Abstract
Context
Estradiol is the primary female sex hormone and plays an important role for skeletal health in both sexes. Several enzymes are involved in estradiol metabolism, but few genome-wide association studies (GWAS) have been performed to characterize the genetic contribution to variation in estrogen levels.
Objective
Identify genetic loci affecting estradiol levels and estimate causal effect of estradiol on bone mineral density (BMD).
Design
We performed GWAS for estradiol in males (n = 147 690) and females (n = 163 985) from UK Biobank. Estradiol was analyzed as a binary phenotype above/below detection limit (175 pmol/L). We further estimated the causal effect of estradiol on BMD using Mendelian randomization.
Results
We identified 14 independent loci associated (P < 5 × 10−8) with estradiol levels in males, of which 1 (CYP3A7) was genome-wide and 7 nominally (P < 0.05) significant in females. In addition, 1 female-specific locus was identified. Most loci contain functionally relevant genes that have not been discussed in relation to estradiol levels in previous GWAS (eg, SRD5A2, which encodes a steroid 5-alpha reductase that is involved in processing androgens, and UGT3A1 and UGT2B7, which encode enzymes likely to be involved in estradiol elimination). The allele that tags the O blood group at the ABO locus was associated with higher estradiol levels. We identified a causal effect of high estradiol levels on increased BMD in both males (P = 1.58 × 10−11) and females (P = 7.48 × 10−6).
Conclusion
Our findings further support the importance of the body’s own estrogen to maintain skeletal health in males and in females.
Keywords: GWAS, estrogen, bone mineral density, mendelian randomization
Estrogen, the primary female sex hormone, is responsible for the development of the female reproductive system and secondary sex characteristics. Estrogen acts primarily as a growth hormone for the reproductive organs and regulates the menstrual cycle in women (1). Additionally, it plays a critical role in male sexual function (2). There are 3 major forms of estrogen: estrone, estradiol, and estriol. Estradiol is the major and most potent form (3). In females, high estrogen levels have been associated with deep vein thrombosis (4) and increased risk for certain cancers, including breast cancer (5) in women. In males, higher levels have been suggested to be associated with reduced risk of type 2 diabetes (T2D) (6). Estrogen is also an important regulator of bone metabolism in both females and males (7). When estrogen levels drop after menopause, the cells that synthesize bone (osteoblasts) are unable to effectively produce bone mass (8). The loss of ovarian estrogens after menopause has therefore, not surprisingly, been associated with reduced bone mineral density (BMD) and higher risk of osteoporosis (9,10).
Previously genome-wide association studies (GWAS) for estrogen have been performed in cohorts of up to 11 000 individuals, mostly of European descent, both male and female and stratified by sex (11-15). In addition, a GWAS was recently performed for sex hormone levels in UK Biobank (UKB), a study that mainly focused on testosterone (16). They identified strong sex-specific genetic effects on serum testosterone levels but did not consider the results from estradiol in women, due to the strong link between estradiol levels and menopausal status/age of menopause.
In addition to GWAS, previous studies have also addressed the causal effect of hormones in relation to disease risk using Mendelian Randomization (MR) (15-17). MR is an approach inspired by randomized controlled studies that uses genetic variants as instrumental variables (IVs) to estimate a causal relationship between an exposure, estradiol in our case, and an outcome (eg, BMD). The underlying reasoning is that if an exposure is directly causing an increased risk of a disease, then the genetic factors increasing the exposure should also indirectly affect disease risk through their effects on the exposure. However, this method requires that some key assumptions are met: (1) the selected IVs are associated with the exposure, (2) the IVs affect the outcome through the exposure, only, and (3) the IVs are not associated with any known or unknown confounders. Because genotypes are assigned at conception, MR can be used to overcome some of the limitations of observational studies, such as reverse causation, where the (alleged) outcome affects the exposure.
For testosterone, many independent genetic variants have been identified and included in previous MR analyses. They concluded that testosterone increases the risk for polycystic ovary syndrome and T2D in women but decreases the risk for T2D in men (16). Even though the number of GWAS significant findings for estradiol has been quite limited in previous studies (11-15), 2 recent MR studies have been performed, using 5 and 2 single-nucleotide polymorphisms (SNPs), respectively (15,17). Both studies identified a beneficial effect of estradiol on BMD in males (15,17). However, no such study has yet been performed in females due to those previous studies have failed to identify genetic variants to be valid instruments for MR analyses.
By using the largest cohort with estradiol levels measured, including almost half a million participants, we have been able to further highlight novel genes, important for regulating estradiol levels. We have stratified the cohort by sex and menopausal status to enable a comparison of genetic effects across strata. Thanks to the larger sample size in UKB compared to previous studies, we were able to identify enough genetic variants associated with estradiol levels to permit causal effect estimation of estradiol on BMD through MR analysis in females, which has not been done previously.
Material and Methods
Sample
UKB is a cross sectional cohort of about 500 000 participants, born between 1939 and 1970 who were recruited between 2006 and 2010. Detailed information was collected on lifestyle and anthropometric traits and on female specific factors that are likely to effect hormone levels, including use of birth control pills, hormone replacement treatments, pregnancies, and menopause. Participants also provided blood samples that were used to genotype approximately 800 000 single nucleotide polymorphisms (SNPs) across the genome and to measure levels of estradiol. The application to use data from UKB has been approved (project number 41143). UKB has an ethics permit from the National Research Ethics Committee (REC reference 11/NW/0382). Phenotype data for this project were extracted from the UKB database in April 2019. All analyses performed in this study are based on the samples and information collected at the participants initial visit to the assessment center. Ethical approval for the analyses performed in this study has also been approved by the Swedish Ethical Review Authority (Dnr: 2020-04415)
Genotype Data
A total of 438 417 participants had been genotyped using the UKB Axiom array, and another 49 994 participants had been genotyped using the similar (95% common marker content) UK BiLEVE array. SNPs had been imputed using UK10K and 1000 genomes phase 3, as reference panels (18,19). For this project we used imputed data from the third release, which contained a total of 93 093 070 SNPs. We only analyzed SNPs with minor allele frequency > 0.01 and removed SNPs deviating from Hardy-Weinberg (P-value < 1 × 10−20) and with markers with more than 5% missing genotype data and/or imputation quality <0.3. After quality control (QC), a total of 7 651 231 autosomal SNPs and 220 486 SNPs on the X chromosome remained. We included Caucasian participants clustering with regards to their genetic principal components. We also excluded first- and second-degree relatives (genetic relationship >0.044) using kinship data. Samples with sex discordance, high heterozygosity/missingness, and/or more than 5% missing genotypes were also excluded. After QC and exclusion, 361 975 unrelated Caucasian participants remained (167 168 males and 194 807 females).
Estradiol Measurements
We only included measurements from the blood samples that were collected during the participants initial visit to the assessment center. Estradiol had been measured by 2-step competitive analysis on a Beckman Coulter Unicel Dxl 800. This assay had a lower detection limit of 175 pmol/L. Normal ranges of serum estradiol levels are known to be 36.7 to 183.6 pmol/L in men, 73.4 to 2753.5 pmol/L in premenopausal women, and 0 to 73.4 pmol/L in postmenopausal women (20). This results in a large fraction of the participants, especially men and postmenopausal women, having levels below detection limit. Among the UKB participants, 76 668 had estradiol measurements above detection limit. Estradiol levels were therefore analyzed as a binary phenotype (above/below detection limit) in our primary analyses, in contrast to a previous GWAS for estradiol, which excluded all individuals below detection limit. However, we also performed sensitivity analyses using the Tobit model where quantitative measures (for those above detection limit) are modeled together with the below-detection-limit samples.
GWAS and Sensitivity Analyses
GWAS was performed in males and females separately, using logistic regression with an additive genetic model implemented in PLINK version 2. The following covariates were included in both male and female analyses: age, body mass index, the first 10 genetic principal components (to adjust for population structures and ethnic origins), and a binary indicator variable for UKB Axiom vs UK BiLEVE genotyping array to adjust for any array differences. For females, hormone replacement therapy (current, ever, or never), oral contraceptive use (current, ever, or never), number of live births, menopausal status, and whether they have undergone hysterectomy were also included. Women who were unsure about menopausal status were excluded. Information for the covariates were collected from the initial visit to the assessment center, which was the same time as the blood sample for the estradiol measurements were taken.
In the GWAS, we applied a P-value threshold of 5 × 10−8, which is commonly used in GWAS when using a minor allele frequency cutoff of 1% (21) and corresponds to a correction of 1 million independent tests (SNPs). Manhattan plots were generated using the R package hudson. We applied the clump function in PLINK 1.90 to define start and end positions for each locus with the main parameters that determines the clumping being set to R2=0.1, p1 = 5 × 10−8 and p2 = 1 × 10−4. We tested all lead SNPs for significant differences in effect sizes between males and females by including an interaction term in the logistic regression model employed by PLINK.
We subsequently performed conditional analyses for each locus, conditioning on the SNP with the lowest P-value from each locus. If there was any significant conditional SNP, this was repeated by also conditioning on the conditional SNP from the previous analyses until no additional significant associations remained.
Several sensitivity analyses were performed for the GWAS. First, we stratified females into being pre- and postmenopausal. Second, we excluded all participants with a cancer diagnosis prior to assessment (when blood samples were taken). Third, we analyzed males and females with testosterone and sex hormone-binding globulin (SHBG) levels included as covariates, in addition to the other covariates, and with age at menopause as a covariate in the female postmenopausal stratum.
In addition, we replicated our results by estimating the effects of our lead SNPs on quantitative estradiol levels. To properly model estradiol levels as a quantitative response with a lower detection limit, we performed censored regression (Tobit-I) modeling (22). By adopting a model with an integrated error term up to the detection limit (175 pmol/L), censored individuals with estradiol levels below the limit could also be included in the estimation. We used the vglm function from the R package VGAM 1.1-2, with the family argument set to “tobit.” Prior to analysis, we transformed estradiol values using the rank-based inverse normal transformation for females and males separately. The corresponding transformed lower detection limit was then given by the maximal (transformed) value for each subset of individuals (males and females) with estradiol levels below 175 pmol/L. The estimated beta coefficient is that of the corresponding latent variable and is here interpreted as the usual additive SNP effect on estradiol level, as if estradiol levels were measured in all individuals. Because it has been recently highlighted that dichotomization of the exposure variable can lead to the IV assumptions in the MR being violated (23), the effect estimates we obtained from this analysis were used for sensitivity MR analyses for males and females separately, as described for the primary analyses in the following discussion.
Annotation of GWAS Lead SNPs
To identify likely target genes for the associated variants, we first identified genes containing the lead SNPs or the closest gene(s) for each individual lead SNP. We also used HaploReg v4.1(24) to identify potential functional effects by the individual lead SNP or any variant in linkage disequilibrium (LD; R2 > 0.8) with the lead SNP. We then checked for overlap between the lead SNPs and expression quantitative trait loci (eQTL) using data from the Genotype-Tissue Expression (GTEx) project (25). We considered our lead SNP to overlap with an eQTL if our lead SNP was in LD (R2 > 0.8) with a lead SNP for an eQTL. To identify overlap with other phenotypes from previously published GWAS and address possible issues with pleiotropy in our MR analyses, we downloaded summary statistics from the GWAS catalog (version 1.0.3 downloaded on November 13, 2020). We defined signals to be overlapping if a lead SNP from a previous GWAS was in LD (R2 > 0.8) with any of our lead SNPs.
Mendelian Randomization
First, we undertook a 1-sample MR approach to investigate the causal effect of estradiol on BMD in UKB in males and females separately. BMD was measured as heel BMD, based on an ultrasound measurement of the calcaneus (heel bone) using the Sahara Clinical Bone Sonometer and automatically converted to T-scores (data field 78 in UKB). The T-score is the number of standard deviations a person’s BMD differs from the mean BMD of their respective sex.
The main MR analyses were performed with the R package gsmr (version 1.0.8) (26). The generalized summary data-based MR (GSMR) approach allows both for heteroskedastic and correlated data via the variance-covariance matrix and identifies and removes pleiotropic outliers prior to causal effect estimation. The default LD threshold was used (R2 = 0.05) and the significance threshold for an SNP being identified as an outlier and therefore not belonging to the set of valid instruments was set to the default value of a = 0.01. GSMR assumes a fixed effects model with the test statistic following a distribution with 1 degree of freedom.
We further performed sensitivity analyses, applying inverse variance weighted, weighted median, and the MR-Egger method, using the TwoSampleMR package implemented in R (27). The MR-Egger intercept was used to assess the possible presence of directional pleiotropy in the set of genetic instruments, where a deviation from zero intercept indicates pleiotropy (28). Both the inverse variance weighted and the MR-Egger methods assume a random effects model, with underdispersion not being allowed. The standard error of the estimate for the weighted median method is estimated using bootstrapping. If heterogeneity is present in the data, these methods would tend to show higher P-values as compared GSMR, as GSMR is expected to remove outliers. To get a reasonable number of instruments to include in the MR analysis for females, we used a less strict P-value cutoff (P < 1 × 10−7), resulting in a total of 4 IVs that were included in the analysis [see Supplementary Table S3 in (29)]. In the GSMR analyses, the HEIDI (heterogeneity in dependent instrument) outlier flag was disabled in females, as there were fewer (<10) SNPs than recommended for this approach to work properly (26).
A 1-sample MR could be sensitive to weak-instrument bias, which could potentially result in bias toward the confounded association (ie, an inflated MR estimate). We therefore also performed a 2-sample MR, which is not affected by weak-instrument bias to further replicate the MR results. For the 2-sample MR, we used GWAS summary statistics for BMD from the GEFOS consortium (30). We used GWAS summary statistics for lumbar spine BMD from the 2012 data release on www.gefos.org (downloaded November 11, 2020), which was the only data set with summary statistics for males and females separately and that did not include samples from UKB. The 2 sets (for males and females) of genetic variants that had been selected in UKB were tested independently using GEFOS BMD summary statistics for males and females, respectively. The overlap, with regards to SNPs (IVs), was not perfect between UKB and GEFOS. We therefore revised the set of SNPs for the MR in GEFOS slightly. For each of our GWAS locus, we selected the most significant SNP in LD with our lead SNP (R2 > 0.6) for which summary statistics were also available in the GEFOS data as a proxy. However, to avoid too weak instruments in the MR, only proxies that were associated with estradiol levels (P < 0.0001) were included in the MR. We harmonized the SNP effects using the harmonize_data function included in the TwoSampleMR package. Finally, we note that any winner’s curse bias in our effect estimates of the genetic variants associated with estradiol levels should, on average, drive the estimates of the causal effect of estrogen on BMD towards the null.
Results
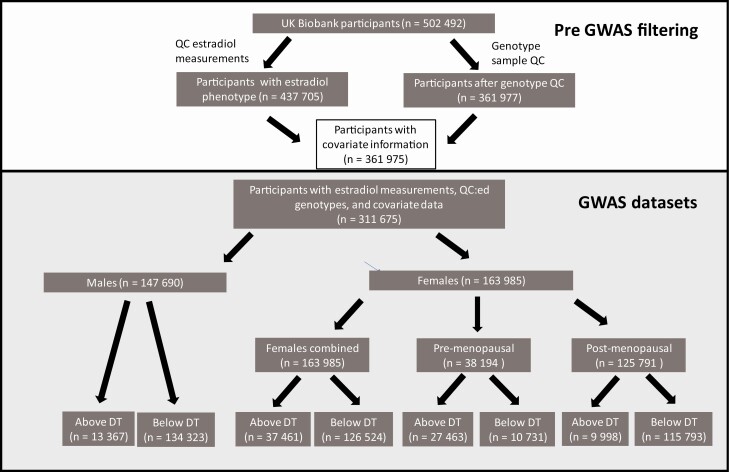
A total of 147 690 male participants had passed our genotype and estradiol QC (Fig. 1; Table 1). Of these, 134 323 had estradiol levels below detection limit (175 pmol/L), while 13 367 individuals had measured estradiol levels above or equal to 175 pmol/L (Table 1). The number of females (n = 163 985) was slightly higher compared to males, with 37 461 and 126 524 having estradiol levels above and below detection limit, respectively. Dramatically smaller fractions of males (9.1%) and postmenopausal women (7.9%) had detectable estrogen levels (Table 1) in comparison to the fraction of premenopausal women (71.9%) with detectable estrogen levels. Number of live births, age, and body mass index were significantly lower among women with detectable estradiol in both post and premenopausal strata (P < 0.05).
Figure 1.
Flowchart of UK Biobank participants included in the four GWAS.
Abbreviations: DT, detection limit (for estradiol measurement); QC, quality control.
Table 1.
Baseline characteristics in the UKB participants included in GWAS
| Males | Postmenopausal females | Premenopausal females | ||||
|---|---|---|---|---|---|---|
| Above DT | Below DT | Above DT | Below DT | Above DT | Below DT | |
| Participants, n (%)a | 13 499 (9.1) | 135 200 (90.1) | 10 090 (7.9) | 117 158 (92.1) | 27 672 (71.9) | 10 807 (28.1) |
| Participants with all covariates, na | 13 367 | 134 323 | 9998 | 115 793 | 27 463 | 10 731 |
| Estradiol levels (pmol/L), mean (Q1-Q3) | 203.8 (188.8-230.8) | NA | 302.2 (221.2-475.2) | NA | 433.7 (291.7-686.5) | NA |
| BMI, mean (Q1-Q3)b | 28.4 (25.2-30.8) | 27.8 (25.0-30.0) | 27.5 (23.6-30.2) | 27.2 (23.7-29.8) | 26.2 (22.7-28.7) | 26.6 (22.9-29.2) |
| Age, mean (Q1-Q3)b | 57.1 (51-64) | 57.1 (51-64) | 55.0 (50-60) | 60.6 (57-65) | 45.9 (43-48) | 47.5 (44-50) |
| Smoking, n (%) | ||||||
| Current | 1179 (8.73) | 11 424 (8.45) | 784 (7.77) | 7445 (6.35) | 1862 (6.73) | 809 (7.49) |
| Never | 6662 (49.3) | 65 993 (48.81) | 5635 (55.85) | 67537 (57.65) | 17 920 (64.76) | 6909 (63.93) |
| Occasional | 423 (3.1) | 4424 (3.27) | 243 (2.41) | 1884 (1.61) | 755 (2.73) | 281 (2.60) |
| Previous | 5175 (38.3) | 52 905 (39.13) | 3399 (33.69) | 39 842 (34.01) | 7083 (25.60) | 2788 (25.80) |
| Had hysterectomy, n (%)c | NA | NA | 4868 (48.25) | 27 841 (23.76) | 79 (0.29) | 76 (0.70) |
| Hormone replacement therapy (never/current/previous), nb | NA | NA | 3474/4586/2024 | 58 112/4606/54 169 | 26 797/403/424 | 10 166/157/463 |
| Oral contraceptives (never/current/previous), nb | NA | NA | 1252/28/8798 | 25 005/87/91 874 | 2442/1090/24 098 | 883/1476/8430 |
| Number of live births, median (Q1-Q3)c | NA | NA | 1.79 (1-2) | 1.9 (1-3) | 1.58 (0-2) | 1.56 (0-2) |
| Age at menarche, median (Q1-Q3)b | NA | NA | 12.62 (12-14) | 12.54 (12-14) | 12.68 (12-14) | 12.59 (12-14) |
| Age at menopause, median (Q1-Q3)b | NA | NA | 44.62 (44-52) | 46.53 (46-53) | NA | NA |
Bold cells are significantly (P < 0.05) associated with binary estradiol phenotype.
Abbreviations: BMI, body mass index; DT, detection limit; NA, not applicable; Q1, 1st quartile; Q3, 3rd quartile.
a These are the actual number of participants analyzed in the GWAS.
b Tested with Student’s t test.
c Tested with Fisher’s exact test.
GWAS Results
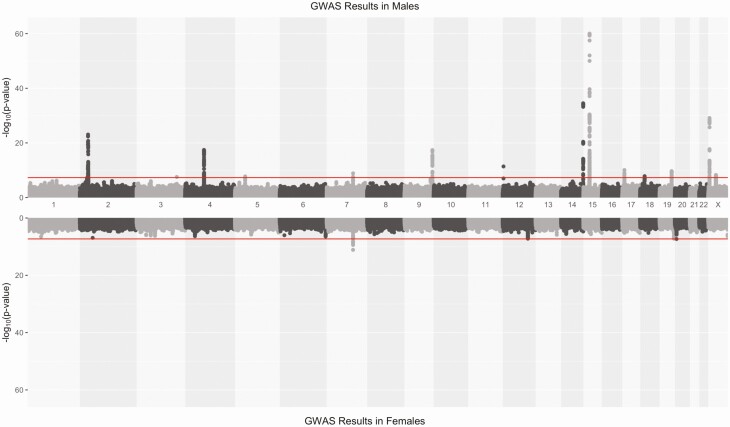
We identified 15 loci, on 14 different chromosomes, to be significantly associated (P < 5 × 10−8) with estradiol in males or females (Fig. 2; Table 2). Of these, 13 were genome-wide significant only in males, 1 only in females (MCM8) and 1 in both males and females (CYP3A7). On chromosome 2, and 15, one conditional SNP was identified per locus, located in the vicinity to the primary lead SNPs (Table 2). Among the lead SNPs identified in males, some were located in or close to genes with a well-known role in estradiol metabolism. Among the most significant loci, we found FAM9A and CYP19A1, which have previously been identified in GWAS for estradiol in males (12,15). One lead SNP, rs45446698, was genome-wide significant in both males and females, mapped to CYP3A7. It is also the most significant eQTL for CYP3A7 in adrenal gland in GTEx (P = 1.2 × 10−12), suggesting that the effect is mediated by the expression level of CYP3A7. The minor (G) allele increases the level of CYP3A7 expression but decreases the estradiol levels. There was also 1 lead SNP in SHBG, which codes for SHBG that had been linked to measurements of several other sex hormones (16). Another lead SNP was located on the X chromosome, within the AR gene that encodes an androgen receptor. We also found a very strong association on chromosome 2, that overlaps with SRD5A2, which codes for 3-oxo-5-alpha-steroid 4-dehydrogenase 2, known to catalyze the conversion of testosterone into androgen or dihydrotestosterone (31). Our most significant SNP is in LD (R2 = 0.75) with rs9282858, which is a missense variant that is predicted to be deleterious (SIFT = 0.03). Here, the minor allele was associated with higher estradiol levels (Table 2). At the same locus on chromosome 2, there was also 1 strong conditional signal, further upstream of the primary signal, closer to MEMO1.
Figure 2.
Hudson plot for the estradiol GWAS in males (upper) and females (lower). The red horizontal line indicated the genome-wide significance threshold (5 × 10−8).
Table 2.
Results of the GWAS including conditional SNPs
| Lead SNP | Chr:position | Primary/conditional | GWAS strata | Gene | Eff allele | MAF | Males | Females | ||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |||||||
| rs112881196a | 2:31982811 | Primary | Males | SRD5A2 | G | 0.039 | 1.353 (1.275-1.435) |
8.52E-24 | 1.106 (1.041-1.175)f |
1.13E-03 |
| rs62142080 | 2:32182528 | Conditional | Males | MEMO1/ SRD5A2 | C | 0.396 | 0.927 (0.903-0.952) |
1.09E-12 | 0.989 (0.965-1.013)f |
1.63E-01 |
| rs57102816 | 3:164506246 | Primary | Males | LINC01324 | TA | 0.257 | 1.084 (1.054-1.115) |
2.87E-08 | 1.008 (0.981-1.036)f |
5.66E-01 |
| rs7662029 | 4:69961912 | Primary | Males | UGT2B7 | G | 0.454 | 0.893 (0.871-0.916) |
3.61E-18 | 0.969 (0.946-0.992)f |
9.45E-03 |
| rs1073548 | 5:36012617 | Primary | Males | UGT3A1 | G | 0.126 | 1.113 (1.072-1.155) |
1.75E-08 | 1.052 (1.015-1.090) |
5.28E-03 |
| rs45446698b | 7:99332948 | Primary | Males/females | CYP3A7 | G | 0.043 | 0.811 (0.758-0.868) |
1.38E-09 | 0.813 (0.767-0.863) |
7.62E-12 |
| rs657152c | 9:136139265 | Primary | Males | ABO | A | 0.340 | 0.887 (0.863-0.911) |
4.14E-18 | 0.955 (0.932-0.980)f |
3.32E-04 |
| rs56196860d | 12:2908330 | Primary | Males | FKBP4 | A | 0.031 | 1.263 (1.183-1.350) |
4.11E-12 | 0.938 (0.877-1.004)f |
6.56E-02 |
| rs11160915 | 14:106514653 | Primary | Males | IGHV3-7 | A | 0.444 | 0.849 (0.827-0.871) |
3.08E-35 | 0.954 (0.931-0.977)f |
1.06E-04 |
| rs28892005 | 15:51519945 | Primary | Males | CYP19A1 | A | 0.350 | 0.795 (0.774-0.817) |
1.01E-60 | 0.962 (0.939-0.986)f |
2.27E-03 |
| rs3751591 | 15:51606710 | Conditional | Males | CYP19A1 | G | 0.167 | 1.125 (1.088-1.163) |
1.73E-10 | 1.004 (0.972-1.036)f |
8.96E-01 |
| rs62059839 | 17:7533015 | Primary | Males | SHBG | T | 0.262 | 1.098 (1.067-1.130) |
8.53E-11 | 1.044 (1.016-1.072) |
1.84E-03 |
| rs113047993 | 18:20585399 | Primary | Males | RBBP8 | T | 0.068 | 0.858 (0.813-0.904) |
1.47E-08 | 0.977 (0.932-1.024)f |
3.37E-01 |
| rs62129966 | 19:48374950 | Primary | Males | SULT2A1 | A | 0.164 | 1.113 (1.077-1.150) |
2.18E-10 | 1.012 (0.981-1.045)f |
4.56E-01 |
| rs16991615 | 20:5948227 | Primary | Females | MCM8 | A | 0.065 | 1.043 (0.992-1.097) |
1.01E-01 | 1.139 (1.087-1.193) |
4.67E-08 |
| rs5933688e | X:8880680 | Primary | Males | FAM9A | G | 0.275 | 1.118 (1.097-1.140) |
7.49E-30 | 1.013 (0.986-1.040)f |
3.48E-01 |
| rs114255570 | X:67005508 | Primary | Males | AR | A | 0.077 | 0.899 (0.867-0.931) |
5.60E-09 | 1.002 (0.959-1.047)f |
9.19E-01 |
a LD with rs9282858, which is missense variant in SRD5A2 (SIFT = 0.03 deleterious, PolyPhen2 = 0.337 benign).
Abbreviation: MAP, minor allele frequency.
b eQTL for CYP3A7.
c rs657152 tags the O blood group.
d rs56196860 is a missense variant in FKBP4.
e rs16991615 is a missense variant in MCM8.
f Significant (P < 0.05) difference in OR between males and females.
There was also an association to rs657152 in ABO, a SNP that is in LD (R2 = 0.98) with rs8176719, which tags the O blood group (32). The A allele of rs657152 tags the non-O blood groups, and we found rs657152-A to be associated with lower levels of estradiol, indicating that individuals with a non-O blood group have lower levels of estradiol compared to individuals with the O blood group (Table 2). Two associations on chromosome 4 and 5 respectively, mapped to UGT2B7 (rs7662029) and UGT3A1 (rs1073548). UGT3A1 and UGT2B7 are glucuronosyltransferases, which are enzymes that catalyze the glucosidation of lipophilic chemicals, facilitating their elimination (33). Another lead SNP mapped to SULT2A1, which encodes a sulfotransferase that is believed to be important for sex-steroid biosynthesis by sulfating hydroxylated steroid hormones and bile acids (34). Another lead SNP on chromosome 14 is a missense variant in FKBP4, which encodes the protein FKBP52 that has been suggested to be involved in the formation of steroid hormone nuclear receptors that regulate hormone levels (35).
The remaining lead SNPs from the GWAS in males were mapped to loci with less clear links to estradiol metabolism. These include 1 lead SNP (and 1 conditional SNP) in a locus with a cluster of genes, including IGHV3-7 and IGHV6-1, that encodes parts of immunoglobulin heavy chains that participates in the antigen recognition. Another lead SNP is located in RBBP8, which encodes Retinoblastoma-binding protein 8 (RBBP8, CtIP), an endonuclease involved in the repair of double-stranded DNA breaks through homologous recombination as well as regulation of G2/M cell-cycle checkpoints as part of the BRCA1-RBBP8 complex (36). Variation in RBBP8 genotype and regulation has been reported in multiple types of cancer (37,38). A third SNP was a missense variant in LINC01324, a long intervening/intergenic noncoding RNA, which has been established as a predictor for melanoma progression (39).
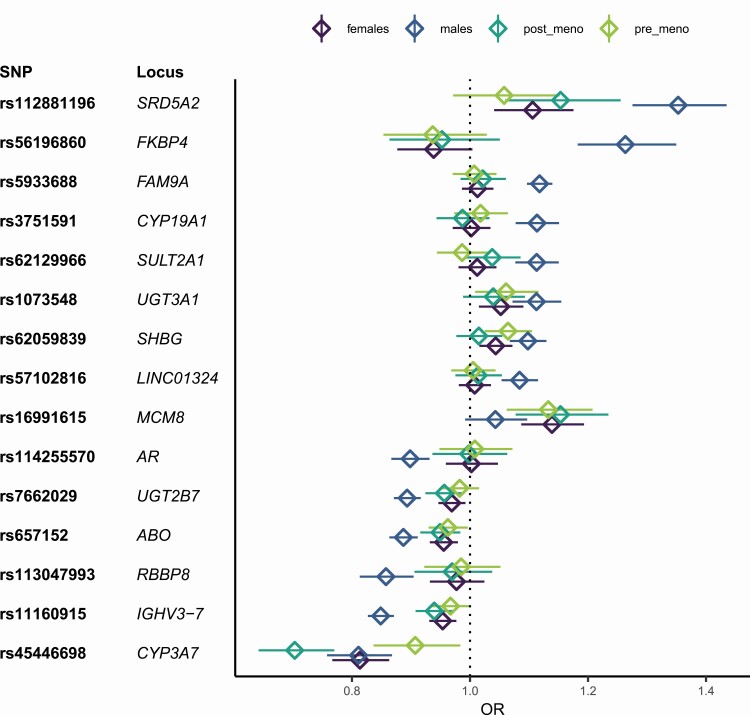
The lead SNP that was only significant in females (rs16991615) (Table 2) is a missense variant located in MCM8. MCM8 codes for a DNA helicase, which is involved in the repair of double-stranded DNA breaks and recombination (40). Even though very few genome-wide significant SNPs were identified in women, 8 out of 14 of the loci that were genome-wide significant in males were at least nominally associated in females as well (P < 0.05) with the same direction of effect (Fig. 3), especially the genes with functions related to hormone metabolism: UGT2B7, UGT3A1, CYP3A7, CYP19A1, SRD5A2, and SHBG replicated in females as well as IGHV3-7 and ABO. However, the effects of all SNPs, except for the SNP in CYP3A7, which achieved genome-wide significance in both sexes, were significantly different between males and females, as identified by significant sex-genotype interactions [see Supplementary Table 1 in (29)].
Figure 3.
ORs and 95% CIs for the lead SNPs in the male and female strata, as well as the 2 female subgroups: pre- and postmenopause. The lead SNP for each locus that reached genome-wide significance in males or females in the primary analyses are included. Non-overlapping confidence intervals typically indicate a significant difference in ORs well below P = 0.05.
Sensitivity Analysis
The primary aim of the sensitivity analyses was to further verify the associations for the lead SNPs from the primary GWAS. The effect size of the lead SNPs from the primary analyses changed somewhat. When stratifying females into pre- and postmenopause, there was 1 locus, CYP3A7, with a significant difference in the odds ratio (OR) between the 2 strata [Fig. 3; also see Supplementary Table 1 in (29)]. For CYP3A7, the largest effect was seen in postmenopausal women [OR = 0.703 (0.642-0.770)], followed by males [OR = 0.811 (0.758-0.868)] and premenopausal [OR = 0.907 (0.837-0.984)].
After removing all participants that had a prior cancer diagnosis at the time of assessment, there was no significant difference in OR compared to the primary GWAS, and all lead SNPs were still genome-wide significant, indicating that a prior cancer diagnosis did not influence our primary results [see Supplementary Table 1 in (29)]. However, when adjusting the estradiol GWAS for testosterone and SHBG levels, the lead SNPs at SHBG and FKBP4 were no longer genome-wide significant in males, and the corresponding effects were significantly lower than the previous signals. UGT3A1 and AR lost genome-wide significance for estradiol when adjusting for testosterone and SHBG levels, but the ORs were not significantly different from the primary analyses. This indicates that the signals at SHBG and FKBP4 might have been completely driven by SHBG and testosterone levels, whereas the other loci are more likely to be estradiol specific loci. We observed the same effect for the female-specific locus MCM8. There was no significant difference in OR between the primary GWAS in postmenopausal women with and without adjustment for age at menopause, suggesting that the results in females are not strongly confounded by age at menopause as has been discussed previously (16). In our quantitative analysis using the Tobit model [see Supplementary Table 1 in (29)], 13 of the lead SNPs were still genome-wide significant (P < 5 × 10−8), while 3 lead SNPs were just below the threshold (5.2 × 10−8, 5.7 × 10−8, and 5.9 × 10−8, respectively).
GWAS Overlap With Estradiol and Related Phenotypes
We did a systematic check for overlap with previous GWAS results that were available in the GWAS catalog [see Supplementary Table 2 in (29)]. For lead SNPs at UGT3A1, LINC01324, and RBBP8, we found no overlap with previous GWAS signals. Several SNPs at the AR locus have previously been associated with male pattern baldness (41), but none of those are in strong LD with our lead SNP (R2 < 0.3). The lead SNP at the other 10 loci all overlapped with at least 1 previous GWAS. There were several overlapping signals with phenotypes that are related to sex hormones. For example, the lead SNPs at SRD5A2 and FAM9A overlap with male pattern baldness as well. The lead SNPs at CYP3A7 and SULT2A1 overlap with GWAS hits for several sex hormones and other metabolites, UGT2B7 overlaps with serum and urinary metabolites, while SHBG overlaps with testosterone levels. Our lead SNPs in FAM9A, CYP19A1, and CYP3A7 all overlap with previous GWAS for BMD. Other loci overlap with GWAS for more distal traits. For example, the lead SNP in ABO overlaps with results from over 100 GWAS, with associations to metabolites, liver enzymes, and proteins as well as cardiovascular diseases, cancer, and T2D. The lead SNP at FKBP4 overlaps with waist-to-hip ratio and lung function. MCM8, the female specific locus, overlaps with traits related to age at menopause. This agrees partly with our sensitivity analyses where the OR decreased slightly from OR = 1.139 (1.087-1.193) in the primary GWAS to OR = 1.126 (1.035-1.225) when adjusted for age at menopause. Still, the OR was significantly different from unity in the sensitivity analyses, suggesting that the locus is probably associated with both age at menopause and estradiol levels.
Mendelian Randomization
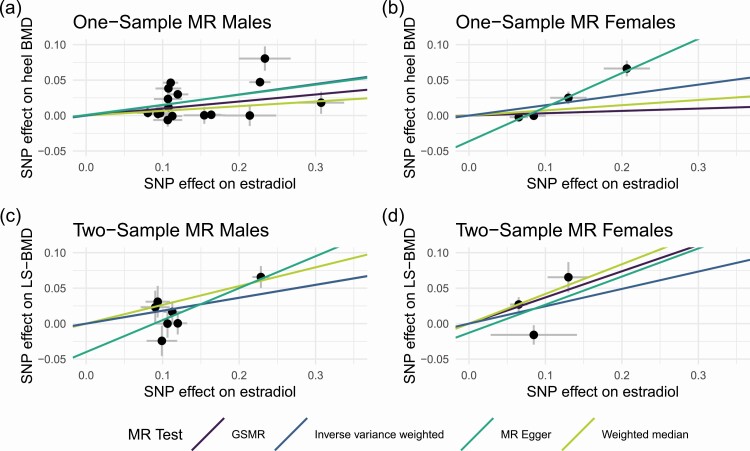
A total of 209 043 participants (96 432 males and 112 611 females) had BMD measurements and covariate information available and were included in the MR analysis. Mean value for T-scores in males was −0.085, and for females −0.58. The below-zero average T-scores agree with the average age in UKB participants being higher than the general population and thereby the BMD being lower compared to the mean BMD of respective sex. The instrument variables for the MR analyses were selected from the lead SNPs in the primary GWAS for males and females, respectively. For males, all significant lead SNPs and conditional SNPs were selected, resulting in 16 instruments (Table 3). The estimated effects on estradiol level [see Supplementary Table 3 in (29)] were taken from the primary GWAS for all instruments and for the SNPs that were identified in the conditional analyses. Hence, the estimates for the conditional SNPs differ slightly from the ones in Table 2. Using GSMR, 3 instruments were identified as pleiotropic and were excluded from the primary analysis in males [see Supplementary Table 3 in (29)]. We identified a causal effect of high estradiol levels on BMD in males (β ≈ 0.10, P = 1.57 × 10−11) (Table 3; Fig. 4A). Similar MR results were obtained using weighted median and MR-Egger, which do not remove pleiotropic SNPs. The MR-Egger intercept was not significantly different from zero, indicating that these results are not strongly influenced by directional pleiotropy.
Table 3.
Results of MR analyses
| Males (1-sample MR: UKB) | Males (2-sample MR:UKB-GEFOS) | Females (1-sample MR: UKB) | Females (2-sample MR:UKB-GEFOS) | |||||
|---|---|---|---|---|---|---|---|---|
| MR method | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value |
| GSMR | 9.89E-02 | 1.57E-11 | 1.82E-01 | 2.08E-04 | 3.27E-02 | 7.48E-06 | 3.69E-01 | 1.67E-03 |
| Weighted median | 6.61E-02 | 2.82E-02 | 2.65E-01 | 7.44E-05 | 7.31E-02 | 1.26E-01 | 4.19E-01 | 3.54E-03 |
| IVW | 1.48E-01 | 5.49E-05 | 1.82E-01 | 3.47E-03 | 1.45E-01 | 9.98E-02 | 2.45E-01 | 2.57E-01 |
| MR-Egger | 1.42E-01 | 2.23E-01 | 4.50E-01 | 2.73E-02 | 4.81E-01 | 2.56E-02 | 3.96E-01 | 7.92E-01 |
| MR-Egger intercept | 8.87E-04 | 6.95E-01 | −4.01E-02 | 1.07E-01 | −3.63E-02 | 1.71E-01 | −1.29E-02 | 9.15E-01 |
Figure 4.
Mendelian Randomization analyses to estimate the causal effect of estradiol on BMD. (A) One-sample MR in UKB males, (B) 1-sample MR in UKB females, (C) 2-sample MR with GEFOS males, and (D) 2-sample MR in GEFOS females. Abbreviation: LS, Lumbar Spine.
To get a reasonable number of instruments to be used for the MR analysis in females, we used a less strict P-value cutoff (P < 1 × 10−7), resulting in a total of 4 IVs [see Supplementary Table 3 in (29)] that were included in the analysis. We identified a causal effect of high estradiol levels on BMD also in women (β = 0.03, P = 7.48 × 10−6) (Fig. 4B). All 3 alternative MR methods showed the same direction of effect, although the null hypothesis of no effect could not be rejected (Table 3). The MR-Egger intercept was relatively large (−3.63 × 10−2), which is an indication of directional pleiotropy but was not significantly different from zero. We investigated each SNP for horizontal pleiotropy using information from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). We did not identify any variants that were in LD with our instruments and associated with correlated traits.
To further validate the MR results, we also performed a 2-sample MR for males and females, respectively, using the instruments selected for estradiol in UKB and the GWAS summary statistics from the BMD GWAS in GEFOS (30). A slightly different set of IVs had to be selected [see Supplementary Table 3 in (29)], since considerably fewer SNPs were analyzed in GEFOS due to their use of the HapMap reference panel for imputations. We included 8 and 4 instruments for the replication in males and females, respectively, none of which were palindromic. The MR estimates were in the same direction as the 1-sample MR (Fig. 4C and 4D), but with a larger effect in both males (β = 0.18, P = 0.00012) and females (β = 0.41, P = 3.49 × 10−5). Similarly to the 1-sample MR, neither MR-Egger intercept was significantly different from zero.
As estradiol was analyzed as a binary phenotype, the unit for the estimate from the MR can be interpreted as standard units increase in T-score associated with having estradiol levels above detection limit. However, our MR results were further replicated using SNP effects on the quantitative estradiol measurements from the Tobit model [see Supplementary Table 4 in (29)]. Here, the P-values were quite comparable to the MR for the binary estradiol exposure. However, the causal estimates are easier to interpreted and indicates that the BMD increases with 0.38 (95% CI 0.30-0.46) and 0.53 (95% CI 0.31-0.74) standard units per 1 standard unit increase in estradiol levels in males and females, respectively.
Discussion
We have presented results from a large GWAS for estradiol levels in males and females. Many of the significant SNPs are annotated to genes with functions that are biologically relevant to estrogen metabolism. For example, by converting and transporting androgens and endogens, synthesis or elimination of steroid hormones. In males, we identified 14 loci to be associated with serum estradiol levels, as well as 2 additional independent SNPs on chromosomes 2 and 15. In females, we identified 2 significant loci. One locus within the CYP3A7 gene overlaps with our findings in males, but the other locus, mapping to MCM8, was specific to females. However, a majority (n = 8) of the male loci, was at least nominally significant in females, although the estimated effect sizes were lower. These results suggest that genetic effects on estradiol levels might differ between males and females. However, in premenopausal women, estradiol levels vary during the menstrual cycle, limiting our ability to estimate the genetic effects on estradiol in females. In postmenopausal women, on the other hand, estradiol levels drop rapidly, and it has previously been suggested that the estradiol levels in females are more likely to be determined by the number of years since menopause (42). For those reasons, we performed sensitivity analyses, stratifying females by menopausal status and adjusting for age at menopause in the postmenopausal strata. The only locus that was significantly different between pre- and postmenopausal women was CYP3A7. The largest effect was seen in postmenopausal women, followed by males and premenopausal women. CYP3A7 encodes cytochrome P450 CYP3A7, which metabolizes dehydroepiandrosterone (DHEA), a precursor of both androgen and estrogen synthesis, and shows catalytic activity for conversion of estrone to hydroxyestrogens (43,44). As much as 75% of the circulating estrogens in premenopausal women and 50% of circulating androgens in men are known to be derived from DHEA. After menopause, DHEA is thought to be the major precursor of androgens and estrogens in females (45). Interestingly, the effect of the CYP3A7 SNP disappeared when adjusting for SHBG and testosterone levels in females, but the effect was unaffected in males. This suggests that CYP3A7 might have different functions in males and in females, as well as in pre- and postmenopausal women. In postmenopausal women CYP3A7 appears to be regulating the amount of available testosterone as a precursor for estrogen.
Several other loci, with previous links to estrogen metabolism were also identified. SULT2A1 encodes a sulfotransferase, which is active on hydroxylated steroid hormones and bile acids (34). Sulfotransferase 2A1, encoded by SULT2A1, mediates the sulfation of a wide range of steroids and sterols, including DHEA, a precursor of estrogen and testosterone. SULT2A1 was only associated with estradiol in males, and this association was similar (slightly stronger) when adjusting for testosterone and SHBG levels. Two of the most well-known loci, which has previously also been associated with estradiol in males in GWAS, are FAM9A and CYP19A1(12,15). Aromatase, encoded by CYP19A1, is a key enzyme in the synthesis of estradiol and estrone from testosterone and androstenedione, respectively (46). The association to CYP19A1 was at least nominally significant, in both the pre- and postmenopausal women, as well as in all sensitivity analyses. The P-values were even numerically slightly lower when adjusting for SHBG and testosterone, which agrees with the function of the protein. FAM9A is exclusively expressed in testis (47), and it is therefore not surprising that there was no association to FAM9A in females. Previously, FAM9A has also been associated with testosterone levels (15), and our analyses indicate that the association was partly driven by associations to testosterone or SHBG levels. We could therefore conclude that the SNP at FAM9A is likely to affect both testosterone and estrogen metabolism. This signal also overlaps with a GWAS signal for male pattern baldness (41), which further supports the importance of this locus. However, FAM9A encodes the protein FAM9A, which has a rather unknown function. Also, the SNP is located between FAM9A and FAM9B, and the annotation of this locus should therefore be interpreted with care.
We identified a strong association between estradiol levels and the SHBG locus. The SHBG gene encodes SHBG that has been linked to measurements of several other sex hormones previously (16). SHBG is a glycoprotein that binds to sex hormones, including testosterone and estrogen, in the bloodstream and thereby regulates the amount of bioavailable hormones (48). Only 1% to 2% of sex hormones are unbound and therefore bioavailable (49-51). The association to SHBG in males completely disappeared after adjusting for SHBG levels. In females, there was a nominally significant association to SHBG in the primary GWAS with the same direction of effect as in males. Interestingly, after adjusting for SHBG and testosterone levels, there was still an association (P = 0.00033), but the effect was in the opposite direction. Similarly, an opposite effect between males and females has previously been seen for total testosterone, where there is a negative genetic correlation between total testosterone and SHBG in females but a positive correlation in males (16). This clearly illustrates the heterogenous effects between males and females for sex hormones. We also identified 2 associations to the X chromosome in males, 1 mapping to the AR gene, which encodes the androgen receptor. In mammals, the action of androgens acts through this single androgen receptor. In different tissues, the androgen receptor is activated by testosterone, or testosterone is first converted into dihydrotestosterone, which is then the activating molecule (52). Therefore, the androgen receptor plays an important role in sex differentiation, mainly through regulating gene expression (53). Interestingly no association (P > 0.05) was seen in females, which might be due to the much lower levels of androgens in females. We also found an association to SRD5A2 which encodes 3-oxo-5-ɑ-steroid 4-dehydrogenase 2 and is located on chromosome 2. This enzyme catalyzes the conversion of testosterone to 5-ɑ-dihydrogentestosterone (31). Thus, the variant in SRD5A2 might influence estradiol levels in men by affecting the amount of testosterone available for conversion to estradiol. The SNP in SRD5A2 is nominally significant in females, an association that is more pronounced in postmenopausal women. There is also a conditional signal in males, 200 kb upstream, close to MEMO1, that encodes the mediator of ErbB2-driven cell motility 1 (MEMO1). In previous studies, it has been shown that MEMO1 regulates the extranuclear function of the estradiol receptor (54), which is a possible link to estradiol levels. It is therefore possible that the GWAS signal on chromosome 2 indeed represents associations with 2 different genes. Another interesting association is with FKBP4, which encodes FKBP52 (FK506-binding protein 4). FKBP52 has been shown to regulate the androgen, glucocorticoid, and progesterone receptor signaling pathways (35) by playing an important role in the formation of steroid hormone nuclear receptors (55). FKBP4 has been shown to regulate androgen receptor transactivation activity and has also been linked to testosterone levels (56). When adjusting for testosterone and SHBG levels, the FKBP4 association was weaker and no longer significant.
Two loci, mapping to UGT2B7 and UGT3A1, were also identified. These 2 loci encode uridine diphosphate (UDP)-glycosyltransferases, which are responsible for the addition of sugars to a broad range of lipophilic molecules to improve the water solubility and thereby play a major role in elimination of potentially toxic xenobiotics and endogenous compounds. Both UGT3A1 and UGT2B7 have been shown to metabolize estrogens by glucuronidation (ie, the transferring of glucuronic acid to the metabolites) (33,57,58). This suggests that UDP-glycosyltransferases might play a major role in estradiol metabolisms. Also, ABO encodes a glycosyltransferase that is known to modify carbohydrates on the red blood cell antigens. ABO is the gene that is responsible for the ABO blood groups (32). The effect allele of our study (the A allele) is in LD with the reference allele of rs8176719 (G), whose alternative allele (a deletion) encodes the O blood group. This indicates that men with a non-O blood type have lower estrogen levels or the other way around—men with blood group O have higher levels. Since ABO and its histoblood group properties have been linked to a vast number of diseases and functions, it is not unlikely that our finding is indicative of an association between ABO antigens and estradiol levels. The most well-known association for ABO is that individuals with the O blood group have lower risk of cardiovascular diseases (59), probably due to lower coagulation activity. There are very limited findings of a clear link between ABO and estrogen levels. However, 1 study found an interaction between ABO and hormone replacement therapy in relation to coagulation activity (47). ABO antigens are present on most epithelial and endothelial cells as well as on T cells, B cells, and platelets and are also detectable in most body fluids such as saliva (60). Recently, ABO was linked to coronavirus disease 2019, where the susceptibility to severe acute respiratory syndrome coronavirus 2 has been suggested to be explained by modulation of sialic acid-containing receptors distributed on host cell surface (61). Coronavirus disease 2019 has a higher mortality in males, which has been suggested to be linked to their lower estrogen levels (62,63). We also identified several loci with less clear links to estrogen metabolism including LINC01324, IGHV3-7, RBBP8, and MCM8. MCM8 is the female-specific locus, where the lead SNP is a missense variant. Genetic variants in MCM8 have previously been associated with premature ovarian failure (64,65), a condition that is characterized by low estrogen levels (55). According to GTEx, MCM8 is highly expressed in both female and male reproductive organs, despite only being significant in the female GWAS cohort, which further support this locus as being involved in sex hormone metabolism.
We performed MR analysis and estimated a causal effect of estradiol on BMD in males and, to our knowledge, for the first time also in females. Compared to previous studies in males, with not more than 5 SNPs as instruments for estradiol (17), we included as many as 16 SNPs in males and 4 in females. Interestingly, the effect estimate was higher for females than males in the 2-sample MR, which could agree with the fact that BMD decreases more rapidly in females by age. Estrogen can be used as a preventative treatment in postmenopausal women with low BMD (66,67) to prevent osteoporosis, and our results were therefore not surprising.
One of the major strengths of this study is the large cohort from UKB with estradiol measurements available. However, only a subset of the participants had estradiol levels above detection limit. To increase the number of participants to analyze, we created a binary phenotype. Since normal levels of estradiol in males and postmenopausal women have a range below or just above the detection limit, we were not able to capture the full spectrum of individual variation in the cohort. However, we also used a second approach, Tobit-I modeling, which takes into account the quantitative measures for samples above detection limit without disregarding the values below detection limit. While this is a well-established method and frequently used (eg, in econometrics), it has not been widely used in genetic association studies. Here we showed that this approach results in similar P-values compared to the binary estradiol variable but provides quantitative effect estimates that are much easier to interpret. Since very few GWAS have been performed for estradiol previously, we only found studies with effects estimates on the quantitative scale for 1 of our GWAS signals: rs28892005 in the CYP19A6 locus [see Supplementary Table 2B in (29)]. In previous studies, only SNPs in LD with our lead variants had been reported, but the absolute value of the effect sizes (β = 0.107, β = 0.065, and β = 0.062) were comparable to our Tobit model estimates for males (β = 0.106) (11,14,15). While many clinical assays have a lower detection limit, we believe that the censored-regression approach using Tobit modeling should be useful for other studies as well. Also, in MR studies, the second MR assumption can be violated by dichotomizing the quantitative phenotype (23). The second MR assumption states that the IVs should be associated with the outcome only through the exposure. However, a change in the underlying quantitative exposure may effect a change in the outcome, without detectable change in the dichotomized, thereby violating this assumption. By re-estimating the causal effects of estradiol on BMD, using the quantitative effect estimates from the Tobit model, we could ensure that a potential violation of the second MR assumption did not dramatically influence our results.
Very few loci were identified to be genome-wide significant in females, which resulted in a low number of IVs available for the MR. This could have been a cause of instability in our MR results, leading to nonsignificant results for some MR approaches. The selection of proxies, for which effect estimates were provided by GEFOS, did not ameliorate this problem. Future studies, where estradiol is measured in an even larger set of women or in women that are more synchronized with regards to timing in relation to menopause and/or ovulation, should be beneficial for identifying additional IVs for females and verifying our MR estimates. In addition, few instruments could also lead to lack of power to detect a significant deviation from zero for the MR-Egger intercept. In fact, we did observe the MR-Egger intercept to deviate from zero, even if it was not a significant deviation, which could possibly indicate that our analyses were affected by directional pleiotropy. However, in a 1-sample MR design, the MR-Egger regression, including the test for directional pleiotropy, has been found to be more strongly affected by weak instrument bias than, for example, the inverse variance weighted method, and it is therefore deemed inappropriate in this setting (68,69). However, through a comparison with the GWAS Catalog (https://www.ebi.ac.uk/gwas/), we did indeed find that many of our GWAS SNPs had previously been associated with different phenotypes. For example, age at menopause, a phenotype that also influences the risk of osteoporosis, is associated with our female-specific locus MCM8. This could potentially have led to pleiotropy (68).
In summary, we have identified biologically relevant genetic loci associated with variation in estradiol levels in males and females and found differences in genetic effects between sexes. In addition, we used an MR approach to show that estradiol levels have a causal effect on good bone health in both males and females, supporting the use of estrogen replacement therapy as a good treatment to prevent osteoporosis and fractures in elderly of both sexes. Our findings support previous research on the synthesis of estradiol and its effects on BMD and provide new insights into the genetic components of estrogen regulation and metabolism.
Acknowledgments
This research was conducted using the UKB Resource under application number 41143, following the restrictions on data availability set up by the UKB. We acknowledge all participants and staff involved in UKB for their valuable contribution. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project sens2017538.
Financial Support : The research was funded by the Swedish Research Council (Å.J. 2019-01497), the Swedish Brain Foundation (Å.J. FO2020-0205 and FO2019-0129), the Swedish Heart Lung Foundation (Å.J. 20200687), as well as the Wiberg (W.E.E.), M Borgström (W.E.E.), Hedströms, K och O F (W.E.E.), A and M Rudbergs (W.E.E.) foundations. The funding sources had no influence and took no part in the design or conduct of this research.
Author Contributions: W.E.E. and Å.J. designed the study; E.B., W.E.E., and D.S. performed the data analysis; D.S. and W.E.E. generated the figures; W.E.E., D.S., and Å.J. wrote the manuscript; W.E.E., T.K., D.S., E.B., Å.J., and J.H. interpreted the data, contributed to, and reviewed the manuscript.
Additional Information
Disclosure Summary: The authors declare no conflict of interests.
Data Availability
Some data generated or analyzed during this study are included in this published article or in the data repositories listed in references. Supplementary data are freely available on Zenodo (https://doi.org/10.5281/zenodo.4926701). GEFOS summary statistics are available for download from the GEFOS Consortium (http://www.gefos.org/). Summary statistics from other GWAS can be downloaded from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). Restrictions apply to the availability of some data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. Individual-level genetic and clinical data are available for bona-fide researchers from UK Biobank (https://www.ukbiobank.ac.uk/) upon filing an application.
References
- 1. Bates GW, Bowling M. Physiology of the female reproductive axis. Periodontol 2000. 2013;61(1):89-102. [DOI] [PubMed] [Google Scholar]
- 2. Hess RA, Bunick D, Lee KH, et al. A role for oestrogens in the male reproductive system. Nature. 1997;390(6659):509-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomas MP, Potter BV. The structural biology of oestrogen metabolism. J Steroid Biochem Mol Biol. 2013;137:27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosendaal FR, Van Hylckama Vlieg A, Tanis BC, Helmerhorst FM. Estrogens, progestogens and thrombosis. J Thromb Haemost. 2003;1(7):1371-1380. [DOI] [PubMed] [Google Scholar]
- 5. Cauley JA, Lucas FL, Kuller LH, Stone K, Browner W, Cummings SR;. Study of Osteoporotic Fractures Research Group. Elevated serum estradiol and testosterone concentrations are associated with a high risk for breast cancer. Ann Intern Med. 1999;130(4 Pt 1):270-277. [DOI] [PubMed] [Google Scholar]
- 6. Vikan T, Schirmer H, Njølstad I, Svartberg J. Low testosterone and sex hormone-binding globulin levels and high estradiol levels are independent predictors of type 2 diabetes in men. Eur J Endocrinol. 2010;162(4):747-754. [DOI] [PubMed] [Google Scholar]
- 7. Manolagas SC, O’Brien CA, Almeida M. The role of estrogen and androgen receptors in bone health and disease. Nat Rev Endocrinol. 2013;9(12):699-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Väänänen HK, Härkönen PL. Estrogen and bone metabolism. Maturitas. 1996;23(Suppl):S65-S69. [DOI] [PubMed] [Google Scholar]
- 9.Manson JE, Bassuk SS. Menopause and Postmenopausal Hormone Therapy. In: Jameson JL, Fauci AS, Kasper DL, Hauser SL, Longo DL, Loscalzo J, eds. Harrison’s Principles of Internal Medicine. 20th ed. New York: McGraw-Hill Education; 2018. [Google Scholar]
- 10. Riggs BL, Khosla S, Melton LJ 3rd. A unitary model for involutional osteoporosis: estrogen deficiency causes both type I and type II osteoporosis in postmenopausal women and contributes to bone loss in aging men. J Bone Miner Res. 1998;13(5):763-773. [DOI] [PubMed] [Google Scholar]
- 11. Pott J, Bae YJ, Horn K, et al. Genetic association study of eight steroid hormones and implications for sexual dimorphism of coronary artery disease. J Clin Endocrinol Metab. 2019;104(11):5008-5023. [DOI] [PubMed] [Google Scholar]
- 12. Chen Z, Tao S, Gao Y, et al. Genome-wide association study of sex hormones, gonadotropins and sex hormone-binding protein in Chinese men. J Med Genet. 2013;50(12):794-801. [DOI] [PubMed] [Google Scholar]
- 13. Liu M, Ingle JN, Fridley BL, et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol. 2013;27(4):657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prescott J, Thompson DJ, Kraft P, et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PloS One. 2012;7(6):e37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eriksson AL, Perry JRB, Coviello AD, et al. Genetic determinants of circulating estrogen levels and evidence of a causal effect of estradiol on bone density in men. J Clin Endocrinol Metab. 2018;103(3):991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruth KS, Day FR, Tyrrell J, et al. ; Endometrial Cancer Association Consortium . Using human genetics to understand the disease impacts of testosterone in men and women. Nat Med. 2020;26(2):252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nethander M, Vandenput L, Eriksson AL, Windahl S, Funck-Brentano T, Ohlsson C. Evidence of a causal effect of estradiol on fracture risk in men. J Clin Endocrinol Metab. 2019;104(2):433-442. [DOI] [PubMed] [Google Scholar]
- 18. Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the wellcome trust case control consortium phase 1 data. Eur J Hum Genet. 2012;20(7):801-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walter K, Min JL, Huang J, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamoto J, Salameh WA, Carlton E. Endocrine Testing. In: Jameson JL, De Groot LJBT, eds. Endocrinology. 6th ed. Philadelphia: Elsevier; 2010:2802-2834. [Google Scholar]
- 21. Xu C, Tachmazidou I, Walter K, Ciampi A, Zeggini E, Greenwood CM; UK10K Consortium . Estimating genome-wide significance for whole-genome sequencing studies. Genet Epidemiol. 2014;38(4):281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tobin J. Estimation of relationships for limited dependent variables. Econometrica. 1958;26(1):24-36. [Google Scholar]
- 23. Glymour MM, Tchetgen Tchetgen EJ, Robins JM. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am J Epidemiol. 2012;175(4):332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930-D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) project. Biopreserv Biobank. 2015;13(5):307-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9(1):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitz D, Ek WE, Berggren E, Höglund J, Karlsson T, Johansson Å. Supplementary data for: Genome-wide association study of estradiol levels, and the causal effect of estradiol on bone mineral density. Zenodo. 10.5281/zenodo.4706671, March 3, 2021, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makridakis NM, di Salle E, Reichardt JK. Biochemical and pharmacogenetic dissection of human steroid 5 alpha-reductase type II. Pharmacogenetics. 2000;10(5):407-413. [DOI] [PubMed] [Google Scholar]
- 32. Ogasawara K, Bannai M, Saitou N, et al. Extensive polymorphism of ABO blood group gene: three major lineages of the alleles for the common ABO phenotypes. Hum Genet. 1996;97(6):777-783. [DOI] [PubMed] [Google Scholar]
- 33. Mackenzie PI, Rogers A, Treloar J, Jorgensen BR, Miners JO, Meech R. Identification of UDP glycosyltransferase 3A1 as a UDP N-acetylglucosaminyltransferase. J Biol Chem. 2008;283(52):36205-36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie.” Drug Metab Dispos. 2009;37(11):2255-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sivils JC, Storer CL, Galigniana MD, Cox MB. Regulation of steroid hormone receptor function by the 52-kDa FK506-binding protein (FKBP52). Curr Opin Pharmacol. 2011;11(4):314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu X, Baer R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J Biol Chem. 2000;275(24):18541-18549. [DOI] [PubMed] [Google Scholar]
- 37. Wong AK, Ormonde PA, Pero R, et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 1998;17(18):2279-2285. [DOI] [PubMed] [Google Scholar]
- 38. Wu M, Soler DR, Abba MC, et al. CtIP silencing as a novel mechanism of tamoxifen resistance in breast cancer. Mol Cancer Res. 2007;5(12):1285-1295. [DOI] [PubMed] [Google Scholar]
- 39. Yang S, Xu J, Zeng X. A six-long non-coding RNA signature predicts prognosis in melanoma patients. Int J Oncol. 2018;52(4):1178-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee KY, Im JS, Shibata E, et al. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat Commun. 2015;6:7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hagenaars SP, Hill WD, Harris SE, et al. Genetic prediction of male pattern baldness. PloS Genet. 2017;13(2):e1006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richardson H, Ho V, Pasquet R, et al. ; MAP.3 Investigators . Baseline estrogen levels in postmenopausal women participating in the MAP.3 breast cancer chemoprevention trial. Menopause. 2020;27(6):693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee AJ, Conney AH, Zhu BT. Human cytochrome P450 3A7 has a distinct high catalytic activity for the 16alpha-hydroxylation of estrone but not 17beta-estradiol. Cancer Res. 2003;63(19):6532-6536. [PubMed] [Google Scholar]
- 44. Ohmori S, Nakasa H, Asanome K, et al. Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells. Biochim Biophys Acta. 1998;1380(3):297-304. [DOI] [PubMed] [Google Scholar]
- 45. Simpson ER, Davis SR. Minireview: aromatase and the regulation of estrogen biosynthesis—some new perspectives. Endocrinology. 2001;142(11):4589-4594. [DOI] [PubMed] [Google Scholar]
- 46. Sohl CD, Guengerich FP. Kinetic analysis of the three-step steroid aromatase reaction of human cytochrome P450 19A1. J Biol Chem. 2010;285(23):17734-17743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guimarães DA, dos Santos MS, Gomes KB, et al. Interaction between oral estrogen plus progestogen therapy and ABO blood groups on coagulation activation in postmenopausal women. Menopause. 2012;19(3):339-345. [DOI] [PubMed] [Google Scholar]
- 48. Hammond GL. Access of reproductive steroids to target tissues. Obstet Gynecol Clin North Am. 2002;29(3):411-423. [DOI] [PubMed] [Google Scholar]
- 49. Coviello AD, Haring R, Wellons M, et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. Plos Genet. 2012;8(7):e1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Somboonporn W, Davis SR; National Health and Medical Research Council . Testosterone effects on the breast: implications for testosterone therapy for women. Endocr Rev. 2004;25(3):374-388. [DOI] [PubMed] [Google Scholar]
- 51. Dunn JF, Nisula BC, Rodbard D. Transport of steroid hormones: binding of 21 endogenous steroids to both testosterone-binding globulin and corticosteroid-binding globulin in human plasma. J Clin Endocrinol Metab. 1981;53(1):58-68. [DOI] [PubMed] [Google Scholar]
- 52. Davison SL, Bell R. Androgen physiology. Semin Reprod Med. 2006;24(2):71-77. [DOI] [PubMed] [Google Scholar]
- 53. Mooradian AD, Morley JE, Korenman SG. Biological actions of androgens. Endocr Rev. 1987;8(1):1-28. [DOI] [PubMed] [Google Scholar]
- 54. Jiang K, Yang Z, Cheng L, et al. Mediator of ERBB2-driven cell motility (MEMO) promotes extranuclear estrogen receptor signaling involving the growth factor receptors IGF1R and ERBB2. J Biol Chem. 2013;288(34):24590-24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hou Q, Gorski J. Estrogen receptor and progesterone receptor genes are expressed differentially in mouse embryos during preimplantation development. Proc Natl Acad Sci U S A. 1993;90(20):9460-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen H, Yong W, Hinds TD Jr, et al. Fkbp52 regulates androgen receptor transactivation activity and male urethra morphogenesis. J Biol Chem. 2010;285(36):27776-27784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dong M, Owens IS, Sheen YY. Cloning and expression of human liver UDP-glucuronosyltransferase cDNA, UDPGTh2. Arch Pharm Res. 1997;20(5):459-464. [DOI] [PubMed] [Google Scholar]
- 58. Kallionpää RA, Järvinen E, Finel M. Glucuronidation of estrone and 16α-hydroxyestrone by human UGT enzymes: the key roles of UGT1A10 and UGT2B7. J Steroid Biochem Mol Biol. 2015;154:104-111. [DOI] [PubMed] [Google Scholar]
- 59. Zhang H, Mooney CJ, Reilly MP. ABO blood groups and cardiovascular diseases. Int J Vasc Med. 2012;2012:641917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schachter H, Michaels MA. A quantitative difference in the activity of blood group A-specific N-acetylgalactosaminyltransferase in serum from A 1 and A 2 human subjects. Biochem Biophys Res Commun. 1971;45(4):1011-1018. [DOI] [PubMed] [Google Scholar]
- 61. Silva-Filho JC, Melo CGF, Oliveira JL. The influence of ABO blood groups on COVID-19 susceptibility and severity: a molecular hypothesis based on carbohydrate-carbohydrate interactions. Med Hypotheses. 2020;144:110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Suba Z. Prevention and therapy of COVID-19 via exogenous estrogen treatment for both male and female patients. J Pharm Pharm Sci. 2020;23(1):75-85. [DOI] [PubMed] [Google Scholar]
- 63. Li Y, Jerkic M, Slutsky AS, Zhang H. Molecular mechanisms of sex bias differences in COVID-19 mortality. Crit Care. 2020;24(1):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. AlAsiri S, Basit S, Wood-Trageser MA, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest. 2015;125(1):258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dou X, Guo T, Li G, Zhou L, Qin Y, Chen ZJ. Minichromosome maintenance complex component 8 mutations cause primary ovarian insufficiency. Fertil Steril. 2016;106(6):1485-1489.e2. [DOI] [PubMed] [Google Scholar]
- 66. Lufkin EG, Wahner HW, O’Fallon WM, et al. Treatment of postmenopausal osteoporosis with transdermal estrogen. Ann Intern Med. 1992;117(1):1-9. [DOI] [PubMed] [Google Scholar]
- 67. Abdi F, Mobedi H, Bayat F, Mosaffa N, Dolatian M, Ramezani Tehrani F. The effects of transdermal estrogen delivery on bone mineral density in postmenopausal women: a meta-analysis. Iran J Pharm Res. 2017;16(1):380-389. [PMC free article] [PubMed] [Google Scholar]
- 68. Bowden J. Misconceptions on the use of MR-Egger regression and the evaluation of the InSIDE assumption. Int J Epidemiol. 2017;46(6):2097-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bowden J, Burgess S, Davey Smith G. Response to Hartwig and Davies. Int J Epidemiol. 2016;45(5):1679-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some data generated or analyzed during this study are included in this published article or in the data repositories listed in references. Supplementary data are freely available on Zenodo (https://doi.org/10.5281/zenodo.4926701). GEFOS summary statistics are available for download from the GEFOS Consortium (http://www.gefos.org/). Summary statistics from other GWAS can be downloaded from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). Restrictions apply to the availability of some data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. Individual-level genetic and clinical data are available for bona-fide researchers from UK Biobank (https://www.ukbiobank.ac.uk/) upon filing an application.