Abstract
Context
Iron overload is a known risk factor for type 2 diabetes (T2D); however, iron overload and iron deficiency have both been associated with metabolic disorders in observational studies.
Objective
Using mendelian randomization (MR), we assessed how genetically predicted systemic iron status affected T2D risk.
Methods
A 2-sample MR analysis was used to obtain a causal estimate. We selected genetic variants strongly associated (P < 5 × 10−8) with 4 biomarkers of systemic iron status from a study involving 48 972 individuals performed by the Genetics of Iron Status consortium and applied these biomarkers to the T2D case-control study (74 124 cases and 824 006 controls) performed by the Diabetes Genetics Replication and Meta-analysis consortium. The simple median, weighted median, MR-Egger, MR analysis using mixture-model, weighted allele scores, and MR based on a Bayesian model averaging approaches were used for the sensitivity analysis.
Results
Genetically instrumented serum iron (odds ratio [OR]: 1.07; 95% CI, 1.02-1.12), ferritin (OR: 1.19; 95% CI, 1.08-1.32), and transferrin saturation (OR: 1.06; 95% CI, 1.02-1.09) were positively associated with T2D. In contrast, genetically instrumented transferrin, a marker of reduced iron status, was inversely associated with T2D (OR: 0.91; 95% CI, 0.87-0.96).
Conclusion
Genetic evidence supports a causal link between increased systemic iron status and increased T2D risk. Further studies involving various ethnic backgrounds based on individual-level data and studies regarding the underlying mechanism are warranted for reducing the risk of T2D.
Keywords: iron, ferritin, transferrin, mendelian randomization, type 2 diabetes
The essential element iron plays a crucial role in many fundamental biological processes, including energy metabolism, redox balance, oxygen delivery, and inflammation. However, iron is also potentially toxic, as excess free iron contributes to the generation of reactive oxygen species and is related to a wide variety of chronic diseases. Indeed, precisely controlled body iron level is essential for maintaining metabolic homeostasis.
The hormone hepcidin (encoded by the HAMP gene) plays a central role in regulating systemic iron and is controlled primarily through the BMP/SMAD (bone morphogenetic protein/mothers against decapentaplegic homolog) signaling pathway in response to iron stimulation. Genetic differences that affect the hepcidin-ferroportin axis are a principal cause of both iron overload and iron deficiency (1). For example, the HFE and TMPRSS6 genes (which encode the hemochromatosis protein HFE and transmembrane serine protease 6, respectively) contain the 3 loci selected in this study and were identified via either monogenic diseases or functional studies. Studies to date have shown that the HFE and TMPRSS6 proteins regulate HAMP transcription primarily via the BMP/SMAD signaling pathway (2). Mutations in the HFE gene cause late-onset (ie, type 1) hemochromatosis (OMIM No. 235200) (3), whereas mutations in the TMPRSS6 gene cause iron-refractory iron deficiency anemia (OMIM No. 206200) (4, 5). Genome-wide association studies (GWASs) revealed that the iron overload‒related single-nucleotide variations (SNVs; formerly single-nucleotide polymorphisms [SNPs]) rs1800562 (C282Y) and rs1799945 (H63D) in the HFE gene are associated with an increased risk of diabetes (6), whereas the iron deficiency‒related SNVs rs855791 (A736V) and rs4820268 (D521) in the TMPRSS6 gene are associated with a decreased risk of diabetes (7, 8).
A case-control study has found a direct correlation between iron stores and the prevalence of type 2 diabetes (T2D, noninsulin-dependent diabetes mellitus), with a lower ratio between the soluble fragment of the transferrin receptor and ferritin being associated with an increased risk of T2D (OR: 2.4; 95% CI, 1.03-5.5) (9). Previous observational studies have also found that higher iron status increases the risk of T2D. In addition, several meta-analyses examined the putative link between systemic iron status and the risk of T2D, suggesting that several iron indices—including ferritin, transferrin saturation, and heme iron intake—are correlated with the risk of diabetes (10-13). We also conducted a meta-analysis previously included 17 studies from 15 publications to evaluate the association between circulating ferritin level (per 100 μg/L) and T2D risk, which also found a positive correlation (relative risk [RR]: 1.22; 95% CI, 1.14-1.31), and a greater effect in women (RR: 1.53; 95% CI, 1.29-1.82) than those in men (RR: 1.21; 95% CI, 1.15-1.27) (14). In contrast, other studies found an association between iron deficiency and obesity. For example, as far back as the early 1960s epidemiology studies suggested a link between obesity and iron deficiency (15, 16). Comprehensive reviews and meta-analyses have since confirmed that obese/overweight participants generally have lower serum iron levels, higher hemoglobin concentration, higher ferritin levels, and lower levels of transferrin saturation (17, 18). Indeed, our previous meta-analysis has revealed the overweight/obese participants had an increased risk of iron deficiency (OR: 1.31; 95% CI, 1.01-1.68) (18). A major limitation with respect to observational studies is the difficulty distinguishing between bona fide causal relationships and spurious associations due to confounding and reverse causation (19, 20). Because genetic variants are determined at conception and have effects that are potentially lifelong, mendelian randomization (MR) studies are less vulnerable to key confounders (21). By using UK Biobank data sets, previous studies have found p.C282Y homozygotes associated with diabetes risk in men (22); however, their following MR analyses have not identified the causal link between genetically instrumented iron status and the risk of T2D (22, 23). Here, we conducted a MR analysis using the summary datasets with a larger sample size from the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) consortium, to elucidate the causal effect of systemic iron status on T2D risk.
Materials and Methods
Study Design
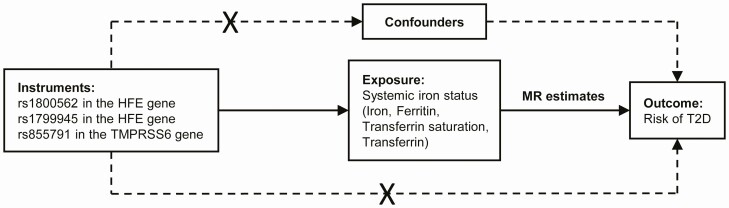
In this 2-sample MR study, we used data from 2 different studies—1 for the exposures and 1 for the outcome—to estimate the effects of exposure on outcome. Essentially, we applied genetic predictors of iron status to extensively genotyped case-control studies of T2D, thereby obtaining estimates of the putative association between iron and T2D risk. The overall study design is depicted graphically in Fig. 1.
Figure 1.
Graphical overview of the 2-sample MR study design. Three SNVs, each of which has a genome-wide significant association with increased serum iron, increased ferritin, increased transferrin saturation, and decreased transferrin levels, were used as instruments for systemic iron status. By using genetic instruments associated with these 4 iron status biomarkers, the MR approach can be used to estimate the causal effect of systemic iron status on the risk of T2D. MR, mendelian randomization; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
Genetic Associations With Systemic Iron Status
A meta-analysis of genome-wide studies was conducted previously by the Genetics of Iron Status (GIS) consortium to obtain association estimates between SNVs and biomarkers of iron status (24). Data from 11 discovery cohorts and 8 replication cohorts were used in the meta-analysis, which combined data obtained from 48 972 European individuals. The descriptive and statistics of the included cohorts in GIS consortium study are shown in Supplementary Table 1 (25). Adjustments were made for age and principal component scores, with analyses performed separately for male and female participants before combining estimates.
The first-stage regression F statistic was used to assess the strength of the instruments and was calculated using the following equation: , where R2 is the proportion of the circulating trace element status variability accounted for by the SNV, k is the number of instruments used in the model, and n is the sample size (26).
Genetic Associations With Type 2 Diabetes
The summary-level GWAS statistics of T2D were obtained from the DIAGRAM study, the aim of which is to characterize genetic information for T2D primarily in samples obtained from participants of European descent. The aggregated GWAS results were included for 32 studies involving 74 124 T2D cases and 824 006 controls of European ancestry. Considering that body mass index (BMI) may mediate the associations between iron status and T2D, we used the estimates for T2D without BMI adjustment in our primary analysis, and then performed a sensitivity analysis by using BMI-adjusted estimates. The descriptive and statistics of the included cohorts in DIAGRAM study are shown in Supplementary Table 2 (25). Imputation was performed using the Haplotype Reference Consortium reference panel for all the included studies, with the exception of the deCODE GWAS, which was imputed using a population-specific reference panel (27). The summary-level data are publicly available at http://diagram-consortium.org/.
Mendelian Randomization Estimates
An inverse variance weighted (IVW) meta-analysis of MR estimates derived using the ratio method was used to generate the main MR estimates for the association between each biomarker of iron status and the risk of T2D (28). The fixed-effect model was employed when using the 3 SNVs associated with all 4 iron status biomarkers. SEs were calculated using the Δ method (29). A threshold of P less than .05 was used to determine statistical significance.
Sensitivity Analysis
To examine whether the instruments for iron status exert effects on T2D risk through pleiotropic pathways that are independent of iron status, thus potentially biasing the results of the MR analysis (30, 31), we used the PhenoScanner database for SNV-phenotype associations (PhenoScanner, http://www.phenoscanner.medschl.cam.ac.uk/phenoscanner) to search for secondary phenotypes associated with the 3 selected instruments at genome-wide significance (P < 5 × 10–8) (32).
To account for potential bias due to unknown pleiotropy, we conducted sensitivity analyses using the simple median, weighted median (33), and MR Egger methods (34), which are more robust to the inclusion of pleiotropic instruments. The simple median and weighted median methods provide consistent estimates even when up to 50% of the information is derived from invalid SNVs (33). MR Egger regression additionally provides an estimate of the true causal effect that is consistent even if all genetic variants are invalid (34); however, MR Egger can be imprecise, particularly if the estimates are similar or if the number of genetic instruments is low. MR Egger intercept tests were conducted to assess the validity of the instrumental variable assumptions, with a non-null intercept indicating that the IVW estimate is biased (34). To verify our main findings in consideration of the horizontal pleiotropy, we employed another MR analysis using mixture-model (MRMix) (35). Given the relatively low statistical power of these approaches compared with the main analysis, they were used solely to confirm a consistent effect estimate compared to that seen in the main IVW estimate, rather than to ascertain statistical significance itself via a P value threshold. Heterogeneity was estimated using the Cochran Q statistic (36, 37).
Since the relatively rare variant rs1800562 (effect allele frequency is 0.067) may produce false-positive findings, we further conducted MR sensitivity analyses using allele scores weighted by various parameters (38), including minor allele frequency (MAF), variance of SNV (Var(SNV)), and the proportion of variance in the risk factor explained by the SNV (R2).
The separately selected SNVs by their genome-wide significant association with each iron status biomarker were used for assessing the consistency in our MR analyses. BMI-adjusted estimates from the DIAGRAM consortium were also used for sensitivity analysis.
Furthermore, to avoid the limitations of logistic regression methods and prioritize the most causally related risk factors, we further conducted a novel analysis based on the Bayesian model averaging approach (39). Briefly, all possible combinations of the 4 biomarkers of iron status were considered and posterior probability (PP) for each specific model was generated. Then, a marginal inclusion probability (MIP) for each iron biomarker was computed, where MIP refers to the sum of the PP over all possible models where the iron biomarker is presented. Furthermore, the model-averaged causal estimate for each iron biomarker by ranking all the iron biomarkers according to the corresponding MIP was computed. Finally, the best models by the PP values (with a PP threshold of 0.02) of the individual models were prioritized. Invalid instruments were detected as outliers with respect to the fit of the linear model using the Q statistic (40). The Cook’s distance was used for quantifying influential observations (41).
Data were analyzed using the TwoSampleMR package (version 0.4.23) in the statistical program R (version 3.6.1; the R Foundation for Statistical Computing).
Results
Genetic Instruments for Systemic Iron Status
Three SNVs including rs1800562 and rs1799945 in the HFE gene and rs855791 in the TMPRSS6 gene were employed for our main analysis. The F statistics for these 3 SNVs ranged from 47 for 2127 among the 4 biomarkers of iron status (Supplementary Table 3) (25), as described previously (42, 43), making significant bias from use of weak instruments unlikely (26). The individual SNV-iron marker estimates are listed in Supplementary Table 3 (25). The SNV-T2D estimates from the summary data without or with BMI adjustment are listed in Supplementary Tables 4 and 5, respectively (25).
Causal Relationship Between Systemic Iron Status and Type 2 Diabetes Risk
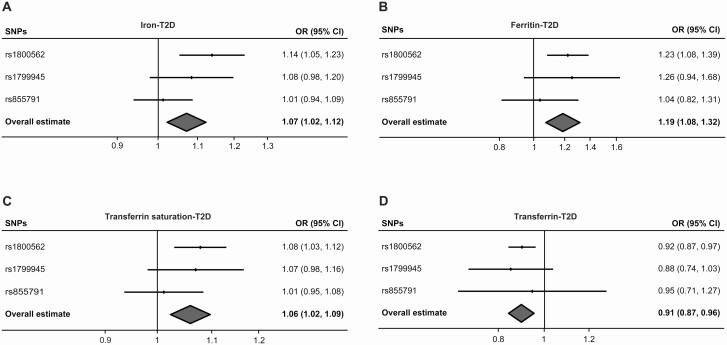
The results of our MR analysis, which are reported as the odds ratio (OR) of T2D per SD unit increase in each iron status biomarker, revealed an association between increased iron status and the risk of T2D, with serum iron (OR: 1.07; 95% CI, 1.02-1.12; P < .01), ferritin (OR: 1.19; 95% CI, 1.08-1.32; P < .01), and transferrin saturation (OR: 1.06; 95% CI, 1.02-1.09; P < .01) having a significant effect. In addition, higher transferrin levels, which are indicative of reduced iron status, were associated with a decreased risk of T2D (OR: 0.91; 95% CI, 0.87-0.96; P < .01). The relationship between each biomarker of iron status and the risk of T2D is shown graphically in Fig. 2.
Figure 2.
Forest plots summarizing the SNV-specific and overall MR estimates for the causal effects (fixed-effect IVW) on T2D without BMI adjustment using the SNVs associated with all four iron biomarkers. The causal effects of serum A, iron; B, ferritin; C, transferrin saturation; and D, transferrin on T2D risk (OR) are estimated. The solid black diamonds represent the estimates of the causal effects for the genetic instruments and the horizontal lines indicate the 95% CIs. The overall MR estimate is indicated by the center of the gray diamond, with the width of the diamond indicating the 95% CI. BMI, body mass index; IVW, inverse variance weighted; MR, mendelian randomization; OR, odds ratio; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
Sensitivity Analysis Provided no Indication of Unknown Pleiotropy
We further examined the biological pleiotropy of these instruments to evaluate the possible biases using the PhenoScanner database (32). As expected, all of these 3 SNVs were also associated with red blood cell traits due to the altered iron status. A potentially protective effect on T2D risk may be contributed by the iron status–increasing allele of rs1800562 with reduced low-density lipoprotein cholesterol and total cholesterol levels (44). Other protective effects on T2D risk may be contributed by all of these 3 SNVs due to their associations with reduced glycated hemoglobin A1c level (45, 46). On the contrary, the 2 SNVs rs1800562 and rs1799945 in HFE region are positively associated with diastolic blood pressure (47-49).
The simple median, weighted median, and MR Egger estimates produced directionally consistent effects as the IVW estimates, albeit with wider CIs (Table 1). The MR Egger intercepts for the 4 biomarkers did not differ significantly from null (P = .30, .51, .41, and .99 for serum iron, ferritin, transferrin saturation, and transferrin, respectively), thus providing no statistical indication of pleiotropy for T2D (see Table 1). The directions of the estimates of causal effects generated by the MRMix approach (θ) are consistent with our previous findings, and the proportions of valid instrumental variables (π 0) are 1 for all biomarkers (see Table 1). Given the relatively low statistical power of these approaches compared with the main analysis, they were used only to confirm an effect estimate directionally consistent to that seen in the main IVW MR, rather than to ascertain statistical significance itself via any given P value threshold. Lastly, for all 4 iron biomarkers, Cochran Q statistics showed low heterogeneities (P = .35, .37, .44, and .56 for serum iron, ferritin, transferrin saturation, and transferrin, respectively) (see Table 1).
Table 1.
Associations between genetically instrumented systemic iron status and type 2 diabetes without body mass index adjustment using the 3 single-nucleotide variations associated with all 4 iron biomarkersa
| IVW-fixed | IVW-random | Simple median | Weighted median | MR Egger | MRMixc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exposureb | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | Q statistic (P) | Intercept (P) | θ | π0 | σ2 |
| Iron | 1.07 | 1.02-1.12 | 1.07 | 1.00-1.15 | 1.08 | 1.00-1.17 | 1.07 | 1.01-1.14 | 1.29 | 1.07-1.56 | 0.88 (.35) | –0.04 (.30) | 0.06 | 1 | 2.64e-04 |
| Ferritin | 1.19 | 1.08-1.32 | 1.19 | 1.08-1.31 | 1.23 | 1.06-1.43 | 1.21 | 1.07-1.36 | 1.29 | 1.07-1.55 | 0.81 (.37) | –0.01 (.51) | 0.125 | 1 | 1.71e-04 |
| Transferrin saturation | 1.06 | 1.02-1.09 | 1.06 | 1.02-1.10 | 1.07 | 1.02-1.12 | 1.07 | 1.03-1.11 | 1.10 | 1.03-1.19 | 0.59 (.44) | –0.01 (.41) | 0.04 | 1 | 2.25e-04 |
| Transferrin | 0.91 | 0.87-0.96 | 0.91 | 0.90-0.93 | 0.92 | 0.83-1.01 | 0.91 | 0.87-0.96 | 0.91 | 0.86-0.97 | 0.33 (.56) | 0.00 (.99) | –0.138 | 1 | 1.42e-04 |
Abbreviations: BMI, body mass index; DIAGRAM, DIAbetes Genetics Replication and Meta-analysis; GIS, Genetics of Iron Status; IVW, inverse variance weighted; MR, mendelian randomization; MRMix, MR analysis using mixture-model; OR, odds ratio; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
a Data source and sample size: T2D case-control (n = 74 124 and 824 006, respectively) study based on the DIAGRAM consortium; genetic instruments were selected based on the GIS consortium study (n = 48 972).
b SNVs rs1800562, rs1799945, and rs855791 associated with all 4 iron status biomarkers at genome-wide significance (P < 5 × 10−8) were used as genetic predictors for systemic iron status.
c θ, the estimates of causal effects generated by MRMix approach; π0, the proportion of valid instrumental variables; and σ2, the unknown variance parameter associated with the invalid instrumental variables.
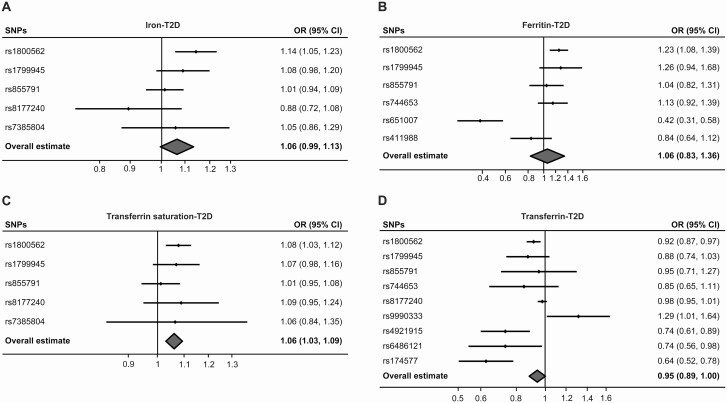
Investigation of BMI-unadjusted T2D risk using the separately selected SNVs by their genome-wide significant association with each iron status biomarker also produced directionally consistent results as shown in Fig. 3 and Table 2. Similarly, the MR Egger intercepts using the separately selected SNVs did not differ significantly from null (P = .21, .16, .69, and .28 for serum iron, ferritin, transferrin saturation, and transferrin, respectively), which providing no statistical indication of pleiotropy for T2D (see Table 2). And using the MRMix approach also derived directionally consistent results with high π 0 values (1, 0.647, 1, and 0.552 for iron, ferritin, transferrin saturation, and transferrin, respectively) (see Table 2). The Cochran Q statistics using the separately selected SNVs showed low heterogeneities for serum iron (P = .21) and transferrin saturation (P = .52); however, they showed significant heterogeneities (P < .01) for ferritin and transferrin (see Table 2). The MR analysis based on T2D summary data with adjustment for BMI derived consistent results as shown in Supplementary Figs. 1 and 2 and Supplementary Tables 6 and 7 (25).
Figure 3.
Forest plots summarizing the SNV-specific and overall MR estimates for the causal effects (random-effect IVW) on T2D without BMI adjustment using the separately selected SNPs associated with each iron status biomarker. The causal effects of serum A, iron; B, ferritin; C, transferrin saturation; and D, transferrin on T2D risk (OR) are estimated. The solid black diamonds represent the estimates of the causal effects for the genetic instruments and the horizontal lines indicate the 95% CIs. The overall MR estimate is indicated by the center of the gray diamond, with the width of the diamond indicating the 95% CI. BMI, body mass index; IVW, inverse variance weighted; MR, mendelian randomization; OR, odds ratio; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
Table 2.
Associations between genetically instrumented iron status and type 2 diabetes without body mass index adjustment using the separately selected single-nucleotide variations associated with each iron biomarkera
| IVW-fixed | IVW-random | Simple median | Weighted median | MR Egger | MRMixc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exposureb | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | Q statistic (P) | Intercept (P) | θ | π0 | σ2 |
| Iron | 1.06 | 1.01-1.11 | 1.06 | 0.99-1.13 | 1.05 | 0.97-1.14 | 1.06 | 1.00-1.13 | 1.14 | 1.02-1.26 | 4.49 (.21) | –0.01 (.21) | 0.035 | 1 | 1.14e-04 |
| Ferritin | 1.06 | 0.98-1.15 | 1.06 | 0.83-1.36 | 1.08 | 0.95-1.24 | 1.17 | 1.05-1.31 | 1.45 | 0.96-2.21 | 25.16 (< .01) | –0.03 (.16) | 0.13 | 0.647 | 1.23e-03 |
| Transferrin saturation | 1.06 | 1.03-1.09 | 1.06 | 1.03-1.09 | 1.07 | 1.01-1.12 | 1.07 | 1.03-1.11 | 1.07 | 1.02-1.13 | 2.28 (.52) | –0.002 (.69) | 0.03 | 1 | 4.95e-05 |
| Transferrin | 0.95 | 0.92-0.97 | 0.95 | 0.89-1.00 | 0.88 | 0.79-0.97 | 0.97 | 0.93-1.00 | 0.98 | 0.90-1.05 | 30.42 (< .01) | –0.008 (.28) | –0.28 | 0.552 | 4.72e-03 |
Abbreviations: BMI, body mass index; DIAGRAM, DIAbetes Genetics Replication and Meta-analysis; GIS, Genetics of Iron Status; IVW, inverse variance weighted; MR, Mendelian randomization; MRMix, MR analysis using mixture-model; OR, odds ratio; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
a Data source and sample size: T2D case-control (n = 74 124 and 824 006, respectively) study based on DIAGRAM consortium; genetic instruments were selected based on GIS consortium study (n = 48 972).
b SNVs associated with serum iron (rs1800562, rs1799945, rs855791, rs8177240, and rs7385804), Ferritin (rs1800562, rs1799945, rs855791, rs744653, rs651007, and rs411988), Transferrin saturation (rs1800562, rs1799945, rs855791, rs8177240, and rs7385804), and Transferrin (rs1800562, rs1799945, rs855791, rs744653, rs8177240, rs9990333, rs4921915, rs6486121, and rs174577) at genome-wide significance (P < 5 × 10−8) were used as genetic predictors for each iron biomarker.
c θ, the estimates of causal effects generated by MRMix approach; π0, the proportion of valid instrumental variables; and σ2, the unknown variance parameter associated with the invalid instrumental variables.
To evaluate the causal effects of the iron biomarkers on T2D risk in consideration of the measured pleiotropy, we conducted MR based on a Bayesian model averaging (MR-BMA) analyses using the SNVs associated with at least one of the biomarkers of iron status. In total, 12 SNVs (including rs1800562, rs1799945, rs855791, rs8177240, rs7385804, rs744653, rs651007, rs411988, rs9990333, rs4921915, rs6486121, and rs174577), 9 SNVs (excluding invalid instruments rs651007, rs174577, and rs4921915 with a Q statistic > 10), or 8 SNVs (further excluding influential instrument rs1800562 with Cook’s distance exceeding the threshold in all 4 best models) were employed for the MR-BMA analyses (Supplementary Tables 8 and 9) (25). The PPs of the best specific models and the MIPs of the risk factors were consistent with the results derived from the logistic regression approaches (Table 3). All the risk factors were then ranked by their MIPs, where the best models were prioritized and ranked by their PPs (see Table 3).
Table 3.
Ranking of risk factors and models (sets of risk factors) for type 2 diabetesa
| Risk factor or model | Ranking by MIP | MIP | CE | Ranking by PP | PP | λ |
|---|---|---|---|---|---|---|
| Model averaging using 12 SNVs (including rs1800562, rs1799945, rs855791, rs8177240, rs7385804, rs744653, rs651007, rs411988, rs9990333, rs4921915, rs6486121, rs174577) | ||||||
| Iron | 4 | 0.177 | 0.008 | 4 | 0.17 | 0.05 |
| Ferritin | 1 | 0.305 | 0.023 | 1 | 0.293 | 0.082 |
| Transferrin saturation | 3 | 0.266 | 0.017 | 3 | 0.254 | 0.061 |
| Transferrin | 2 | 0.27 | –0.015 | 2 | 0.264 | –0.056 |
| Model averaging using 9 SNVs (excluding invalid instruments rs651007, rs174577, rs4921915 with Q statistic > 10) | ||||||
| Iron | 4 | 0.089 | 0.004 | 4 | 0.08 | 0.052 |
| Ferritin | 1 | 0.532 | 0.074 | 1 | 0.519 | 0.14 |
| Transferrin saturation | 2 | 0.268 | 0.015 | 2 | 0.257 | 0.055 |
| Transferrin | 3 | 0.131 | –0.006 | 3 | 0.125 | –0.043 |
| Model averaging using 8 SNVs (excluding influential instrument rs1800562 with Cook’s distance exceeding the threshold) | ||||||
| Iron | 4 | 0.15 | 0.002 | 4 | 0.143 | 0.017 |
| Ferritin | 1 | 0.488 | 0.04 | 1 | 0.477 | 0.083 |
| Transferrin saturation | 2 | 0.214 | 0.008 | 2 | 0.206 | 0.035 |
| Transferrin | 3 | 0.164 | –0.004 | 3 | 0.158 | –0.026 |
Abbreviations: MIP, marginal inclusion probability; MR, mendelian randomization; MR-BMA, MR based on Bayesian model averaging; PP, posterior probability; SNV, single-nucleotide variation (formerly single-nucleotide polymorphism [SNP]); T2D, type 2 diabetes.
a Results were generated using the MR-BMA approach. In total, 4 genetically instrumented biomarkers of systemic iron status were assessed as risk factors. All the risk factors and the best individual models with a PP value greater than 0.02 were presented. A negative causal estimate (MACE or λ) indicates a protective effect as suggested by the model, whereas a positive value indicates a risk factor. λ is the causal effect estimate for a specific model and MACE is the model averaged causal effect of a risk factor.
Furthermore, when using MAF, Var(SNV), and R2-weighted allele scores for MR sensitivity analyses, the R2-weighted analysis still derived statistically consistent results (P < .05), whereas the results of MAF- and Var(SNV)-weighted analyses were only directionally consistent (Supplementary Table 10) (25).
Discussion
Here, we report the first evidence that several genetically determined markers of systemic iron status are associated with the risk of T2D. Our assumption in MR is that the instruments (SNVs) should be associated with the outcome of interest (T2D) only via the exposure (systemic iron status as reflected by the 4 iron biomarkers). We used a 2-sample study design that can cost-effectively produce unbiased estimates to determine the effects of systemic iron status on T2D risk. In addition, our use of samples generally pertaining to European individuals with appropriate genomic control (27) reduced the likelihood of bias due to concealed genetic associations. Our main MR analysis using the SNVs associated with all 4 iron biomarkers revealed that higher systemic iron status is associated with an increased risk of T2D based on IVW (with fixed effects, given that we included only 3 SNVs). Finally, simple median, weighted median, MR Egger, MRMix, weighted allele scores, and MR-BMA approaches yielded directionally consistent results as IVW.
Iron overload and iron deficiency both are associated with metabolic disorders, and perturbations in iron homeostasis can have a plethora of effects on T2D (50). For example, iron overload is a potent risk factor for diabetes, whereas iron deficiency is associated with obesity and insulin resistance (51). Although the pathogenic mechanism still needs further investigation, various tissues may have reflected the deleterious effects of iron overload on glycemic control. First, the elevated iron level could impair the function of pancreatic β cells both in humans (52) and high-fat diet–fed mice (53). Second, dietary iron overload could elevate adenosine monophosphate–activated protein kinase C activity and impair insulin signaling in skeletal muscle and liver in mice (54). Third, elevated iron status could decrease adiponectin secretion and insulin sensitivity of adipocytes in humans and high-iron diet–fed mice (55). Finally, breakdown of heme into carbon monoxide, biliverdin, and free iron by the enzyme heme oxygenase-1 could also promote chronic metabolic inflammation and insulin resistance in hepatocytes and macrophages (56).
To examine the causal relationship between iron status and T2D, we used an MR approach and found that higher systemic iron is associated with an increased risk of T2D. Increased systemic iron status has been associated with increased levels of serum iron, transferrin saturation, and ferritin, as well as decreased levels of transferrin (57). Thus, genetic variants were selected as instrumental variables based on their genome-wide significant association with these 4 biomarkers of iron status (ie, increased serum iron, increased ferritin, increased transferrin saturation, and decreased transferrin levels) (21). The aforementioned GIS consortium study identified 11 loci related to these biomarkers of iron status with genome-wide significance (P < 5 × 10–8) (24). Of these 11 loci, 3 (rs1800562 and rs1799945 in the HFE gene, and rs855791 in the TMPRSS6 gene) were associated with all 4 iron status biomarkers at genome-wide significance (P < 5 × 10–8), with low linkage disequilibrium between the rs1800562 and rs1799945 SNPs in the HFE gene (linkage disequilibrium: r2 < 0.01) (24).
Nutritional factors are causally associated with many chronic diseases (58, 59); however, such effects would be difficult to study in observational studies as these factors are correlated and often co-occur. In this respect, MR is valuable for identifying risk factors that could serve as potential targets for clinical and/or behavioral interventions (60, 61). Our use of genetically instrumented serum iron, ferritin, transferrin saturation, and transferrin enabled us to avoid potential confounding, therefore distinguishing the effects of iron status. Given that most genetic variants explain only a small portion of the variation in a risk factor, a relatively large sample size is required for an MR study with sufficient power (62). Using cross-trait meta-analysis of GWAS on a certain disease or health status could identify novel genetic loci (63). Several consortia containing large numbers of participants, including the GIS consortium for studying iron status (24) and the DIAGRAM consortium for studying T2D (27), have published data and have publicly available information regarding the association between genetic variants and either risk factors or disease status, thus providing precise estimates of genetic associations and enabling us to obtain causal estimates based on a well-powered MR study using a cost-effective approach (28). Previous MR studies have revealed that increased iron levels are causally associated with increased risk of stroke (particularly cardioembolic stroke) (43), and decreased risk of coronary artery disease (42) and Parkinson disease (64). No causal relationship has been identified previously between instrumented iron status and diabetes using the MR approach (22, 23). Thus, we suggested a causal link between systemic iron status and T2D for the first time.
A previous study using UK Biobank data sets has found that male p.C282Y homozygotes had a higher prevalence of diagnosed diabetes mellitus (predominantly T2D but including type 1; OR: 1.53; 95% CI, 1.16-1.98), compared with no p.C282Y mutations (irrespective of H63D status) (22). However, the following MR analysis showed no, but directionally consistent, association (β = .006; P = .06) between genetically instrumented transferrin saturation and diabetes in men (22). Another MR study evaluated the causal links between 25 predominantly metabolic traits and liver iron content and found a causative effect of central obesity, as measured by higher waist-to-hip ratio (adjusted for BMI), on elevated liver iron content (β = .162; P = .003) (65). However, T2D showed no causative effect on liver iron content (β = –0.021; P = .393) (65). Interestingly, a MR–phenome-wide association study (MR–PheWAS) using data from the UK Biobank also found a directionally consistent trend of the association (β = .06; P = .11) between the 3 SNVs (rs1800562, rs1799945, and rs855791) and instrumented serum iron and T2D risk (23). Actually, there are 32 cohorts (including the UK Biobank) used in the DIAGRAM study in our analysis. Our findings provide the first evidence that all 4 genetically determined biomarkers (serum iron, ferritin, transferrin saturation, and transferrin) of systemic iron status are significantly associated with the risk of T2D.
There are several limitations to this study. First, our analyses were conducted at the summary level, which made it impossible to conduct stratified analysis. Second, although the MR Egger intercepts and MRMix results suggested no statistical evidence of horizontal pleiotropy, the PhenoScanner database showed that the instrumental variables used in our MR analyses were truly associated with total cholesterol, low-density lipoprotein cholesterol, glycated hemoglobin A1c, and diastolic blood pressure, which may introduce some pleiotropy bias. Finally, the data sets used in our analyses were mainly derived from European individuals, which may hamper their translational relevance to other racial groups.
The findings of this study provided evidence that higher systemic iron status plays a causal role in the pathogenesis of T2D, which is consistent with a previous recognized association between iron and T2D based on observational studies. We have conducted meta-analyses previously and found that the disturbances of iron homeostasis are associated with obesity (18) and T2D (14). Indeed, a positive correlation has been established between heme iron intake and the risk of T2D (11).
Conclusions
Using an MR approach, we examined the putative hypothesis that systemic iron status has a causal effect on T2D risk. Specifically, we performed a 2-sample MR study based on iron status data measured in 48 972 individuals in the general population and T2D data obtained from 74 124 T2D cases and 824 006 controls. We then used 3 SNVs as instruments in order to increase statistical power by combining their MR estimates and to investigate the possible presence of pleiotropy. In summary, our results provide the first evidence that systemic iron status could be a causal factor in T2D development. Future studies should focus on examining this causal relationship in various populations with different ethnic backgrounds based on individual-level data, as well as the possible underlying mechanism, thereby providing new insights into potential strategies designed to prevent T2D.
Acknowledgments
The publicly available summary data on type 2 diabetes were provided by the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) investigators and were downloaded from http://diagram-consortium.org/. The authors thank all the investigators for sharing these data.
Financial Support: This work was supported by the Information Technology Center, Zhejiang University; the National Key R&D Program of China (grant Nos. 2018YFA0507801 to J.M., 2018YFA0507802 to F.W., and 2020YFC2008002 to M.X.); the National Natural Science Foundation of China (grant Nos. 31600953 to X.W., 31900835 to X.F., 31530034 and 31930057 to F.W., and 31570791 and 31970689 to J.M.); the Fundamental Research Funds for the Central Universities (grant No. 2020QNA7019 to X.W.); and the China Postdoctoral Science Foundation (grant No. 2019M650139 to X.F.).
Author Contributions: X.W., X.F., M.X., J.M., and F.W. conceived and designed the study; X.W., X.F., and W.Z. conducted the research; X.W., X.F., W.Z., J.Z., and Z.S. collected and analyzed the data; X.W., X.F., W.Z., J.Z., M.X., J.M., and F.W. wrote and revised the paper; and all authors read and approved the final manuscript.
Glossary
Abbreviations
- BMI
body mass index
- BMP
bone morphogenetic protein
- DIAGRAM
DIAbetes Genetics Replication and Meta-analysis
- GIS
Genetics of Iron Status
- GWAS
genome-wide association study
- HFE
hemochromatosis protein
- IVW
inverse variance weighted
- MAF
minor allele frequency
- MIP
marginal inclusion probability
- MR
mendelian randomization
- MR-BMA
mendelian randomization based on Bayesian model averaging
- MRMix
mendelian randomization analysis using mixture model
- OR
odds ratio
- PP
posterior probability
- RR
relative risk
- SMAD
mothers against decapentaplegic homolog
- SNV
single-nucleotide variation
- TMPRSS6
transmembrane serine protease 6
- T2D
type 2 diabetes
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112(2):219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117(17):4425-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399-408. [DOI] [PubMed] [Google Scholar]
- 4. Finberg KE, Heeney MM, Campagna DR, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40(5):569-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. An P, Wu Q, Wang H, et al. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet. 2012;21(9):2124-2131. [DOI] [PubMed] [Google Scholar]
- 6. Qi L, Meigs J, Manson JE, et al. HFE genetic variability, body iron stores, and the risk of type 2 diabetes in U.S. women. Diabetes. 2005;54(12):3567-3572. [DOI] [PubMed] [Google Scholar]
- 7. Gan W, Guan Y, Wu Q, et al. Association of TMPRSS6 polymorphisms with ferritin, hemoglobin, and type 2 diabetes risk in a Chinese Han population. Am J Clin Nutr. 2012;95(3):626-632. [DOI] [PubMed] [Google Scholar]
- 8. He M, Workalemahu T, Manson JE, Hu FB, Qi L. Genetic determinants for body iron store and type 2 diabetes risk in US men and women. PloS One. 2012;7(7):e40919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salonen JT, Tuomainen TP, Nyyssönen K, Lakka HM, Punnonen K. Relation between iron stores and non-insulin dependent diabetes in men: case-control study. BMJ. 1998;317(7160):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Z, Li S, Liu G, et al. Body iron stores and heme-iron intake in relation to risk of type 2 diabetes: a systematic review and meta-analysis. PloS One. 2012;7(7):e41641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bao W, Rong Y, Rong S, Liu L. Dietary iron intake, body iron stores, and the risk of type 2 diabetes: a systematic review and meta-analysis. BMC Med. 2012;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunutsor SK, Apekey TA, Walley J, Kain K. Ferritin levels and risk of type 2 diabetes mellitus: an updated systematic review and meta-analysis of prospective evidence. Diabetes Metab Res Rev. 2013;29(4):308-318. [DOI] [PubMed] [Google Scholar]
- 13. Orban E, Schwab S, Thorand B, Huth C. Association of iron indices and type 2 diabetes: a meta-analysis of observational studies. Diabetes Metab Res Rev. 2014;30(5):372-394. [DOI] [PubMed] [Google Scholar]
- 14. Jiang L, Wang K, Lo K, et al. Sex-specific association of circulating ferritin level and risk of type 2 diabetes: a dose-response meta-analysis of prospective studies. J Clin Endocrinol Metab. 2019;104(10):4539-4551. [DOI] [PubMed] [Google Scholar]
- 15. Wenzel BJ, Stults HB, Mayer J. Hypoferraemia in obese adolescents. Lancet. 1962;2(7251):327-328. [DOI] [PubMed] [Google Scholar]
- 16. Seltzer CC, Mayer J. Serum iron and iron-binding capacity in adolescents. II. Comparison of obese and nonobese subjects. Am J Clin Nutr. 1963;13:354-361. [DOI] [PubMed] [Google Scholar]
- 17. Cheng HL, Bryant C, Cook R, O’Connor H, Rooney K, Steinbeck K. The relationship between obesity and hypoferraemia in adults: a systematic review. Obes Rev. 2012;13(2):150-161. [DOI] [PubMed] [Google Scholar]
- 18. Zhao L, Zhang X, Shen Y, Fang X, Wang Y, Wang F. Obesity and iron deficiency: a quantitative meta-analysis. Obes Rev. 2015;16(12):1081-1093. [DOI] [PubMed] [Google Scholar]
- 19. Didelez V, Meng S, Sheehan NA. Assumptions of IV methods for observational epidemiology. Stat Sci. 2010;25(1):22-40. [Google Scholar]
- 20. Wang X, Jiao X, Tian Y, et al. Shanghai Birth Cohort Study . Associations between maternal vitamin D status during three trimesters and cord blood 25(OH)D concentrations in newborns: a prospective Shanghai birth cohort study. Eur J Nutr. Published online March 4, 2021. doi:10.1007/s00394-021-02528-w [DOI] [PubMed] [Google Scholar]
- 21. Taylor AE, Davies NM, Ware JJ, VanderWeele T, Smith GD, Munafò MR. Mendelian randomization in health research: using appropriate genetic variants and avoiding biased estimates. Econ Hum Biol. 2014;13:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pilling LC, Tamosauskaite J, Jones G, et al. Common conditions associated with hereditary haemochromatosis genetic variants: cohort study in UK Biobank. BMJ. 2019;364:k5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gill D, Benyamin B, Moore LSP, et al. Associations of genetically determined iron status across the phenome: a mendelian randomization study. PloS Med. 2019;16(6):e1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benyamin B, Esko T, Ried JS, et al. ; InterAct Consortium . Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X, Fang X, Zheng W, et al. Data from: Genetic support of a causal relationship between iron status and type 2 diabetes: a Mendelian randomization study—Supplementary material. Figshare. 2021. Deposited May 7. 2021. 10.6084/m9.figshare.14552463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palmer TM, Lawlor DA, Harbord RM, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21(3):223-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG; EPIC- InterAct Consortium . Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson JR, Minelli C, Del Greco MF. Mendelian randomization using public data from genetic consortia. Int J Biostat. 2016;12(2):20150074. [DOI] [PubMed] [Google Scholar]
- 30. Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29(2):66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PloS Med. 2008;5(8):e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi G, Chatterjee N. Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat Commun. 2019;10(1):1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10(1):101-129. [Google Scholar]
- 37. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177-188. [DOI] [PubMed] [Google Scholar]
- 38. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuber V, Colijn JM, Klaver C, Burgess S. Selecting likely causal risk factors from high-throughput experiments using multivariable Mendelian randomization. Nat Commun. 2020;11(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cook RD. Influential observations in linear regression. J Am Stat Assoc. 1979;74(365):169-174. [Google Scholar]
- 42. Gill D, Del Greco M F, Walker AP, Srai SKS, Laffan MA, Minelli C. The effect of iron status on risk of coronary artery disease: a mendelian randomization study—brief report. Arterioscler Thromb Vasc Biol. 2017;37(9):1788-1792. [DOI] [PubMed] [Google Scholar]
- 43. Gill D, Monori G, Tzoulaki I, Dehghan A. Iron status and risk of stroke. Stroke. 2018;49(12):2815-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soranzo N, Sanna S, Wheeler E, et al. ; WTCCC . Common variants at 10 genomic loci influence hemoglobin A1 (C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wheeler E, Leong A, Liu CT, et al. ; EPIC-CVD Consortium; EPIC-InterAct Consortium; Lifelines Cohort Study . Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PloS Med. 2017;14(9):e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wain LV, Vaez A, Jansen R, et al. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 2017;70(3):e4-e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Surendran P, Drenos F, Young R, et al. ; CHARGE-Heart Failure Consortium; EchoGen Consortium; METASTROKE Consortium; GIANT Consortium; EPIC-InterAct Consortium; Lifelines Cohort Study; Wellcome Trust Case Control Consortium; Understanding Society Scientific Group; EPIC-CVD Consortium; CHARGE+ Exome Chip Blood Pressure Consortium; T2D-GENES Consortium; GoT2DGenes Consortium; ExomeBP Consortium; CHD Exome+ Consortium . Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48(10):1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ehret GB, Munroe PB, Rice KM, et al. ; International Consortium for Blood Pressure Genome-Wide Association Studies; CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium . Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simcox JA, McClain DA. Iron and diabetes risk. Cell Metab. 2013;17(3):329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X, Fang X, Wang F. Pleiotropic actions of iron balance in diabetes mellitus. Rev Endocr Metab Disord. 2015;16(1):15-23. [DOI] [PubMed] [Google Scholar]
- 52. Utzschneider KM, Largajolli A, Bertoldo A, et al. Serum ferritin is associated with non-alcoholic fatty liver disease and decreased Β-cell function in non-diabetic men and women. J Diabetes Complications. 2014;28(2):177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng K, Ho K, Stokes R, et al. Hypoxia-inducible factor-1alpha regulates beta cell function in mouse and human islets. J Clin Invest. 2010;120(6):2171-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang J, Simcox J, Mitchell TC, et al. Iron regulates glucose homeostasis in liver and muscle via AMP-activated protein kinase in mice. FASEB J. 2013;27(7):2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gabrielsen JS, Gao Y, Simcox JA, et al. Adipocyte iron regulates adiponectin and insulin sensitivity. J Clin Invest. 2012;122(10):3529-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jais A, Einwallner E, Sharif O, et al. Heme oxygenase-1 drives metaflammation and insulin resistance in mouse and man. Cell. 2014;158(1):25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wish JB. Assessing iron status: beyond serum ferritin and transferrin saturation. Clin J Am Soc Nephrol. 2006;1(Suppl 1):S4-S8. [DOI] [PubMed] [Google Scholar]
- 58. Chen J, Zhao X, Cui L, et al. Genetic regulatory subnetworks and key regulating genes in rat hippocampus perturbed by prenatal malnutrition: implications for major brain disorders. Aging (Albany NY). 2020;12(9):8434-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng S, Zhao T, Yuan S, et al. Immunodeficiency promotes adaptive alterations of host gut microbiome: an observational metagenomic study in mice. Front Microbiol. 2019;10:2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang F, Baranova A, Zhou C, et al. Causal influences of neuroticism on mental health and cardiovascular disease. Hum Genet. Published online May 11, 2021. doi:10.1007/s00439-021-02288-x [DOI] [PubMed] [Google Scholar]
- 61. Zhang F, Rao S, Cao H, et al. Genetic evidence suggests posttraumatic stress disorder as a subtype of major depressive disorder. J Clin Invest. Published online April 27, 2021. doi:10.1172/JCI145942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schatzkin A, Abnet CC, Cross AJ, et al. Mendelian randomization: how it can—and cannot—help confirm causal relations between nutrition and cancer. Cancer Prev Res (Phila). 2009;2(2):104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu Y, Cao H, Baranova A, et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl Psychiatry. 2020;10(1):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pichler I, Del Greco M F, Gögele M, et al. ; PD GWAS Consortium; International Parkinson’s Disease Genomics Consortium; Wellcome Trust Case Control Consortium 2; Genetics of Iron Status Consortium . Serum iron levels and the risk of Parkinson disease: a Mendelian randomization study. PloS Med. 2013;10(6):e1001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilman HR, Parisinos CA, Atabaki-Pasdar N, et al. ; IMI DIRECT Consortium . Genetic studies of abdominal MRI data identify genes regulating hepcidin as major determinants of liver iron concentration. J Hepatol. 2019;71(3):594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”