Abstract
Context
Anaplastic thyroid cancer (ATC) is a rare, aggressive, and deadly disease. Robust preclinical thyroid cancer models are needed to adequately develop and study novel therapeutic agents. Patient-derived xenograft (PDX) models may resemble patient tumors by recapitulating key genetic alterations and gene expression patterns, making them excellent preclinical models for drug response evaluation.
Objective
We developed distinct ATC PDX models concurrently with cell lines and characterized them in vitro and in vivo.
Methods
Fresh thyroid tumor from patients with a preoperative diagnosis of ATC was surgically collected and divided for concurrent cell line and PDX model development. Cell lines were created by generating single cells through enzymatic digestion. PDX models were developed following direct subcutaneous implantation of fresh tumor on the flank of immune compromised/athymic mice.
Results
Six ATC PDX models and 4 cell lines were developed with distinct genetic profiles. Mutational characterization showed one BRAF/TP53/CDKN2A, one BRAF/CDKN2A, one BRAF/TP53, one TP53 only, one TERT-promoter/HRAS, and one TERT-promoter/KRAS/TP53/NF2/NFE2L2 mutated phenotype. Hematoxylin-eosin staining comparing the PDX models to the original patient surgical specimens show remarkable resemblance, while immunohistochemistry stains for important biomarkers were in full concordance (cytokeratin, TTF-1, PAX8, BRAF). Short tandem repeats DNA fingerprinting analysis of all PDX models and cell lines showed strong concordance with the original tumor. PDX successful establishment rate was 32%.
Conclusion
We have developed and characterized 6 novel ATC PDX models with 4 matching cell lines. Each PDX model harbors a distinct genetic profile, making them excellent tools for preclinical therapeutic trials.
Keywords: anaplastic thyroid carcinoma, BRAF mutation, TP53 mutation, patient-derived xenograft
Anaplastic thyroid cancer (ATC), which represents less than 2% of all thyroid cancers, has an overall historic median survival of 4 months (1) and causes more than 50% of annual thyroid related mortality (2). Patients with ATC usually present with locoregionally invasive disease, while having metastatic lesions more than 50% of the time (2). This renders surgical management extremally difficult, with these patients historically treated with palliative approaches. As such, conducting translational studies have been limited by access to tumor specimens, while clinical trials are continuously challenged by the short length of survival of patients to adequately evaluate therapeutic options. Although we have recently reported significant improvement in ATC outcomes (3), clearly preclinical models remain the foundation for therapeutic advances.
Over the past 2 decades, several ATC cell lines have become available for research (4-6). Furthermore, several research groups have reported the genomic profile of ATC using whole-exome sequencing and next-generation sequencing (NGS) to shed light on the genomic landscape of ATC (7-9). With human cancer-derived cell lines established, using them in an in vitro and in vivo setting becomes critical to study the cancer’s molecular characteristics and rudimentary response to various therapeutic options. However, it is becoming apparent that using cell lines in an animal model setting, whether they are injected orthotopically or subcutaneously, leads to a significant persistent limitation due to the loss of tumor heterogeneity, tumor microenvironment, and inability to mirror patients’ responses to therapy. With ATC being a very diverse type of cancer, expanding models to mirror this heterogeneity is needed to improve translational studies in identifying novel targeted therapies.
Patient-derived xenograft (PDX) models have been shown to recreate human tumor biology, specifically in terms of gene expression, tumor heterogeneity (10), and drug responses, making them a robust in vivo preclinical model for new drug development. PDX models have been successfully established in many types of cancer, including lung (11, 12), breast (13), and head and neck (6, 14-16) cancers. In this study, we report the development and genetic characterization of 6 novel ATC PDX models and their associated cell lines, each having key genetic distinctions that are pathologically and clinically relevant to the development of novel ATC-targeted therapeutics.
Materials and Methods
This study was approved by the institutional review board at The University of Texas MD Anderson Cancer Center (MDACC).
Patient Demographics
Six patients (4 female and 2 male) with histopathologically confirmed diagnosis of ATC between October 2015 and March 2019 were included in this study. Mean age of diagnosis of ATC was 65 years, ranging from 51 to 75 years. None of the patients remain alive. Median follow-up time from diagnosis was 0.64 years (234 days), ranging from 0.05 to 1.43 years. Two patients included received BRAF-directed therapy prior to tumoral tissue collection. Five of 6 patients had NGS on their tumors ranging from 50 to 134 gene analyses.
Patient-Derived Xenograft Models
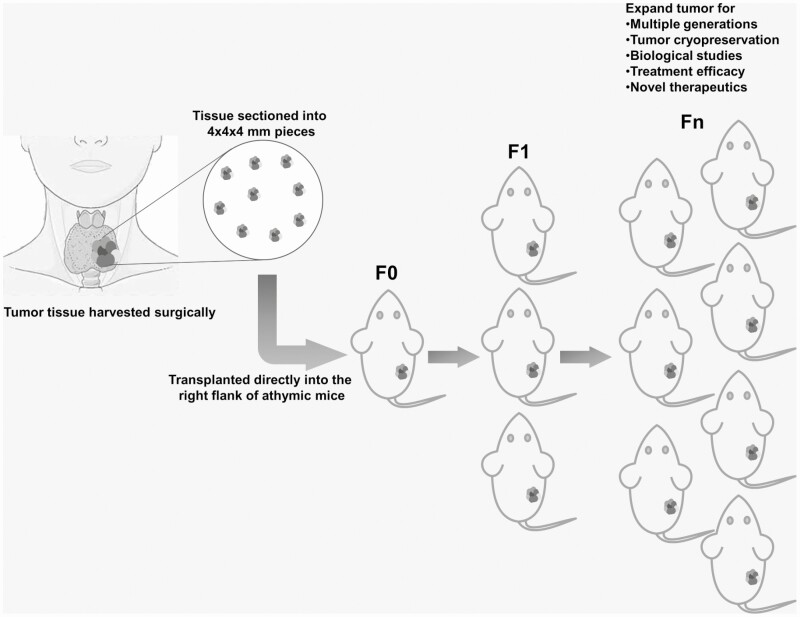
All experimental procedures and care for mice were approved by the institutional animal care and use committee and the department of veterinary medicine at our institution. Following our groups’ past successful experience with PDX models in head and neck squamous cell carcinoma (16), we used the patient tumoral tissue collected and prepared 4 × 4 × 4-mm pieces (Fig. 1) scalpels and implanted them in the flank of 6-week-old immunodeficient athymic nu/nu female mice (Envigo, No. 69; founder generation: F0) under 5% inhaled isoflurane anesthesia, followed by buprenorphine injection for pain management. Mice were then followed for tumor growth in vivo by recording tumor volume (height × length × width) weekly using a caliper. Once tumors reached a minimum of 1000 mm3 volume, the mice were euthanized, and the tumor was retrieved, weighed, measured, and sectioned into pieces. Tumor pieces were then used for PDX model expansion (F1, 1:5), snap-freezing in liquid nitrogen for short tandem repeat (STR) analysis, formalin fixation for histological evaluation, and tumor library storage by cryogenically freezing in 90% fetal bovine serum (FBS, Sigma-Aldrich F0926) plus 10% dimethyl sulfoxide (Sigma-Aldrich, No. D2650). PDX models were deemed established when expansion was completed successfully from F0 to F3, which represents a total of 4 generations, as well as the ability to expand the tumor from a previously cryogenically frozen sample. Established PDX ATC models were named with MDA-ATC numbers on engraftment.
Figure 1.
Fresh patient tumor specimen is prepared in 4 × 4 × 4-mm pieces to implant into the right flank of immune compromised/athymic nu/nu mice.
Cell Line Generation
All tumor specimens were obtained from patients who underwent a surgical intervention at our institution and provided written informed consent. Cell lines were generated and passaged as previously described (17, 18). Briefly, fresh tumor specimens from patients were minced using sterile scalpel blades and incubated in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich No. R8758) containing 2 mg/mL collagenase type I (Sigma-Aldrich No. C0130) and 0.002% DNase I (Fisher Scientific No. NC9082558) for 2 hours. Following incubation, single cells were filtered through a 40-μm nylon mesh strainer (Fisher Scientific No. 087711), incubated in hemolysis buffer (ammonium chloride solution; STEMCELL Technologies No. 07850) to remove the red blood cells, and washed with phosphate-buffered saline. Cells were then cultured in RPMI 1640 medium supplemented with 10% FBS (Sigma-Aldrich No. F0926), 2 mM L-glutamine, and 2 mg/mL Primocin (Fisher Scientific No. NC9392943) in an incubator supplied with 95% air and 5% CO2 at 37 °C. Fibroblasts were labeled with antihuman fibroblast antibody attached to microbeads (Miltenyi Biotec No. 130-050-601) and removed by passing through an LS+ column (Miltenyi Biotec No. 130-042-401) under a magnetic field. Cells were then cultured in RPMI 1640 supplemented with 10% FBS, nonessential amino acid mixture (Cambrex BioScience No. MT25025CI), and 1-mM sodium pyruvate (Cambrex BioScience MT25000CI), and counted as passage 1 after fibroblast removal. A total of 20 passages and at least 6 months in culture were deemed adequate to characterize a cell line as established.
Cell line doubling time was calculated using 1 × 105 cells in 6-well plates in triplicate. Cell numbers were determined 24 or 48 hours later by hemocytometer. Cell doubling time (Roth V. 2006 Doubling Time Computing, http://www.doubling-time.com/compute.php) was calculated using the following formula: DoublingTime = duration*log (2)/[log(FinalConcentration) − log(InitalConcentration)].
The doubling time experiment was repeated 3 times for accuracy.
Short Tandem Repeat DNA Fingerprinting and Mutational Analysis
Genomic DNA was extracted from frozen tissue using the Gentra Puregene kit (Qiagen). STR analysis for each cell line, PDX model, and its matching patient tissue was performed at the Characterized Cell Line Core Facility at MDACC using Promega PowerPlex 16 HS (catalog No. DC2100). These STR profiles were then compared with those in the ATCC (RRID:SCR_001672), the DSMZ (RRID:SCR_001711), the JCRB, the RIKEN (SCR_003250), Cellosaurus using the CLASTR tool (RRID:SCR_013869), and institutional databases for possible matches. Polymerase chain reaction (PCR)-based sequencing was performed using an NGS platform on genomic DNA to screen for mutations and copy number amplifications in the coding sequences of genes in the Clinical Laboratory Improvement Amendments–certified molecular diagnostics laboratory at MDACC. The human genomic reference sequence used was GRCh37 hg19. Percentage of STR identity was determined using patient frozen tissue as reference and the following formula: Percentage match = (number shared loci × 2)/(total 13 loci in the questioned profile + total 13 loci in the patient frozen tissue). All 13 loci were used in calculation and only 8 loci were reported in this study according to MDACC policy.
Genomic DNA from PDX models and cell lines were used in mutational analyses to confirm the mutations detected in patient tumors by whole-exome and/or Sanger sequencing. Whole-exome sequencing was conducted at the Human Genome Sequencing Center at Baylor College of Medicine (19). For Sanger sequencing, KAPA HiFi hotstart DNA polymerase (Fisher Scientific No. 501965229) was used for all PCRs, and PCR products were either treated with ExoSAP-IT PCR Product Cleanup Reagent to eliminate unincorporated primers and deoxynucleoside triphosphates (Fisher Scientific No. 78200) or were gel-purified before sequencing. PCR conditions and primers (Integrated DNA Technologies) used in PCR for BRAF (20), TP53 (21), CDKN2A (22), HRAS (20), KRAS (20), NF2 (23), NFE2L2 (24), and telomerase reverse transcriptase (TERT) promoter (17) were as previously described, except the antisense primer for NF2 was 5′-GATGTCACTGTGTGGTCAG. Allele frequency was determined from whole-exome sequencing data (25). Allele frequency was not determined from Sanger sequencing; instead, each mutation was indicated by homozygous or heterozygous nucleotide change.
Tumorigenesis Analysis of Cell Lines in Immunodeficient Mice
The orthotopic thyroid carcinoma model in athymic nude (nu/nu) mice (Envigo/Harlan Labs, No. 069) using established cell line has been previously described (26). Briefly, 1 × 106 or 4 × 106 MDA-T187 cells were injected into the thyroid or subcutaneously, respectively. Tumor growth was monitored daily and mice were euthanized when neck tumors hindered breathing or significantly affected the mouse’s general health. Subcutaneous tumor volumes were measured multiple times a week. Cells were not injected with Matrigel.
Histologic Characterization and Immunohistochemistry
The mice were euthanized and tumor specimens were collected and fixed in 10% neutral buffered formalin. Formalin fixed tissues were processed into 5-μm thick sections, stained with hematoxylin and eosin, and examined microscopically by head and neck pathologists (D.B., M.D.W.) using a BX41 Olympus microscope and an Aperio (Leica Biosystems) digital image scanner. Morphology, degree of differentiation (growth pattern, cytologic features, formation of keratin), and extent of inflammation were initially evaluated. Immunohistochemistry (IHC) analyses were performed against human-specific pancytokeratin (CK) AE1/AE3 (Dako; 1:100 dilution), thyroid transcription factor-1 (TTF-1) (catalog No. IS05630-2, Dako; 1:200 dilution), paired box gene 8 (PAX8) (No. 379, Biocare; 1:100 dilution), BRAFV600E (catalog No. E19290, clone VE1, Spring Biosciences; 1:50 dilution) and tumor protein TP53 (catalog No. GA61661-2, clone DO-7, Dako; 1:100 dilution). Diaminobenzidine was used as a chromogen for antigen localization. BRAFV600E-cytoplasmic and membranous staining was scored as positive or negative, positive score being defined as a greater than 90% strongly or weakly positive homogeneous stain for anti-BRAFV600E. TTF1, Pax8, or TP53-nuclear staining in the tumor cells was scored on a sliding scale of 0 to 4+ according to the percentage of reactive cells (0, negative; 1+, 1%-25%; 2+, 26%-50%; 3+, 51%-75%; 4+, 76%-100%). The staining intensity was graded as weak, intermediate, or strong. Cytoplasmic staining alone was scored as negative. PDX specimens were compared intergenerationally, as well as with their respective patient source surgical specimen.
Statistical Analyses
Pairwise t test with unequal variances was used to analyze continuous variables, while a Fisher’s exact test was used to analyze categorical variables. The Kaplan-Meier method was used to estimate overall survival (OS), while log-rank test was used to assess between-group differences in OS. A Cox proportional-hazards model and Efron’s method of tie handling were used to assess relative risk (hazard) of death, adjusting for age, sex, disease stage, and BRAF status. All statistical analyses were performed using Stata 13.0 (StataCorp LP).
Results
Development of Patient-Derived Xenograft Models
Nineteen patients with histopathologically confirmed ATC had tumor specimens transplanted into athymic nu/nu mice, of which 8 (42%) showed successful engraftment, defined as successful tumor growth in F0 and transplanted to F1. From the latter group, 6 were successfully established (32% overall) (Table 1). MDA-ATC1 originated from a T4bN1bM1 (stage IVC) tumor, harboring a BRAFV600E, and TP53K132N mutations. MDA-ATC2 originated from a T4aN1aM0 (stage IVB) tumor, wild-type (no mutation) for BRAFV600E, harboring a TP53T253P mutation. MDA-ATC3 originated from a T4aN1aM0 (stage IVB) tumor harboring BRAFV600E, TP53D208V, and CDKN2AE88* mutations, previously treated with dabrafenib and trametinib. MDA-ATC4 originated from a T4aN1bM0 (stage IVB) tumor with focally associated papillary thyroid cancer (PTC) harboring BRAFV600E, TP53R280T, and CDKN2AD74N mutations, previously treated with atezolizumab, vemurafenib, and cobimetinib. MDA-ATC5 originated from a T4aN0M1 (stage IVC) tumor, wild-type for TP53 and BRAFV600E mutations, but harboring TERT promoter –124C > T, and HRASG13R mutations. MDA-ATC6 originated from a T4aN0M1 (stage IVC) tumor with focally associated PTC, wild-type for BRAFV600E, but harboring TERT promoter –124C > T, TP53G783T, 731del, KRASG12R, NF2E317*, and NFE2L2D27Y mutations. The time required for generation-to-generation expansion represents the time needed for a tumor to grow from implant size (65 mm3) to successful expansion size (1000 mm3). Doubling times for the PDX models are also summarized in Table 1. A summary of the 13 tumor specimens that failed to generate PDX models can be seen in Table 2. There was no significant difference in clinocopathological features between the 6 patients from whom successful PDX models were developed and the 13 that failed.
Table 1.
Patient-derived xenograft model characteristics
| PDX model name/tumor ID | TNM | Disease stage | Diagnosis | Treatment prior to tissue collection | Last PDX generation | Expansion of PDX from frozen | Final generation-to-generation expansion time, d | PDX tumor doubling time, d | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MDA-ATC1 | T4bN1bM1 | IVC | ATC | No | F5 | F4 | 32 | 5-7 |
| 2 | MDA-ATC2 | T4aN1aM0 | IVB | ATC | No | F4 | F4 | 28 | 5-6 |
| 3 | MDA-ATC3 | T4aN1M0 | IVB | Multifocal clusters of viable carcinoma consistent with residual ATC | Dabrafenib, trametinib | F4 | F4 | 14 | 4-5 |
| 4 | MDA-ATC4 | T4aN1bM1 | IVB | ATC arising associated with PTC | Atezolizumab, vemurafenib, cobimetinib | F5 | F3 | 14 | 3-5 |
| 5 | MDA-ATC5 | T3aN0M0 | IVC | ATC | No | F5 | F2 | 14 | 3-4 |
| 6 | MDA-ATC6 | T4aN0M0 | IVC | ATC focal associated PTC | No | F6 | F6 | 14 | 3-4 |
Abbreviations: ATC, anaplastic thyroid cancer; d, days; ID, identification; PDX, patient-derived xenograft; PTC, papillary thyroid cancer.
Table 2.
Patient tumor specimens that were unsuccessful in generating patient-derived xenograft models
| Tumor ID | TNM | Diagnosis | Treatment prior to tissue collection | Current status | Platform used to detect mutations | BRAF | TP53 | Other mutations | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P178 | T4bN1bM0 | ATC | None | Deceased | IHC | WT | Yes | None |
| 2 | P184 | T4bN1bM1 | ATC | None | Deceased | 50-Gene somatic mutation analysis | WT | WT | None |
| 3 | P215 | T4bN1bM1 | ATC | None | Deceased | IHC and solid tumor genomics assay | WT | R248W | DNMT3A |
| 4 | P220LN | T4aN1aM1 | ATC with extensive squamous differentiation, LN | Dabrafenib, trametinib | Deceased | IHC and solid tumor genomics assay | V600E | WT | PIK3CA |
| 5 | P221 | T3NxM0 | ATC arising from a follicular neoplasm | None | Deceased | IHC | WT | WT | None |
| 6 | P225 | T4aN1bM0 | PTC with anaplastic (undifferentiated) component (~ 10%) | None | Alive | IHC and solid tumor genomics assay | WT | R280K | CDH1 CDKN2A NF1 PIK3CA |
| 7 | P239 | T4N1bM0 | ATC and PTC | Abraxane, dabrafenib, trametinib | Deceased | IHC and solid tumor genomics assay | V600E | Q331a | None |
| 8 | P244 | T4aN1bM0 | ATC, squamous type | None | Alive | IHC and solid tumor genomics assay | WT | E56a | TERT RB1 |
| 9 | P245 | T4aN1bM0 | ATC (40%) with PDTC | None | Deceased | Solid tumor genomics assay | WT | WT | None |
| 10 | P258 | T4bN1bM0 | ATC | Dabrafenib, trametinib | Deceased | IHC and solid tumor genomics assay | V600E | WT | NF2 TERT |
| 11 | P264 | T4bN1bM1 | ATC | Radiation | Deceased | IHC and solid tumor genomics assay | D594N | WT | NRAS TERT |
| 12 | P265 | T4bN1bM1 | ATC | Lenvatinib, dabrafenib, trametinib | Alive | IHC and solid tumor genomics assay | V600E | WT | PIK3CA TERT |
| 13 | P267 | T4aN0M0 | ATC | None | Alive | Solid tumor genomics assay | WT | R273C | ATM NF1 NOTCH3 RET |
Abbreviations: ATC, anaplastic thyroid cancer; ID, identification; IHC, immunohistochemistry; LN, lymph node; PDTC, poorly differentiated thyroid cancer; TERT, TERT promoter; WT, wild-type.
a Nonsense mutation.
PDX models were analyzed by whole-exome and/or Sanger sequencing to confirm mutations found in patient tumors (Table 3). We did not detect the mutation TP53T253P (that was seen in the patient’s cell-free DNA analysis) in MDA-ATC2; instead, we detected a possible exon 4 splice site mutation (c. 376-1G > A) in TP53 for that PDX model. Also, mutations in NF2E317* and TP53G244fs*, c.783-1G > T splice were detected in the patient tumor, but not in MDA-ATC6. All remaining mutations in patient tumors were confirmed in their respective PDX models. For example, Sanger sequencing of the PDX models for the presence or absence of the BRAFV600E mutation showed the expected GTG → GAG transversion in MDA-ATC1, 3, and 4, which were the same BRAF mutations found in patients’ tumors.
Table 3.
Summary of mutations in patient’s tumor, patient-derived xenograft, and cell lines
| Patient tumor | PDX | Cell linea | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDX name | Cell line name (RRID No.) | Platform used for patient tumor | BRAF | TP53 | Other mutations | BRAF (VAF %) | TP53 (VAF %) | Other mutations | BRAF (VAF %) | TP53 (VAF %) | Other mutations (VAF %) |
| MDA-ATC1 | MDA-T187 (CVCL_A1CS) | IHC | V600E | WT | None | V600E (57)a | K132N (5)a | None | V600E (94) | K132N (100) | None |
| MDA-ATC2 | NA | IHC, liquid biopsy | WT | T253Pc | None | WT | c. 376-1G → Ab,e | None | NA | NA | NA |
| MDA-ATC3 | NA | 50-Gene somatic mutation analysis | V600Ed | D208Vd | CDKN2A_E88*d | V600Eb1 | D208Vb,e | CDKN2A_E88*b,f | NA | NA | NA |
| MDA-ATC4 | MDA-T248 (CVCL_A1CT) | Solid tumor genomics assay | V600Ec,d | R280Tc | CDKN2A_D74Nc,d | V600Eb,e | R280Tb,e | CDKN2A_D74Nb,e | V600E (59) | c.994-1G > C (96) | CDKN2A_D74N (100) |
| MDA-ATC5 | MDA-T269 (CVCL_A1CU) | Solid tumor genomics assay | WT | WT | pTERT-124C > Td, HRAS_G13Rd | WT | WT | pTERT-124C > Tb,f, HRAS_G13Rb,f | WT | WT | pTERT-124C > Tb,fHRAS_G13Rb,f |
| MDA-ATC6 | MDA-T273 (CVCL_A1CV) | Solid tumor genomics assay, liquid biopsy | WT | G244fs*d, c.783-1G > Td, R175Hc | pTERT-124C > Td, KRAS_G12Rd, NF2_E317* d, NFE2L2_D27Yd | WT | R175Hb,e | pTERT-124C > Tb2, KRAS_G12Rb,eNFE2L2_D27Yb,e | WT | R175H (100) | pTERT-124C > Tb,f, KRAS_G12R (100), NFE2L2_D27Y (100) |
Abbreviations: ATC, anaplastic thyroid cancer; IHC, immunohistochemistry; NA, not available; PDX, patient-derived xenograft; VAF, variant allele frequency; WT, wild-type.
a Mutation detected by whole-exome sequencing.
b Mutation detected by Sanger sequencing.
c Mutation detected in cell-free DNA.
d Mutation detected by next-generation sequencing.
e Homozygous mutation.
f Heterozygous mutation.
Expansion from F0 to F1 occurred at a median 59 days (range, 47-244 days), 31 days (range, 20-147 days) for F2, 50 days (range, 25-219 days) for F3, 25 days (range, 10-70 days) for F4, 23 days (range, 14-32 days) for F5, and one model (MDA-ATC6) was expanded to F6, which took 14 days to reach required tumor volume of 1000 mm3. All models demonstrated tumorigenecity using frozen tissue stock. Once models were successfully established, additional expansions were 84% successful as demonstrated in a recent preclinical trial (27).
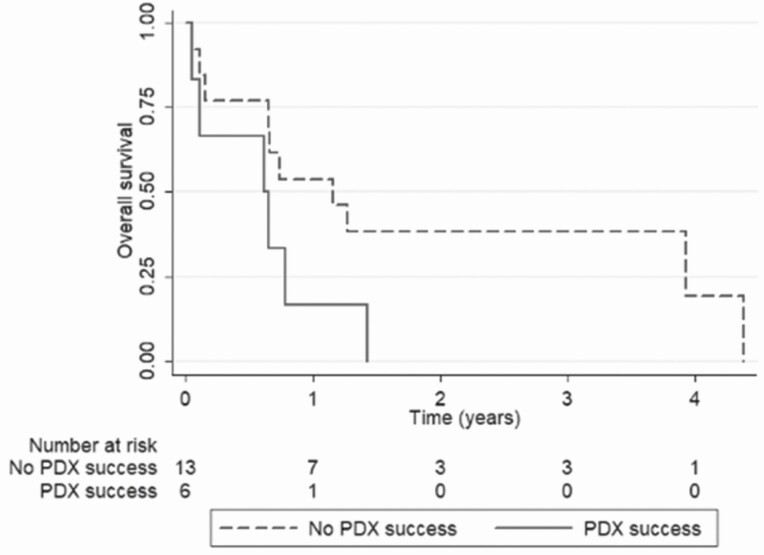
Median survival in patients for whom PDX models (n = 6) were successfully established was 0.64 years (range, 0.05-1.43 years), while the remaining patients (n = 13) had a median survival of 1.16 years (range, 0.05-4.38 years) (P = .065), with an age, sex, disease stage, and BRAF status–adjusted hazard ratio of 2.276 (95% CI, 0.666-7.784; P = .190) (Fig. 2).
Figure 2.
Overall survival (OS) of anaplastic thyroid cancer patients according to successful patient-derived xenograft (PDX) establishment or not. Patients for whom PDX models were not successfully established (blue curve; n = 13) had a median OS of 1.16 years (range, 0.05-4.38 years), whereas medial OS for patients with successful PDX models (red curve; n = 6) was 0.64 years (range, 0.05-1.43 years). Age, sex, disease stage, BRAF status–adjusted hazard ratio was 2.276 (95% CI, 0.666-7.784; P = .190)
Histopathological Evaluation
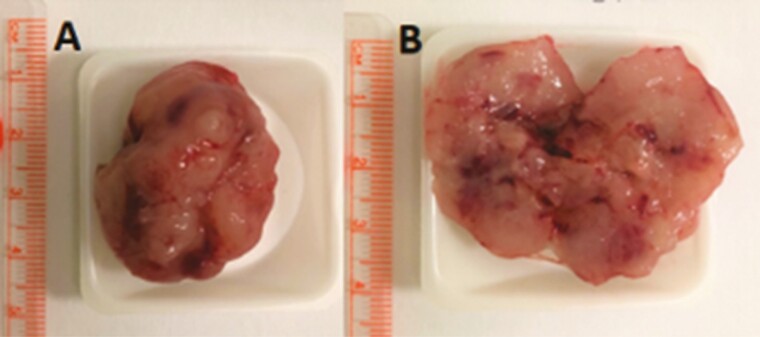
Gross evaluation of the PDX models demonstrated large, solid tumors with admixed necrosis, granularity, hemorrhage, and a marbled, tan-gray mucoid surface (Fig. 3), similar in description to their original patients’ surgical specimens.
Figure 3.
Gross evaluation of the patient-derived xenograft models demonstrates A, large solid tumors with B, admixed necrosis, granularity, hemorrhage, and a marbled tan-gray mucoid surface.
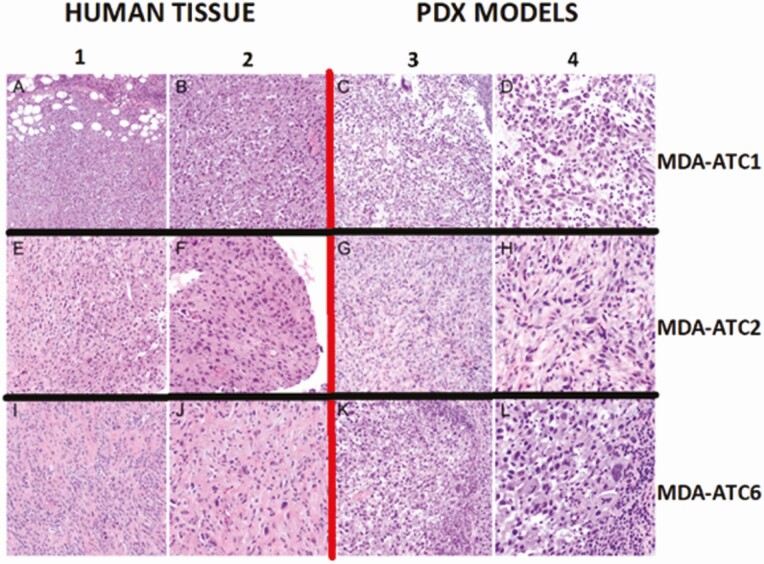
Microscopic evaluation of the models shows a variety of patterns classically described in ATC pathologies. Globally, all models had microscopic histologic evidence of necrosis, hemorrhage, high-grade mitoses, malignant spindle cells, and variable-sized nucleoli (Fig. 4). Specifically, MDA-ATC1 demonstrated a lymphoepithelioma-like aspect with prominent germinal centers, and stained diffusely positive for BRAFV600E and TP53, while being negative for TTF-1. MDA-ATC2 demonstrated a hypocellular, collagenous, spindle-variant with geographical type necrosis, and stained positive for TP53, while being negative for TTF-1 and BRAFV600E. MDA-ATC3 stained diffusely positive for TP53, while being weakly positive for BRAFV600E, comparable to the patient’s core biopsy where there were rare BRAFV600E-positive viable tumor cells. MDA-ATC4 stained diffusely positive for BRAFV600E and TP53, while being negative for TTF-1. MDA-ATC5 stained negative both for BRAFV600E and TP53. MDA-ATC6 stained partially positive for cytokeratin, diffusely for TP53, and minimally (2%) for PAX8, while being negative for TTF-1 and BRAFV600E. All microscopic histologic and IHC evaluations were very similar, if not undistinguishable to the human specimen slides, as demonstrated in the models depicted in Fig. 4 and 5. Of note, patient 3’s slides show significant inflammatory infiltration (small nuclei), which dilutes the number of tumor cells evaluated; however, the tumor cells remain strongly positive both for the TP53 and BRAFV600E stains.
Figure 4.
Comparative evaluation of histology and morphology of human specimens (columns 1 and 2) and associated patient-derived xenograft (PDX) models (columns 3 and 4). Columns 1 and 3 are in low-power field, while columns 2 and 4 are in high-power field.
Figure 5.
Comparative evaluation of immunohistochemistry (IHC) stainings of human specimens and associated patient-derived xenograft (PDX) models. IHC analyses were performed against human-specific tumor protein TP53, BRAFV600E, thyroid transcription factor-1 (TTF-1), paired box gene 8 (PAX8), and pancytokeratin (CK).
Anaplastic Thyroid Cancer Cell Lines
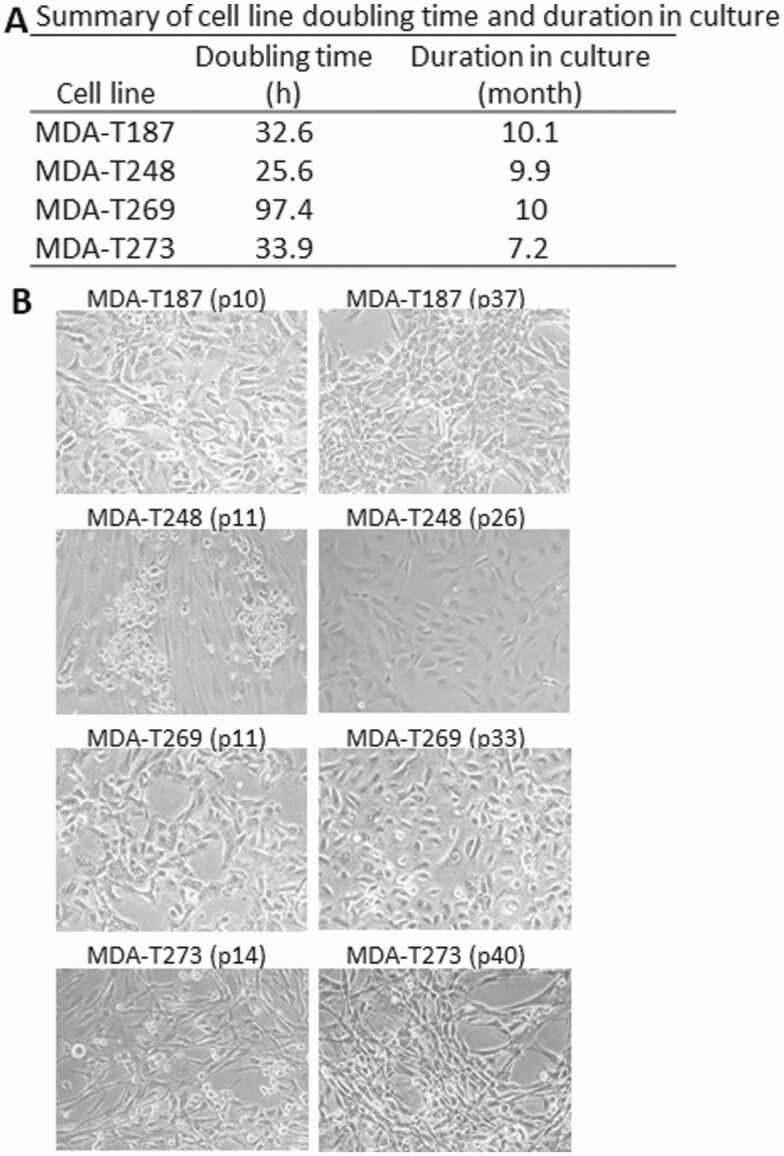
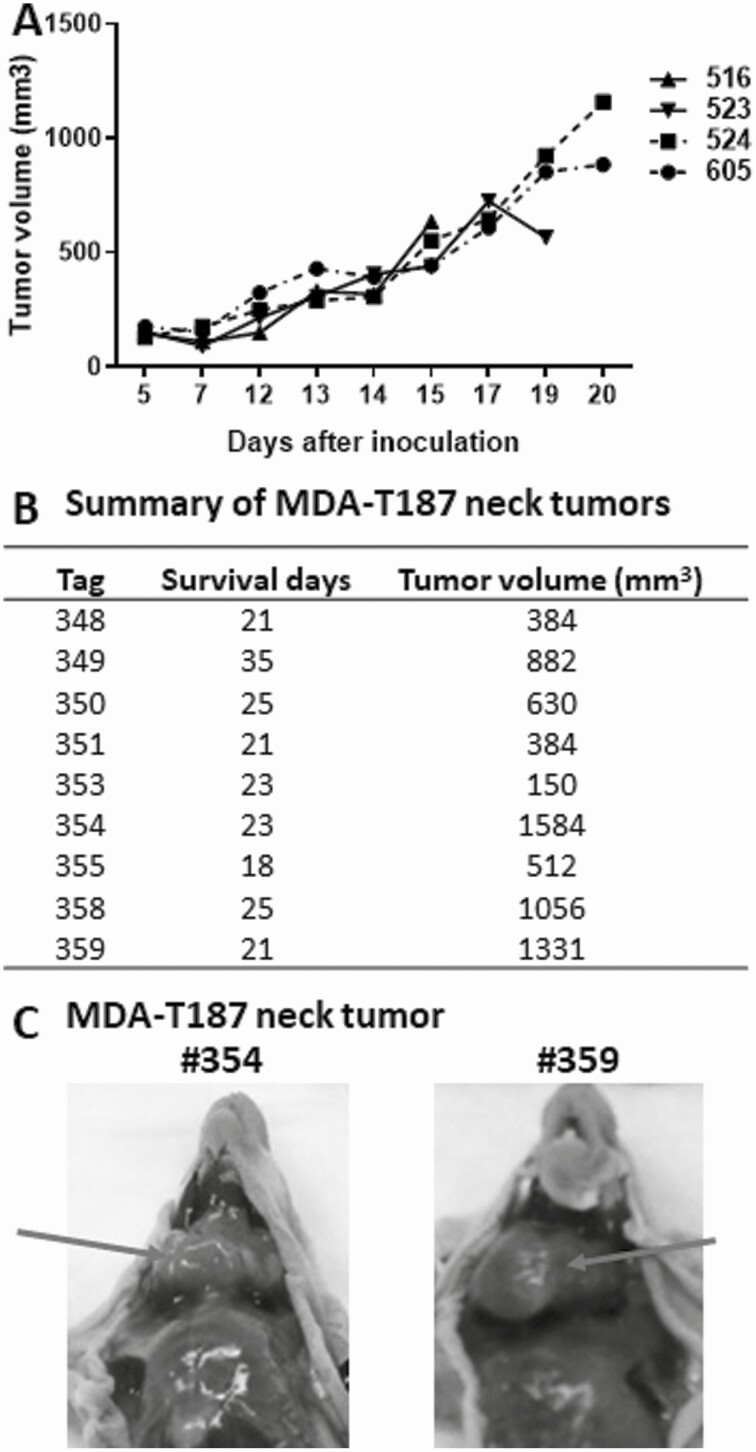
ATC cell lines were established in 4 of 6 patient tumor specimens. MDA-ATC2 did not have a human ATC cell line counterpart because the cells stopped growing after the sixth passage. Single cells corresponding to MDA-ATC3 reached 20 passages but lost adequate ATC cell morphology and cell-marker phenotype. The remaining 4 cell lines reached at least 20 passages and remained in culture for more than 6 months to be ensured as a successful cell line (Fig. 6A). The morphology of cells during development appeared to be similar when comparing cells at early and late passages (Fig. 6B). Cell doubling time for each cell line was determined (see Fig. 6A). MDA-T269 cells took much longer to double (97.4 hours) than the other 3 cell lines (25.6-33.9 hours). Tumorigenicity was demonstrated in cell line analyses using the MDA-ATC1–associated line, MDA-T187, in an in vivo setting by injecting cells subcutaneously and orthotopically. Growth curves of subcutaneous tumor are shown in Fig. 7A. Growth curves for orthotopic tumors are not available as tumors are not as evident as subcutaneous tumors. The final tumor volumes of orthotopic tumors are provided in Fig. 7B. Examples of 2 orthotopic tumors (from mouse tags No. 354 and No. 359) are shown in Fig. 7C as large neck tumors on day 23 and 21 after injection, respectively. Tumors successfully grew within 7 days of injection, while mouse euthanasia and tumor harvest were required starting day 15 for subcutaneous tumors and day 18 for orthotopic tumors, indicative of the aggressivity of the cell line. Overall, the tumorigenic take rates of MDA-T187 among orthotopic and subcutaneous groups were 100% (10/10) and 90% (9/10), respectively.
Figure 6.
A, Doubling time of cell lines. B, morphology of cell lines in early and late stages of cell line development. Average doubling time for each cell line was determined as described in “Materials and Methods” from 3 independent assays. Morphology of cell lines were taken by a Nikon camera using a microscope at different passages of cells during cell line development.
Figure 7.
Growth curve of subcutaneous tumors (A) and final tumor volume of orthotopic tumors (B). A, Subcutaneous tumors were measured after tumors were established 5 days after injection. Tumor volume was measured by caliper as for patient-derived xenograft models and calculated using the formula (V = length × width × depth). B, tumor volume for orthotopic tumors was measured using the same formula as for subcutaneous tumors after mice were euthanized. The survival days were counted using the injection day as day 0. C, examples of images of orthotopic tumors were indicated by arrow. Mouse tag No. 354 was euthanized on day 23 and No. 359 on day 21 after injection.
Genetic alterations in cell lines were determined both by whole exome and/or Sanger sequencing to confirm the mutations found in patient tumors (see Table 3). Most mutations found in patient tumors were also found in their resultant cell lines, except for the following: (1) The TP53K132N mutation was detected in MDA-T187 cells but not in the patient’s tumor. (2) TP53R280T was not detected in MDA-T248 cells, but was detected in the patient’s tumor and corresponding PDX model (MDA-ATC4). A mutation in the splice site of intron 9 and exon 10 junction of TP53 was found in MDA-T248 cells (c.994-1G > C), which may result in early termination. This mutation has been reported in Li-Fraumeni syndrome (28). (3) TP53 mutations G244fs* and c.783-1G > T splice, and the NF2E317* mutation detected in the patient tumor were not found in the PDX model (MDA-ATC6) and MDA-T273 cells.
STR DNA fingerprinting was performed on all models and cell lines and showed concordance with the original frozen patient tissue as indicated by the percentage of identity (Table 4). All STR from the PDX models were above 90%, except for MDA-ATC3, which demonstrated 73.1% identity to the reference tissue. No new polymorphisms were noted in MDA-ATC3, simply consistent losses relative to the original patient tumor. Electropherograms of STR for all samples can be found on an online data-sharing platform (29).
Table 4.
Short-tandem repeats of patient tissues, cell lines, and patient-derived xenograft models
| Sample (sex) | STR source (passage or generation) | AM | CSF1PO | D5S818 | D7S820 | D13S317 | D16S539 | TH01 | TPOX | vWA | Identitya (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MDA-ATC1 | Frozen tissue | X | 10,12 | 11,14 | 9,12 | 11,12 | 11 | 9.3 | 8 | 16,18 | 100 |
| (Female) | MDA-T187 cell line (p28) | X | 12 | 14 | 9,12 | 12 | 11 | 9.3 | 8 | 16,18 | 88.5 |
| PDX (F1) | X | 10,12 | 11,14 | 12 | 12 | 11 | 9.3 | 8 | 16,18 | 92.3 | |
| MDA-ATC2 | Frozen tissue | X,Y | 12,13 | 9,12 | 10,12 | 11,12 | 11,12 | 6,9.3 | 8,12 | 17,18 | 100 |
| (Male) | PDX (F3) | X,Y | 12,13 | 9,12 | 10,12 | 12 | 11 | 6,9.3 | 8,12 | 17,18 | 92.3 |
| MDA-ATC3 | Frozen tissue | X | 10,12 | 12 | 9,12 | 11,12 | 8,10 | 6,9.3 | 8,12 | 14,15 | 100 |
| (Female) | PDX (F3) | X | 12 | 12 | 12 | 11 | 8,10 | 6,9.3 | 12 | 14 | 73.1 |
| MDA-ATC4 | Frozen tissue | X | 12 | 9,11 | 11 | 11,12 | 11,13 | 6,8 | 9,10 | 14,18 | 100 |
| (Female) | MDA-T248 cell line (p25) | X | 12 | 9,11 | 11 | 11 | 11,13 | 6,8 | 9,10 | 14,18 | 96.2 |
| PDX (F2) | X | 12 | 9,11 | 11 | 11 | 11,13 | 6,8 | 9,10 | 14,18 | 96.2 | |
| MDA-ATC5 | Frozen tissue | X,Y | 11,14 | 12 | 8,10 | 11,12 | 11,13 | 6 | 8,11 | 14,17 | 100 |
| (Male) | MDA-T269 cell line (p20) | X | 11,14 | 12 | 10 | 11,12 | 11,13 | 6 | 8,11 | 14,17 | 96.2 |
| PDX (F3) | X | 11,14 | 12 | 10 | 11,12 | 11,13 | 6 | 8,11 | 14,17 | 96.2 | |
| MDA-ATC6 | Frozen tissue | X | 11,12 | 12 | 10,11 | 9,11 | 11 | 7,9 | 8,11 | 14,17 | 100 |
| (Female) | MDA-T273 cell line (p21) | X | 11,12 | 12 | 10,11 | 9 | 11 | 7,9 | 8,11 | 14 | 92.3 |
| PDX (F3) | X | 11,12 | 12 | 10,11 | 9 | 11 | 7,9 | 8,11 | 14 | 92.3 |
Abbreviations: PDX, patient-derived xenograft; STR, short-tandem repeat.
a Percentage identity was determined by Tanabe algorithm based on 13 loci. Percentage match = (number shared loci × 2)/(total number of loci in the questioned profile + total number of loci in the reference profile). Reference profile was frozen tissue.
Discussion
The first reported ATC PDX model dates back to 2013, when Wunderlich et al (30) demonstrated the feasibility of not only transplanting patient tumor to athymic mice, but also using it as a preclinical model for drug testing. Since then, 7 other ATC PDX models have been reported (6, 31), although none have been used for drug evaluation. In this study, we report 6 new ATC PDX models that we have established in a robust manner, demonstrating very strong histopathologic concordance with original patient specimens, and favorable and timely generation-to-generation expansion rates. In addition, we noted a difference in median OS in patients from whom we successfully generated a PDX model, compared to those who were unsuccessful. Our success rate for PDX model establishment of 32% is higher than the reported 15% for all thyroid cancers (6); however, when comparing it to ATC-specific PDX models, Marlow et al had a success rate of 75% (6). Overall, the success rate remains quite elevated, which is associated with the aggressiveness of the disease and ATC’s dedifferentiated state. Furthermore, once established, tumor expansion from one mouse to several for drug trial purposes was almost 85% successful (27), truly making these strong preclinical tools.
ATC tumors have a high mutation burden and often harbor multiple genetic alterations, which are involved in different signaling pathways (32). Each mutated gene may play a different role in ATC progression and thus gives ATC tumors unique biologic properties. Comprehensively analyzing the mutational status of ATC tumors is a key factor in understanding the disease and the basis of molecular targeted therapy development.
Landa et al (9) analyzed the genomic backgrounds of 33 ATC patients using NGS and found that 45% had a BRAF mutation, whereas 24% had a RAS mutation. Similar to differentiated thyroid cancer (33), these 2 types of mutations were mutually exclusive, which suggest they may be early events and driver mutations during tumor evolution. Other common genetic alterations in ATC were TERT promoter (73%), TP53 (73%), PIK3CA (18%), PTEN (15%), and EIF1AX (9%) mutations (9). These mutations were concurrent with the driver mutations, and it is suggested they play an important role in tumor progression. In this study, findings support the stepwise dedifferentiation model that ATC was derived from differentiated thyroid cancer through the accumulation of important genetic alterations (9). The division of these driver mutations into BRAF- or RAS-dependent drivers (8, 34) is supported in our study, as none of the PDX models contained both the RAS and BRAF mutations.
Preclinical models play a major role in the analysis of disease progression. Cancer cell lines are a well-defined tool to obtain reproducible data both for in vitro and in vivo work. Although ATC cell lines have been previously generated (5, 35, 36), developing novel ATC cell lines is important as prior ATC cell lines have been found to harbor various cross-contaminations (37, 38). We confirmed concordance of the STR results of the newly established cell lines and PDX models with the corresponding tumor tissues from patients.
ATC harbors a vast library of molecular alteration combinations; adding to the current library of cell lines will help further characterize this disease, especially when the cell line has a matching PDX model. In this study, we developed and characterized 4 new ATC cell lines with their corresponding PDX model, while 2 additional PDX models (MDA-ATC2 and MDA-ATC3) were established without a companion cell line. Although the specific cause for failure to generate a cell line is unclear, MDA-ATC2 was hypocellular and collagenous, which may have contributed to fibroblast overgrowth. Nevertheless, both PDX models were successfully propagated until F4, with an average generation-to-generation expansion rate of 36 days. PDX establishment for MDA-ATC2 and MDA-ATC3, as opposed to their cell line counterparts, may be due to the preservation of the tumor heterogeneity and gene expression when transplanted as a solid single piece of tumor. Overall, we had a strong engraftment rate, which is testament to the aggressive and dedifferentiated nature of ATC, compared to the more indolent nature of PTC, which has a reported engraftment rate of 14% (6). By generating additional robust ATC PDX models, we are enriching the library of PDX thyroid cancer models for future preclinical drug therapies. To date, our group has shown successful use of our PDX models in such preclinical trials, evaluating known and novel treatments for ATC (27). Future work on our PDX models will include the evaluation of novel drug combinations, analysis of resistance patterns, and serving as platforms for novel treatments that have yet to be tested in patients.
In conclusion, we have developed and characterized 6 novel ATC PDX models, and their cell line counterparts in 4 of the 6 models, that contain unique histologic and genetic variations that may serve as reliable preclinical models for drug development and therapeutic trials.
Acknowledgments
We thank Xuesong Li and Keri Sherman for providing STR profiling; Viju Varghese for Sanger-based DNA sequencing; and Yan Cai, Cynthia Steward, Deborah Rodriguez, Kareem Metwalli, Bridget Reeves, Maitrayee Goswami, and Barbara Deleon for technical support. We also wish to acknowledge the contributions of Dr Richard Gibbs, Donna Muzny, and the rest of the production team at the BCM-HGSC.
Financial Support: This work was partly supported by the Cancer Prevention and Research Institute of Texas (CPRIT) (grant No. RP170366; principal investigator: S.Y.L.), an institutional Multi-investigator Research Program grant, and National Cancer Institute Cancer Center Support (CORE) Grant P30CA016672 for media production and cell line authentication. A.M. received a research grant from the Fonds de Recherche du Québec-Santé (FRQS) for ATC PDX model development. Donations were generously provided through Marty Schaffel, Kevin Weinrich, the Anaplastic Thyroid Cancer Research Fund, the Michael A. O’Bannon Endowment for Cancer Research, and the Petrick ATC Research Fund.
Glossary
Abbreviations
- ATC
anaplastic thyroid cancer
- FBS
fetal bovine serum
- IHC
immunohistochemistry
- MDACC
The University of Texas MD Anderson Cancer Center
- NGS
next-generation sequencing
- OS
overall survival
- PCR
polymerase chain reaction
- PDX
patient-derived xenograft
- PTC
papillary thyroid cancer
- RPMI
Roswell Park Memorial Institute
- STR
short tandem repeats
- TTF-1
thyroid transcription factor-1
Additional Information
Disclosures: A.M., Y.C.H., H.H., S.P., Y.Y.C., Y.J., S.J., M.C., D.B., M.C.H., S.E.S., D.A.W., and S.Y.L. declare no competing interests; M.Z.: principal investigator for clinical trial funded by Merck; M.E.C.: research funding from Eisai, Exelixis, Kura Oncology, and Genentech, and participated in advisory boards for LOXO and Ignyta in the past 2 years; R.D.: research funding from Merck, Eisai, Exelixis, AstraZeneca, and Bayer; N.L.B.: research funding from Novartis through the National Comprehensive Cancer Network (NCCN); F.M.J.:research funding from PIQUR Therapeutics and Trovagene.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.” All 4 cell lines have been registered with RRID/Celllosaurus, CVCL_A1CS for MDA-T187, CVCL_A1CT for MDA-T248, CVCL_A1CU for MDA-T269, and CVCL_A1CV for MDA-T273.
References
- 1. Lin B, Ma H, Ma M, et al. . The incidence and survival analysis for anaplastic thyroid cancer: a SEER database analysis. Am J Transl Res. 2019;11(9):5888-5896. [PMC free article] [PubMed] [Google Scholar]
- 2. Smallridge RC, Ain KB, Asa SL, et al. ; American Thyroid Association Anaplastic Thyroid Cancer Guidelines Taskforce . American Thyroid Association guidelines for management of patients with anaplastic thyroid cancer. Thyroid. 2012;22(11):1104-1139. [DOI] [PubMed] [Google Scholar]
- 3. Maniakas A, Dadu R, Busaidy NL, et al. . Evaluation of overall survival in patients with anaplastic thyroid carcinoma, 2000-2019. JAMA Oncol. 2020;6(9):1397-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schweppe RE, Klopper JP, Korch C, et al. . Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93(11):4331-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marlow LA, D’Innocenzi J, Zhang Y, et al. . Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab. 2010;95(12):5338-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marlow LA, Rohl SD, Miller JL, et al. . Methodology, criteria, and characterization of patient-matched thyroid cell lines and patient-derived tumor xenografts. J Clin Endocrinol Metab. 2018;103(9):3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kasaian K, Wiseman SM, Walker BA, et al. . The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer. 2015;15:984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kunstman JW, Juhlin CC, Goh G, et al. . Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24(8):2318-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Landa I, Ibrahimpasic T, Boucai L, et al. . Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126(3):1052-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bousquet G, Janin A. Patient-derived xenograft: an adjuvant technology for the treatment of metastatic disease. Pathobiology. 2016;83(4):170-176. [DOI] [PubMed] [Google Scholar]
- 11. Gebauer F, Wicklein D, Tachezy M, et al. . Establishment and characterization of a pair of patient-derived human non-small cell lung cancer cell lines from a primary tumor and corresponding lymph node metastasis. Anticancer Res. 2016;36:1507-1518. [PubMed] [Google Scholar]
- 12. Hao C, Wang L, Peng S, et al. . Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015;357(1):179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moon HG, Oh K, Lee J, et al. . Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast Cancer Res Treat. 2015;154(1):13-22. [DOI] [PubMed] [Google Scholar]
- 14. Karamboulas C, Bruce JP, Hope AJ, et al. . Patient-derived xenografts for prognostication and personalized treatment for head and neck squamous cell carcinoma. Cell Rep. 2018;25:1318-1331.e1314. [DOI] [PubMed] [Google Scholar]
- 15. Hsu CL, Lui KW, Chi LM, et al. . Integrated genomic analyses in PDX model reveal a cyclin-dependent kinase inhibitor Palbociclib as a novel candidate drug for nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2018;37(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng S, Creighton CJ, Zhang Y, et al. . Tumor grafts derived from patients with head and neck squamous carcinoma authentically maintain the molecular and histologic characteristics of human cancers. J Transl Med. 2013;11:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henderson YC, Ahn SH, Ryu J, et al. . Development and characterization of six new human papillary thyroid carcinoma cell lines. J Clin Endocrinol Metab. 2015;100(2):E243-E252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahn SH, Henderson YC, Williams MD, Lai SY, Clayman GL. Detection of thyroid cancer stem cells in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2014;99(2):536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Ni X, Covington KR, et al. . Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47(12):1426-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henderson YC, Shellenberger TD, Williams MD, et al. . High rate of BRAF and RET/PTC dual mutations associated with recurrent papillary thyroid carcinoma. Clin Cancer Res. 2009;15(2):485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lehman TA, Bennett WP, Metcalf RA, et al. . p53 Mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51(15):4090-4096. [PubMed] [Google Scholar]
- 22. Harland M, Goldstein AM, Kukalizch K, et al. . GenoMEL, the Melanoma Genetics Consortium . A comparison of CDKN2A mutation detection within the Melanoma Genetics Consortium (GenoMEL). Eur J Cancer. 2008;44(9):1269-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andujar P, Pairon JC, Renier A, et al. . Differential mutation profiles and similar intronic TP53 polymorphisms in asbestos-related lung cancer and pleural mesothelioma. Mutagenesis. 2013;28(3):323-331. [DOI] [PubMed] [Google Scholar]
- 24. Kim YR, Oh JE, Kim MS, et al. . Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220(4):446-451. [DOI] [PubMed] [Google Scholar]
- 25. Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling somatic SNVs and indels with Mutect2, biorXiv, 2019. https://www.biorxiv.org/content/10.1101/861054v1.
- 26. Ahn SH, Henderson Y, Kang Y, et al. . An orthotopic model of papillary thyroid carcinoma in athymic nude mice. Arch Otolaryngol Head Neck Surg. 2008;134(2):190-197. [DOI] [PubMed] [Google Scholar]
- 27. Maniakas A MA, Henderson YC, Hei H, et al. . In vivo drug response evaluation in anaplastic thyroid cancer patient-derived tumor xenografts following high-throughput screening [abstract]. Paper presented at: 111th Annual Meeting of the American Association for Cancer Research; April 24-29, 2020,. Philadelphia, PA.
- 28. Verselis SJ, Rheinwald JG, Fraumeni JF Jr, Li FP. Novel p53 splice site mutations in three families with Li-Fraumeni syndrome. Oncogene. 2000;19(37):4230-4235. [DOI] [PubMed] [Google Scholar]
- 29. Maniakas A, Henderson Y, Lai SY. STR electropherogram for novel ATC PDX models and cell lines. figshare 2021. Figure. Deposited 18 July 2021. 10.6084/m9.figshare.14567736.v2. [DOI]
- 30. Wunderlich A, Khoruzhyk M, Roth S, et al. . Pretherapeutic drug evaluation by tumor xenografting in anaplastic thyroid cancer. J Surg Res. 2013;185(2):676-683. [DOI] [PubMed] [Google Scholar]
- 31. Schweppe RE, Pozdeyev N, Pike LA, et al. . Establishment and characterization of four novel thyroid cancer cell lines and PDX models expressing the RET/PTC1 rearrangement, BRAFV600E, or RASQ61R as drivers. Mol Cancer Res. 2019;17(5):1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bible KC, Ryder M. Evolving molecularly targeted therapies for advanced-stage thyroid cancers. Nat Rev Clin Oncol. 2016;13(7):403-416. [DOI] [PubMed] [Google Scholar]
- 33. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikiforov YE. Thyroid cancer in 2015: molecular landscape of thyroid cancer continues to be deciphered. Nat Rev Endocrinol. 2016;12(2):67-68. [DOI] [PubMed] [Google Scholar]
- 35. Garg M, Okamoto R, Nagata Y, et al. . Establishment and characterization of novel human primary and metastatic anaplastic thyroid cancer cell lines and their genomic evolution over a year as a primagraft. J Clin Endocrinol Metab. 2015;100(2):725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Onoda N, Nakamura M, Aomatsu N, Noda S, Kashiwagi S, Hirakawa K. Establishment, characterization and comparison of seven authentic anaplastic thyroid cancer cell lines retaining clinical features of the original tumors. World J Surg. 2014;38(3):688-695. [DOI] [PubMed] [Google Scholar]
- 37. American Type Culture Collection Standards Development Organization Workgroup ASN-0002 . Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10(6):441-448. [DOI] [PubMed] [Google Scholar]
- 38. Zhao M, Sano D, Pickering CR, et al. . Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17(23):7248-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.” All 4 cell lines have been registered with RRID/Celllosaurus, CVCL_A1CS for MDA-T187, CVCL_A1CT for MDA-T248, CVCL_A1CU for MDA-T269, and CVCL_A1CV for MDA-T273.