other strains
| Parameter | No. of patients infected with indicated strain type
|
||
|---|---|---|---|
| Type A | Other types | P valuea | |
| Total case number | 25 | 50 | |
| Age (yr) | 49.5 ± 19.3 | 51.4 ± 18.1 | 0.48 |
| Male | 15 | 38 | 0.24 |
| Risk factorb | 14 | 16 | 0.27 |
| Mechanical ventilation | 14 | 23 | 0.13 |
| Nosocomial | 6 | 6 | 0.15 |
| Deceased | 8 | 12 | 0.64 |
No significant differences were found between patients infected by type A and patients infected by other types (χ2 test with Yates’ correction or Fisher’s exact test).
Patients with at least one risk factor: malignancy, organ transplantation, immunosuppressive therapy, or AIDS.
Environmental isolates.
To identify a possible environmental source, we applied the same molecular methods to 64 environmental strains isolated in the Paris area between 1986 and 1997 (Table 2). AP-PCR with primer B1245 gave the type A pattern with 25 isolates. AP-PCR with the other two primers, and PFGE after SfiI digestion, confirmed that 16 isolates collected from 15 sites over an 11-year period had the type A pattern. There was no evidence of geographic clustering (Fig. 1). L. pneumophila isolates belonging to type A were regularly recovered from Paris environmental samples (from zero to five times a year, with no obvious peak incidence) (Fig. 2B) in hot water supplies, cooling towers, and a swimming pool (Table 2).
TABLE 2.
Sources of environmental isolates
| Source | No. of isolates with:
|
|
|---|---|---|
| Type A profile | Other profiles | |
| Hospitalsa | ||
| Hot water supply | 9 | 25 |
| Cooling tower | 2 | 3 |
| Other buildings | ||
| Hot water supply | 2 | 8 |
| Cooling tower | 2 | 4 |
| Swimming pool | 1 | 3 |
| Whirlpool spa | 0 | 2 |
| Miscellaneous | 0 | 3 |
| Total | 16 | 48 |
The type A profile was found in 10 different hospitals.
Comparison of the type A PFGE profile with profiles in the data bank of the French National Reference Center for Legionellosis (containing more than 250 PFGE profiles of clinical L. pneumophila serogroup 1 strains, 33 of which were isolated in the Paris area) indicated that the type A profile was not found outside the Paris area.
DISCUSSION
Various molecular methods based on genomic DNA polymorphisms of L. pneumophila serogroup 1 can be used to discriminate between epidemic and nonepidemic isolates (9, 10, 13, 14, 16, 18, 21, 22, 25–27). The finding that 25 sporadic clinical L. pneumophila isolates from the Paris area had identical AP-PCR patterns led us to suspect that the method lacked discriminatory power. However, a second molecular method, PFGE, which analyzes different portions of the Legionella genome, confirmed the strict identity of the 25 isolates. Unpublished results obtained by one of us (J.E.) show that PFGE, used to type more than 250 clinical L. pneumophila serogroup 1 isolates, has a discriminatory power of over 95% and that strains with the same pulsotype are most often epidemiologically related. The assumption, based on investigations of Legionnaires’ disease outbreaks, that clinical L. pneumophila isolates with identical PFGE types come from a single source should thus be reconsidered if the cases occur in large cities such as Paris. Indeed, 33% of our L. pneumophila serogroup 1 clinical isolates and 25% of the Paris environmental isolates had the same pattern (designated type A) by AP-PCR, with three different primers, and by PFGE. Six cases were considered definitely nosocomial but occurred in six different hospitals, further suggesting that a common environmental localized source was not involved. In the 19 cases considered to be community-acquired, no retrospective epidemiological study was undertaken to identify a possible common source. The latter cases were not clustered geographically or chronologically but occurred in various parts of Paris, and the incidence remained stable over the 10-year study period, suggesting that the infection was endemic rather than epidemic and that contamination of water sources is widespread in Paris. These results were confirmed by the presence of type A strains in various sites throughout the Paris area over a long period. This is the first time, to our knowledge, that dissemination of a single clone of L. pneumophila serogroup 1 throughout a water distribution network has been described. The fact that apparently unrelated isolates of L. pneumophila can display the same PFGE pattern has already been reported (21); such isolates can be found at sites separated by up to 60 km (14). It has also been shown, in Los Angeles County, that environmental strains exhibiting the same molecular markers as strains isolated from patients can be found at sites distinct from the site of infection (3). Our study emphasizes the need for molecular epidemiologic studies to include a large number of control strains to establish the validity of a given marker in a given area. It also suggests that molecular epidemiologic studies can only be used in conjunction with clinical epidemiologic findings and cannot stand on their own.
Moreover, it is also known that a single type of L. pneumophila can persist in the same water supply system for at least 10 years (4). Our results and other published data indicate that certain types of L. pneumophila are better suited to persistence in the environment (20). The similar proportions of type A strains among clinical and environmental isolates in this study suggest that the high incidence of type A infections is due more to the abundance of this strain in the environment than to greater pathogenicity. Taken together, our results show that type A is ubiquitous and constantly present in the L. pneumophila population of the Paris water supply network. The spread of this type from a water catchment or storage site is one possible explanation. Molecular analysis of all isolates from patients with legionellosis acquired in large cities should throw new light on the diversity of L. pneumophila.
ACKNOWLEDGMENT
This work was supported by a grant from the Programme Hospitalier de Recherche Clinique (AOM 96134).
REFERENCES
- 1.Bhopal R. Source of infection for sporadic Legionnaires’ disease: a review. J Infect. 1995;30:9–12. doi: 10.1016/s0163-4453(95)92665-8. [DOI] [PubMed] [Google Scholar]
- 2.Breiman R F. Mode of transmission in epidemic and nonepidemic Legionella infection: direction for further study. In: Barbaree J M, Breiman R F, Dufour A P, editors. Legionella: current status and emerging perspectives. Washington, D.C: American Society for Microbiology; 1993. pp. 30–35. [Google Scholar]
- 3.Buchholz U, Peterson C, Kool J, Mascola L, Brown E, Benson R, Pruckler J, Fields B, Sturgeon J, Lehnkering E, Butler J. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. 1-800-DNA-TYPE: strengths and limitations of molecular subtyping in a community outbreak of Legionnaires’ disease in Los Angeles County, abstr. L-116; p. 583. [Google Scholar]
- 4.Chang F, Jacobs S L, Colodny S M, Stout J E, Yu V L. Nosocomial Legionnaires’ disease caused by Legionella pneumophila serogroup 5: laboratory and epidemiologic implications. J Infect Dis. 1996;174:1116–1119. doi: 10.1093/infdis/174.5.1116. [DOI] [PubMed] [Google Scholar]
- 5.Commission de Normalisation. Norme AFNOR T90-431. Essais des eaux. Recherche et dénombrement des Legionella et L. pneumophila. Méthodes générales par ensemensement direct et filtration sur membrane. Paris, France: Association Française de Normalisation; 1993. [Google Scholar]
- 6.Edelstein P H, Nakahama C, Tobin J O, Calarco K, Beer K B, Joly J R, Selander R K. Paleoepidemiologic investigation of Legionnaires disease at Wadsworth Veterans Administration hospital by using three typing methods for comparison of legionellae from clinical and environmental sources. J Clin Microbiol. 1986;23:1121–1126. doi: 10.1128/jcm.23.6.1121-1126.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelstein P H. Legionnaires’ disease. Clin Infect Dis. 1993;16:741–749. doi: 10.1093/clind/16.6.741. [DOI] [PubMed] [Google Scholar]
- 8.Fraser D W, Tsai T R, Orenstein W, Parkin W E, Beecham H J, Sharrar R G, Harris J, Mallison G F, Martin S M, McDade J E, Shepard C C, Brachman P S. Legionnaires’ disease. Description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Luz P, Fields B S, Benson R F, Martin W T, O’Connor S P, Black C M. Comparison of arbitrarily primed polymerase chain reaction, ribotyping, and monoclonal antibody analysis for subtyping Legionella pneumophila serogroup 1. J Clin Microbiol. 1993;31:1940–1942. doi: 10.1128/jcm.31.7.1940-1942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grattard F, Berthelot P, Reyrolle M, Ros A, Etienne J, Pozzetto B. Molecular typing of nosocomial strains of Legionella pneumophila by arbitrarily primed PCR. J Clin Microbiol. 1996;34:1595–1598. doi: 10.1128/jcm.34.6.1595-1598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jernigan D B, Hofmann J, Cetron M S, Genese C A, Pekka Nuorti J, Fields B S, Benson R F, Carter R L, Edelstein P H, Guerrero I C, Paul S M, Lipman H B, Breiman R F. Outbreak of Legionnaires’ disease among cruise ship passengers exposed to a contaminated whirlpool spa. Lancet. 1996;347:494–499. doi: 10.1016/s0140-6736(96)91137-x. [DOI] [PubMed] [Google Scholar]
- 12.Keller D W, Hajjeh R, DeMaria A, Jr, Fields B S, Pruckler J M, Benson R S, Kludt P E, Lett S M, Mermel L A, Giorgio C, Breiman R F. Community outbreak of Legionnaires’ disease: an investigation confirming the potential for cooling towers to transmit Legionella species. Clin Infect Dis. 1996;22:257–261. doi: 10.1093/clinids/22.2.257. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence C, Ronco E, Dubrou S, Leclercq R, Nauciel C, Matsiota-Bernard P. Molecular typing of Legionella pneumophila serogroup 1 isolates from patients and the nosocomial environment by arbitrarily primed PCR and pulsed-field gel electrophoresis. J Med Microbiol. 1999;48:327–333. doi: 10.1099/00222615-48-4-327. [DOI] [PubMed] [Google Scholar]
- 14.Lück P C, Helbig J H, Günter U, Assmann M, Blau R, Koch H, Klepp M. Epidemiologic investigation by macrorestriction analysis and by using monoclonal antibodies of nosocomial pneumonia caused by Legionella pneumophila serogroup 10. J Clin Microbiol. 1994;32:2692–2697. doi: 10.1128/jcm.32.11.2692-2697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher W E, Para M F, Plouffe J F. Subtyping of Legionella pneumophila serogroup 1 isolates by monoclonal antibody and plasmid techniques. J Clin Microbiol. 1987;25:2281–2284. doi: 10.1128/jcm.25.12.2281-2284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamolen M, Breiman R F, Barbaree J, Gunn R A, Stone K M, Spika J S, Dennis D T, Mao S H, Vogt R L. Use of multiple molecular subtyping techniques to investigate a Legionnaires’ disease outbreak due to identical strains at two tourist lodges. J Clin Microbiol. 1993;31:2584–2588. doi: 10.1128/jcm.31.10.2584-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston B J, Lipman H B, Breiman R F. Surveillance for Legionnaires’ disease. Risk factor for morbidity and mortality. Arch Intern Med. 1994;154:2417–2422. [PubMed] [Google Scholar]
- 18.Matsiota-Bernard P, Thierry D, Guesdon J-L, Nauciel C. Molecular epidemiology of Legionella pneumophila serogroup 1 by ribotyping with a non-radioactive probe and PCR fingerprinting. FEMS Immunol Med Microbiol. 1994;9:23–27. doi: 10.1111/j.1574-695X.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 19.Nolte F S, Conlin C A, Roisin A J M, Redmond S R. Plasmids as epidemiological markers in nosocomial Legionnaires’ disease. J Infect Dis. 1984;149:251–256. doi: 10.1093/infdis/149.2.251. [DOI] [PubMed] [Google Scholar]
- 20.Plouffe J F, Para M F, Maher W E, Hackman B, Webster L. Subtypes of Legionella pneumophila serogroup 1 associated with different attack rates. Lancet. 1983;2:649–650. doi: 10.1016/s0140-6736(83)92531-x. [DOI] [PubMed] [Google Scholar]
- 21.Pruckler J M, Mermel L A, Benson R F. Comparison of Legionella pneumophila isolates by arbitrarily primed PCR and pulsed-field gel electrophoresis: analysis from seven epidemic investigations. J Clin Microbiol. 1995;33:2872–2875. doi: 10.1128/jcm.33.11.2872-2875.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–1498. doi: 10.1128/jcm.30.6.1491-1498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stout J E, Yu V L, Muraca P, Joly J, Troup N, Tompkins L S. Potable water as a cause of sporadic cases of community-acquired Legionnaires’ disease. N Engl J Med. 1992;326:151–155. doi: 10.1056/NEJM199201163260302. [DOI] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tram G, Simonet M, Nicolas M-H, Offredo C, Grimont F, Lefevre M, Ageron E, Debure A, Grimont P A. Molecular typing of nosocomial isolates of Legionella pneumophila. J Clin Microbiol. 1990;28:242–245. doi: 10.1128/jcm.28.2.242-245.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Belkum A, Struelens M, Quint W. Typing of Legionella pneumophila strains by polymerase chain reaction-mediated DNA fingerprinting. J Clin Microbiol. 1993;31:2198–2200. doi: 10.1128/jcm.31.8.2198-2200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitney C G, Hofmann J, Prukcler J M, Benson R F, Fields B S, Bandyopadhyay U, Donnally E F, Giorgio-Almonte C, Mermel L A, Boland S, Matyas B T, Breiman R F. The role of arbitrarily primed PCR in identifying the source of an outbreak of Legionnaires’ disease. J Clin Microbiol. 1997;35:1800–1804. doi: 10.1128/jcm.35.7.1800-1804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]