Abstract
Background
The genomic and transcriptomic landscape of widely invasive follicular thyroid carcinomas (wiFTCs) and Hürthle cell carcinoma (HCC) are poorly characterized, and subsets of these tumors lack information on genetic driver events.
Objective
The aim of this study was to bridge this gap.
Methods
We performed whole-genome and RNA sequencing and subsequent bioinformatic analyses of 11 wiFTCs and 2 HCCs with a particularly poor prognosis, and matched normal tissue.
Results
All wiFTCs exhibited one or several mutations in established thyroid cancer genes, including TERT (n = 4), NRAS (n = 3), HRAS, KRAS, AKT, PTEN, PIK3CA, MUTYH, TSHR, and MEN1 (n = 1 each). MutSig2CV analysis revealed recurrent somatic mutations in FAM72D (n = 3, in 2 wiFTCs and in a single HCC), TP53 (n = 3, in 2 wiFTCs and a single HCC), and EIF1AX (n = 3), with DGCR8 (n = 2) as borderline significant. The DGCR8 mutations were recurrent p.E518K missense alterations, known to cause familial multinodular goiter via disruption of microRNA (miRNA) processing. Expression analyses showed reduced DGCR8 messenger RNA expression in FTCs in general, and the 2 DGCR8 mutants displayed a distinct miRNA profile compared to DGCR8 wild-types. Copy number analyses revealed recurrent gains on chromosomes 4, 6, and 10, and fusion gene analyses revealed 27 high-quality events. Both HCCs displayed hyperploidy, which was fairly unusual in the FTC cohort. Based on the transcriptome data, tumors amassed in 2 principal clusters.
Conclusion
We describe the genomic and transcriptomic landscape in wiFTCs and HCCs and identify novel recurrent mutations and copy number alterations with possible driver properties and lay the foundation for future studies.
Keywords: follicular, thyroid, carcinoma, cancer, mutation, whole-genome
Thyroid cancer is the most common malignancy in endocrine organs and the eleventh most common malignancy worldwide. Remarkably, thyroid cancer has shown a steep increase in incidence for the last few decades (1, 2). Although the prognosis of thyroid cancer in general is quite favorable, the second most common type, follicular thyroid carcinoma (FTC), shows a 5-year survival of around 90% and drops to 80% after 10 years (3). In 2017, the World Health Organization revised the subclassifications of FTC based on the grade of capsular and vascular invasion where the widely invasive FTC (wiFTC) is the most aggressive form, which is reflected in its poor prognosis (4, 5). FTCs with more than 75% oncocytic features were also separated into a distinct tumor entity denoted Hürthle cell carcinoma (HCC) (5). FTCs and HCCs rarely metastasize to neck lymph nodes but predominantly spread to the lungs and bone, causing significant morbidity (6).
The diagnostic procedure includes a fine-needle aspiration biopsy from the tumor; however, a preoperative diagnosis cannot rely solely on this procedure because it is morphologically impossible to distinguish FTC and HCC cells from its benign counterpart follicular thyroid adenoma (FTA) and Hürthle cell adenoma, respectively (5). Therefore, a diagnostic hemithyroidectomy is performed for a conclusive histopathological diagnosis, and if the specimen fulfils the criteria of an FTC or HCC (capsular or vascular invasion) the patient undergoes a completion thyroidectomy, during which the other lobe is removed, providing conditions for a more successful subsequent treatment and follow-up (7). The treatment regimen is based on surgery, radioiodine therapy, and subsequent thyrotropin suppression with thyroxine, where the dose of radioiodine and the length of suppression is based on clinicopathological variables (7). Thus, so far, no molecular alterations have been widely implemented in the diagnostic, predictive and prognostic workup.
Most FTCs are characterized either by point mutations in the RAS gene family or by PAX8-PPARγ rearrangements, and they appear to occur mutually exclusively (8). The frequency of RAS gene family mutations and PAX8-PPARγ rearrangements in FTC ranges from 30% to 60% and 10% to 60%, respectively (9). Other genetic alterations that include activation of the PI3K/protein kinase B (AKT) pathway are also common in FTC, and are together with the RAS gene family mutations and PAX8-PPARγ rearrangements the only widely accepted cancer-driver events in sporadic FTC, despite also being present to a lesser extent in FTA (9-15). Mechanisms involving TERT alterations, including point mutations in the TERT promoter (TERTp), have frequently been reported in FTCs and are associated with a worse clinical outcome. However, although they contribute to thyroid cancer progression, it is unclear whether the activating TERTp mutations can act as a cancer driver alone (16-21). Despite these advances in surveying follicular thyroid tumorigenesis, a substantial proportion of FTCs lacks an identifiable driver. HCCs (previously known as “oxyphilic follicular thyroid carcinomas”) show mutations affecting the mitogen-activated protein kinase pathway, PI3/Akt pathway, TERTp, and PAX8-PPARγ rearrangements but to a lesser extent than FTCs (5). In contrast to FTCs, HCCs display mitochondrial DNA mutations possibly related to its mitochondrial rich cytoplasm (5).
A few recent attempts have been made to further elucidate possible cancer drivers and prognostic markers in FTC by using various next-generation sequencing (NGS) methods. These studies have used either whole-exome sequencing (WES) or targeted NGS. However, very few novel recurrent genetic events have been identified, especially in wiFTC (22-27). Furthermore, to our knowledge, no study has evaluated HCCs using pangenomic sequencing. Whole-genome sequencing (WGS) provides comprehensive insight into all types of genomic alterations, including an unbiased detection of somatic mutations and a superiority over WES in detecting copy number alterations (CNAs) (28). In this study, our aim was to characterize the genomic and transcriptomic landscape in wiFTC and HCC with a particularly poor prognosis. We first characterized the somatic genomic landscape by applying the Sarek (29) workflow using WGS data from 11 wiFTCs and 2 HCCs with matched adjacent normal thyroid tissue, and the transcriptional profile from RNA sequencing of the 13 tumors and 2 unrelated normal thyroid tissues.
Materials and Methods
Clinical Characteristics of Follicular Thyroid Carcinomas and Hürthle Cell Carcinomas
The clinicopathological characteristics of the 11 wiFTC and 2 HCC patients are summarized in Table 1 and visualized in Fig. 1. Detailed information is available as supplementary data (30). The cohort consisted of 11 wiFTCs and 2 HCCs retrieved through a thyroid lobectomy and subsequently diagnosed by histopathological examination at the Department of Pathology and Cytology, Karolinska University Hospital in Stockholm, Sweden. All tumors were assessed using the most recent World Health Organization criteria (5), all tumors were large, grossly invasive tumors displaying multiple foci both with capsular and vascular invasion. All cases were also reviewed for the so-called Turin criteria (solid, trabecular and/or insular growth, increased mitotic count, and/or the presence of tumor necrosis), in order to exclude the presence of a poorly differentiated thyroid carcinoma (PDTC). We deliberately aimed to include fresh-frozen tumors from patients with adverse clinical parameters, such as metastasized disease (n = 8) and/or disease-specific fatal outcomes (n = 5), and for the remaining nonmetastasized cases we selectively collected larger tumors (> 40 mm, n = 5). As matched constitutional tissues for the pangenomic analysis, we used histologically normal thyroid tissue.
Table 1.
Clinicopathologic characteristics of the 11 follicular thyroid carcinomas and 2 Hürthle cell carcinomas examined by whole-genome and RNA sequencing
| Parameter | ||
|---|---|---|
| FTC, case IDs 101, 102, 201, 104, 105, 106, 203, 204, 205, 206, and 207 | ||
| n (%) | ||
| Age, y | ≥ 55 | 8 (73%) |
| < 55 | 3 (27%) | |
| Sex | Female | 7 (63%) |
| Male | 4 (37%) | |
| Recurrence/Metastasis | No | 7 (63%) |
| Yes | 4 (37%) | |
| AJCC staging a | I | 4 (36%) |
| II | 6 (55%) | |
| III | 0 (0%) | |
| IVA | 0 (0%) | |
| IVB | 1 (9%) | |
| Median (min-max) | ||
| Tumor size, mm | 50 (35-100) | |
| Ki-67 index, % | 2 (1-11) | |
| HCC, case IDs 202 and 208 | ||
| n (%) | ||
| Age, y | ≥ 55 | 1 (50%) |
| < 55 | 1 (50%) | |
| Sex | Female | 1 (50%) |
| Male | 1 (50%) | |
| Recurrence/Metastasis | No | 1 (50%) |
| Yes | 1 (50%) | |
| AJCC staging a | I | 1 (50%) |
| II | 1 (50%) | |
| III | 0 (0%) | |
| IVA | 0 (0%) | |
| IVB | 0 (0%) | |
| Median (min-max) | ||
| Tumor size, mm | 45.5 (45-46) | |
| Ki-67 index, % | 4 (4) |
Abbreviations: FTC, follicular thyroid carcinoma; HCC, Hürthle cell carcinoma; ID, identification; max, maximum; min, minimum.
a American Joint Committee on Cancer, eighth edition.
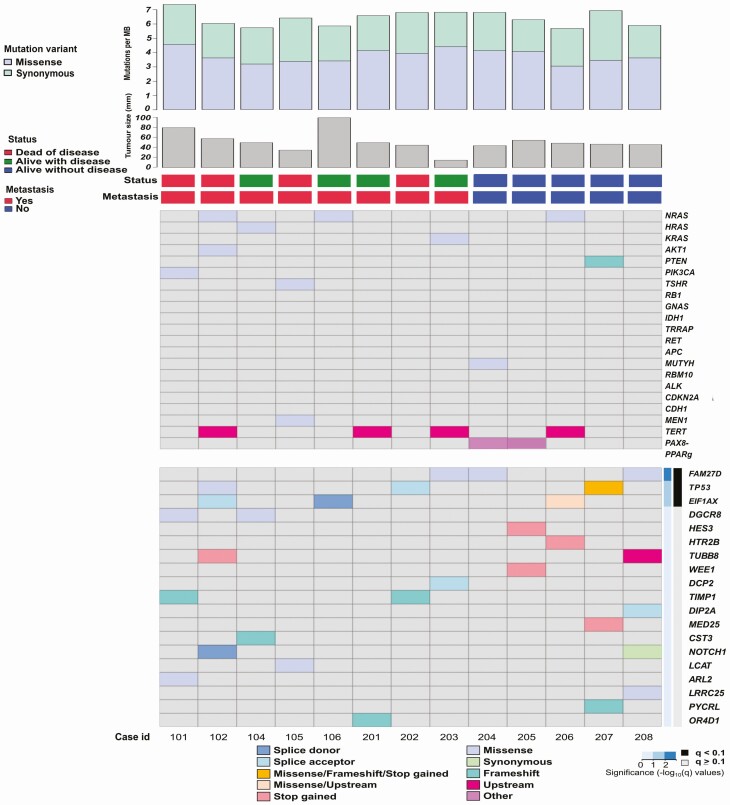
Figure 1.
Heat map of the somatic mutational landscape across the whole-genome sequenced tumor cohort. Each column represents one patient/tumor. Each row represents a mutated gene and the color code represents the mutation type. The top grid displays thyroid-related genes (top 20 COSMIC mutated genes), while the bottom grid displays top mutations called by MutSigCV2 sorted by P value. Cases 202 and 208 are Hürthle cell carcinomas; the remaining 11 cases are widely invasive follicular thyroid carcinomas (wiFTCs).
Tissue Representativity Testing
To verify adequate and representative cell contents for all tumors and normal thyroid specimens subsequently submitted for nucleic acid extraction, a small piece of the tissue was dissected using a sterile scalpel, fixated in formalin, embedded in paraffin, cut at 4 μm, mounted on slides, and stained with hematoxylin-eosin. An endocrine pathologist (C.C.J.) verified high tumor cell percentages (> 90%) for all included tumors and histologically normal thyroid tissue for the constitutional DNA extractions (data not shown).
DNA and RNA Extraction
For all included samples, genomic DNA was extracted from fresh-frozen tissue using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. Total RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. MicroRNA (miRNA) was extracted using the mirVana miRNA isolation kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Whole-Genome Sequencing and an Overview of the Associated Bioinformatics
WGS was performed at the National Genomics Infrastructure Sweden (SciLifeLab, Stockholm, Sweden). Library preparation was performed using Illumina TruSeq polymerase chain reaction (PCR)-free libraries (350 bp) from 13 tumors and 13 corresponding normal samples. Subsequent sequencing was performed using the Illumina HighSeq X with a coverage of 60× for tumors and 30× for normal samples. The library preparation, sequencing, and data processing are validated methods under International Standards Organization accreditation 17025:2005. WGS bioinformatics processing and analysis was performed at the National Bioinformatics Infrastructure Sweden by 2 of the authors (N.R. and S.D.L.). Briefly, WGS data of 13 matched tumor/control samples were analyzed. The reads were realigned to reference genome GRCh38 before analysis, and the subsequent analyses were divided into 2 parts: small-variant analyses and large-variant analyses. For the small-variant analyses, a panel of normal and selected high-quality variants were generated for downstream analyses. The identified variants were then annotated for their functional effect on genes. Results were compared with other publicly available databases such as The Cancer Genome Atlas and the Catalogue of Somatic Mutations in Cancer (COSMIC; both accessed October 16, 2019). For large structural variation analyses, the genome was screened for CNA and gene fusions.
Analyses of Somatic DNA Alterations
Following quality controls, Genome Analysis Toolkit (GATK) best practices were followed as implemented in the Sarek pipeline, version 2.3, developed at National Bioinformatics Infrastructure Sweden and National Genomic Infrastructure (29). Sarek uses nextflow DSL, in this study version 19.03.0-SNAPSHOT, as a computational framework. In this project, the following steps were performed: aligning the reads with BWA (31), marking duplicate reads with GATK4 MarkDuplicates, base quality recalibration with GATK4 BQSR, calling germline small variants with GATK4 HaplotypeCaller (32), calling somatic small variants with GATK4 Mutect2 (33), calling somatic structural variation with Manta (34), somatic CNA and ploidy with ASCAT (35), annotation both of small variants and structural variants with snpEff (36) and VEP (37), and quality control with MultiQC (38).
To extract high-quality small somatic variants (single-nucleotide variations [SNVs] and insertion-deletion mutations [indels]), we created a panel of normals based on the Mutect2 recommendations. The panel of normals was used to remove additional normal variations, and the remaining variants were used for downstream analyses. To identify significantly mutated genes, we used MutSig2CV version 3.11 (39). Because this tool is hard coded to hg19, the VCF files for each sample were lifted over to the hg19 genome reference using picard LiftOverVCF, version 2.10.3. The variants were filtered in 2 additional steps before MutSig2CV analysis, first using the GATK4 FilterMutectCalls version 4.1.0.0, which became standard best practice for somatic short mutation calling after the Sarek pipeline was run, and second, using the R software version 3.5 to apply custom filters. In the second filtering step using R, filters were inspected and customized. The filters were as follows: 1) filter intergenic region and unknown gene variants; 2) filter by the population database gnomAD (≥ 1%); 3) filter out low-impact variants; 4) filter out variants deemed benign both by PolyPhen and SIFT annotations; 5) filter out low-coverage variants (< 10× in tumor); 6) filter out upstream and downstream gene variants; 7) filter out intronic variants (nonregulatory regions like splice sites excluded); and 8) filter out custom clustered variant regions with several low AF variants. All code and supplementary plots of which variants were affected are available on request.
Structural Genomic Variations
To analyze somatic CNAs, calls from ASCAT were used to identify gain and loss events across the genome. We calculated gain and loss using reported ploidy by ASCAT based on the following formula: Gain = #copies > ploidy + 0.6, Loss = #copies < ploidy + 0.6, Deletion= 0 #copies is the sum of major and minor allele reported by ASCAT. For visualization, we summarized the events along each chromosome based on their location in cytobands and their size, filtering away CNAs less than 1 kb. For some cytobands, we observed more than one CNA event and selected the largest CNA per region.
Genomic Rearrangements
For genomic rearrangements, we extracted the gene_fusion term from snpEff annotation of structural variation detected by Manta. We selected events passing the filtering step (only PASS) and classified them based on reported rearrangement by Manta (translocation, inversion, duplication, and deletion). The filtered list was then visualized by the circlize R package (40). We parsed the data and visualized them using R.
RNA Sequencing
The transcriptomes of 13 tumor samples and 2 normal thyroid samples were sequenced. Library construction was performed with the Illumina TruSeq Stranded messenger RNA (mRNA) kit, followed by Poly-A selection and clustering using “cBot.” Libraries were sequenced on HiSeq2500 (HiSeq Control Software 2.2.58/RTA 1.18.64) with a 2 × 126 setup using “HiSeq SBS Kit v4” chemistry. The Bcl to FastQ conversion was performed using bcl2fastq_v2.19.1.403 from the CASAVA software suite. The quality scale used was Sanger/phred33/Illumina 1.8+. Reads were aligned using STAR (41) and gene counts were generated with featureCounts (42). National Genomic Infrastructure performed read alignment and extracted read counts by using nextflow/rnaseq pipeline (available in the GitHub repository https://github.com/nf-core/rnaseq).
Differential Expression Analysis and Gene Set Enrichment Analysis
For gene set enrichment analysis in R, normalized counts were used to identify gene sets significantly associated with patient survival, tumor metastasis, and TERTp mutations. Individual genes in the significantly enriched pathways were visualized using pheatmap package in R. Next, differential expression with an individual false discovery rate of less than 0.01 was calculated between normal and tumors using the DESeq2 (43) package in R. Differentially expressed genes were visualized on a heat map and samples were clustered using unsupervised hierarchal clustering and visualized with pheatmap and the Enhanced volcanoplot package (v 1.8.0, Blighe, 2020) in R. Upregulated genes from the differential expression analysis were analyzed for gene ontology using Enrichr (v 2.1, Jawaid, 2019).
Expression of Genes in Minimal Region of Amplification
Canonical genes in the cytoband 10q11.21 were extracted using the BioMart tool through Ensembl. Tumors with gain and neutral/loss in the minimal region of amplification (MRA) were analyzed for differential expression using DESeq2.
Sanger Sequencing Validation
The NGS cohort of 13 tumors and an additional 120 thyroid carcinomas (37 HCCs, 33 minimally invasive FTCs, 26 wiFTCs, 20 PDTCs, and 7 encapsulated angioinvasive FTCs were sequenced to validate and screen for the selected mutation in the DGCR8 gene. Exon 7 of the DGCR8 gene was amplified using F primer 5′-TGTGATGTGTTGAGGGCATG and R primer 5′-CCTTTCCTCCCGTTCCAGT and Sanger sequenced in both directions. Mutational calling including visual inspection of each chromatogram was performed using CodonCode Aligner (CodonCode Corp).
Quantitative Real-Time Polymerase Chain Reaction
Total mRNA was used for complementary DNA synthesis using the High-Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR was performed in 65 FTCs using the QuantStudio 6 Real-Time PCR system (Thermo Fisher Scientific) with TaqMan Gene expression assay (Hs00377897_m1) to investigate DGCR8 expression. The 18S ribosomal RNA expression was used as an endogenous reference (Hs99999901_s1). A total of 42 FTAs served as controls. Relative expression was calculated as 2–Δct.
MicroRNA Profiling
In total, 100 ng of total RNA from fresh-frozen tissue was used for miRNA profiling using the direct molecular barcoding with digital detection method NanoString nCounter platform (NanoString Technologies). The panel includes expression of 827 human miRNAs and 6 positive mRNA controls, 8 negative mRNA controls, 3 ligation-positive miRNA controls, 3 ligation-negative miRNA controls, 5 mRNA reference controls, and 5 spike-in controls. Samples were hybridized with probes containing unique barcodes and counted using the nCounter platform. The data were analyzed with nSolver v.4 (NanoString Technologies) and Rosalind (Rosalind Inc, https://rosalind.onramp.bio/). Normalization, fold changes, and P values were calculated using criteria provided by NanoString. Background was subtracted based on POS A probe correction factors. Normalization was performed by calculating the geometric mean from positive controls and code-set mRNA housekeeping genes. An unsupervised clustering heat map was generated as part of the QC step including the top 50 most variable miRNAs. Differential expression between DGCR8-mutated (n = 2) FTC and DGCR8 wild-type (n = 9) FTC cases was performed with fold change as 1.5 and the adjusted P value threshold set to .05.
Results
Patient Characteristics and Sequencing Quality Parameters
The clinical characteristics of the included patients are summarized in Table 1 and detailed in Supplementary Table S1 (30). In total, 26 samples were successfully sequenced using WGS and the Illumina HiSeq X platform, including 11 primary wiFTCs, 2 primary HCCs, and corresponding constitutional tissues. The average genome coverage for tumors and normals was 68.4× and 34.0×, respectively, and the average alignment to the reference human genome was 98.0% and 98.6%, respectively. The aggregated percentage of bases that had a quality score higher than Q30 was 90% on average, ranging from 87.06% to 92.19%. Q30 is equal to an inferred base call accuracy of 99.9%.
Somatic Mutational Overview
By using the previously described Sarek workflow, we found a total of 63 855 genomic, SNVs, and indels in 17 184 genes. These alterations are schematically depicted in Supplementary Fig. S1 (30). After extensive filtering (as outlined in “Materials and Methods”), 889 SNVs remained for subsequent analysis with MutSig2CV. The mutational burden and main events are illustrated in Fig. 1. The complete list of somatic SNVs in all tumors and the final MutSig2CV input list are available as Supplementary Table S2 and Supplementary Table S3, with MutSig2CV genes ranked by P value in Supplementary Table S4 (30).
Mutations in Thyroid Cancer-Associated Genes
To highlight mutational events in established thyroid cancer genes, we compared our complete list of somatic mutations with the top 20 genes mutated in FTCs as reported by the COSMIC database (see Fig. 1, Table 2). All wiFTCs exhibited one or several mutations with predicted functional impact in established genes ranked as the top 20 mutated in FTC, including the TERTp (n = 4), NRAS (n = 3), HRAS, KRAS, AKT, PTEN, PIK3CA, TSHR, MUTYH, and MEN1 (n = 1 each) (see Table 2). The 2 HCCs did not display any coding mutations among the top 20 thyroid-related mutated genes (see Table 2, case identifications 202 and 208), but several alterations in other cancer-related genes not commonly associated with thyroid cancer (see Fig. 1).
Table 2.
List of top 20 thyroid-related mutated genes (COSMIC) present in the whole-genome sequencing cohort
| Gene symbol | Location (GRCh37) | Transcript | HGVSc | HGVSp | Mutation classification | Hot spot (COSMIC) | Case ID | Allele frequency, % |
|---|---|---|---|---|---|---|---|---|
| NRAS | Chr1:115256529 | ENST00000369535 | c.182A > G | p.Gln61Arg | Missense | Yes | 102 | 39 |
| 106 | 30 | |||||||
| Chr1:115256530 | ENST00000369535 | c.181C > A | p.Gln61Lys | Missense | Yes | 206 | 30 | |
| HRAS | Chr11:533874 | ENST00000451590 | c.182A > G | p.Gln61Arg | Missense | Yes | 104 | 42 |
| KRAS | Chr12:25380276 | ENST00000256078 | c.182A > G | p.Gln61Arg | Missense | Yes | 203 | 39 |
| AKT1 | Chr14:105246551 | ENST00000554581 | c.49G > A | p.Glu17Lys | Missense | Yes | 102 | 46 |
| PTEN | Chr10:89624274 | ENST00000371953 | c.49dup | p.Gln17ProfsTer27 | Frameshift | No | 207 | 86 |
| PIK3CA | Chr3:178952085 | ENST00000263967 | c.3140A > G | p.His1047Arg | Missense | Yes | 101 | 20 |
| TSHR | Chr14:81609860 | ENST00000541158 | c.1458C > G | p.Ile486Met | Missense | No | 105 | 42 |
| MUTYH | Chr1:45332926 | ENST00000450313.5 | c.496T > C | p.Ser166Pro | Missense | 204 | 5 | |
| MEN1 | Chr11:64574544 | ENST00000337652 | c.866C > A | p.Ala289Glu | Missense | No | 105 | 53 |
| TERT | Chr5:1295228 | ENST00000310581 | Upstream | Yes | 102 | 61 | ||
| 201 | 56 | |||||||
| 203 | 43 | |||||||
| 206 | 58 |
Abbreviations: COSMIC, Catalogue of Somatic Mutations in Cancer; Gene symbol, HUGO gene symbol; HGVSc, human genome variation society coding sequence; HGVSp, human genome variation society protein sequence; ID, identification; Location (GRCh37), chromosome location according to GRCh37 assembly; Transcript, ensembl transcript.
Novel Genes of Interest
In addition, a separate analysis using MutSig2CV was performed (see Fig. 1). From the 889 SNVs that remained after initial filtering, 564 were run in MutSig2CV. In-depth analyses of these variants discovered somatic mutations in 3 genes annotated as significant according to MutSig2CV with a q value of less than 0.1; FAM72D (4 mutations in 3 cases), TP53 (3 cases), and EIF1AX (3 cases), with DGCR8 (2 cases) as borderline significant (see Fig. 1). The properties of these mutations are detailed in Table 3. Of these genes, TP53 and EIF1AX have previously been reported as recurrently mutated in FTC, whereas FAM72D and DGCR8 mutations are unusual in this context (with a prevalence of 0.67% and 0.29% in unspecified thyroid cancer specimen respectively when consulting the COSMIC database). The TP53 and EIF1AX mutations were distinct from each other. In 3 patients, an indel (c.295_296delinsGA) in FAM72D caused an amino acid change at codon 99 (p.Arg99Glu) and the 2 samples with a DGCR8 mutation exhibited an identical c.1552G > A (p.Glu518Lys) missense alteration. The FAM72D mutation p.Arg99Glu has not been reported in the COSMIC database (accessed December 2020), and was considered as benign using in silico analyses, with low/no impact of protein structure and function (PolyPhen2 score of 0.001). The DGCR8 mutation p.Glu518Lys, however, was considered probably damaging (PolyPhen2 score of 1).
Table 3.
List of top MutSig2CV somatic mutations in the whole-genome sequencing cohort of 13 widely invasive follicular thyroid carcinomas
| Gene | P/q | Location (GRCh37) | Transcript | HGVSc | HGVSp | Mutation classification | Hot spot (COSMIC) | Case ID | Allele frequency, % |
|---|---|---|---|---|---|---|---|---|---|
| FAM72D | < .001/0.007 | Chr1:143906071 | ENST00000400889 | c.295_296delinsGA | p.Arg99Glu | Missense | Not reported | 203 | 38 |
| Chr1:143906086 | ENST00000400889 | p.Leu94Pro | p.L94P | Missense | Not reported | 203 | 38 | ||
| Chr1:143906071 | ENST00000400889 | c.295_296delinsGA | p.Arg99Glu | Missense | Not reported | 204 | 20 | ||
| Chr1:143906072 | ENST00000400889 | c.295_296delinsGA | p.Arg99Glu | Missense | Not reported | 208 | 13 | ||
| TP53 | < .001/0.032 | Chr17:7675070 | ENST00000635293 | c.425G > A | p.Arg142His | Missense | No | 102 | 9 |
| Chr17:7577156 | ENST00000269305 | c.783-1G > T | p.X261_splice | Splice acceptor | No | 202 | 93 | ||
| Chr17:7577120 | ENST00000269305 | c.818G > T | p.Arg273Leu | Missense | Yes | 207 a | 23 | ||
| Chr17:7578253 | ENST00000269305 | c.596G > A | p.Gly199Glu | Missense | No | 207 a | 11 | ||
| Chr17:7579366 | ENST00000269305 | c.320dup | p.Tyr107Ter | Nonsense | No | 207 a | 9 | ||
| EIF1AX | < .001/0.032 | Chr23:20148726 | ENST00000379607 | c.338-1G > T | p.X113_splice | Splice acceptor | No | 102 | 97 |
| Chr23:20148633 | ENST00000379607 | c.429 + 1G > A | p.X143_splice | Splice donor | No | 106 | 86 | ||
| Chr23:20156732 | ENST00000379607 | c.25G > C | p.Gly9Arg | Missense | Yes | 206 | 38 | ||
| DGCR8 | < .001/0.802 | Chr22:20079439 | ENST00000351989 | c.1552G > A | p.Glu518Lys | Missense | Yes | 101 | 88 |
| Chr22:20079439 | ENST00000351989 | c.1552G > A | p.Glu518Lys | Missense | Yes | 104 | 48 |
Abbreviations: COSMIC, Catalogue of Somatic Mutations in Cancer; ID, identification.
a The genomic coordinates in the MutSig2CV analysis are based on hg19 and the genomic coordinates in the heat map (Fig. 1) are based on hg38.
Gene Ontology of Top Mutated Genes
Since most of the top genes in the MutSig2CV analysis are unknown in thyroid and most are not curated cancer census genes, we performed a gene ontology analysis using Enrichr. The top Mutsig2CV genes showed a significant enrichment in the gene ontology (GO) Cellular components “RNAi effector complex” and “RISC complex,” P value equal to .009 for both (data not shown). Both GO Cellular components are essential components in the miRNA machinery and downstream effects.
Copy Number Landscape and Overall Ploidy
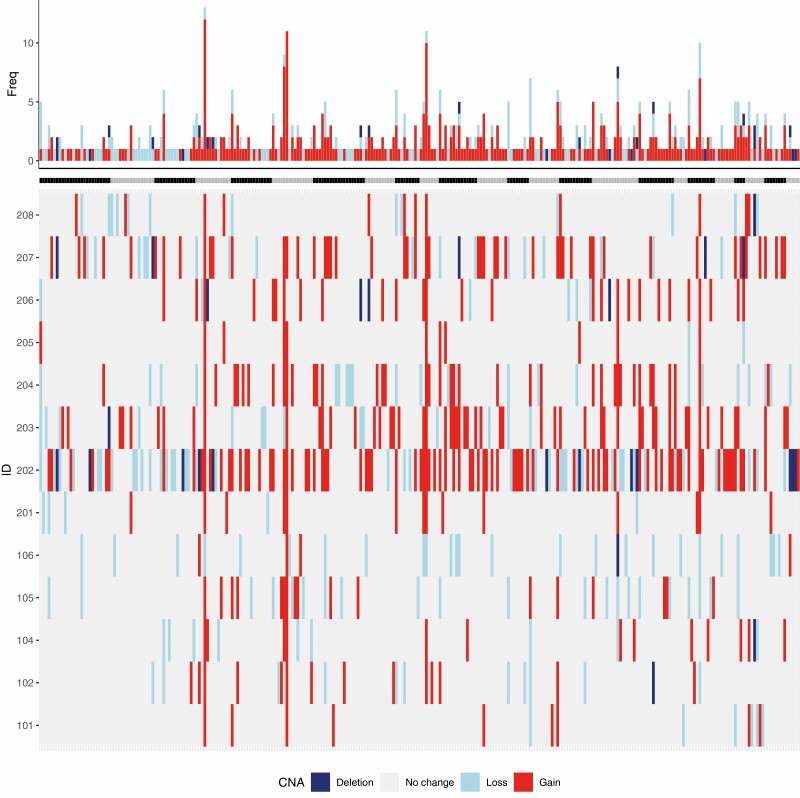
One of the benefits of using WGS as opposed to the more common WES is that it also contains noncoding regions, which allows for a more detailed interrogation of global CNAs. By analyzing our data with ASCAT, we present the genomic copy number landscape in wiFTCs and HCCs, and depict recurrent loci displaying gain and loss events (Fig. 2). A subset of tumors was particularly amassed in terms of CNA events: samples 202, 203, 204, and 207, but without any clear correlation to underlying genetics or patient outcome (see Fig. 1, Supplementary Table S1) (30). When consulting loci with recurrent CNAs, chromosomes 4, 6, and 10 were particularly enriched for copy number gains. The MRA was pinpointed to regions 4p11, 6p21.32, and 10q11.21, in which gain was noted in 12, 11, and 10 samples respectively out of the 13 tumors investigated. Candidate genes for all 3 loci and their implications for cancer in general are detailed in Supplementary Table S5 (30). Of particular interest, chromosome 4p11 harbors TEC, SLAIN2, FRYL, and OCIAD1, all of which have been associated with cancer development. Chromosome 6p21.32 contains DAXX, a well-established cancer-associated gene with tumor suppressive and oncogenic properties, while chromosome 10q11.21 harbors the thyroid oncogene RET.
Figure 2.
The landscape of copy number alterations (CNAs) in the whole-genome sequenced cohort. Summarized gain and loss events across the genome (columns represent cytobands) for each patient/tumor (rows). The bar plot on top shows the total number of CNA across each chromosome. Note the frequent gain of regions 4p11, 6p21.32, and 10q11.21 across the cohort. Cases 202 and 208 are Hürthle cell carcinomas; the remaining 11 cases are widely invasive follicular thyroid carcinomas (wiFTCs).
For 12 out of 13 tumors, the ploidy could be safely estimated, and these results are detailed in Supplementary Table S6 (30). In short, 8 informative samples were denoted as diploid, whereas 2 cases are assumed to be triploid (case 202, an HCC; and case 207, a wiFTC) and 2 cases were tetraploid (case 203, a wiFTC; and case 208, an HCC). Three out of the four hyperploid samples were among the 4 tumors displaying markedly increased copy number gains as compared to the remaining samples.
Genomic Rearrangements
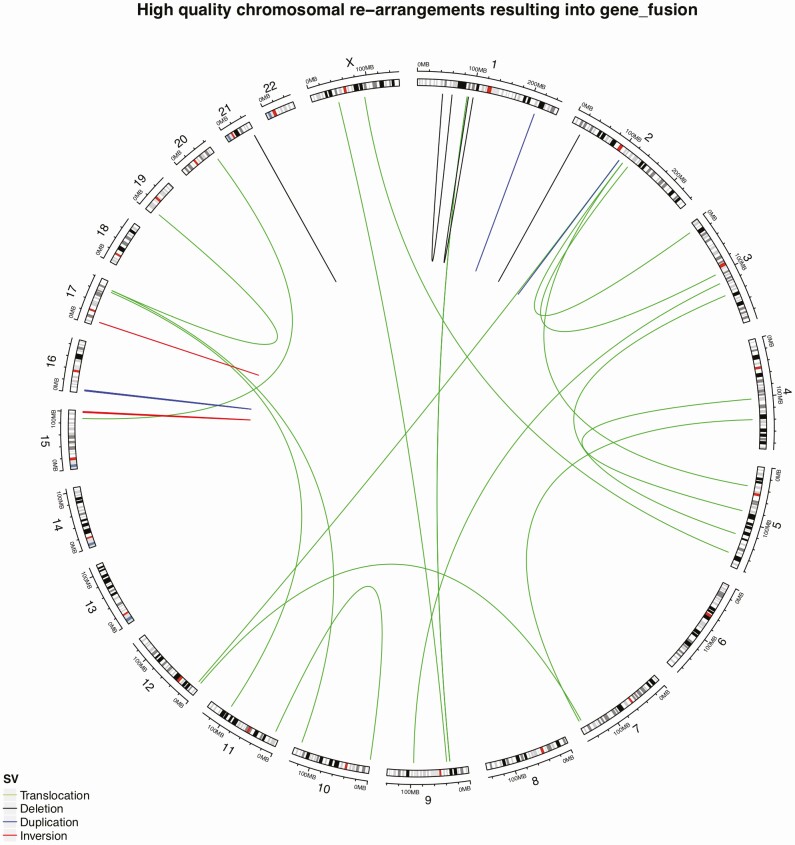
To analyze structural variants, we selected fusion events from the snpEff output corresponding to high-quality events reported by Manta (34). In total, we identified 268 structural events, of which 27 were denoted as high-confidence events resulting in bona fide gene fusions as shown in Fig. 3. Samples FTC 105 and HCC 208 had the highest number of rearrangements resulting in gene fusions (n = 4 each). When manually scrutinizing these 28 high-confidence events, 2 tumors (case 204 FTC and case 205 FTC) were found to exhibit the established PAX8-PPARγ fusion (see Figs. 1 and 3). All high-confidence fusion events are detailed in Supplementary Table S7 (30).
Figure 3.
Gene fusion events across the tumor genome. Lines represent the location of gene fusion and colors correspond to structural variation type. 28 high-confidence events were observed. Cases 204 and 205 were found to exhibit the established PAX8-PPARγ fusion (between chromosomes 2q13 and 3p25).
DGCR8 Loss of Heterozygosity Analyses
Following the identification of a recurrent p.E518K DGCR8 mutation in 2 out of 13 tumors, we further investigated this gene. Five tumors displayed LOH at the DGCR8 locus, including the 2 DGCR8-mutated cases, an additional 2 FTCs, and 1 HCC. Following this finding, the transcriptome data were analyzed to investigate the expression pattern of DGCR8 in relation to loss of heterozygosity (LOH) status. After normalizing the expression values using edgeR (44), a pairwise comparison between samples with and without LOH was performed, revealing no significant expression difference between tumors with or without LOH of the DGCR8 locus (P = .13, Supplementary Fig. S2) (30).
Targeted DGCR8 Analyses
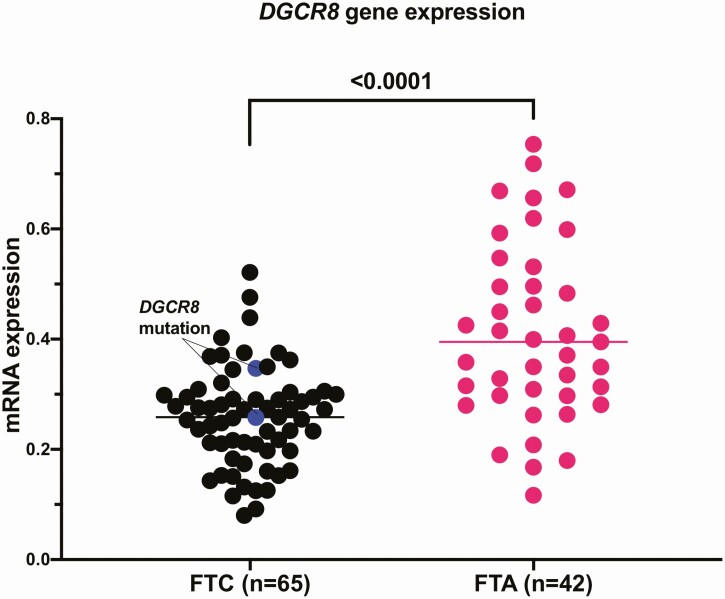
Direct sequencing of the p.E518 region of DGCR8 in 123 thyroid carcinomas (7 encapsulated angioinvasive FTCs, 37 HCCs, 33 minimally invasive FTCs, 26 wiFTCs, and 20 PDTCs) was performed. All FTCs and HCCs were successfully sequenced. Seventeen out of 20 PDTC were successfully sequenced, whereas 3 gave no product, possibly because of loss of genetic material. No additional mutations were found. In all, the frequency of this recurrent mutation in FTCs would correspond to 8% in wiFTCs and less or nonexistent in other subtypes and PDTCs. Moreover, by quantitative real-time PCR, DGCR8 mRNA expression was analyzed in 107 follicular thyroid tumors (65 FTCs, 42 FTAs). In general, FTAs displayed significantly higher expression than FTCs (P < .001, Fig. 4).
Figure 4.
DGCR8 messenger RNA (mRNA) expression in an extended follicular tumor cohort. The DGCR8 mRNA expression is significantly lower in follicular thyroid carcinoma (FTC) compared to follicular thyroid adenoma (FTA). The DGCR8-mutated cases are marked with blue. These cases displayed near median expression.
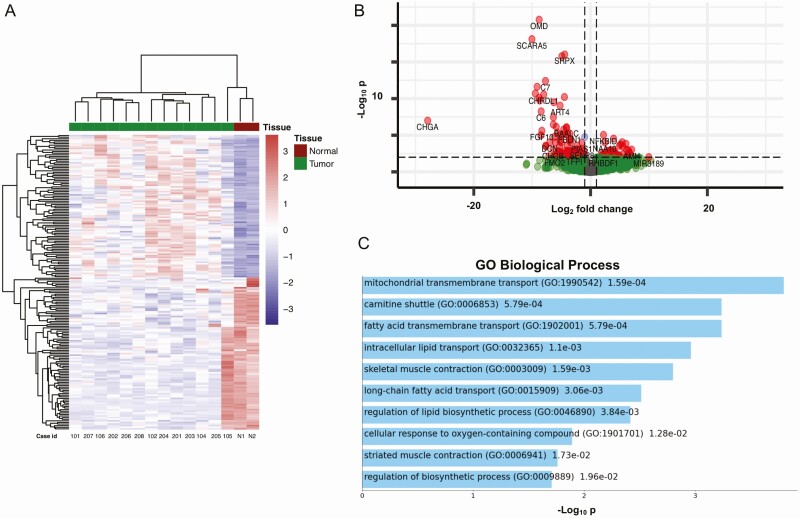
Transcriptome Profiling
The aggregated percentage of bases that had a quality score more than the Q30 value was 91.8%. DESeq2 identified 184 differentially expressed genes between the tumor and normal samples, as visualized by a heat map (Fig. 5A). Unsupervised clustering revealed 3 main clusters: the 2 normal samples clustered together with a single FTC (case 105), whereas the remaining tumors aggregated in 2 different clusters; 4 wiFTCs and both HCCs in one cluster and 6 wiFTCs in the other. Three out of 4 samples with the TERTp mutation (102, 201, and 203) clustered together. One of the 2 clusters (containing the TERTp-mutated cases) was defined by more pronounced expression in the significantly upregulated genes (Fig. 5B). GO analysis showed significant enrichment in the GO Biological Processes mitochondrial transmembrane transport, carnitine shuttle, and fatty acid transmembrane transport (Fig. 5C). The complete list of differentially expressed genes is available as Supplementary Table S8 (30). The first main tumor cluster showed a tendency to shorter survival compared with the second cluster; however, it was not significant (overall survival, disease-specific survival, and disease-free survival, with P = .28, P = .28, and P = .64, respectively) (data not shown). Our data suggest that wiFTCs and HCCs adhere to 2 principal clusters. Furthermore, a DESeq2 analysis was also performed in the DGCR8-mutated tumors and compared to the wild type cases. Despite the low number of cases, 4 genes were significantly downregulated with enrichment in GO cellular component “bicellular tight junctions” (data not shown).
Figure 5.
Transcriptome analyses of follicular thyroid carcinoma, Hürthle cell carcinoma, and normal thyroid tissue. A, Unsupervised cluster analysis of tumor and normal (N) tissue. Three main clusters are seen; the 2 normal samples clustered together with a single follicular thyroid carcinoma (FTC) (case 105), whereas the remaining tumors aggregated in 2 principal clusters. Three out of 4 samples with the TERT promoter (TERTp) mutation (102, 201, and 203) clustered together. The 2 Hürthle cell carcinomas (cases 202 and 208) appeared in the same cluster as 4 widely invasive FTCs. B, Volcano plot displaying significantly upregulated and downregulated genes. Genes labeled with red dots show a fold change greater than 1 and a P value of less than .01 One of the 2 tumor clusters (containing the majority of the TERTp-mutated cases) was defined by more pronounced expression in the significantly upregulated genes. C, Enrichment analysis of upregulated genes revealed associations with genes associated with mitochondrial transmembrane transport, carnitine shuttle, and fatty acid transmembrane transport.
Sample 105 clustered with the 2 normal thyroid tissues. This tumor has been histologically confirmed to exhibit a tumor purity of 90%, making contamination with normal tissue highly unlikely. Moreover, this case exhibited somatic MEN1 and TSHR gene mutations (see Fig. 1), and furthermore displayed CNAs on par with many other FTCs in this series (see Fig. 2), supporting the tumorous nature of this sample.
MicroRNA Profiling
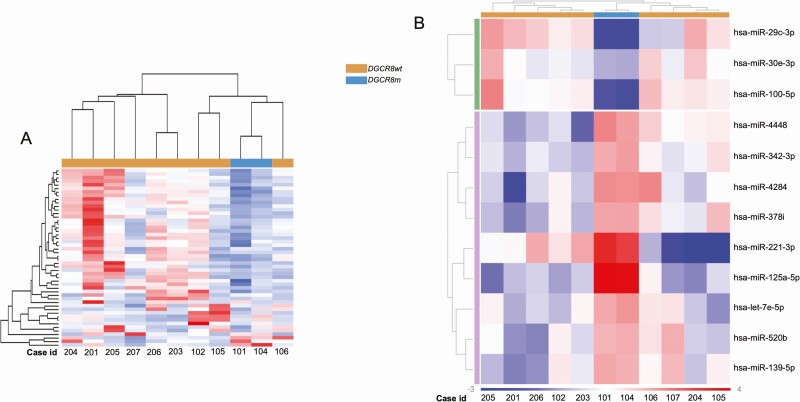
After the identification of missense mutation and LOH of the DGCR8 gene, we sought to explore the consequences on miRNA expression. The 11 wiFTCs from the genomic and transcriptomic analyses (2 harboring the DGCR8 p.E518K mutation and 9 the DGCR8 p.E518 wild type) were successfully profiled for 827 miRNAs (NanoString Human v3 miRNA assay). The 2 DGCR8-mutated cases clustered together in the unsupervised clustering analysis of the top 50 most variable miRNAs (Fig. 6A). An additional 2 cases showed similar general downregulation of miRNAs and clustered in close proximity to the mutated cases. These cases (105 and 106) displayed low DGCR8 mRNA expression in the transcriptomic analysis (see Supplementary Fig. S2). Case 102 showed remarkably higher miRNA expression compared to the other cases. This case displayed the highest level of DGCR8 mRNA expression, similar to the levels in normal tissue (see Supplementary Fig. S2). Differential expression analysis of the DGCR8-mutated cases compared to the wild-type cases revealed that only 12 miRNAs were significantly differentially expressed (Fig. 6B).
Figure 6.
MicroRNA (miRNA) profiling in follicular thyroid carcinoma (FTC). A, Unsupervised clustering of the top 50 most variable miRNAs in follicular thyroid carcinoma (FTC) (n = 11). The annotation on top indicates the DGCR8 p.E518K–mutated cases, which clustered together and show a general downregulation of miRNA. Nearby cases 105 and 106 both showed low DGCR8 messenger RNA (mRNA) expression levels from the transcriptomic data. Case 201 displayed augmented upregulation compared to other cases. This case showed DGCR8 mRNA expression levels similar to that of normal thyroid tissues. B, Differential expression analysis in DGCR8 p.E518K–mutated cases (n = 2) compared to DGCR8 p.E518 wild-type cases (n = 9). Only 12 miRNAs were differentially expressed between groups, possibly because of a general downregulation of miRNA in cases 105 and 106. Fold change cutoff was set to –1.5 and 1.5, and the threshold for adjusted P value was set to .05.
10q11.21 Cytoband Gene Expression
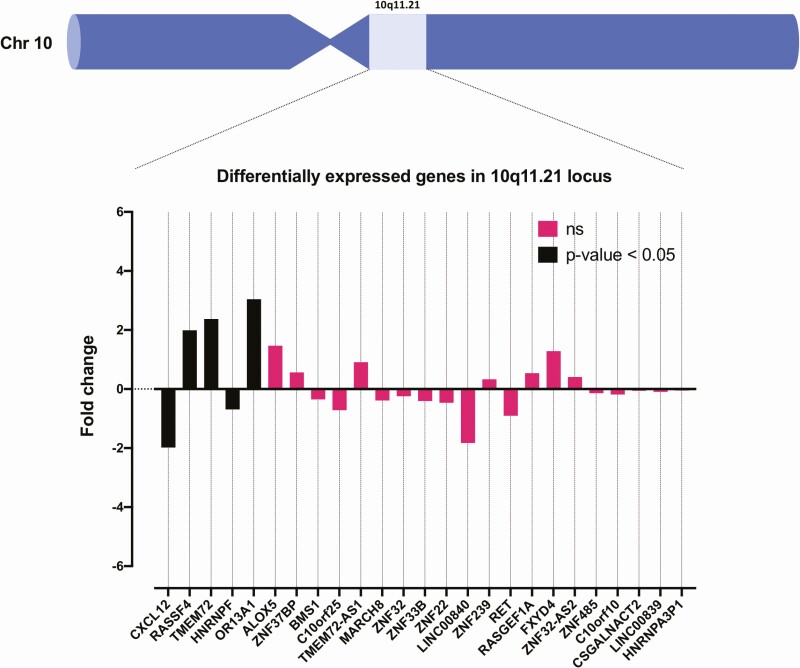
Given the detection of recurrent gain at chromosome 10q11.21, we specifically analyzed the gene expression of genes in the MRA (10q11.21) in gain vs neutral/loss tumors (n = 10 and n = 3, respectively). In this cytoband, RASSF4, TMEM72, and OR13A1 were significantly upregulated (Fig. 7).
Figure 7.
Tumors with gain vs no gain in the 10q11.21 cytoband were analyzed for differentially expressed genes in the same cytoband. RASSF4, TMEM72, and OR13A1 were all significantly upregulated in the tumor cohort when analyzing RNA sequencing data, suggesting that the augmented expression of one or several of these genes might be associated with recurrent 10q11.21 gains observed in the cohort.
Subanalysis of TERTp Mutated Cases
Given the established role of TERTp mutations in FTCs with exceptionally poor outcome, we also analyzed the transcriptome of TERTp-mutated wiFTCs (cases 102, 201, 203, and 206) compared to that of the TERTp wild-type cases using gene set enrichment analysis in hallmark pathways. We found that TERTp-mutated cases had significant (normalized P value < .01 and a false discovery rate < 0.1) enrichment in gene sets associated with adipogenesis, genes responding to androgens, and genes encoding the peroxisome (Supplementary Fig. S3) (30). These differences suggest that the TERTp mutation may be associated with altered metabolic activity.
Discussion
Although recent efforts with focused NGS panels as well as exomewide analyses have helped elucidate the mutational landscape of FTCs, few have tried to tackle this tumor entity via comprehensive WGS. By selecting FTCs and HCCs with histological and clinical features coupled to aggressiveness, the aim of this study was to characterize unchartered mutational, gross chromosomal, and transcriptome events to identify novel markers of prognostic and therapeutic significance in poor-prognosis cases.
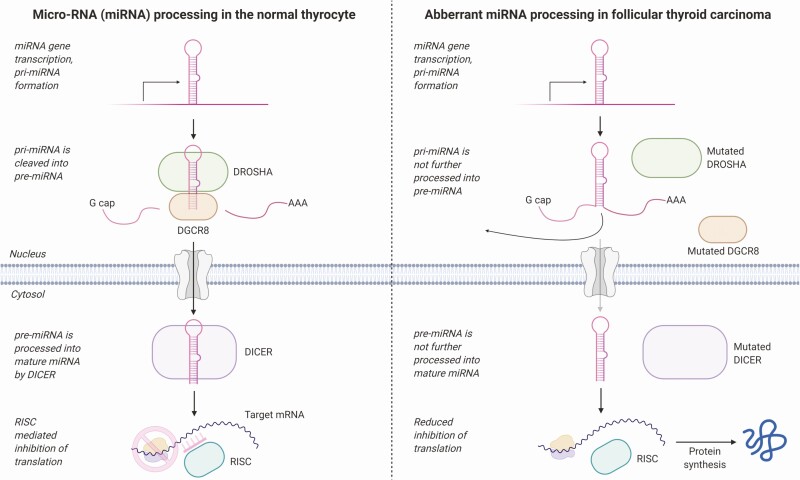
We identified a recurrent, heterozygous, and somatic p.E518K mutation in the miRNA master processor DGRC8 in 2 out of 13 tumors. The specific p.E518K variant in combination with LOH of the WT DGCR8 allele was recently proposed as causing a familial syndrome characterized by multinodular goiter and schwannoma (45). This combination of a p.E518K mutation and LOH of the WT DGCR8 allele has also been demonstrated in approximately 2% to 3% of Wilms tumors (46). The mutation has also been demonstrated in thyroid cancer, however, only in a small subset of papillary thyroid carcinomas (PTCs) (and follicular variant PTCs)—but not previously reported in FTCs (47). The mutation also led to loss of function in the miRNA processing causing downregulation of canonical miRNAs (45)—a phenomenon that has been shown to enhance cellular transformation and tumorigenesis (48). This is in line with our results wherein the DGCR8-mutated cases appeared in the same cluster from the global miRNA profiling (Fig. 6). Of particular interest from a thyroid perspective, miR139-5p is reported as aberrantly expressed in thyroid cancer, and this miRNA was also significantly dysregulated in the 2 DGRC8 mutants compared to DGCR8 wild-type cases (49, 50). Moreover, 2 DGCR8 wild-type cases displayed a similar global miRNA expression pattern as the 2 mutants. Both these cases displayed low DGCR8 mRNA expression, suggesting that not only mutations but also mRNA downregulation potentially could alter the miRNA machinery. The cause of DGCR8 downregulation in our tumor cohort (see Fig. 4) is unknown. We have previously shown that DICER1, another essential component in the miRNA machinery, is downregulated in FTC and is downregulated by the transcription factor GABPA (51). However, whether epigenetic dysregulation is at play also at the DGRC8 locus remains to be established. For example, the transcription factor YY1 has been found to influence DGCR8 gene output in vitro, and YY1 has been found expressed exclusively at invasive fronts of FTCs (52, 53).
In our data, the gene expression pattern of the DGCR8-mutated cases showed an enrichment of downregulated genes in bicellular tight junctions. The miRNA processing RISC complex where DGCR8 is included has recently been shown to be associated with epithelial cell junctions in colon cancer and, when lost, by various mechanisms might promote cell transformation (54). Also, knockdown of DGCR8 in breast cancer cells increased the expression of the invasion-essential urokinase-type plasminogen activator (uPA) (55). Dysregulation of the miRNA machinery and a possible activation of such enzymes could thus play a role in thyroid cancer invasion. Indeed, previous results have observed a marked increase in expression of the uPA receptor, especially in FTC (56). The finding of DGCR8 mutations is also of immediate interest given the established association between FTCs and inactivating mutations in DICER1 and DROSHA, 2 master miRNA processing genes (51, 57). Our findings thus strengthen the association between abnormal miRNA processing and the development of FTC and underline a central mechanism in which subsets of these tumors probably arise as a direct consequence of aborted miRNA maturation (Fig. 8). However, the driver properties of the p.E518K DGCR8 mutation in thyroid cancer remain to be established, as the 2 cases with this mutation also carried a somatic HRAS and PIK3CA mutation, respectively. Therefore, it is possible that the DGCR8 mutation found in these tumors influenced the tumor progression or invasive behavior without driving the tumor formation per se. However, recent studies have found recurrent DICER1 mutations in various forms of thyroid cancer, of which most cases did not display established driver-gene alterations, vaguely suggesting that aberrant miRNA expressional patterns could be at play even at the level of tumor initiation (58, 59).
Figure 8.
Schematic overview of microRNA (miRNA) regulators in wild-type thyrocytes and mutated follicular thyroid carcinoma (FTCs). The left aspect depicts normal miRNA processing, in which miRNAs are transcribed, forming a pri-miRNA, which is subsequently targeted by Drosha and DGCR8. This molecular complex cleaves the pri-miRNA into pre-miRNA. The pre-miRNA is transported to the nucleus, where it is processed into mature miRNA by DICER. This mature miRNA sequence interacts with the RNA-induced silencing complex (RISC) and inhibits translation of target messenger RNAs (mRNAs), thereby regulating gene expression. The right aspect displays the potential functional consequences of DROSHA, DGCR8, and DICER1 mutations in FTCs. As the mutations in theory lead to the inactivation of the corresponding proteins, a defect in miRNA processing would affect gene expression output. Image created using BioRender.com.
We did not observe the mutation in extended material of 103 FTCs and 17 PDTCs, suggesting that the variant is rarely seen overall. However, when analyzing DGCR8 expression in an extended panel, we observed significant downregulation in FTCs when compared to FTAs, suggesting the importance of DGCR8 in FTC development. The GO for the top MutSig2CV genes in the WGS cohort was significantly associated with the GO cellular component “RISC complex,” which further strengthens the hypothesis that the miRNA machinery plays a vital role in these tumors. In all, future studies might help elucidate whether downregulation of DGCR8 in any way orchestrates invasive behavior in follicular thyroid neoplasms.
Two additional wiFTCs and one HCC carried a recurrent somatic alteration in the Family with Sequence Similarity 72 Member D (FAM72D) gene. Located on chromosome 1q21.1, this gene encompasses a region frequently amplified in myeloma (60), and FAM72 paralogs are upregulated in glioblastoma (61). The former study also suggests that FAM72D interact with the FOXM1 transcription factor network controlling cell proliferation (60), but little is known regarding its putative function in the thyroid gland. The mutation detected in this study was not believed to alter the protein function, and the putative roles of this alteration will need to be functionally linked to a cellular phenotype before they can be considered of importance in the development of FTC and HCC. Moreover, 2 out of 3 cases with a FAM72D mutation already exhibited established genetic events (case 203, mutations in KRAS and TERTp; case 204, MUTYH mutation and PAX8-PPARγ fusion)—thereby somewhat diminishing the chance of the FAM72D alteration being a true driver. From a clinical standpoint, all 3 tumors with this aberration were relapse free, but no clear-cut associations to other clinical parameters could be made.
Moreover, we highlighted a set of novel high-confidence fusions in our cohort, although only the established PAX8-PPARγ fusion was present in 2 cases (cases 204 and 205). It is also interesting to note that both HCCs displayed hyperploidy, and 1 of the 2 HCC cases also displayed among the highest number of structural variants in the entire cohort. As both HCCs did not carry pathogenic mutational events in bona fide thyroid-related genes, the notion that gross chromosomal alterations were overrepresented in these cases compared to conventional FTCs might indicate an association worthy of future exploration. Strikingly in terms of structural alterations, recurrent gains at chromosomes 4p11, 6p21.32, and 10q11.21 were observed across our cohort. To our knowledge, these regions have not been previously reported as recurrently amplified in FTCs, and this could be because we used high-resolution WGS as opposed to more blunt technology such as PCR of microsatellite markers or comparative genomic hybridization. The regions discussed earlier contain multiple genes with associations with cancer development, of which several could be of great interest for further studies. Most notably, chromosome 10q11.21 harbors the thyroid oncogene RET, which is mutated on the somatic and constitutional level in medullary thyroid carcinoma and fusions involving this gene are a recurrent feature in PTC, but there is no known association with any of these genetic aberrancies and FTCs. By our analyses, RET gene expression was not associated with gain of this gene locus when consulting RNA sequencing data (Fig. 7). Instead, we found that RASSF4, TMEM72, and OR13A1 were the only 3 genes in the region with significant overexpression. Although not all genes within a region of gain will be overexpressed, we lack functional evidence linking the observed overexpression with the actual chromosomal gain, and our results are therefore merely observational in nature.
This study cohort is overrepresented in terms of poor-prognosis cases (large tumors, history of tumor recurrences, and several have succumbed to disease). Therefore, it comes as little surprise that 4 of 13 cases (31%) exhibited TERTp mutations, as this genetic event is tightly coupled to adverse clinical outcome in patients with thyroid cancer. The roles of TERT in thyroid cancer is only partly understood, but the mutations themselves are not considered driver alterations—which is mirrored by the fact that 3 of 4 wiFTCs in our cohort with a TERTp mutation also exhibited driver-gene mutations in NRAS (n = 2) or KRAS (n = 1) (see Fig. 1). To elucidate how TERT affects the transcriptome profile output in wiFTCs and HCCs, we analyzed the RNA-sequencing data stratified with regards to TERTp mutations and found remarkable differences in the expression of genes responsible for metabolic pathways, such as adipogenesis, androgen response, and peroxisome-related pathways. This was also true when performing an unsupervised clustering of all tumors included, in which the majority of TERTp mutants aggregated (see Fig. 5). It is tempting to speculate that one or several of these metabolic pathways are crucial for the metastatic potential or disseminated growth so often seen in this tumor cohort, and will be a highly relevant topic for future studies.
We conclude that subsets of FTCs display recurrent DGCR8 mutations, adding yet another player to the growing palette of miRNA-related gene mutations in this disease. Moreover, we identified recurrent gain of chromosome 10q11.21, and identified overexpression of RASSF4, TMEM72, and OR13A1 as potential candidates for further studies. Finally, we identified 2 principal expressional clusters by global RNA sequencing, which differed in terms of TERTp mutations and metabolic pathway gene expression. Future analyses in extended series will possibly help elucidate whether these clusters exhibit differences in clinical outcomes.
Acknowledgments
The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, UPPMAX, and SNIC for providing assistance in massive parallel sequencing and computational infrastructure. The authors also acknowledge the support from the Knut and Alice Wallenberg Foundation and the Swedish Research Council. The authors would also like to acknowledge the KI Gene core facility at Karolinska Institutet for providing assistance with the NanoString analysis.
Financial Support: This work was supported by the Swedish Cancer Society (CAN 2016/1126), the Swedish Society for Medical Research, the Cancer Research Funds of Radiumhemmet, the Swedish Society of Medicine, the Lisa and Johan Grönberg Foundation, the Stockholm City Council, and Karolinska Institutet.
Author Contributions: C.C.J. and J.Z. conceived and supervised the project. J.O.P. and C.C.J. designed and performed the research. N.R. and S.D.L. performed genomic bioinformatic analyses and parts of the transcriptomic analyses. Y.C., F.H., and J.O. P. performed transcriptomic analyses. J.O.P. performed the miRNA analysis and subsequent bioinformatics. J.Z. and J.O.P. extracted clinical information from patient medical charts. J.O.P. performed molecular wet lab analyses. J.O.P. and C.C.J. compiled the data and wrote the manuscript with input and consent from all authors.
Glossary
Abbreviations
- CNA
copy number alteration
- FTA
follicular thyroid adenoma
- FTC
follicular thyroid carcinoma
- GATK
Genome Analysis Toolkit
- GO
gene ontology
- HCC
Hürthle cell carcinoma
- indel
insertion-deletion mutation
- LOH
loss of heterozygosity
- MRA
minimal region of amplification
- mRNA
messenger RNA
- miRNA
microRNA
- NGS
next-generation sequencing
- PCR
polymerase chain reaction
- PDTC
poorly differentiated thyroid carcinoma
- PTC
papillary thyroid carcinoma
- SNV
single-nucleotide variation
- TERTp
TERT promoter
- uPA
urokinase-type plasminogen activator
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
- wiFTC
widely invasive follicular thyroid carcinoma
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
The data sets analyzed during the present study are available to a variable extent. Gene expression data sets and miRNA data sets will be available upon reasonable request. Genomic raw data can be available upon reasonable request. All supplementary materials including the complete mutect2 list and figures are located in a digital repository https://doi.org/10.5878/6fcv-1795. The codes for the genomic analyses are available at GitHub https://github.com/NBISweden/SMS_4472_19_Thy_Cancer.
References
- 1. Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12(11):646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 3. James BC, Aschebrook-Kilfoy B, Cipriani N, Kaplan EL, Angelos P, Grogan RH. The incidence and survival of rare cancers of the thyroid, parathyroid, adrenal, and pancreas. Ann Surg Oncol. 2016;23(2):424-433. [DOI] [PubMed] [Google Scholar]
- 4. Jin M, Kim ES, Kim BH, et al. Clinical implication of World Health Organization classification in patients with follicular thyroid carcinoma in South Korea: a multicenter cohort study. Endocrinol Metab (Seoul). 2020;35(3):618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lloyd RV, Osamura RY, Klöppel G, Rosai J.. WHO Classification of Tumours of Endocrine Organs. WHO/IARC Classification of Tumours. 4th ed. Vol. 10. Lyon: International Agency for Research on Cancer; 2017. [Google Scholar]
- 6. Ruegemer JJ, Hay ID, Bergstralh EJ, Ryan JJ, Offord KP, Gorman CA. Distant metastases in differentiated thyroid carcinoma: a multivariate analysis of prognostic variables. J Clin Endocrinol Metab. 1988;67(3):501-508. [DOI] [PubMed] [Google Scholar]
- 7. Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26(1):1-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nikiforova MN, Lynch RA, Biddinger PW, et al. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88(5):2318-2326. [DOI] [PubMed] [Google Scholar]
- 9. Jeong SH, Hong HS, Kwak JJ, Lee EH. Analysis of RAS mutation and PAX8/PPARγ rearrangements in follicular-derived thyroid neoplasms in a Korean population: frequency and ultrasound findings. J Endocrinol Invest. 2015;38(8):849-857. [DOI] [PubMed] [Google Scholar]
- 10. Hou P, Liu D, Shan Y, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13(4):1161-1170. [DOI] [PubMed] [Google Scholar]
- 11. Kim CS, Vasko VV, Kato Y, et al. AKT activation promotes metastasis in a mouse model of follicular thyroid carcinoma. Endocrinology. 2005;146(10):4456-4463. [DOI] [PubMed] [Google Scholar]
- 12. Saito J, Kohn AD, Roth RA, et al. Regulation of FRTL-5 thyroid cell growth by phosphatidylinositol (OH) 3 kinase-dependent Akt-mediated signaling. Thyroid. 2001;11(4):339-351. [DOI] [PubMed] [Google Scholar]
- 13. Lemoine NR, Mayall ES, Wyllie FS, et al. High frequency of ras oncogene activation in all stages of human thyroid tumorigenesis. Oncogene. 1989;4(2):159-164. [PubMed] [Google Scholar]
- 14. Namba H, Rubin SA, Fagin JA. Point mutations of ras oncogenes are an early event in thyroid tumorigenesis. Mol Endocrinol. 1990;4(10):1474-1479. [DOI] [PubMed] [Google Scholar]
- 15. Nikiforov YE, Carty SE, Chiosea SI, et al. Impact of the multi-gene ThyroSeq next-generation sequencing assay on cancer diagnosis in thyroid nodules with atypia of undetermined significance/follicular lesion of undetermined significance cytology. Thyroid. 2015;25(11):1217-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23(3):R143-R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu T, Wang N, Cao J, et al. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas. Oncogene. 2014;33(42):4978-4984. [DOI] [PubMed] [Google Scholar]
- 18. Melo M, da Rocha AG, Vinagre J, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014;99(5): E754-E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paulsson JO, Mu N, Shabo I, et al. TERT aberrancies: a screening tool for malignancy in follicular thyroid tumours. Endocr Relat Cancer. 2018;25(7):723-733. [DOI] [PubMed] [Google Scholar]
- 20. Wang N, Liu T, Sofiadis A, et al. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer. 2014;120(19):2965-2979. [DOI] [PubMed] [Google Scholar]
- 21. Borah S, Xi L, Zaug AJ, et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347(6225):1006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicolson NG, Murtha TD, Dong W, et al. Comprehensive genetic analysis of follicular thyroid carcinoma predicts prognosis independent of histology. J Clin Endocrinol Metab. 2018;103(7):2640-2650. [DOI] [PubMed] [Google Scholar]
- 23. Yoo SK, Lee S, Kim SJ, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PloS Genet. 2016;12(8):e1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoo SK, Song YS, Lee EK, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun. 2019;10(1):2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swierniak M, Pfeifer A, Stokowy T, et al. Somatic mutation profiling of follicular thyroid cancer by next generation sequencing. Mol Cell Endocrinol. 2016;433:130-137. [DOI] [PubMed] [Google Scholar]
- 26. Duan H, Liu X, Ren X, Zhang H, Wu H, Liang Z. Mutation profiles of follicular thyroid tumors by targeted sequencing. Diagn Pathol. 2019;14(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borowczyk M, Szczepanek-Parulska E, Debicki S, et al. Differences in mutational profile between follicular thyroid carcinoma and follicular thyroid adenoma identified using next generation sequencing. Int J Mol Sci. 2019;20(13):3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X, Li X, Cheng Y, et al. Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma. Hum Genomics. 2015;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia M, Juhos S, Larsson M, et al. Sarek: a portable workflow for whole-genome sequencing analysis of germline and somatic variants. F1000Res. 2020;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paulsson J. Supplementary data for: Whole-genome and transcriptome sequencing of widely invasive follicular thyroid carcinomas reveal recurrent mutations of the microRNA regulator DGCR8, Swedish National Data Service (SND), Dataset. Date uploaded June 24, 2021. 10.5878/6fcv-1795. [DOI]
- 31. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220-1222. [DOI] [PubMed] [Google Scholar]
- 35. Van Loo P, Nordgard SH, Lingjærde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107(39):16910-16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811-2812. [DOI] [PubMed] [Google Scholar]
- 41. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [DOI] [PubMed] [Google Scholar]
- 43. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rivera B, Nadaf J, Fahiminiya S, et al. DGCR8 microprocessor defect characterizes familial multinodular goiter with schwannomatosis. J Clin Invest. 2020;130(3):1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gadd S, Huff V, Walz AL, et al. A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet. 2017;49(10):1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673-677. [DOI] [PubMed] [Google Scholar]
- 49. Chi J, Zheng X, Gao M, et al. Integrated microRNA-mRNA analyses of distinct expression profiles in follicular thyroid tumors. Oncol Lett. 2017;14(6):7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Montero-Conde C, Graña-Castro O, Martín-Serrano G, et al. Hsa-miR-139-5p is a prognostic thyroid cancer marker involved in HNRNPF-mediated alternative splicing. Int J Cancer. 2020;146(2):521-530. [DOI] [PubMed] [Google Scholar]
- 51. Paulsson JO, Wang N, Gao J, et al. GABPA-dependent down-regulation of DICER1 in follicular thyroid tumours. Endocr Relat Cancer. 2020;27(5):295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shan X, Ren M, Chen K, Huang A, Tang H. Regulation of the microRNA processor DGCR8 by hepatitis B virus proteins via the transcription factor YY1. Arch Virol. 2015;160(3):795-803. [DOI] [PubMed] [Google Scholar]
- 53. Nicolson NG, Paulsson JO, Juhlin CC, Carling T, Korah R. Transcription factor profiling identifies spatially heterogenous mediators of follicular thyroid cancer invasion. Endocr Pathol. 2020;31(4):367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nair-Menon J, Daulagala AC, Connor DM, et al. Predominant distribution of the RNAi machinery at apical adherens junctions in colonic epithelia is disrupted in cancer. Int J Mol Sci. 2020;21(7):2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S. Impaired MicroRNA processing facilitates breast cancer cell invasion by upregulating urokinase-type plasminogen activator expression. Genes Cancer. 2011;2(2):140-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim SJ, Shiba E, Taguchi T, et al. uPA receptor expression in benign and malignant thyroid tumors. Anticancer Res. 2002;22(1A):387-393. [PubMed] [Google Scholar]
- 57. Paulsson JO, Backman S, Wang N, et al. Whole-genome sequencing of synchronous thyroid carcinomas identifies aberrant DNA repair in thyroid cancer dedifferentiation. J Pathol. 2020;250(2):183-194. [DOI] [PubMed] [Google Scholar]
- 58. Juhlin CC, Stenman A, Zedenius J. Macrofollicular variant follicular thyroid tumors are DICER1 mutated and exhibit distinct histological features. Histopathology. Published online May 18, 2021. doi:10.1111/his.14416 [DOI] [PubMed] [Google Scholar]
- 59. Chong AS, Nikiforov YE, Condello V, et al. Prevalence and spectrum of DICER1 mutations in adult-onset thyroid nodules with indeterminate cytology. J Clin Endocrinol Metab. 2021;106(4):968-977. [DOI] [PubMed] [Google Scholar]
- 60. Chatonnet F, Pignarre A, Sérandour AA, et al. The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation. Haematologica. 2020;105(3):774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rahane CS, Kutzner A, Heese K. A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J Neurooncol. 2019;141(1):57-70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets analyzed during the present study are available to a variable extent. Gene expression data sets and miRNA data sets will be available upon reasonable request. Genomic raw data can be available upon reasonable request. All supplementary materials including the complete mutect2 list and figures are located in a digital repository https://doi.org/10.5878/6fcv-1795. The codes for the genomic analyses are available at GitHub https://github.com/NBISweden/SMS_4472_19_Thy_Cancer.