Abstract
Context
Intrauterine growth restriction (IUGR) is an immediate outcome of an adverse womb environment, exposing newborns to developing cardiometabolic disorders later in life.
Objective
This study investigates the cardiac metabolic consequences and underlying mechanism of energy expenditure in developing fetuses under conditions of IUGR.
Methods
Using an animal model of IUGR characterized by uteroplacental vascular insufficiency, mitochondrial function, gene profiling, lipidomic analysis, and transcriptional assay were determined in fetal cardiac tissue and cardiomyocytes.
Results
IUGR fetuses exhibited an upregulation of key genes associated with fatty acid breakdown and β-oxidation (Acadvl, Acadl, Acaa2), and mitochondrial carnitine shuttle (Cpt1a, Cpt2), instigating a metabolic gene reprogramming in the heart. Induction of Ech1, Acox1, Acox3, Acsl1, and Pex11a indicated a coordinated interplay with peroxisomal β-oxidation and biogenesis mainly observed in females, suggesting sexual dimorphism in peroxisomal activation. Concurring with the sex-related changes, mitochondrial respiration rates were stronger in IUGR female fetal cardiomyocytes, accounting for enhanced adenosine 5′-triphosphate production. Mitochondrial biogenesis was induced in fetal hearts with elevated expression of Ppargc1a transcript specifically in IUGR females. Lipidomic analysis identified the accumulation of arachidonic, eicosapentaenoic, and docosapentaenoic polyunsaturated long-chain fatty acids (LCFAs) in IUGR fetal hearts, which leads to nuclear receptor peroxisome proliferator-activated receptor α (PPARα) transcriptional activation in cardiomyocytes. Also, the enrichment of H3K27ac chromatin marks to PPARα-responsive metabolic genes in IUGR fetal hearts outlines an epigenetic control in the early metabolic energy switch.
Conclusion
This study describes a premature and sex-related remodeling of cardiac metabolism in response to an unfavorable intrauterine environment, with specific LCFAs that may serve as predictive effectors leading to IUGR.
Keywords: fetal growth retardation, energy metabolism, cardiomyocyte, LCFAs, seahorse analysis, peroxisome
Cardiovascular diseases are the leading cause of mortality globally, and their prevalence is increasing in several developing countries. Including sedentary lifestyle and genetically determined conditions as predisposing factors, a suboptimal womb environment also leads to cardiometabolic complications later in life, as originally proposed by Barker et al (1). Indeed, increasing evidence has indicated that individuals who have developed in an adverse in utero environment are predisposed to an increased risk of disease in adulthood, including insulin resistance, diabetes, hypertension, chronic lung disease, and cardiovascular defects (2-8). Exposure of fetuses to subnormal intrauterine conditions also largely results in perinatal complications of pulmonary hypertension, hypoxic-ischemic conditions, and unregulated glucose, which are associated with a high incidence of neonatal morbidity (9, 10). Hence, prenatal exposure to a suboptimal environment leads to immediate metabolic complications as well as to programming fetuses with an increased risk of cardiovascular and metabolic diseases later in life. However, the molecular mechanism underlying the fetal adaptation to an adverse uterine environment remains to be better elucidated.
Intrauterine growth restriction (IUGR) is an immediate consequence of an inadequate fetal environment, where the fetus cannot reach its full growth potential resulting in a bodyweight less than the tenth percentile for their gestational age. When excluding factors such as maternal undernourishment and fetal malformation, abnormal placental function characterized by a reduction in vascularization and impaired fetomaternal exchange of substrates becomes the leading cause of IUGR (8, 11, 12).
Human studies of IUGR have reported several postnatal complications, such as altered skeletal muscle development, brain damage, dysregulation of glucose metabolism, predisposition to type 2 diabetes, hypertension, adiposity, and impaired cardiovascular function (4, 13-15). Various animal models of IUGR have been developed to recapitulate the clinical conditions of placental vascular dysfunction and adverse uterine conditions and to better understand the link with adult diseases. These models using either small or large animals are based on diverse experimental approaches to induce fetal growth retardation, including caloric or protein restriction, hypoxic exposure, ligation of uterine artery, and prenatal exposure to dexamethasone (16-22). Under these conditions, IUGR offspring have shown to exhibit problems of kidney development, renal insufficiency, impaired lung function, altered cerebellar development, insulin resistance, late-onset diabetes, and sensitivity to postischemic injury, exemplifying the pathologic consequences of suboptimal fetoplacental exchange. Also, alterations in fetal cardiac maturation and function associated with reduced heart growth, and impaired cardiomyocyte proliferation, apoptosis, and signaling, have been described in IUGR models (19, 23-25).
We have developed an animal model of IUGR based on a low-sodium diet given to dams in the last week of gestation. Such a decrease in sodium intake was shown to alter the expansion of maternal plasma volume due to deficient renal sodium reabsorption and to reduce placental perfusion, supporting the onset of placental insufficiency (26-28). Studies in humans have determined that early pregnancy mean serum sodium and maternal volume expansion were lower in IUGR pregnancies and defective maternal hemodynamic adaptation is strongly associated with fetal growth restriction pregnancies, highlighting the crucial role of maternal plasma volume expansion and remodeling of uterine vessels in fetal growth (29-32). In our model, adult IUGR animals exhibit a higher arterial blood pressure and sex-dependent alterations of the renin-angiotensin-aldosterone system and renal function, demonstrating future metabolic disturbances derived from adverse conditions in fetal life (33). In addition, we have shown that the fetal brain and heart were increased in size relative to body weight in IUGR, suggesting a redistribution of cardiac output to these organs (33). Also, at adult age, elevated depth and volume and diminished contractility were observed in cardiomyocytes from left ventricles of IUGR females (34). Altogether, these findings suggest critical alterations in fetal heart physiology occurring in response to inadequate placental perfusion and IUGR. However, the molecular mechanism underlying such deleterious changes in cardiac metabolism is unknown.
Here we show that IUGR is associated with alterations in lipid metabolic gene programming, changes in long-chain fatty acid (LCFA) profile, and increased peroxisomal FA oxidation markers in fetal cardiac tissue, with more prominent effects in females. We also identified nuclear receptor peroxisome proliferator-activated receptor α (PPARα) as a key responsive factor to accumulating FAs observed in the metabolic adaptation to adverse intrauterine conditions and demonstrate critical changes in mitochondrial function and energy production. Our findings delineate the molecular basis of a premature metabolic remodeling in heart energy expenditure as part of the adaptive process taking place in developing fetuses in response to suboptimal uterine conditions.
Materials and Methods
Animals and Tissue Preparation
All animal experiments were reviewed and approved by the animal care committee of Université de Montréal (No. 18-089) and conform to the Canadian Council on Animal Care and the US National Institute of Health guidelines. Female Sprague-Dawley rats (Charles River Canada) aged 12 to 14 weeks and weighing 225 to 250 g were mated with known fertile males. Based on a protocol developed in the laboratory (28), IUGR was induced by giving an isocaloric low-sodium diet containing 0.03% (w/w) sodium and 0.85% potassium (low-sodium diet 5881; PMI Feed Inc, Ren’s Feed and Supplies) with demineralized water from day 15 to 22 of gestation. The control group was fed a normal diet containing 0.20% sodium and 0.40% potassium (normal diet 5755; PMI Feed Inc) and tap water. The composition of both diets was similar in terms of protein (19%), carbohydrate (60.6%), and fat (10%) content, and animals were randomly assigned to each diet. On day 22 of gestation (1 day before term), animals were anesthetized with 4% isoflurane mixed with oxygen at a rate of 1 L/min in a plexiglass chamber and then euthanized (8-9 am) by decapitation. Fetuses were removed from the womb, fetal annexes (placenta, umbilical cord, amnios) were discarded, and fetuses were sexed and weighed, while kept on a warm heating pad and protected from light. Fetuses were beheaded, hearts were quickly collected and either processed immediately for cardiomyocyte isolation or snap-frozen in liquid nitrogen and stored at –80 °C.
RNA Isolation and Quantitative Polymerase Chain Reaction
Total RNA was extracted with TRIzol (Thermo) and complementary DNA was prepared and subjected to quantitative polymerase chain reaction (qPCR) analysis as described previously (35). For tissues, approximately 20 mg of fetal heart was used per each group for RNA isolation. Each value is derived from one fetus from a separate litter (one fetus of each sex per litter per group). For cells, values are derived from at least 3 independent experiments. In each case, values were normalized to ribosomal protein RPLP0 expression.
Mitochondrial Biogenesis
The relative number of mitochondria was determined by measuring the ratio of mitochondrial DNA to nuclear DNA from fetal hearts obtained from IUGR and normal fetuses. DNA was isolated and subjected to qPCR using specific primers for respectively mitochondrial cytochrome b (mt-Cytb) and the nuclear gene Pgk2.
Fetal Cardiomyocyte Isolation
Fetal cardiomyocytes were isolated as previously described (36) with the following modifications. Approximately 80 mg of fetal heart tissue were excised quickly and placed in calcium-free S-MEM medium (Gibco BRL) supplemented with 24-mM NaHCO3, 0.6-mM MgSO4, and 1-mM dl-carnitine; pH 7.4. The atria were removed, and the ventricles were carefully minced and incubated at 37 °C for 5 minutes with the addition of 23-mM taurine, 10% bovine serum albumin, and 0.23-mg/mL collagenase (Yakult). Supernatants were collected and inactivated in M199 medium (Sigma) containing 10-mM N-2-hydroxyethylpiperazine-N′-2-ethane sulfonic acid (HEPES), pH 7.4, 26-mM NaHCO3, 30% fetal bovine serum (FBS), 1% penicillin/streptomycin (P/S), and 1.25-U/mL insulin. Cells were then resuspended in M199 medium supplemented with 10-mM HEPES, pH 7.4, 26-mM NaHCO3, 15% FBS, 1% P/S, and 1.25-U/mL insulin) and plated in culture dishes for 30 minutes at 37 °C and 5% CO2 to let the fibroblasts adhere. Cardiomyocytes were then collected and resuspended in M199 medium supplemented as described earlier at the same cell density for each group and prepared in the same conditions and the same day (synchronized pregnancies) to minimize intergroup disparities.
Oxygen Consumption Rate
An equal number of isolated cardiomyocytes from control and IUGR fetuses were plated (70%-80% confluency) in a collagen- or laminin-coated 96-well plate in Dulbecco’s modified Eagle’s medium 5030 supplemented with 5.5-mM glucose, 1-mM sodium pyruvate, and 4-mM glutamine. Plates were preincubated at 37 °C to let the cardiomyocytes adhere and analyzed for oxygen consumption rate (OCR) measurements in a Seahorse Xfe96 apparatus. OCR was monitored on serial injections of 1-µM oligomycin, 1-µM (optimized) carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP), and 1-µM antimycin A/rotenone (AA/rot) mixture. The maximal respiration rate was determined by subtracting the AA/rot minimum reading from the maximal respiratory capacity following FCCP treatment. The spare respiratory capacity (reserve capacity) is the difference between the basal respiration rate and the maximal respiratory capacity. Adenosine 5′-triphosphate (ATP) production (ATP-linked oxygen consumption) is the difference between basal oxygen consumption and OCR readings after ATPase inhibition with oligomycin. The proton leak (nonmitochondrial oxygen utilization) is defined as the basal respiration rate less the AA/rot minimum rate. Data presented were corrected for nonmitochondrial respiration in each case.
Lipidomics
Snap-frozen hearts were processed for quantitative profiling of fatty acid derivatives by gas chromatography–mass spectrometry according to Pinçon and colleagues (37). For gas chromatography–mass spectrometry, 2 hearts from the same sex and same litter were pooled and considered as 1 sample (n = 1). In total, 12 samples derived in each case from a different litter were analyzed.
Cell Culture and Treatments
Rat H9C2 cardiomyocytes (ATCC), a cell lineage derived from female rat fetal ventricular cardiomyocytes, were cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS and 1% P/S, in a 5% CO2-humidified incubator at 37 °C. H9C2 cells were grown at a maximum of 70% confluency. Treatments with arachidonic acid (ARA), eicosapentaenoic acid (EPA), and docosapentaenoic acid (DPA) (Cayman Chemical) were performed in culture medium for 16 to 24 hours.
Luciferase Reporter Gene Assay
Luciferase assays were conducted as previously described (38). Briefly, cells were transfected with a Gal4-PPARα fusion construct in the presence of a UAStkLuc reporter, using polyethyleneimine (Gibco). Values were normalized to the β-galactosidase activity and expressed as fold compared to control cells. Data were obtained from at least 3 independent experiments performed in triplicate.
Fluorescence Microscopy
H9C2 cardiomyocytes were seeded in Lab-Tek coverglass chambers (Nalge Nunc) and treated or not (vehicle) with 100-µM ARA or 100-µM EPA in culture medium for 72 hours. Live cells were then rinsed with phosphate-buffered saline and labeled at 37 °C for 15 minutes with 10-µg/mL rhodamine-123, a mitochondrial-specific fluorochrome (Sigma), as described previously (38). Mitochondria were visualized by fluorescence microscopy (Leica DMI 8) with an excitation at 488 nm and emission at 525 nm. Signal intensity was measured with a CLARIOstar reader using the same excitation/emission wavelengths. Data were obtained from 3 independent experiments performed in duplicate and normalized to protein content.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (39). Approximately 80 mg of snap-frozen fetal heart tissue were pulverized in liquid nitrogen and cross-linked with 1% formaldehyde. Then, 0.125-M glycine was added, and samples were further homogenized in 10-mM Tris, pH 7.4 containing 3-mM CaCl2, 3-mM MgCl2, 1-mM phenylmethylsulfonyl fluoride, and protease inhibitors. ChIP was performed using antibodies against PPARα (sc-398394, Santa-Cruz Biotech) and H3K27ac (ab4729, Abcam).
Statistical Analysis
Experiments were routinely repeated at least 3 times with replicate determinations, and the repeat number was increased according to effect size or sample variation. Animals were randomly assigned for diets (IUGR; control) and one fetus per sex per litter was considered n = 1. All values are expressed as the mean ± SEM, except for the qPCR data, which are expressed as median, minimum, and maximum (box plot). For the experiments with animal tissues, multiple comparisons were performed by 2-way analysis of variance for the fetal environment (PIUGR), sex (Psex), and interaction (Pint), and significant differences were analyzed with Bonferroni post hoc test using GraphPad Prism, version 8 (GraphPad Software Inc). If an interaction (Pint) is presented, differences between groups were determined by t test. Elsewhere, single comparisons between 2 groups were determined by t test. P values less than .05 were considered significant.
Results
Key Genes of Fatty Acid Breakdown and β-Oxidation Are Upregulated in the Intrauterine Growth Restriction Fetal Heart
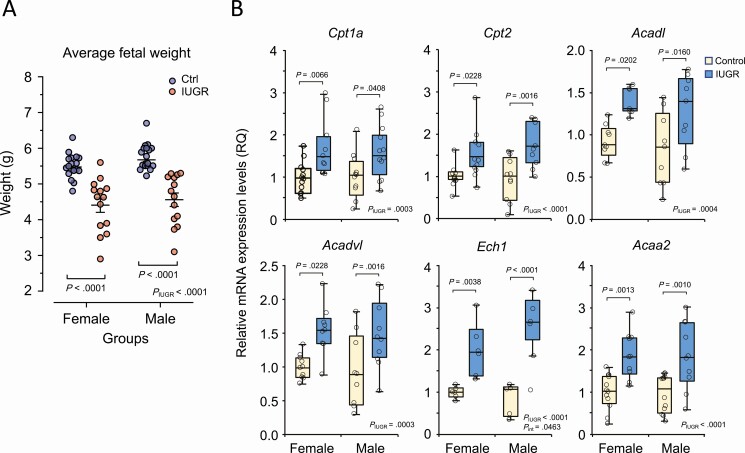
We have previously reported that giving a low-sodium diet to dams in the last week of gestation resulted in reduced uterine blood flow, placental under-perfusion, and intrauterine growth restriction, establishing an animal model of IUGR (27, 40). Here, under the same conditions, we show that the average body weight was indeed clearly reduced both in female and male fetuses from the IUGR group (Fig. 1A), without significantly affecting the litter size and sex distribution (Supplementary Fig. S1A and S1B) (41). Based on our previous findings of structural remodeling of adult cardiomyocytes in these conditions (34), we thus addressed fetal heart metabolism in the context of IUGR by first profiling key genes involved in FA and energy metabolism. Gene expression levels of carnitine palmitoyl-transferases Cpt1a and Cpt2, 2 mitochondrial transporters that allow LCFAs to cross the mitochondrial outer and inner membrane, were significantly increased in the IUGR group from both sexes (Fig. 1B). As CPT1a and CPT2 control the rate-limiting step in the process of LCFA β-oxidation (42), we next analyzed the expression levels of critical genes involved in the various steps of FA β-oxidation. Interestingly, genes for acyl-CoA dehydrogenase Acadl and Acadvl, enoyl-CoA hydratase 1 (Ech1), and acetyl-CoA acyltransferase 2 (Acaa2) were significantly upregulated in female and male IUGR fetal hearts compared to controls (see Fig. 1B). Given that Acadvl and Acadl are primarily involved in the initial and unidirectional steps of LCFA oxidation, and that Ech1 and Acaa2 contribute to the final steps, our results suggest an increase in the overall β-oxidation process in IUGR fetal hearts.
Figure 1.
Upregulation of fatty acid metabolic genes in the intrauterine growth restriction (IUGR) fetal heart. A, Distribution of fetal body weight of the IUGR and control group at 22 days of gestation. The transversal bars represent the mean ± SEM. B, Box plot representation of gene expression levels of key metabolic genes of fatty acid uptake and metabolism in IUGR compared to control fetal hearts collected at 22 days of gestation. Results are expressed as relative messenger RNA (mRNA) levels (RQ; relative quotient) after normalization to Rplp0 expression. Data were analyzed using 2-way analysis of variance (PIUGR, Psex, and Pint) and Bonferroni post hoc test derived from independent samples (fetus/sex/litter; n = 6-12) from each group. Statistical significance (P value, post test) is indicated between control and IUGR for each sex.
Genes of Fatty Acid Uptake and Peroxisomal β-Oxidation Are Regulated in a Sex-related Manner in Intrauterine Growth Restriction
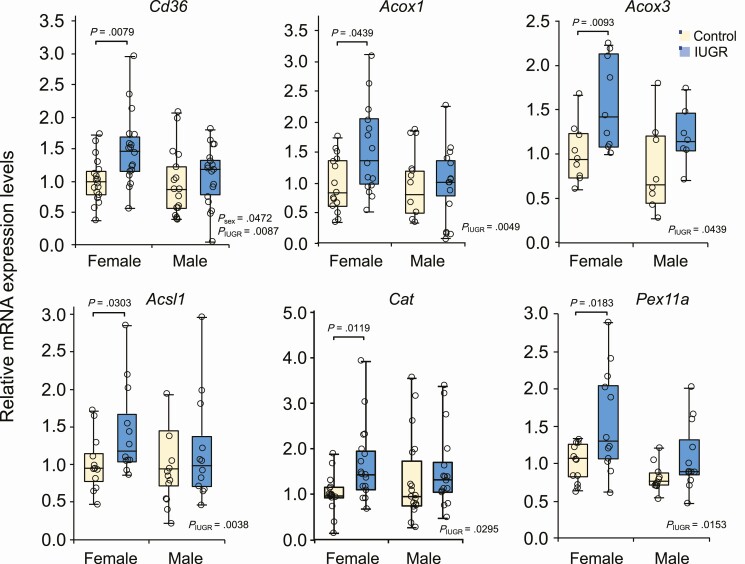
Scavenger receptor CD36 is known as an FA translocase that promotes the uptake of LCFAs, a process that is crucial for adequate fetal development but also associated with lipotoxic cardiomyopathy in adults (43, 44). We found an increase in Cd36 expression levels in female IUGR fetal hearts, whereas no significant changes were observed in males (Fig. 2). To better support these variations, we also found that gene expression levels of acyl-CoA oxidases Acox1 and Acox3, involved in the first and rate-limiting step in the peroxisomal β-oxidation pathway, were significantly increased in IUGR female hearts, whereas no major changes were observed in male fetuses. Similar results were also observed for Acsl1, responsible for the esterification of free LCFAs to fatty acyl-CoA esters to enter β-oxidation, Cat (catalase), encoding a peroxisomal enzyme of the redox pathway, and Pex11a, a marker of peroxisomal biogenesis (see Fig. 2). These results suggest a sex-related regulation of LCFA uptake and peroxisomal β-oxidation as part of an adaptive mechanism taking place during IUGR.
Figure 2.
Key genes of fatty acid uptake and peroxisomal β-oxidation are upregulated in a sex-related fashion in the intrauterine growth restriction (IUGR) fetal heart. Box plot representation of relative gene expression levels of metabolic genes involved in fatty acid (FA) uptake and peroxisomal FA breakdown and biogenesis. Female and male IUGR and control fetal hearts were collected at 22 days of gestation and analyzed by quantitative polymerase chain reaction. Gene expression levels were normalized to Rplp0 expression. Data were analyzed using 2-way analysis of variance (PIUGR, Psex, and Pint) and Bonferroni post hoc test derived from independent samples (fetus/sex/litter; n = 9-18) from each group.
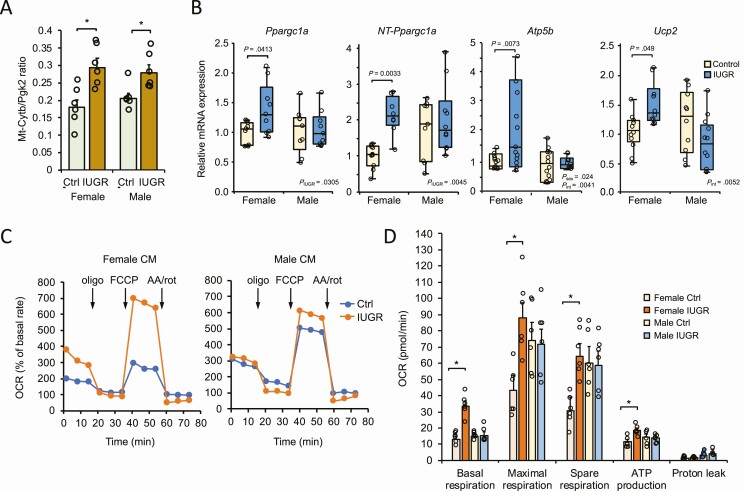
Cardiac Mitochondrial Biogenesis Is Enhanced in Intrauterine Growth Restriction Fetuses
The extent to which numerous genes involved in LCFA metabolism and oxidation are upregulated in IUGR fetal cardiac tissue suggests an increase in mitochondrial function. We thus measured the ratio of mtDNA-encoded cytochrome b (mt-Cytb) gene compared to nuclear DNA-encoded phosphoglycerate kinase 2 (Pgk2) levels as an indication of mitochondrial biogenesis in cardiac tissue of normal and IUGR fetuses. A higher ratio of mtDNA relative to nuclear DNA was observed in IUGR, supporting an increased number of mitochondria fetal cardiac tissues in the context of IUGR (Fig. 3A). The thermogenic and transcriptional nuclear receptor coregulator peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) is an essential component regulating mitochondrial biogenesis and FA β-oxidation in metabolic tissues, including the heart (45). We found that PGC-1α (Ppargc1a) gene expression was significantly induced in female IUGR fetal hearts but not in males compared to their respective controls (Fig. 3B). To further investigate such a response, we addressed whether splicing variants of PGC-1α could be differently regulated in IUGR, given that alternative 3′ splicing events affecting exon 6 to 7 boundaries of the Ppargc1a gene produces an N-truncated form of PGC-1α (referred to as NT-PGC-1α) shown to exhibit shared features with PGC-1α (46). Interestingly, we found the NT-PGC-1α spliced transcript to be expressed in both sexes, suggesting a functional role in fetal cardiac metabolism (see Fig. 3B). However, the NT-Ppargc-1α transcript was only significantly augmented in IUGR females, supporting a sex-dependent role of the variant.
Figure 3.
Intrauterine growth restriction (IUGR) is associated with a sex-specific increase in mitochondrial biogenesis and respiration in fetal cardiac cells. A, Gene expression analysis of mitochondrial Cytb and nuclear Pgk2 from female and male control and IUGR fetal cardiac tissues. Results are expressed as a ratio representative of mitochondrial enrichment. Data are expressed as mean ± SEM (n = 6). *P < .05 vs respective control group. B, Box plots showing expression levels of Ppargc1a, NT-Ppargc1a, Atp5b, and Ucp2 transcripts in fetal cardiac tissues from each group. NT-Ppargc1a represents the N-terminal truncated PGC-1α transcript. Data were normalized to the Rplp0 gene and are derived from independent litters (one fetus per litter; n = 9-12) from each group. C, Single trace of oxygen consumption rate (OCR) analysis was performed on isolated cardiomyocytes (CM) from control and IUGR fetuses. Profiles are shown using female (left panel) and male (right panel) cardiomyocytes. For group comparisons, OCR was expressed as a percentage (%) relative to baseline measured before the addition of mitochondrial effectors oligomycin, carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP), and antimycin A/rotenone (AA/rot) where indicated. D, OCR was monitored for basal, maximal, and spare (reserve) respiration capacity, as well as adenosine 5′-triphosphate (ATP) production and proton leak from each group. Data were corrected for nonmitochondrial respiration and represented as mean ± SEM from 6 independent experiments (fetus/sex/litter). *P less than .05 relative to the respective same-sex control group.
The increases in PGC-1α expression and mitochondrial content in cardiac tissue of IUGR fetuses indicate a requirement for mitochondrial activity and energy production. Consistent with such upregulation, gene markers of enhanced mitochondrial activity such as Atp5b, a subunit from the F1 catalytic core of ATP synthase, and the uncoupling protein Ucp2 were significantly increased in IUGR females, but not in males, compared to their respective controls (see Fig. 3B). A significant interaction between sex and IUGR (Pint) was observed for the 2 genes, strengthening the sex-specific regulation of these 2 genes in IUGR. Overall, these results suggest that fetal hearts might promote an adaptive mechanism implicating mitochondrial biogenesis and activation associated with IUGR. Such adaptation appears to be more efficient in female fetuses.
Increased Mitochondrial Respiration and Energy Production in Intrauterine Growth Restriction Fetal Myocardiac Cells
Our results suggest sex-dependent and adapted alterations in fetal cardiac energy metabolism in conditions of IUGR. To further investigate whether the metabolic adaptation was associated with changes in mitochondrial function, we performed OCR analysis using a Seahorse flux analyzer on female and male primary cardiomyocytes prepared from control and IUGR fetuses (Fig. 3C). Measurement analysis indicates that both basal respiration and maximal respiration rates were increased more than 2-fold in IUGR female fetal cardiomyocytes compared to normal fetal cells (Fig. 3D). A marked increase in the spare respiration capacity was also observed in female IUGR cardiomyocytes, which is consistent with an increase in the reserve capacity of mitochondria. These results suggest a rise in oxygen consumption in female cardiomyocytes to possibly adapt to the context of the adverse fetal environment by inducing their mitochondrial oxidation capacity. No significant changes in terms of maximal respiration rate or spare respiration capacity were observed in male fetal cells, further indicating that male fetuses exhibit an impaired or delayed adaptative potential.
Cellular respiration is usually tightly coupled to ATP synthesis, but the increase in UCP2 expression in the female IUGR fetal heart might also allow uncoupling of fuel oxidation from ATP production (Fig. 3B). However, OCR readings obtained in the presence of the ATP synthase inhibitor oligomycin indicate an enhanced respiration-coupled ATP production in female IUGR cardiomyocytes that was not present in male cells (see Fig. 3D). Although more elevated in male than in female cells, alterations of uncoupled respiration as revealed by proton leak values were not significant. These results uncover the strong potential of mitochondrial oxidative function in IUGR female cardiac cells to provide readily available ATP as an adaptative mechanism to respond to energy expenditure.
C16-C22 Long-Chain Fatty Acids Accumulate in Intrauterine Growth Restriction Fetal Cardiac Tissue
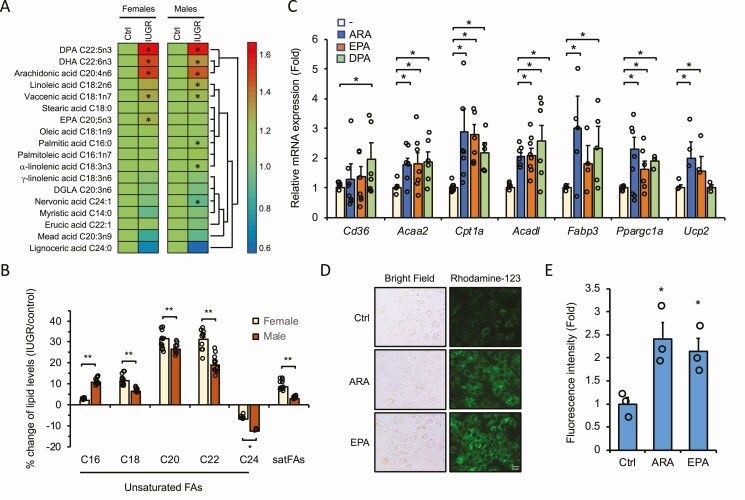
Anaerobic glycolysis with transition to oxidative phosphorylation are the preferred pathways adopted by the fetal heart for energy production during gestation, and maturation to the predominant use of FAs as energy substrates occurs generally in the postnatal period (47, 48). However, our findings on the induced expression of mitochondrial and peroxisomal β-oxidation genes and enhanced mitochondrial respiration in IUGR fetal hearts suggest that FAs may contribute prematurely to energy needs. We thus performed mass spectrometry–based lipidomic analysis using fetal hearts of normal and IUGR groups to identify relevant changes in FA metabolites. We found a significant accumulation of several polyunsaturated LCFA (LCPUFA) derivatives in the IUGR fetal heart when compared with normal fetuses (Fig. 4A). Polyunsaturated C22:5 DPA (P < .001), C20:4 ARA (P < .001), and C22:6 docosahexaenoic acid (DHA; P < .05) were increased by more than 40% both in female and male IUGR fetal cardiac tissues relative to their respective controls. Other derivatives were also significantly elevated in a sex-specific fashion, such as C18:2 linoleic acid (P = .02) in males and C20:5 EPA (P = .04) in females. When comparing FAs in terms of respective carbon chain lengths, the percentage changes of C18, C20, and C22 derivatives were stronger in IUGR female compared to male cardiac tissues (Fig. 4B). Such enhanced availability of unsaturated LCFAs in the IUGR fetal heart might indicate an adaptive mechanism taking place to ensure the appropriate supply of energetic FA metabolites and signaling effectors readily available for cellular needs.
Figure 4.
Long-chain fatty acid (LCFA) derivatives accumulate in intrauterine growth restriction (IUGR) fetal hearts and upregulate metabolic genes and mitochondrial activity. A, Lipidomic analysis reveals accumulation of LCFA derivatives in IUGR fetal hearts. Heat map representation and cluster analysis of LCFA derivatives measured by mass spectrometry analysis of fetal cardiac tissue prepared from each group. Results represent the mean ± SEM of fold changes of each LCFA concentration (nmol/mg total protein) from IUGR compared to respective controls (Ctrls) set at 1.0. Data are derived from 12 independent samples, each representing a unique pool of 2 fetal hearts (sex-matched) from the same litter (12 litters in total). *P less than .05 relative to the same-sex control group. DPA, docosapentaenoic acid; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; DGLA, dihomo-γ-linolenic acid. B, Percentage change of total accumulation of polyunsaturated LCFAs and saturated FAs clustered by carbon-chain length in female and male IUGR fetal hearts compared to control. Data are derived from results presented in A and are expressed as mean ± SEM. C, Upregulation of key metabolic genes by LCFAs. Quantitative polymerase chain reaction analysis of H9C2 cardiomyocytes treated with 100 μM ARA, 100 μM EPA, or 60 μM DPA in ethanol (1:1000, v:v) for 24 hours. Data were normalized to Rplp0 expression and are expressed as fold change (mean ± SEM) over vehicle-treated cells set at 1.0. *P less than .05 relative to vehicle-treated cells. D, Representative images of H9C2 cells stained with mitochondria-specific rhodamine-123 dye. Cells were treated with vehicle (Ctrl), 100-μM ARA, or 100-µM EPA for 72 hours. E, Quantitative determination of fluorescence signals of cells treated as in D. Data were normalized to protein content and expressed as fold change (mean ± SEM; n = 3) over vehicle-treated cells set at 1.0. *P less than .05 relative to vehicle-treated cells.
Long-Chain Polyunsaturated Fatty Acids Induce Expression of Key Fatty Acid Metabolic Genes in H9C2 Cardiomyocytes
To get more insight into the molecular mechanism underlying the metabolic adaptation observed in IUGR fetal hearts and address the effect of specific LCFAs in transactivation assays, we used H9C2 cells derived from rat fetal ventricular cardiomyocytes. The LCFAs used to treat H9C2 cells were chosen based on their upregulated pattern from the lipidomic analysis (Fig. 4A) as an attempt to replicate the in vivo condition of LCFA excess. Interestingly, when exposed to ARA, DPA, or EPA, H9C2 cells exhibited an increased expression of key genes of FA metabolism and mitochondrial function in a manner comparable to the profile obtained in IUGR fetal cardiac tissue. More specifically, genes such as Acaa2, Cpt1a, Acad1, Fabp3, Ppargc1a, and Ucp2 were upregulated in response to LCPUFAs in H9C2 cardiomyocytes (Fig. 4C). These results are consistent with the expression profile we observed in tissues (Figs. 1 and 2) and further support the adaptive response to LCPUFA signaling. We next determined whether the changes in mitochondrial gene expression correlated with mitochondrial activity by staining H9C2 cells with rhodamine-123, a nontoxic fluorescent dye that labels active mitochondria. We found a significant increase in mitochondrial staining in cells treated with LCPUFAs ARA and EPA with densely packed structures to some extent when compared to untreated cells (Fig. 4D and 4E). This indicates that LCPUFAs promote mitochondrial activity in H9C2 cardiomyocytes.
Regulation of Nuclear Receptor Peroxisome Proliferator-Activated Receptor α in the Metabolic Adaptation to Intrauterine Growth Restriction
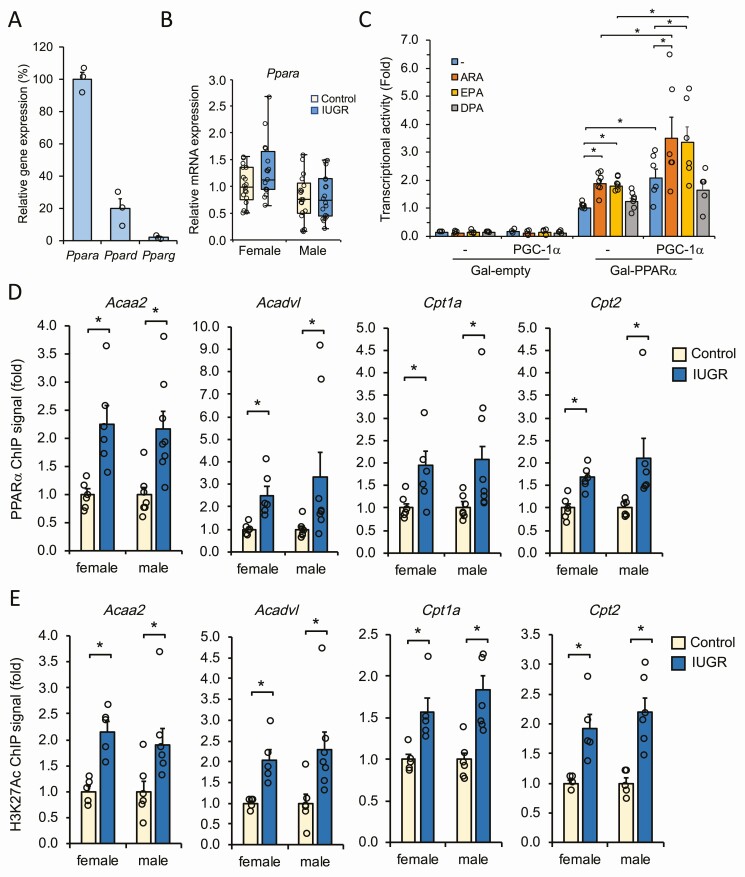
The PPAR members of nuclear receptors comprise PPARα, PPARβ/δ, and PPARγ, which are essential transcriptional regulators of key genes controlling lipid and glucose homeostasis in metabolic tissues including the heart (49, 50). Gene expression profiling of the 3 PPAR genes has indicated that Ppara is highly expressed in fetal cardiac tissues compared to the Ppard and Pparg genes (Fig. 5A), in support of its pivotal role in heart metabolism. Also, although not reaching significance (P = .06), the Ppara gene was upregulated in the IUGR female fetal heart compared to males (Fig. 5B) and compared to Ppard and Pparg genes that remained mostly unchanged in the context of IUGR (Supplementary Fig. S2) (41). Therefore, we tested the effect of ARA, EPA, and DPA on the transcriptional activity of PPARα using the luciferase gene reporter assay in H9C2 cells. Cells exposed to the LCPUFAs showed an increase in PPARα activity, with ARA and EPA being the more potent (Fig. 5C). Also, transient expression of PGC-1α in H9C2 cells further increased the transcriptional response of PPARα to ARA and EPA, indicating the contribution of PGC-1α coactivator to promote LCFA responsiveness (see Fig. 5C). Although slightly less affected, PPARβ/δ and PPARγ were also activated by ARA and EPA under the same conditions, not to exclude their possible implication in responding to LCPUFAs in the fetal heart (Supplementary Fig. S3) (41). These results suggest that in the condition of excess LCPUFAs such as the one observed in IUGR fetal cardiac tissues, transcriptional activation of the PPARs, with a more prevalent contribution of PPARα, might ensue to mediate the cardiac metabolic remodeling to adverse intrauterine environment.
Figure 5.
Peroxisome proliferator-activated receptor α (PPARα) is involved in the metabolic adaptation of the fetal heart to an adverse intrauterine environment. A, Relative expression levels of Ppara, Ppard, and Pparg genes in fetal cardiac tissues. Expression was determined by quantitative polymerase chain reaction analysis and normalized to Rplp0 expression. Data represent mean ± SEM (n = 3) of percentage expression relative to Ppara set at 100%. B, Box plot representation of Ppara gene expression in the fetal heart from female and male IUGR and control groups. Data were derived from independent samples (fetus/sex/litter) from each group and were normalized to Rplp0 expression. C, Transcriptional activity of PPARα and contribution of peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) in response to long-chain fatty acid derivatives. H9C2 cardiomyocytes were transfected with Gal4-PPARα fusion in the presence of a UAStkLuc luciferase reporter with or without PGC-1α expression construct. Cells were then treated with arachidonic acid (ARA; 100 μM), eicosapentaenoic acid (EPA; 100 μM), or docosapentaenoic acid (DPA, 60 μM) for 16 hours. Data were normalized to β-galactosidase and expressed as fold response compared to vehicle-treated PPARα-transfected cells set at 1.0. Negative control cells were transfected with an empty Gal4 plasmid. *P less than .05 vs control group. D, Chromatin immunoprecipitation (ChIP) analysis of PPARα binding to metabolic target genes. PPARα enrichment to peroxisome proliferator response element (PPRE)-containing genes was measured from fetal cardiac tissues prepared from female and male intrauterine growth restriction (IUGR) and control groups. Data are presented as mean ± SEM (n = 6) of fold changes compared to same-sex control set at 1.0. *P less than .05 vs control. E, ChIP mapping of transcriptionally active histone H3K27ac mark relative to each PPARα binding sites analyzed in E. Data are derived using the same samples as in D.
Peroxisome Proliferator-Activated Receptor α Recruitment to Promoters of Target Metabolic Genes Is Enhanced in to Intrauterine Growth Restriction Fetal Heart
Considering that several metabolic genes found to be upregulated in IUGR fetal hearts (see Figs. 1 and 2) are recognized targets of PPARs and based on the upregulation of Ppara in fetal cardiac tissue (see Fig. 5A), we addressed whether PPARα recruitment to target gene promoters was affected in IUGR using the ChIP assay. ChIP assay was thus performed on fetal heart tissue derived from normal and IUGR groups. Considering that no ChIP data are yet available for PPARα on rat heart tissue, we based our ChIP procedure mainly on described peroxisome proliferator response element (PPRE) sequences from other species and available datasets in mice (51, 52), to help us identifying responsive rat PPARα binding sites. Among the genes analyzed, we found an enhanced enrichment of PPARα binding to Acaa2, Acadvl, Cpt1a, and Cpt2 promoters, indicating greater recruitment of PPARα to these genes in the condition of IUGR (Fig. 5D). To further distinguish between active and less active PPREs, we performed ChIP for transcriptionally active histone H3K27ac marks at each respective PPRE. Significant enrichment of H3K27ac marks was associated mostly with each PPRE found to associate with PPARα, correlating with transcriptional gene activation (Fig. 5E). These results demonstrate a direct role of PPARα in avidly interacting with key cardiac metabolic gene promoters in fetuses subjected to IUGR. Such enrichment of PPARα was coupled to increased transcriptional histone marks, thus favoring the establishment of an active chromatin landscape through epigenetic changes as part of a metabolic attempt adopted by IUGR fetuses to adapt from an adverse in utero environment. A proposed model outlining the cellular pathways involved in such fetal cardiac metabolic adaptation is depicted in Fig. 6.
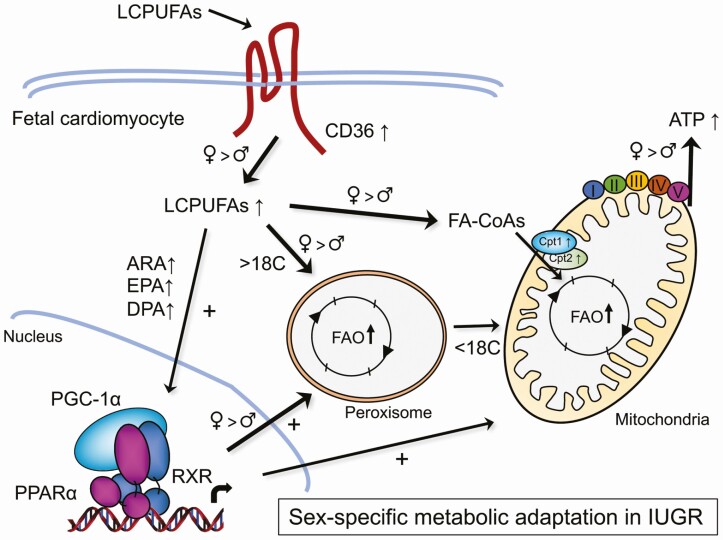
Figure 6.
Proposed model of fetal cardiac metabolic adaptation to adverse intrauterine environment. Uteroplacental vascular dysfunction leading to intrauterine growth restriction (IUGR) induces metabolic stress to the fetal heart, resulting in an early and sex-adaptive shift in fatty acid (FA) metabolism to adjust for energy expenditure. The increase in specific LCPUFA derivatives in fetal cardiomyocytes leads to PPARα transcriptional activation and PGC-1α coregulation, resulting in upregulation of key genes in lipid metabolism including key markers of peroxisomal and mitochondrial β-oxidation and transport. These effects are also supported by increased peroxisomal and mitochondrial biogenesis, contributing to the adapted mechanism to mobilize LCPUFAs for adenosine 5′-triphosphate (ATP) production and energy expenditure. While each arrow represents the metabolic pathways occurring both in IUGR female and male fetuses, thicker arrows highlight the preferred pathways observed in females. The model outlines a sex-dependent metabolic attempt adopted by IUGR fetuses to adapt from an adverse in utero environment. ARA, arachidonic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid; FA-CoA, fatty acyl-CoA; FAO, fatty acid oxidation; LCPUFAs, long-chain polyunsaturated fatty acids; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α; PPARα, peroxisome proliferator-activated receptor α.
Discussion
In this study, we describe the mechanistic adaptation of the fetal heart to achieve cellular energy requirements using an animal model of IUGR. In normal conditions, the fetal heart uses predominantly carbohydrates for energy production principally due to the anaerobic womb environment at early stages of development with transition to oxidative phosphorylation (47, 48, 53). After birth the aerobic environment, increasing workload, and ingestion of fat-rich maternal milk, contribute to the transition of metabolic fluxes to use lipids for energy requirements (48, 54). Our findings demonstrate an accumulation and increased metabolic effects of LCPUFAs that occur prenatally in fetal hearts, suggesting an early shift toward lipid use for energy needs to adapt to the adverse intrauterine environment. Such lipid surge was associated with an increase in specific LCPUFAs, including DPA, ARA, DHA, and EPA. LCPUFA derivatives have been shown to exert anti-inflammatory and cardioprotective effects, such as recently demonstrated for omega-3 DHA- and EPA-derived oxylipins (55). Here we show that specific LCPUFAs might contribute to the metabolic reprogramming in the IUGR fetal heart by mostly acting as β-oxidation substrates and activating effectors of nuclear receptor PPARα. The accumulation of LCPUFAs is present both in females and males but seems to be more prominent in females, with a greater accumulation of C18, C20, and C22 derivatives compared to shorter FAs. Also, the increased expression levels of the Cd36 gene involved in LCFA uptake by cells and of LCPUFA metabolic genes, such as Cpt1a, Cpt2, and several β-oxidation genes, suggest that the IUGR fetal heart is adapting to a higher surge of LCPUFAs, likely derived from an enhanced cellular entry, rather than a dysfunctional oxidation capacity. Because the fetus is unable to meet its own needs in LCPUFA synthesis, the majority is usually transferred from the mother to the fetal circulation through the placenta (43). Accordingly, the existence of a placental metabolic pool, buffering the FA transfer from the maternal to the fetal circulation, would allow the preferential transport of LCPUFAs to the fetus (56). However, there is evidence that the maternal metabolic status alters placental lipid metabolism and transporters during adverse fetal environment leading to IUGR and that such alterations affect FA metabolism and placental genes in a sex-dependent manner (57-60). Consistent with this, we have reported a placental underperfusion and increased clustering of glycogen cells at the junctional zone of the placenta in our IUGR model, supporting alterations in uterine vascular bed remodeling and normal uteroplacental circulation (27). Hence, our results indicate that IUGR fetuses may trigger an adaptive mechanism to switch the metabolic needs for energy by promoting an efficient entry and use of LCPUFAs by the heart.
The expression pattern of several LCPUFA metabolic genes suggest an increase in FA uptake and β-oxidation that seems more prevalent in the female IUGR fetal heart. For instance Cd36 and peroxisomal genes are involved at key steps of LCPUFA mobilization into the β-oxidation process (61-63), and their sex-related regulation might be indicative of the favored propensity of LCPUFAs to serve as energy fuel in the female IUGR fetal heart compared to IUGR males. Interestingly, female sex hormones have been proposed to accelerate cardiac LCFA uptake kinetics and intracellular trafficking of acyl CoA using ex vivo adult mouse heart perfusion (64). Myocardial FA utilization and oxidation rates were higher in women compared to men at the onset of diabetic conditions when FAs are more abundant, possibly related to the effect of estrogen in inducing myocardial β-oxidation (65, 66). Although estrogen production is minimal at the fetal stage because of immaturity of gonad function, expression of estrogen receptors may contribute to the observed sex differences in the gene profile, especially given that sex-specific interactions between estrogen signaling pathways and PPARα target genes have been reported (67). Whether such a mechanism may underlie the sex-dependent response of PPAR-regulated genes in the fetal heart deserves further investigation.
Peroxisomes preferably oxidize very-long-chain FAs (VLCFAs) without completing the β-oxidation process, thus feeding mitochondria with shorter-chain FAs (68). Our results on the apparent sex-related regulation of peroxisomal markers, such as Acox1, Acox3, and Pex11a, suggest a sexually dimorphic response of peroxisomal function. Not much is known about fetal cardiac sex dimorphism and peroxisomal dysfunction occurring during IUGR. Sexual dimorphism has been observed with dysregulated epigenetic patterns in cord blood stem cells of IUGR male newborns (69), and sex-specific expression of fetal cardiac microRNAs was reported in a baboon model of IUGR (70). Our results suggest an enhanced peroxisomal FA oxidation in female fetuses, whereas this process appears less effective in males. Impaired peroxisomal assembly and activity is known to be associated with metabolic and developmental problems (71). In our model, the mobilization of peroxisomes is thus envisioned as a mechanism to achieve a more supportive breakdown of VLCFAs, especially in female fetuses, to fulfill the critical demand for energy production in the IUGR condition. Although the exact mechanism to explain the sex differences in peroxisomal response remains unclear, we suspect that the low response of key peroxisomal genes in male hearts may result from an impaired or at least delayed peroxisomal function in the context of IUGR. Peroxisomal ACOX1 deficiency leads to VLCFA accumulation and patients are characterized by severe developmental delay, neurological regression, and poor survival past early childhood (72, 73). Also, alterations of peroxisomal gene expression have been reported in the pancreas of IUGR fetal rats, although the animal sex was not specified (74), further supporting peroxisomal dysfunction to affect several metabolic organs during IUGR.
The increased mitochondrial biogenesis in the IUGR group supports an enhanced adaptation toward lipid metabolism in the fetal heart, outlining a premature shift in energy substrates. During heart development, the mitochondrial density increases with the maturation of cardiomyocytes, especially during the perinatal period, correlating with the metabolic shift to use lipids instead of carbohydrates as the main energy source in the postnatal period (54, 75). Enhanced mitochondrial density was also reported in the fetal blood cord of human IUGR babies and placentas from IUGR pregnancies (76, 77). Our observation of enhanced mitochondrial biogenesis is also consistent with the upregulation of the Ppargc1a gene. PGC-1α is an essential biogenic regulator of mitochondrial function and a transcriptional coactivator of several nuclear receptors (78). Interestingly, the NT-Ppargc1a transcript was also induced in the female IUGR heart. NT-PGC-1α is often coexpressed with PGC-1α in metabolically active tissues, such as the liver, muscle, and fat, and its expression is highly dependent on the energy status (46, 79). Our results suggest a shared role of NT-PGC-1α in heart metabolism and adaptation to cardiac metabolic alterations and IUGR.
Real-time analysis of OCRs has revealed that IUGR females adapted with an improved bioenergetics profile by elevating steady-state basal respiration, maximal respiration, and spare respiratory capacity in isolated fetal cardiomyocytes. Also, the increase in ATP production in fetal cardiac cells indicates that elevated aerobic mitochondrial oxidation is tightly coupled to ATP synthesis, therefore responding to energy needs. In contrast, male IUGR cardiomyocytes failed to exhibit improved levels of oxygen consumption in the same conditions, revealing an altered response in cellular respiration capacity and ATP-linked production despite already increased maximal and spare respiration rates under normal conditions. Elevated respiration capacity in males has also been reported in other tissues, such as lung and brain sections, revealing a sex-specific mitochondrial respiration (80-82). Although the exact mechanism remains unclear, such enhanced cellular respiration in males would not be as protective in certain conditions, being highly sensitive to impaired mitochondrial function (81). In our study, the sex-specific regulation of Atp5b encoding a subunit of the F1 catalytic complex of ATP synthase might therefore impose an impaired or at least delayed response in the electron transport chain process, providing a possible mechanism for the sex-specific metabolic adaptation of IUGR cardiomyocytes. As the Atp5b gene has been reported as a direct PGC-1–regulated gene (83), alterations in Atp5b expression would reflect the differential expression pattern of PGC-1 seen in male and female fetal cardiomyocytes. Another interesting observation reinforcing sex-divergent effects is the significant upregulation of UCP2 seen only in the female. UCP2 is the principal heart isoform of the UCP family of uncoupling proteins classically referred to as specific proton carriers that dissipate the proton gradient as heat in brown fat (84). Our data on proton leaks suggest that some uncoupling in energy production may occur more in male fetal cardiomyocytes compared to females, suggesting that these differences may relate to UCP2 expression. Enhanced UCP2 expression in the heart was recently shown to promote FA oxidation, resulting in a beneficial effect on mitochondrial function without affecting proton leak kinetics and uncoupling activity (85), consistent with a broader role of UCP2 (86, 87). In line with the sex-related bioenergetics profile observed during IUGR, we propose that such a beneficial role of UCP2 might be part of the adapted mechanism to adjust heart metabolism to an adverse in utero environment.
We found the genes for PPARα, PPARβ/δ, and PPARγ to be expressed in the fetal heart, consistent with their essential role in energy metabolism (49, 50, 88). However, the increased expression pattern of several established PPARα target genes, such as Ctp1a, Acaa2, and Acad family members in fetal hearts and cultured cardiomyocytes, and changes in their epigenetic active histone marks argue for a predominant role of PPARα-mediated transcriptional events in governing the metabolic adaptation in the IUGR fetal heart. Overexpression of Ppara has been reported to upregulate genes involved in FA uptake and oxidation, with a concomitant decrease in genes of glucose uptake and utilization in cardiac-specific PPARα transgenic mice (89). Perinatal upregulation of PPARα hepatic genes also prepares neonates for adaptation to nutrients and milk intake (90, 91). In our IUGR model, the buildup in specific LCPUFAs that can serve as PPARα-signaling lipids and the increase in PGC-1α expression are thought to prime PPARα sensitive gene activation in the IUGR fetal heart.
In this study, we show that IUGR fetal heart metabolism is adaptive to an adverse fetal environment by increasing the peroxisomal and mitochondrial β-oxidation process, elevating the LCPUFA intracellular pool, adjusting mitochondrial biogenesis, and enhancing cellular respiratory capacity and ATP production. As depicted in Fig. 6, such metabolic buildup is suggested to drive substantial energy expenditure, underlying an attempt to meet the high energy demand of an unfavorable in utero environment and placental dysfunction. However, the reduced response of key genes involved in LCFA use and the resulting difficulty in elevating cellular mitochondrial respiration capacity and ATP production might compromise the ability of IUGR male fetuses to properly respond to energy needs associated with cardiac metabolic adaptation. Our findings therefore suggest a molecular basis to the evolving concept of sex-related adaptation in heart metabolism with FA derivatives serving as signaling effectors that may predict metabolic consequences to develop chronic diseases later in life.
Acknowledgments
We thank members of the A.T. laboratory for critical and useful comments. We acknowledge the useful help and support of Matthieu Ruiz for the lipidomics experiment; Céline Fiset, Eli Louwagie, and Michelle Baack for the cardiomyocyte isolation; and Emily Heckel and Jean-Sébastien Joyal for the OCR experiments.
Financial Support: This work was supported by the Canadian Institutes of Health Research to M.B. (CIHR; grant No. MOP136790) and A.T. (CIHR; grant No. MOP152870). L.M. is supported by a doctoral award from the Fonds de recherche du Québec–Santé (FRQS).
Glossary
Abbreviations
- AA/rot
antimycin A/rotenone
- ARA
arachidonic acid
- ATP
adenosine 5′-triphosphate
- ChIP
chromatin immunoprecipitation
- DHA
docosahexaenoic acid
- DPA
docosapentaenoic acid
- EPA
eicosapentaenoic acid
- FA
fatty acid
- FA-CoA
fatty acyl-CoA
- FAO
fatty acid oxidation
- FBS
fetal bovine serum
- FCCP
carbonylcyanide p-trifluoromethoxyphenylhydrazone
- IUGR
intrauterine growth restriction
- LCFA
long-chain fatty acid
- LCPUFA
long-chain polyunsaturated fatty acid
- mtDNA
mitochondrial DNA
- NT-PGC-1α
N-truncated form of peroxisome proliferator-activated receptor-γ coactivator 1α
- OCR
oxygen consumption rate
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator 1α
- PPARα
peroxisome proliferator-activated receptor α
- PPRE
peroxisome proliferator response element
- P/S
penicillin/streptomycin
- qPCR
quantitative polymerase chain reaction
- VLCFA
very-long-chain fatty acid.
Author Contributions
L.M., M.B., and A.T. designed the research; L.M., B.S., and V.C. performed the experiments; L.M., B.S., V.C., M.B., and A.T. analyzed the data; L.M. wrote the manuscript, and M.B. and A.T. supervised the project and edited the manuscript.
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
The data supporting the findings of this study are available within this article and its supplementary materials.
References
- 1. Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341(8850):938-941. [DOI] [PubMed] [Google Scholar]
- 2. Victora CG, Adair L, Fall C, et al. ; Maternal and Child Undernutrition Study Group . Maternal and child undernutrition: consequences for adult health and human capital. Lancet. 2008;371(9609):340-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85(2):571-633. [DOI] [PubMed] [Google Scholar]
- 4. Crispi F, Miranda J, Gratacós E. Long-term cardiovascular consequences of fetal growth restriction: biology, clinical implications, and opportunities for prevention of adult disease. Am J Obstet Gynecol. 2018;218(2S):S869-S879. [DOI] [PubMed] [Google Scholar]
- 5. Chatmethakul T, Roghair RD. Risk of hypertension following perinatal adversity: IUGR and prematurity. J Endocrinol. 2019;242(1):T21-T32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simeoni U, Armengaud JB, Siddeek B, Tolsa JF. Perinatal origins of adult disease. Neonatology. 2018;113(4):393-399. [DOI] [PubMed] [Google Scholar]
- 7. Cianfarani S, Agostoni C, Bedogni G, et al. Effect of intrauterine growth retardation on liver and long-term metabolic risk. Int J Obes (Lond). 2012;36(10):1270-1277. [DOI] [PubMed] [Google Scholar]
- 8. Malhotra A, Allison BJ, Castillo-Melendez M, Jenkin G, Polglase GR, Miller SL. Neonatal morbidities of fetal growth restriction: pathophysiology and impact. Front Endocrinol (Lausanne). 2019;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nardozza LM, Caetano AC, Zamarian AC, et al. Fetal growth restriction: current knowledge. Arch Gynecol Obstet. 2017;295(5):1061-1077. [DOI] [PubMed] [Google Scholar]
- 10. Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371(9608):261-269. [DOI] [PubMed] [Google Scholar]
- 11. Burton GJ, Jauniaux E. Pathophysiology of placental-derived fetal growth restriction. Am J Obstet Gynecol. 2018;218(2S):S745-S761. [DOI] [PubMed] [Google Scholar]
- 12. Zhang S, Regnault TR, Barker PL, et al. Placental adaptations in growth restriction. Nutrients. 2015;7(1):360-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleiss B, Wong F, Brownfoot F, et al. Knowledge gaps and emerging research areas in intrauterine growth restriction-associated brain injury. Front Endocrinol (Lausanne). 2019;10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Năstase L, Cretoiu D, Stoicescu SM. Skeletal muscle damage in intrauterine growth restriction. Adv Exp Med Biol. 2018;1088:93-106. [DOI] [PubMed] [Google Scholar]
- 15. Carducci B, Bhutta ZA. Care of the growth-restricted newborn. Best Pract Res Clin Obstet Gynaecol. 2018;49:103-116. [DOI] [PubMed] [Google Scholar]
- 16. Garg M, Thamotharan M, Dai Y, et al. Early postnatal caloric restriction protects adult male intrauterine growth-restricted offspring from obesity. Diabetes. 2012;61(6):1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beauchamp B, Thrush AB, Quizi J, et al. Undernutrition during pregnancy in mice leads to dysfunctional cardiac muscle respiration in adult offspring. Biosci Rep. 2015;35(3):e00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu Y, Williams SJ, O’Brien D, Davidge ST. Hypoxia or nutrient restriction during pregnancy in rats leads to progressive cardiac remodeling and impairs postischemic recovery in adult male offspring. FASEB J. 2006;20(8):1251-1253. [DOI] [PubMed] [Google Scholar]
- 19. O’Sullivan L, Cuffe JS, Paravicini TM, et al. Prenatal exposure to dexamethasone in the mouse alters cardiac growth patterns and increases pulse pressure in aged male offspring. PLoS One. 2013;8(7):e69149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khazaee R, McCaig LA, Yamashita C, Hardy DB, Veldhuizen RAW. Maternal protein restriction during perinatal life affects lung mechanics and the surfactant system during early postnatal life in female rats. PLoS One. 2019;14(4):e0215611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nüsken E, Fink G, Lechner F, et al. Altered molecular signatures during kidney development after intrauterine growth restriction of different origins. J Mol Med (Berl). 2020;98(3):395-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lane SL, Doyle AS, Bales ES, Lorca RA, Julian CG, Moore LG. Increased uterine artery blood flow in hypoxic murine pregnancy is not sufficient to prevent fetal growth restriction. Biol Reprod. 2020;102(3):660-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wadley GD, McConell GK, Goodman CA, Siebel AL, Westcott KT, Wlodek ME. Growth restriction in the rat alters expression of metabolic genes during postnatal cardiac development in a sex-specific manner. Physiol Genomics. 2013;45(3):99-105. [DOI] [PubMed] [Google Scholar]
- 24. Botting KJ, McMillen IC, Forbes H, Nyengaard JR, Morrison JL. Chronic hypoxemia in late gestation decreases cardiomyocyte number but does not change expression of hypoxia-responsive genes. J Am Heart Assoc. 2014;3(4):e000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Botting KJ, Wang KC, Padhee M, et al. Early origins of heart disease: low birth weight and determinants of cardiomyocyte endowment. Clin Exp Pharmacol Physiol. 2012;39(9):814-823. [DOI] [PubMed] [Google Scholar]
- 26. Bédard S, Sicotte B, St-Louis J, Brochu M. Modulation of body fluids and angiotensin II receptors in a rat model of intra-uterine growth restriction. J Physiol. 2005;562(Pt 3):937-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bibeau K, Sicotte B, Béland M, et al. Placental underperfusion in a rat model of intrauterine growth restriction induced by a reduced plasma volume expansion. PLoS One. 2016;11(1):e0145982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roy-Clavel E, Picard S, St-Louis J, Brochu M. Induction of intrauterine growth restriction with a low-sodium diet fed to pregnant rats. Am J Obstet Gynecol. 1999;180(3 Pt 1):608-613. [DOI] [PubMed] [Google Scholar]
- 29. Duvekot JJ, Cheriex EC, Pieters FA, Menheere PP, Schouten HJ, Peeters LL. Maternal volume homeostasis in early pregnancy in relation to fetal growth restriction. Obstet Gynecol. 1995;85(3):361-367. [DOI] [PubMed] [Google Scholar]
- 30. St-Louis J, Paré H, Sicotte B, Brochu M. Increased reactivity of rat uterine arcuate artery throughout gestation and postpartum. Am J Physiol. 1997;273(3 Pt 2):H1148-H1153. [DOI] [PubMed] [Google Scholar]
- 31. Salas SP, Marshall G, Gutiérrez BL, Rosso P. Time course of maternal plasma volume and hormonal changes in women with preeclampsia or fetal growth restriction. Hypertension. 2006;47(2):203-208. [DOI] [PubMed] [Google Scholar]
- 32. Osol G, Moore LG. Maternal uterine vascular remodeling during pregnancy. Microcirculation. 2014;21(1):38-47. [DOI] [PubMed] [Google Scholar]
- 33. Battista MC, Oligny LL, St-Louis J, Brochu M. Intrauterine growth restriction in rats is associated with hypertension and renal dysfunction in adulthood. Am J Physiol Endocrinol Metab. 2002;283(1):E124-E131. [DOI] [PubMed] [Google Scholar]
- 34. Battista MC, Calvo E, Chorvatova A, Comte B, Corbeil J, Brochu M. Intra-uterine growth restriction and the programming of left ventricular remodelling in female rats. J Physiol. 2005;565(Pt 1):197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanchez M, Picard N, Sauvé K, Tremblay A. Coordinate regulation of estrogen receptor β degradation by Mdm2 and CREB-binding protein in response to growth signals. Oncogene. 2013;32(1):117-126. [DOI] [PubMed] [Google Scholar]
- 36. Larsen TD, Sabey KH, Knutson AJ, et al. Diabetic pregnancy and maternal high-fat diet impair mitochondrial dynamism in the developing fetal rat heart by sex-specific mechanisms. Int J Mol Sci. 2019;20(12):3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pinçon A, De Montgolfier O, Akkoyunlu N, et al. Non-alcoholic fatty liver disease, and the underlying altered fatty acid metabolism, reveals brain hypoperfusion and contributes to the cognitive decline in APP/PS1 mice. Metabolites. 2019;9(5):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodrigue-Way A, Caron V, Bilodeau S, et al. Scavenger receptor CD36 mediates inhibition of cholesterol synthesis via activation of the PPARγ/PGC-1α pathway and Insig1/2 expression in hepatocytes. FASEB J. 2014;28(4):1910-1923. [DOI] [PubMed] [Google Scholar]
- 39. Benhadjeba S, Edjekouane L, Sauvé K, Carmona E, Tremblay A. Feedback control of the CXCR7/CXCL11 chemokine axis by estrogen receptor α in ovarian cancer. Mol Oncol. 2018;12(10):1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bigonnesse E, Sicotte B, Brochu M. Activated NO pathway in uterine arteries during pregnancy in an IUGR rat model. Am J Physiol Heart Circ Physiol. 2018;315(2):H415-H422. [DOI] [PubMed] [Google Scholar]
- 41. Maréchal L, Sicotte B, Caron V, Brochu M, Tremblay A. Supplementary data for Fetal cardiac lipid sensing triggers an early and sex-related metabolic energy switch in intrauterine growth restriction.2021. Posted May 19, 2021. doi: 10.6084/m9.figshare.14619111.v1 [DOI] [PMC free article] [PubMed]
- 42. Schlaepfer IR, Joshi M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 2020;161(2):bqz046. [DOI] [PubMed] [Google Scholar]
- 43. Cetin I, Alvino G, Cardellicchio M. Long chain fatty acids and dietary fats in fetal nutrition. J Physiol. 2009;587(Pt 14):3441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J, Sambandam N, Han X, et al. CD36 deficiency rescues lipotoxic cardiomyopathy. Circ Res. 2007;100(8):1208-1217. [DOI] [PubMed] [Google Scholar]
- 45. Arany Z, He H, Lin J, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1(4):259-271. [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y, Huypens P, Adamson AW, et al. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284(47):32813-32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56(2):130-140. [DOI] [PubMed] [Google Scholar]
- 48. Porter GA Jr, Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS. Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol. 2011;31(2):75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289-312. [DOI] [PubMed] [Google Scholar]
- 50. Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355-361. [DOI] [PubMed] [Google Scholar]
- 51. Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soltis AR, Motola S, Vernia S, et al. Hyper- and hypo- nutrition studies of the hepatic transcriptome and epigenome suggest that PPARα regulates anaerobic glycolysis. Sci Rep. 2017;7(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Girard J, Ferré P, Pégorier JP, Duée PH. Adaptations of glucose and fatty acid metabolism during perinatal period and suckling-weaning transition. Physiol Rev. 1992;72(2):507-562. [DOI] [PubMed] [Google Scholar]
- 55. O’Connell TD, Mason RP, Budoff MJ, Navar AM, Shearer GC. Mechanistic insights into cardiovascular protection for omega-3 fatty acids and their bioactive lipid metabolites. Eur Heart J Suppl. 2020;22(Suppl J):J3-J20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perazzolo S, Hirschmugl B, Wadsack C, Desoye G, Lewis RM, Sengers BG. The influence of placental metabolism on fatty acid transfer to the fetus. J Lipid Res. 2017;58(2):443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dubé E, Gravel A, Martin C, et al. Modulation of fatty acid transport and metabolism by maternal obesity in the human full-term placenta. Biol Reprod. 2012;87(1):14, 1-1. [DOI] [PubMed] [Google Scholar]
- 58. Brass E, Hanson E, O’Tierney-Ginn PF. Placental oleic acid uptake is lower in male offspring of obese women. Placenta. 2013;34(6):503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. O’Tierney-Ginn PF, Gillingham M, Fowler J, Brass E, Marshall NE, Thornburg KL. Maternal weight gain regulates omega-3 fatty acids in male, not female, neonates. Reprod Sci. 2017;24(4):560-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Phuthong S, Reyes-Hernandez CG, Rodríguez-Rodríguez P, et al. Sex differences in placental protein expression and efficiency in a rat model of fetal programming induced by maternal undernutrition. Int J Mol Sci. 2020;22(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Avallone R, Demers A, Rodrigue-Way A, et al. A growth hormone-releasing peptide that binds scavenger receptor CD36 and ghrelin receptor up-regulates sterol transporters and cholesterol efflux in macrophages through a peroxisome proliferator-activated receptor gamma-dependent pathway. Mol Endocrinol. 2006;20(12):3165-3178. [DOI] [PubMed] [Google Scholar]
- 62. Maréchal L, Laviolette M, Rodrigue-Way A, et al. The CD36-PPARγ pathway in metabolic disorders. Int J Mol Sci. 2018;19(5):1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ellis JM, Mentock SM, Depetrillo MA, et al. Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs fatty acid oxidation and induces cardiac hypertrophy. Mol Cell Biol. 2011;31(6):1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goldenberg JR, Wang X, Lewandowski ED. Acyl CoA synthetase-1 links facilitated long chain fatty acid uptake to intracellular metabolic trafficking differently in hearts of male versus female mice. J Mol Cell Cardiol. 2016;94:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lyons MR, Peterson LR, McGill JB, et al. Impact of sex on the heart’s metabolic and functional responses to diabetic therapies. Am J Physiol Heart Circ Physiol. 2013;305(11):H1584-H1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Peterson LR, Saeed IM, McGill JB, et al. Sex and type 2 diabetes: obesity-independent effects on left ventricular substrate metabolism and relaxation in humans. Obesity (Silver Spring). 2012;20(4):802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Djouadi F, Weinheimer CJ, Saffitz JE, et al. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J Clin Invest. 1998;102(6):1083-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sargsyan Y, Thoms S. Staying in healthy contact: how peroxisomes interact with other cell organelles. Trends Mol Med. 2020;26(2):201-214. [DOI] [PubMed] [Google Scholar]
- 69. Delahaye F, Wijetunga NA, Heo HJ, et al. Sexual dimorphism in epigenomic responses of stem cells to extreme fetal growth. Nat Commun. 2014;5:5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Muralimanoharan S, Li C, Nakayasu ES, et al. Sexual dimorphism in the fetal cardiac response to maternal nutrient restriction. J Mol Cell Cardiol. 2017;108:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wanders RJA, Vaz FM, Waterham HR, Ferdinandusse S. Fatty acid oxidation in peroxisomes: enzymology, metabolic crosstalk with other organelles and peroxisomal disorders. Adv Exp Med Biol. 2020;1299:55-70. [DOI] [PubMed] [Google Scholar]
- 72. Ferdinandusse S, Denis S, Hogenhout EM, et al. Clinical, biochemical, and mutational spectrum of peroxisomal acyl-coenzyme A oxidase deficiency. Hum Mutat. 2007;28(9):904-912. [DOI] [PubMed] [Google Scholar]
- 73. Morita A, Enokizono T, Ohto T, et al. Novel ACOX1 mutations in two siblings with peroxisomal acyl-CoA oxidase deficiency. Brain Dev. 2021;43(3):475-481. [DOI] [PubMed] [Google Scholar]
- 74. Liu X, Guo Y, Wang J, Gao L, Liu C. Label-free proteomics of the fetal pancreas identifies deficits in the peroxisome in rats with intrauterine growth restriction. Oxid Med Cell Longev. 2019;2019:1520753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Piquereau J, Ventura-Clapier R. Maturation of cardiac energy metabolism during perinatal development. Front Physiol. 2018;9:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mandò C, De Palma C, Stampalija T, et al. Placental mitochondrial content and function in intrauterine growth restriction and preeclampsia. Am J Physiol Endocrinol Metab. 2014;306(4):E404-E413. [DOI] [PubMed] [Google Scholar]
- 77. Novielli C, Mandò C, Tabano S, et al. Mitochondrial DNA content and methylation in fetal cord blood of pregnancies with placental insufficiency. Placenta. 2017;55:63-70. [DOI] [PubMed] [Google Scholar]
- 78. Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361-370. [DOI] [PubMed] [Google Scholar]
- 79. Chang JS, Jun HJ, Park M. Transcriptional coactivator NT-PGC-1α promotes gluconeogenic gene expression and enhances hepatic gluconeogenesis. Physiol Rep. 2016;4(20):e13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Arias-Reyes C, Losantos-Ramos K, Gonzales M, Furrer D, Soliz J. NADH-linked mitochondrial respiration in the developing mouse brain is sex-, age- and tissue-dependent. Respir Physiol Neurobiol. 2019;266:156-162. [DOI] [PubMed] [Google Scholar]
- 81. Zemskova M, Kurdyukov S, James J, McClain N, Rafikov R, Rafikova O. Sex-specific stress response and HMGB1 release in pulmonary endothelial cells. PLoS One. 2020;15(4):e0231267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Djordjevic J, Thomson E, Chowdhury SR, et al. Brain region- and sex-specific alterations in mitochondrial function and NF-κB signaling in the TgCRND8 mouse model of Alzheimer’s disease. Neuroscience. 2017;361:81-92. [DOI] [PubMed] [Google Scholar]
- 83. Schreiber SN, Emter R, Hock MB, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101(17):6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chouchani ET, Kazak L, Spiegelman BM. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 2019;29(1):27-37. [DOI] [PubMed] [Google Scholar]
- 85. Kukat A, Dogan SA, Edgar D, et al. Loss of UCP2 attenuates mitochondrial dysfunction without altering ROS production and uncoupling activity. PLoS Genet. 2014;10(6):e1004385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cadenas S. Mitochondrial uncoupling, ROS generation and cardioprotection. Biochim Biophys Acta Bioenerg. 2018;1859(9):940-950. [DOI] [PubMed] [Google Scholar]
- 87. Nicholls DG. Mitochondrial proton leaks and uncoupling proteins. Biochim Biophys Acta Bioenerg. 2021;1862(7):148428. [DOI] [PubMed] [Google Scholar]
- 88. Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44(6):968-975. [DOI] [PubMed] [Google Scholar]
- 89. Finck BN, Lehman JJ, Leone TC, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109(1): 121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rando G, Tan CK, Khaled N, et al. Glucocorticoid receptor-PPARα axis in fetal mouse liver prepares neonates for milk lipid catabolism. Elife. 2016;5:e11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bowman CE, Selen Alpergin ES, Cavagnini K, Smith DM, Scafidi S, Wolfgang MJ. Maternal lipid metabolism directs fetal liver programming following nutrient stress. Cell Rep. 2019;29(5):1299-1310.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available within this article and its supplementary materials.