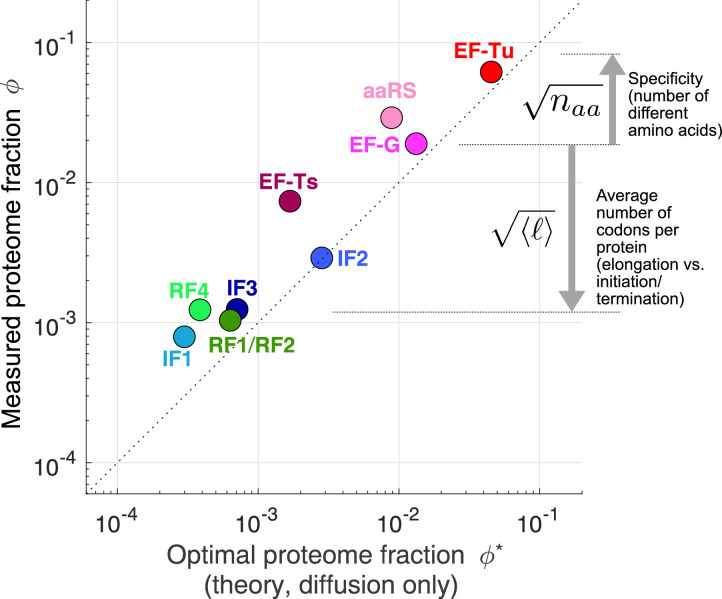
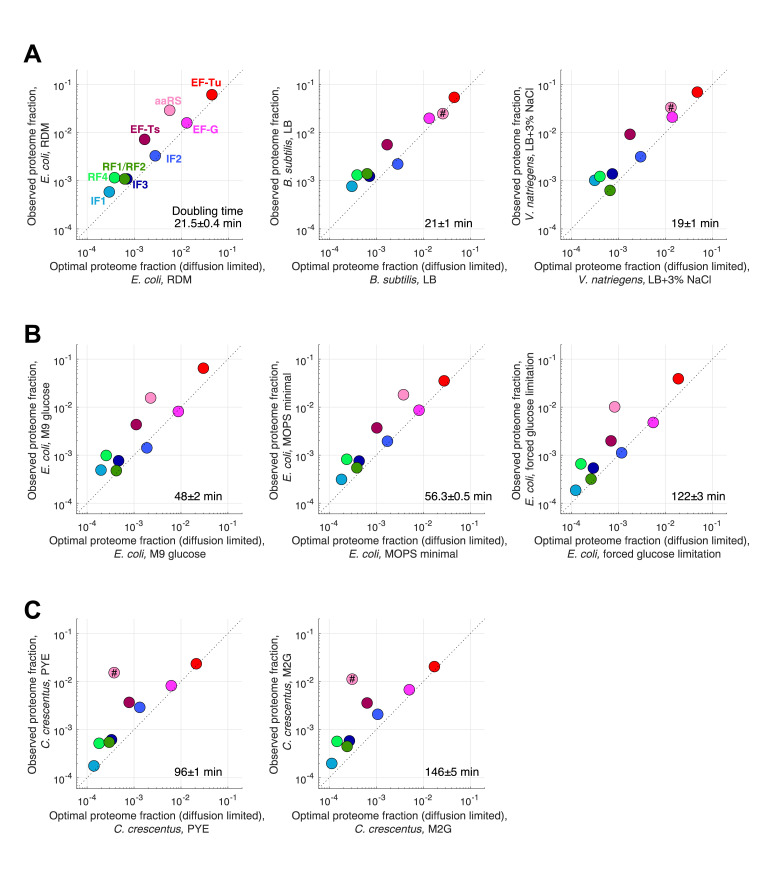
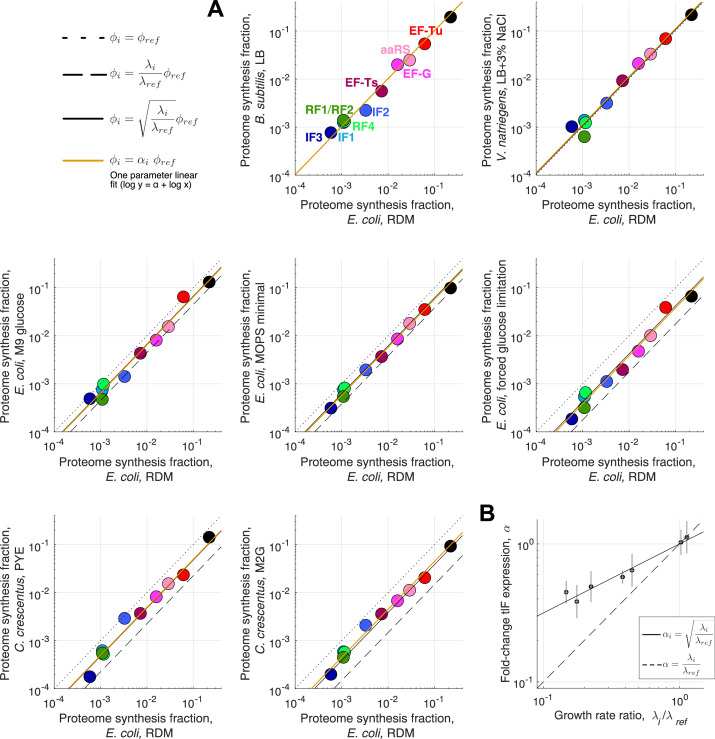
Expression stoichiometry of core translation factors in different species and at different growth rates. (
A) Comparison of measured (ribosome profiling) proteome fraction for core translation factors across different species and growth conditions (same conditions as
Figure 4—figure supplement 1). All conditions are compared to the
E. coli RDM dataset (reference:
, condition of interest:
). Dotted line correspond to
, dashed line to
and full black line to
(the parameter free prediction from the binding-limited regime of the model, optimal abundance
). Orange line corresponds to the one parameter fit
(excluding aaRS, not expected to follow the square root scaling, and ribosomes), corresponding to the scaling of all factor’s abundance. (
B) Best one-parameter fit
(scale factor) from (
A) as a function of the growth rate ratio
. Square root scaling: full line. Linear scaling: dashed line. Uncertainties on the growth ratio are propagated from uncertainties of the respective growth rates. Uncertainties in
are 95% confidence interval from the linear fits in (
A).