Abstract
A recent cluster of reports have considerably deepened our understanding of the transcriptional diversity of serotonin neurons of the dorsal raphe nucleus (DR). In this commentary a subset of implications from these studies is highlighted such as: serotonin neurons in the lateral wings have a newly discovered close relationship with those in rostral and dorsal locations and that cre-lines may be just as likely to cut across several transcriptional subtypes as to define a single subtype. To evolve understanding of DR organization, it may be prudent to correlate transcriptional snapshots in time with other known features of DR neurons. Here we bring together new and old information on serotonin neuron diversity with the goal of developing increasingly useful schemes of DR organization.
The first things that students seem to learn about serotonin are the misconceptions that serotonin neurons are homogenous, project nonspecifically over the brain and monotonically signal. It has been obvious almost from the discovery of serotonin neurons that serotonin neurons are highly organized. The problem is that serotonin neurons, although few in number, have far reaching functional impact on behavior with a complex structure to match. Suffice it to say that some of the logic underpinning the organization of the serotonin system has been elusive and the diffuse and nonspecific stereotype has been hard to shake.
In the last several years the idea that there may be distinct functional modules of serotonin neurons has taken hold of the field and the race to define those modules pursued. Historically serotonin neurons have been divided by their nuclear and subnuclear cytoarchitecture with increasing granularity. Different groups of neurons with interesting features and selective projection features have been defined in this way. In addition, early trends were identified in differential gene expression, such as serotonin neurons that express substance P in the medulla tend to innervate the dorsal but not ventral horn of the spinal cord (Johansson et al., 1981). In the age of Cre-lox and related tools such transcriptional differences have become key to both identifying and manipulating serotonin neurons.
Next generation sequencing has brought serotonin subtype identification to a new level. A cadre of recent studies have considerably refined understanding of the transcriptional profile and profile variance of serotonin neurons, mostly of the dorsal raphe nucleus (DR) but to some extent also the median raphe (MR) (Huang et al., 2019; Okaty et al., 2020; Ren et al., 2019). These studies again evolve our understanding of serotonin neuron organization. Here these new data are reconciled with old, to facilitate ways of thinking about the DR. By necessity this process involves some broad generalizations and speculation since mostly indirect evidence relates transcriptional cell-type to previous literature on other characteristics such as subnuclear location and projections. The reader should be cautioned that there are few clear-cut rules visible in the data but rather many trends and lines of circumstantial evidence.
Here we refer to transcriptional cell types as those defined by the most recent and detailed analysis (Okaty et al. 2020) as Okaty groups 1–14. Within Okaty et al. there is a description of how those groups correspond to groups defined in the studies by Ren et al., and Huang et al. While it is useful to define 14 subtypes as they allow a refined look at the transcriptional data, for some purposes it may be helpful to take a step back and consider groupings of the 14 subtypes that would be more approachable, easier to remember and useful for other purposes. The following discussion develops towards that goal while highlighting specific insights from the data.
Focusing on rostral 2/3 of the DRN for the moment, this area is populated, mostly, by what can be thought of as perhaps 4 groups (Table 1, Fig. 1). The first are those that have more markers for GABA neurotransmission than glutamate (Okaty groups 2, 3 and 4). Since the evidence for co-transmission of serotonin with GABA within these neurons is skant (Okaty et al., 2019), we call these “less Vglut3 5-HT neurons”. The reciprocal group has more markers of glutamate neurotransmission than GABA (Okaty groups 9, 10 and 11) or the “more Vglut3 5-HT neurons”. There is an in-between or “transitional” group (Okaty 5, 6) as well as some “scattered” neurons (Okaty 1).
Table 1.
Grouping of transcriptional subtypes and alignment with a mnemonic nickname, general location and likely projection targets. Currently group 13, NK1-Vglut3 neurons would not be identified as serotonergic by any other means and thus we call them ‘non-5-HT’, although they do express low levels of genes could enable serotonin neurotransmission
| Okaty et al. Group | Nickname | Location Bias | Projection Bias |
|---|---|---|---|
| 1 | Scattered | Scattered | |
| 2 | Less Vglut3 | Lateral, Rostral and Dorsal | Subcortical |
| 3 | |||
| 4 | |||
| 5 | Transitional | Rostral>Caudal | |
| 6 | |||
| 7 | MR-like | Caudal | Septohippocampal |
| 8 | |||
| 9 | More Vglut3 | Middle Ventral | Cortical |
| 10 | |||
| 11 | |||
| 12 | Met+ | Caudal Dorsal | Ventricular walls |
| 13 | NK1-Vglut3 (non-5-HT) | Middle Central | VTA |
| 14 | Atypical | Mixed/Caudal | Distinctive |
Figure 1:
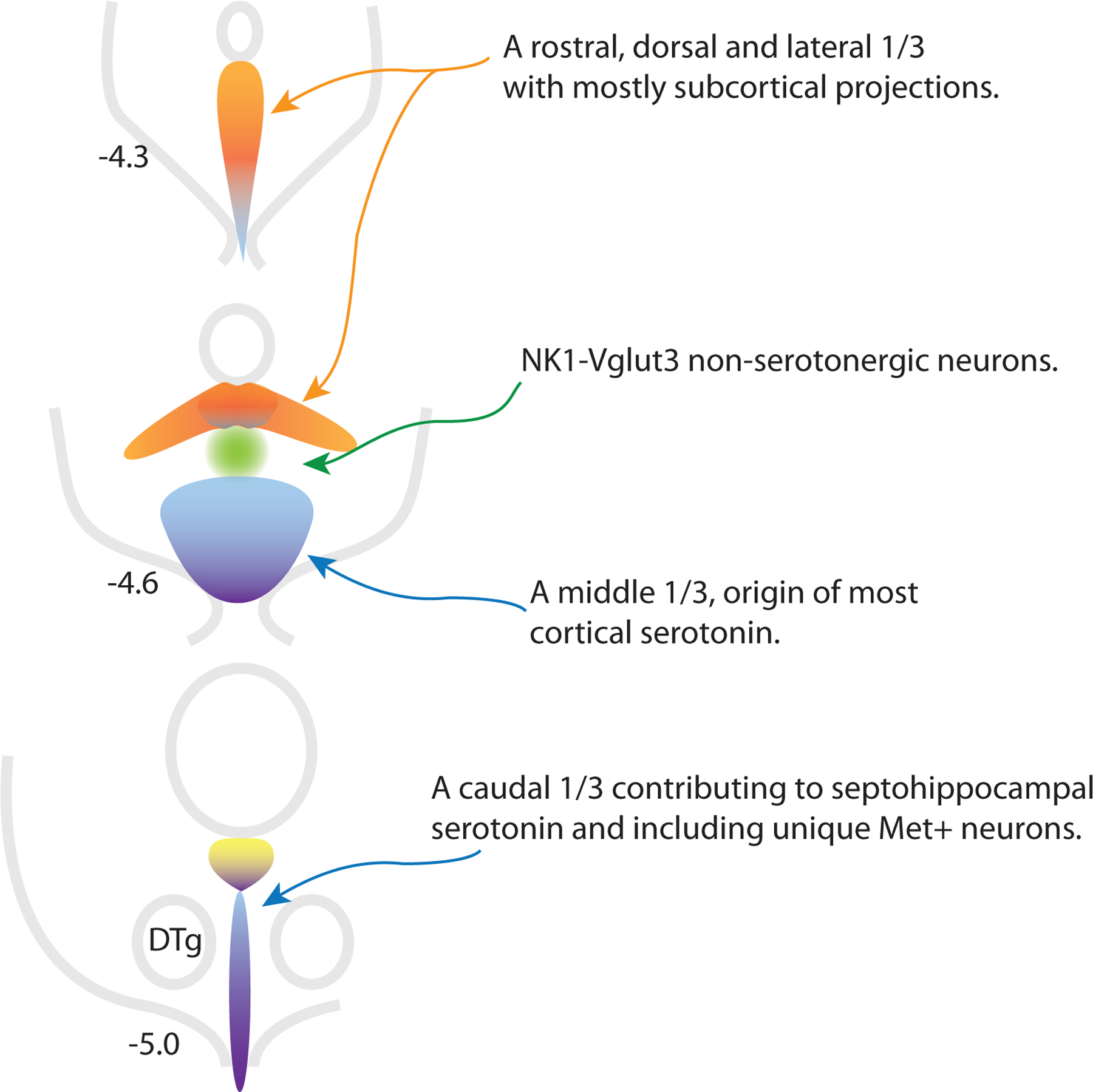
Schematic representation of different zones of the dorsal raphe nucleus from rostral (top) to caudal with distance from Bregma noted. New data link the lateral wings of the DR with neurons in rostral and dorsal locations in the nucleus, these areas may be more heavily populated by less Vglut3 and transitional neurons (groups 2–6, orange). NK1-Vglut3 neurons (group 13, green) have Pet1 lineage but express low levels of genes associated with serotonergic neurotransmission, but would not be identified as serotonergic by other criteria. These neurons may help coordinate ascending serotonin function with dopaminergic activity. The largest cluster of serotonin neurons likes in the middle-ventral part of the nucleus and is the major source of cortical serotonin (blue, more Vglut3 neurons including groups 9–11). The caudal third of the DR (purple, probably more heavily groups 7, 8, and 14) resembles the MR in connectivity, although Met+ neurons at the base of the aqueduct have many unique characteristics (yellow, group 12). Dorsal tegmental nucleus of Gudden (DTg).
Relating these to an anatomic distribution, less Vglut3 5-HT neurons tend to have a biased distribution lateral as well as rostral and dorsal. Our recent observations suggest that this is because these neurons share developmental origin from progenitors that express Fgf8 later in development (Guajardo et al., in press). The observation of a fundamental commonality between the lateral wings of the DR and neurons that skew rostral and dorsal is a major insight because previously neurons in the lateral wings were seen as unique and indeed their cytoarchitecture is very distinctive (Crawford et al., 2010; Kirifides et al., 2001). In retrospect there is a common theme in projections from these areas is that they tend to be subcortical (Muzerelle et al., 2014). Lateral neurons were well characterized as preferentially targeting sensory-related areas while projection targets of the rostral DR seem to show a preference for the ventral limbic forebrain (Kirifides et al., 2001; Muzerelle et al., 2014).
In contrast more Vglut3 5-HT neurons may be the major contributors to the largest single cluster of neurons in the DR located ventrally at mid-rostrocaudal levels. This area is known to be the major source of cortical serotonin (O’Hearn and Molliver, 1984; Waterhouse et al., 1986). Indeed evidence identifies Vglut3-expressing serotonin neurons as projecting cortically (Prouty et al., 2017; Ren et al., 2019). Mapping information would place transitional 5-HT neurons, which seem to share aspects with both former groups, generally more rostral in the nucleus than caudal (Okaty, et al., 2020).
There is no bright line between more Vglut3 5-HT neurons and neurons in Okaty group 8. Group 8 neurons were among those sampled from the caudal DR and they seem to overlap with neurons Ren et al sampled from the MR, suggesting the idea that these neurons are ‘MR like’ and located in the caudal DR. Group 7 was also sampled within the caudal DR and with transcriptional similarities to group 8, it may be worth considering these two together as ‘MR-like’.
While there is no direct evidence on the projections of these neurons, if they are transcriptionally similar to MR neurons and have a biased distribution in the caudal DR, it would certainly be reasonable to speculate that they contribute to serotoninergic innervation of the septohippocampal system. Interestingly MR-like neurons are not transcriptionally distant to more Vglut3 neurons and in different clustering schemes Okaty group 10 neurons switch between sorting with groups 7 and 8 vs. 9 and 11 (Okaty, et al., 2020). We speculate that all these groups share a common bias in their projections in that they favor cortical structures.
Even though transcriptionally defined cell types may correlate to some extent with location and projection pattern, all of the previously mentioned groups are fairly similar to each other. In fact further simplification could be argued using different criteria. For example, if Vglut3 expression was deemed particularly meaningful, one might envision a ‘larger less Vglut3’ group encompassing Okaty groups 1–6 and part of 14, vs. a ‘larger more Vglut3’ group including Okaty groups 7–12.
This propensity of serotonin neuron transcriptional profile to be fairly similar leads to a major implication of the recent studies. That is, a given Cre-line, even when restricted to a subset of serotonin neurons, may not define a single identified cell type as defined by gene expression holistically. For example, neurons identified by the Npyr2-cre that are located rostral likely belong to Okaty group 6 whereas those caudally may belong to groups 7 or possibly 10 or 14 (Okaty, et al., 2020). This is an important factor to consider when an effort is made to map function onto groups of neurons.
Thus most serotonin neurons seem remarkably similar to each other and this rule is emphasized by it’s exception, that is Okaty group 12 neurons, or ‘Met-5-HT neurons’. And now for something completely different. These neurons are very unique and have been identified and highlighted in previous studies, although Okaty et al., add another interesting insight in the definition of a cre-line (P2ry1) that selectively captures this population revealing new characteristics of these neurons as well as facilitating future studies.
Several unique characteristics of neurons Met-5-HT neurons that mostly reside in a densely packed cluster at the base of the aqueduct in the caudal pole of the DR have been described in the literature. Their projections are well characterized and they provide the major contribution to the supraependymal plexus, implicated in regulating cerebral spinal fluid as well as proliferation in the subventricular zone (Tong et al., 2014). Met-5-HT neurons may also innervate the hippocampus (Kast et al., 2017). Met-5-HT neurons seem to have very high colocalization of Vglut3 and 5-HT in their axon terminals, more so than many other 5-HT neurons, even those in the greater more Vglut3 population (Commons, 2009).
Some nuances with respect to anatomy of Met- 5-HT neurons should be pointed out however. While Met-5HT neurons cluster at the base of the aqueduct in a distinctive group, that anatomical group may not be homogenous and other transcriptional types may reside there. Further Met neurons may be located at other locations as well. Neurons that project to the aqueduct and separately Met-expressing 5-HT neurons have been identified ventrally (Mikkelsen et al., 1997; Okaty et al., 2015). Thus like other serotonin neurons, the relationship between transcription subtypes and anatomic location should be viewed as selective but not specific, even for these neurons that have both strikingly unique anatomical characteristics and transcriptional profile (Kast et al., 2017; Wu and Levitt, 2013).
An interesting group identified by using the Pet1-cre is a Vglut3 expressing population of neurons (Okaty group 13), which although they truly have a Pet1 lineage, have never been historically identified as serotonergic. It seems as though these neurons express very low levels of the genes that would confer the characteristic of serotonin neurotransmission and have been referred to as “low serotonin”. However it remains an open question if these neurons use serotonin as a transmitter, or if they might be plastic with the potential to use serotonin at some developmental time or under some unique environmental or behavioral circumstance. Until evidence of their use of serotonin as a transmitter develops more fully, we would characterize these as non-serotonergic Pet-1 lineage neurons.
A significant portion of Okaty group 13 neurons express neurokinin 1 or NK1 receptor (Tacr1) and Vglut3 and are located in the middle third of the nucleus between the dorsal and ventral clusters of serotonin neurons, thus these very likely overlap heavily with a group of neurons that have been fairly well described (Soiza-Reilly and Commons, 2011). The area occupied by NK1-Vglut3 neurons in the DR is associated with unique projections. It is specifically targeted by the rostromedial tegmental nucleus (RMTg) (Sego et al., 2014) and sends a glutamatergic projection to the ventral tegmental area (VTA) (Qi et al., 2014). Consistent with this circuitry, they play a role in reward processing (McDevitt et al., 2014; Qi et al., 2014).
NK1-Vglut3 neurons also seem to play a unique role in regulating network 5-HT activity (Soiza-Reilly and Commons, 2011). When these neurons are activated by substance P (SP), they release glutamate to activate a subpopulation of 5-HT neurons and these in turn produced a 5-HT1A-receptor dependent inhibition of other 5-HT neurons (Valentino et al., 2003). That is, they produce a mixed excitation/inhibition of other serotonin neurons. If NK1-Vglut3 neurons are functionally disabled, there is a characteristic adaptation of DR serotonin neurons in that 5-HT1A receptors desensitize (Amilhon et al., 2010; Froger et al., 2001). Thus in addition to influencing VTA dopaminergic function, these NK1-Vglut3 neurons probably have a specific role in coordinating serotonin neuron activity in the DR.
Related to NK1-Vglut3 neurons and Met-5-HT neurons are Okaty group 14, that are somewhat mysterious and perhaps deserve the moniker “atypical”. While their expression of genes conferring serotonergic neurotransmission is variable and may be low, for the moment we assume these would be largely identified as serotonergic using other means. In comparison to other serotonin neurons, atypical neurons express distinctive heparan sulfate proteoglycans, which are most similar to NK1-Vglut3 neurons. Therefore it may be speculated that these neurons project to areas different from other serotonin neurons, and a tenable hypothesis might be similar to those NK1-Vglut3 neurons.
As a final point, a spotlight was shown on neurons that do not cluster within a subregion of DR but are scattered through the area, the scattered group (Okaty group 1). Thus another interesting finding of the transcriptomic studies in that there are certain populations of serotonin neurons that have generally escaped prior study. Another very large group that remains largely unexplored is B9, even though in the rat there are in fact more serotonin neurons located in B9 than in the MR (Vertes and Crane, 1997).
In conclusion we might ask how many subtypes of serotonin neurons are there? The most comprehensive transcriptomics is able to resolve as many as 14. Subjectively these could be viewed as 7 groups plus NK1-Vglut3 neurons (Table 1). However, these 7 groups populate a DR that from an anatomical perspective might be roughly divided in thirds (Fig. 1), a more rostral/dorsal/lateral third, a ventral third and a caudal third that resembles the MR. Note that previously a division of the DR in thirds would link lateral wing neurons with midline neurons within the same coronal section but an emerging understanding is that these probably should be considered as more closely related to rostrally-located midline neurons. Each of these thirds may be further divided by various criteria.
It’s possible that we still lack information about some important features of serotonin neuron function that would better clarify meaningful groups, such as aspects of their individual phasic and tonic firing characteristics. In addition, the transcriptional analyses so far give us a snapshot in the transcriptional life of these neurons and the level of behavioral and developmental plasticity of their transcriptional repertoire is unknown. For example transitional 5-HT neurons could be a dynamic population that should shift transcriptional profile depending on the circumstances. Thus the number of useful groups may still evolve. Moreover the number of useful groups may depend on the purpose, and therefore the criteria used for grouping.
On the other hand, if a question can have many different answers, maybe the question itself is at fault. Imagine if the serotonin system were analogous to a piano, each serotonin neuron a different key. We might find ways to group the keys, by color (cytoarchtecture), register (connectivity) or note (transcriptional type) in useful ways. But it might not help us identify the major or minor key of a song, the rhythm, the genre or the song itself. Taking this analogy a step further, we can imaging that with gain and loss of function we could manipulate certain keys and selectively impact different songs. But would that mean the function of a particular set of keys maps to a particular song? That analogy might deserve some thought as the serotonin system is an instrument that regulates our emotions, whether we’re feeling the blues or ready to rock ‘n roll. That is to say, the evolution of questions may lead to better answers.
Acknowledgments
Funding from DA021801.
Footnotes
Conflict of interest: KGC received consulting fee from Zogenix, Inc.
References
- Amilhon B, Lepicard E, Renoir T, Mongeau R, Popa D, Poirel O, Miot S, Gras C, Gardier AM, Gallego J, Hamon M, Lanfumey L, Gasnier B, Giros B, El Mestikawy S, 2010. VGLUT3 (vesicular glutamate transporter type 3) contribution to the regulation of serotonergic transmission and anxiety. J Neurosci 30, 2198–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commons KG, 2009. Locally collateralizing glutamate neurons in the dorsal raphe nucleus responsive to substance P contain vesicular glutamate transporter 3 (VGLUT3). J Chem Neuroanat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LK, Craige CP, Beck SG, 2010. Increased intrinsic excitability of lateral wing serotonin neurons of the dorsal raphe: a mechanism for selective activation in stress circuits. Journal of neurophysiology 103, 2652–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froger N, Gardier AM, Moratalla R, Alberti I, Lena I, Boni C, De Felipe C, Rupniak NM, Hunt SP, Jacquot C, Hamon M, Lanfumey L, 2001. 5-hydroxytryptamine (5-HT)1A autoreceptor adaptive changes in substance P (neurokinin 1) receptor knock-out mice mimic antidepressant-induced desensitization. J Neurosci 21, 8188–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guajardo H, Hatini PG, Commons KG, In preparation. The Dorsal Raphe Nucleus as Understood by Temporal Fgf8 Lineage Analysis. [DOI] [PMC free article] [PubMed]
- Huang KW, Ochandarena NE, Philson AC, Hyun M, Birnbaum JE, Cicconet M, Sabatini BL, 2019. Molecular and anatomical organization of the dorsal raphe nucleus. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson O, Hokfelt T, Pernow B, Jeffcoate SL, White N, Steinbusch HW, Verhofstad AA, Emson PC, Spindel E, 1981. Immunohistochemical support for three putative transmitters in one neuron: coexistence of 5-hydroxytryptamine, substance P- and thyrotropin releasing hormone-like immunoreactivity in medullary neurons projecting to the spinal cord. Neuroscience 6, 1857–1881. [DOI] [PubMed] [Google Scholar]
- Kast RJ, Wu HH, Williams P, Gaspar P, Levitt P, 2017. Specific Connectivity and Unique Molecular Identity of MET Receptor Tyrosine Kinase Expressing Serotonergic Neurons in the Caudal Dorsal Raphe Nuclei. ACS chemical neuroscience 8, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirifides ML, Simpson KL, Lin RC, Waterhouse BD, 2001. Topographic organization and neurochemical identity of dorsal raphe neurons that project to the trigeminal somatosensory pathway in the rat. J Comp Neurol 435, 325–340. [DOI] [PubMed] [Google Scholar]
- McDevitt RA, Tiran-Cappello A, Shen H, Balderas I, Britt JP, Marino RAM, Chung SL, Richie CT, Harvey BK, Bonci A, 2014. Serotonergic versus nonserotonergic dorsal raphe projection neurons: differential participation in reward circuitry. Cell reports 8, 1857–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen JD, Hay-Schmidt A, Larsen PJ, 1997. Central innervation of the rat ependyma and subcommissural organ with special reference to ascending serotoninergic projections from the raphe nuclei. J Comp Neurol 384, 556–568. [PubMed] [Google Scholar]
- Muzerelle A, Scotto-Lomassese S, Bernard JF, Soiza-Reilly M, Gaspar P, 2014. Conditional anterograde tracing reveals distinct targeting of individual serotonin cell groups (B5-B9) to the forebrain and brainstem. Brain Struct Funct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hearn E, Molliver ME, 1984. Organization of raphe-cortical projections in rat: a quantitative retrograde study. Brain Res Bull 13, 709–726. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Commons KG, Dymecki SM, 2019. Embracing diversity in the 5-HT neuronal system. Nat Rev Neurosci 20, 397–424. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Freret ME, Rood BD, Brust RD, Hennessy ML, deBairos D, Kim JC, Cook MN, Dymecki SM, 2015. Multi-Scale Molecular Deconstruction of the Serotonin Neuron System. Neuron 88, 774–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sturrock N, Lozoya YE, Chang Y, Senft R, Lyon K, Alekseyenko OV, Dymecki SM, 2020. A single-cell transcriptomic and anatomic atlas of mouse dorsal raphe Pet1 neurons. eLife in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty EW, Chandler DJ, Waterhouse BD, 2017. Neurochemical differences between target-specific populations of rat dorsal raphe projection neurons. Brain Res 1675, 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Zhang S, Wang HL, Wang H, de Jesus Aceves Buendia J, Hoffman AF, Lupica CR, Seal RP, Morales M, 2014. A glutamatergic reward input from the dorsal raphe to ventral tegmental area dopamine neurons. Nat Commun 5, 5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Isakova A, Friedmann D, Zeng J, Grutzner SM, Pun A, Zhao GQ, Kolluru SS, Wang R, Lin R, Li P, Li A, Raymond JL, Luo Q, Luo M, Quake SR, Luo L, 2019. Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sego C, Goncalves L, Lima L, Furigo IC, Donato J Jr., Metzger M, 2014. Lateral habenula and the rostromedial tegmental nucleus innervate neurochemically distinct subdivisions of the dorsal raphe nucleus in the rat. J Comp Neurol 522, 1454–1484. [DOI] [PubMed] [Google Scholar]
- Soiza-Reilly M, Commons KG, 2011. Glutamatergic drive of the dorsal raphe nucleus. J Chem Neuroanat 41, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong CK, Chen J, Cebrian-Silla A, Mirzadeh Z, Obernier K, Guinto CD, Tecott LH, Garcia-Verdugo JM, Kriegstein A, Alvarez-Buylla A, 2014. Axonal control of the adult neural stem cell niche. Cell Stem Cell 14, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino RJ, Bey V, Pernar L, Commons KG, 2003. Substance P Acts through local circuits within the rat dorsal raphe nucleus to alter serotonergic neuronal activity. J Neurosci 23, 7155–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes RP, Crane AM, 1997. Distribution, quantification, and morphological characteristics of serotonin-immunoreactive cells of the supralemniscal nucleus (B9) and pontomesencephalic reticular formation in the rat. J Comp Neurol 378, 411–424. [DOI] [PubMed] [Google Scholar]
- Waterhouse BD, Mihailoff GA, Baack JC, Woodward DJ, 1986. Topographical distribution of dorsal and median raphe neurons projecting to motor, sensorimotor, and visual cortical areas in the rat. J Comp Neurol 249, 460–476, 478–481. [DOI] [PubMed] [Google Scholar]
- Wu HH, Levitt P, 2013. Prenatal expression of MET receptor tyrosine kinase in the fetal mouse dorsal raphe nuclei and the visceral motor/sensory brainstem. Developmental neuroscience 35, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


