Abstract
Dysbiosis of gut microbiota is associated with many pathologies, yet host factors modulating microbiota remain unclear. Interstitial cystitis/bladder pain syndrome (IC/BPS) is a debilitating condition of chronic pelvic pain often with comorbid urinary dysfunction and anxiety/depression, and recent studies find fecal dysbiosis in patients with IC/BPS. We identified the locus encoding acyloxyacyl hydrolase, Aoah, as a modulator of pelvic pain severity in a murine IC/BPS model. AOAH-deficient mice spontaneously develop rodent correlates of pelvic pain, increased responses to induced pelvic pain models, voiding dysfunction, and anxious/depressive behaviors. Here, we report that AOAH-deficient mice exhibit dysbiosis of gastrointestinal (GI) microbiota. AOAH-deficient mice exhibit an enlarged cecum, a phenotype long associated with germ-free rodents, and a “leaky gut” phenotype. AOAH-deficient ceca showed altered gene expression consistent with inflammation, Wnt signaling, and urologic disease. 16S sequencing of stool revealed altered microbiota in AOAH-deficient mice, and GC-MS identified altered metabolomes. Cohousing AOAH-deficient mice with wild-type mice resulted in converged microbiota and altered predicted metagenomes. Cohousing also abrogated the pelvic pain phenotype of AOAH-deficient mice, which was corroborated by oral gavage of AOAH-deficient mice with stool slurry of wild-type mice. Converged microbiota also alleviated comorbid anxiety-like behavior in AOAH-deficient mice. Oral gavage of AOAH-deficient mice with anaerobes cultured from IC/BPS stool resulted in exacerbation of pelvic allodynia. Together, these data indicate that AOAH is a host determinant of normal gut microbiota, and dysbiosis associated with AOAH deficiency contributes to pelvic pain. These findings suggest that the gut microbiome is a potential therapeutic target for IC/BPS.
Keywords: acyloxyacyl hydrolase, gut dysbiosis, interstitial cystitis, microbiome, pelvic pain
INTRODUCTION
Interstitial cystitis/bladder pain syndrome (IC/BPS or “IC”) is a chronic condition characterized by bladder discomfort and moderate-to-severe pelvic pain affecting as many as eight million patients in the United States (1, 2). Patients suffering from IC/BPS commonly exhibit comorbid conditions including voiding dysfunction of increased urinary frequency and urgency and anxiety/depression (3–5). The etiology of IC/BPS is unknown and its pathophysiological mechanisms remain unclear, so IC/BPS is a clinical challenge with no widely effective therapy. Because of the profound impact of chronic pelvic pain, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) established the Multi-Disciplinary Approaches to Chronic Pelvic Pain (MAPP) Research Network in an effort to study urologic chronic pelvic pain syndromes at the clinical, epidemiological, and basic levels. In our MAPP studies, we previously conducted a genetic screen for loci modulating pelvic pain severity and identified Aoah, the locus encoding acyloxyacyl hydrolase, as a modulator of pelvic pain in a murine model of IC/BPS (6).
Acyloxyacyl hydrolase (AOAH) is a host enzyme best known for detoxifying bacterial lipopolysaccharides (LPS) by selectively removing secondary acyl chains from the lipid A moiety, mitigating the host response to bacterial infection and attenuating sustained inflammation (7–10). Previous studies in our laboratory demonstrated that mice deficient for AOAH spontaneously develop pelvic allodynia consistent with pelvic pain and exhibit heightened response in induced pelvic pain models (6). AOAH-deficient mice also have voiding dysfunction (11) and display rodent correlates of anxious/depressive behaviors (12). Expressed along the bladder-brain axis, AOAH functions as an arachidonyl acyl transferase, and AOAH-deficient mice exhibit defects in central nervous system (CNS) arachidonic acid homeostasis and increased accumulation of PGE2, contributing to pelvic pain (13). Moreover, corticotropin-releasing factor (CRF) is the initiator of the hypothalamic-pituitary-adrenal (HPA) axis and a modulator of pain responses, and AOAH-deficient mice have increased Crf expression and increased serum corticosterone consistent with HPA axis dysregulation (12). Patients with IC/BPS also have aberrant diurnal cortisol fluctuations indicating HPA axis dysregulation (14), another similarity with AOAH-deficient mice. Thus, AOAH-deficient mice recapitulate several key aspects of IC/BPS.
Gut dysbiosis has been increasingly recognized as a key player in visceral pain, and recent studies have indicated altered fecal microbiota in women with IC/BPS and in male patients suffering an analogous condition known as chronic prostatitis/chronic pelvic pain syndrome (15–17). Visceral convergence of sensory inputs that drives pelvic organ cross talk and the gut-brain axis provides mechanisms by which gut dysbiosis may influence IC/BPS urologic and cognitive symptoms (18–21). Given the strong parallels between phenotypes of AOAH-deficient mice and IC/BPS, we hypothesized that AOAH-deficient mice would exhibit gut dysbiosis that mediates pelvic pain and other comorbidities. We observed multiple phenotypes supporting this hypothesis including altered cecal morphology and transcriptomes, fecal and cecal dysbiosis, and a role for gut microbiota in pelvic pain behaviors.
METHODS
Animals
Ten- to twelve-week-old male and female wild-type (WT) C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Aoah−/− mice (B6.129S6-Aoahtm1Rsm/J) were obtained as a generous gift from Dr. Robert Munford of National Institute of Allergy and Infectious Diseases (NIAID) and maintained on a 12 h:12 h light:dark cycle as previously described (12). All animals were maintained at the Center for Comparative Medicine at Northwestern University and used for experimentation under Northwestern Institutional Animal Care and Use Committee (IACUC) approved protocols.
GI Characterization
Animals were euthanized and placed on a pan balance to obtain body weight. The cecum and bladder were excised and weighed. The cecum was then dissected, and cecal stool was removed and weighed.
Transepithelial Electrical Resistance
Cecum and bladder permeability were measured as transepithelial electrical resistance (TEER) as previously described for bladders in Aguiniga et al. (11). Excised organs were bisected and immediately placed in Ringer solution at room temperature. After rinsing, ceca and bladders were mounted onto cassettes with apertures of 0.126 cm2 and 0.04 cm2, respectively. Cassettes were inserted between two-halves of an Ussing chamber (EM-CSYS-2, Physiologic Instruments, San Diego, CA). KCl saturated salt-bridge electrodes were inserted. The chambers were filled with Ringer solution and bubbled with carbogen (95% O2-5% CO2) and maintained at 37°C until preliminary resistance became stabilized, typically at least 1 h. Voltage was clamped and current was passed every 3 s at 30 s intervals using a VCC MC2 multichannel voltage-current clamp amplifier controlled with Acquire and Analyze software v2.3.4 (Physiologic Instruments).
Hematoxylin and Eosin Staining
All hematoxylin and eosin (H&E) staining in wild-type and AOAH-deficient ceca was performed by Northwestern University Mouse Histology & Phenotyping Laboratory (MHPL, Chicago, IL).
Cecum Staining
Animals were euthanized and ceca were dissected. The cecum was flushed first with PBS and then with 10% neutral buffered formalin (NBF). Tissue was Swiss-rolled and placed in 15 mL of 10% NBF before embedding in paraffin. Tissue was then sectioned at 4 μm. Tissue was deparaffinized with a series of washes as follows: two times for 3 min with 100% xylene, three times for 3 min with 100% ethanol, once for 3 min with 94% ethanol, once for 3 min with 70% ethanol, once for 3 min with 50% ethanol, and once for 3 min with H2O. Antigen retrieval was performed in a pressure cooker using Target Retrieval Solution (S1699, DAKO, Santa Clara, CA) at 100°C for 10 min, 90°C for 10 min, and then washed in PBS for 3 min. Tissue was blocked in FC Receptor Block (NB309, Innovex Biosciences, Richmond, CA) for 10 min and Background Buster (NB306, Innovex Biosciences) for 30 min. Tissue was incubated with AOAH primary antibody (ab130594, Abcam, Cambridge, UK) at 1:100 and Peptide YY (PYY) primary antibody (10 R-8025, clone RPY-B12, Fitzgerald Industries, Acton, MA) at 1:60 in Antibody Dilution Buffer (S0809, DAKO) overnight at 4°C. Tissue was then washed in 1X PBS three times for 5 min followed by incubation with goat anti-mouse (A11029, Invitrogen, Carlsbad, CA) or goat anti-rabbit (4412S, Cell Signaling, Danvers, MA) Alexa Fluor 488 secondary antibody or goat anti-rabbit (4413S, Cell Signaling) Alexa Fluor 555 secondary antibody diluted to 1:500 in Antibody Dilution Buffer (DAKO) for 2 h at room temperature. Secondary was washed in 1X PBS three times for 5 min, and nuclei were stained using DAPI followed by imaging using Volocity 5 (Improvision v5.3.1, PerkinElmer Incorporated, Downers Grove, IL).
Gut Microbiome Analyses
Fecal and cecal gut microbiota were analyzed as previously reported in Braundmeier-Fleming et al. (16). Briefly, DNA was extracted from fecal pellets or cecal stool by QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) by homogenizing with a Mini-Beadbeater and 0.1 mm zirconia/silica beads (Biospec, Bartlesville, OK) in 1.4 mL ASL stool lysis buffer (QIAGEN). Phylotype profiles of microbiota were generated by deep amplicon sequencing of the 16S rRNA V3–V5 hypervariable region. Barcoding samples followed by MiSeq tag sequencing yielded ∼50,000 reads/sample, detecting dominant and poorly represented taxa. The V3–V5 hypervariable region was selectively amplified through 30 cycles with primers 357F ( CCTACGGGAGGCAGCAG) and 926R ( CCGTCAATTCMTTTRAGT). Amplicon pools were quantified on a Qubit fluorometer, and fragment sizes were determined by Agilent bioanalyzer High Sensitivity DNA LabChip (Agilent Technologies, Wilmington, DE) followed by dilution to 10 nM. To accurately calculate matrix, phasing, and prephasing, amplicons were spiked with a PhiX control library to 20%. Mixtures were sequenced on an Illumina MiSeq V2 (250 nt from each end). To define operational taxonomic unit (OTU) abundance profiles and phylogenic relationships, sequence reads were binned at 97% identity using Quantitative Insights Into Microbial Ecology (QIIME) and Galaxy.
Predictive Functional Analysis
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was implemented using 16S rRNA sequence data from fecal samples to predict function as previously described (16, 22).
Fecal Metabolomics
Analyses of fecal and cecal gut metabolomes were analyzed as previously reported in Braundmeier-Fleming et al. (16). Briefly, duplicate samples were dried, and one was derivatized (23) with minor modifications: 90 min at 50°C with 80 μL of methoxyamine hydrochloride in pyridine (20 mg/mL) followed by incubating 60 min at 50°C with 80 μL N-methyl-N-trimethylsilytrifluoroacetamide. A 5-μL aliquot of a C31 fatty acid internal standard was added to each; in derivatized samples, this occurred before trimethylsilylation. Then 1 mL samples were injected with a split ratio of 7:1 into an Agilent GC-MS system (Agilent Inc, Palo Alto, CA) consisting of a 7890A gas chromatograph, a 5975C mass-selective detector, and a 7683B autosampler. GC was performed on 60-m HP-5MS columns (0.25 mm inner diameter, 0.25 mm film thickness; Agilent Inc) at 250°C injection and interface temperatures and 230°C ion source and helium carrier flow rate of 1.5 mL/min. A 5-min isothermal heating program at 70°C was followed by a 50°C/min increase to 310°C and finally 20 min at 310°C. The mass spectrometer was operated in positive electron impact mode at 69.9 eV ionization energy in a scan range of m/z 30–800. All resulting chromatogram were compared with mass spectrum libraries NIST08 (NIST, Gaithersburg, MD), WILEY08 (Palisade Corporation, Ithaca, NY), and a University of Illinois Metabolomics Center (Urbana, IL) custom library. All data were normalized to internal standards to facilitate direct comparisons, and chromatograms and mass spectra were evaluated using MSD ChemStation (Agilent Inc) and Automated Mass Spectral Deconvolution and Identification System (AMDIS; NIST), where retention time and mass spectra were implemented within the AMDIS method formats.
Cecum RNA Preparation and Microarray
The cecum was dissected from mice immediately following euthanasia by cervical dislocation and stored at −80°C. Cecal epithelium was homogenized in ice-cold TRIzol with a homogenizer, and total RNA was purified according to the manufacturer’s instructions (Invitrogen). Gene expression was quantified using a Affymetrix Mouse Genome 430 2.0 array that contain 45,101 probes and measure the expression level of 20,022 unique National Center for Biotechnology Information (NCBI) Entrez-identified genes. The data sets were preprocessed with normalization, variance stabilization, and log2 transformation. Student’s t tests were used to identify genes significantly differentially expressed (P < 0.05 and 2-fold) between WT and AOAH-deficient mice. Average difference values were normalized to median over the arrays. The data were filtered so that only those genes that were adequately measured on 75% of the arrays were included. A class comparison protocol was used to identify genes whose degree of expression differed significantly by twofold or more among the three groups. To visualize whole genome expression level by function and pathway, the microarray data were analyzed with MetaCore Analysis (Ingenuity Pathway Analysis, IPA; Ingenuity Systems, Redwood City, CA). IPA analysis identified canonical pathways differentially expressed (P < 0.05) between WT and AOAH-deficient mice.
Cohousing and Fecal Microbiota Transfer Experiments
For cohousing experiments, two WT and two AOAH-deficient mice were housed in the same cage (4 mice/cage) for at least 1 wk before experimentation. For fecal microbiota transfer (FMT) experiments, mice from both groups received oral FMT from either WT or AOAH-deficient mice. Fecal suspensions were prepared from three FMT donor mice by suspending one fecal pellet in 1-mL sterile PBS using a syringe and 18-gauge needle (Becton Dickinson, Vernon Hills, IL) until the entire volume could pass through the needle. We allowed particulates to settle for 30 s before drawing supernatant into a syringe attached to a 22-gauge gavage needle (Fine Science Tools, Foster City, CA). We pooled together 500 μL of the supernatants from each of the three 1 mL suspensions and deposited 200 μL of this pooled supernatant into the stomach of each recipient mouse by gavage. Each FMT recipient was gavaged every other day for 7 days. We cleaned gavage needles with 70% ethanol between mice, using different gavage needles for each treatment group.
FMT of Human Samples
Human stool samples from healthy patients and patients with IC/BPS were placed in PBS with 0.1% cysteine (PBSc). Diluted (10−4) samples were plated on 10-cm plates and grown under anaerobic conditions at 37°C. Once plates were dense, they were scraped into PBSc. A 100-μL of sample was gavaged into male mice three times for 3 days. After 7 days, mice were tested for tactile allodynia to assess pelvic pain.
Patients were enrolled using standard pelvic pain inclusion and exclusion criteria. The inclusion criteria for healthy control patients are “No” answers to the inclusion and exclusion questions for patients with pelvic pain. Pelvic pain inclusion criteria are must be at least 18-yr old; participant reports an unpleasant sensation of pain, pressure, or discomfort, perceived to be related to the bladder and/or pelvic region, associated with lower urinary tract symptoms (LUTS); symptoms have been present for the majority of the time during any 3 mo in the previous 6 mo; and symptoms have been present for the majority of the time during the most recent 3 mo. Pelvic pain exclusion criteria are the following: under the age of 18 yr; participant has an on-going symptomatic urethral stricture; participant has an on-going neurological disease or disorder affecting the bladder or bowel fistula; participant has a history of cystitis caused by tuberculosis, radiation therapy, or Cytoxan/cyclophosphamide therapy; participant has augmentation cystoplasty or cystectomy; participant has an active autoimmune or infectious disorder [such as Crohn’s disease or ulcerative colitis, lupus, rheumatoid arthritis, multiple sclerosis, or human immunodeficiency virus (HIV)]; participant has a history of cancer (with the exception of skin cancer); participant has any psychiatric or medical comorbidities that would interfere with study participation (e.g., dementia, psychosis, upcoming major surgery, lupus, active heart failure, diabetes, etc.); participant currently has a urinary tract infection and/or has had a positive urine culture in the past 6 wk; and participant is currently taking antibiotics or has in the past 3 mo.
Pelvic Allodynia
Mice were tested for pelvic allodynia using a von Frey filament stimulation to the pelvic region, as previously described in Rudick et al. (24). Briefly, before cohousing, baseline pelvic allodynia was measured for WT and AOAH-deficient mice. Mice were cohoused for 28 days followed by testing. After cohousing, mice were placed in a test chamber and allowed to acclimate for 5 min. Starting with the lowest force filament, five von Frey filaments were applied 10 times to the pelvic region. A response was considered to be painful if the animal jumped, lifted, and shook the hind paws, or excessively licked the pelvic region.
Urinary Bladder Distention Evoked Visceromotor Response
Mice were anesthetized with 2% isoflurane. Silver wire electrodes were placed on the superior oblique abdominal muscle and subcutaneously across the abdominal wall (as a ground) to allow differential amplification of the abdominal visceromotor response (VMR) signals. A lubricated angiocatheter was inserted into the bladder via the urethra for bladder distention. After completion of the surgical preparation, isoflurane anesthesia was reduced to ∼0.875% until a flexion reflex response was present (evoked by pinching the paw), but spontaneous escape behavior and righting reflex were absent. The animals were not restrained in any fashion. Body temperature was maintained using an overhead radiant light and monitored throughout the experiment. The conditions were optimized to establish a stable depth of anesthesia and consistent baseline VMR activity. Phasic bladder distention with PBS was then used to evoke bladder nociception. The PBS pressure was controlled by an automated distention control device custom made in the Washington University School of Medicine Electronic Shop (St. Louis, MO). The distention stimulus applied 20–80 mmHg pressure for 20 s every 2 min. The VMR signal was relayed in real-time using a Grass CP511 preamplifier to a PC via a WinDaq DI-720 module. Data were exported to Igor Pro 6.05 software (Wavemetrics, Portland, OR). Using a custom script, VMR signals were subtracted from the baseline, rectified and integrated over 20 s to quantify the area under the curve, and presented in arbitrary units. The investigator who quantified the VMR was blinded to treatment.
Defensive Burying
To measure anxiety-like behavior, AOAH-deficient and WT mice were singly placed in a novel holding cage with 7 cm of fresh bedding for 15 min. Anxiety-like behavior was recorded as latency of first (measured in seconds) and number of burying behaviors. Burying behavior was defined as the mouse aggressively sifting through bedding with both front paws and with their head facing down.
Statistical Analysis
Results were analyzed by Student’s t test or one-way/two-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison tests with the use of Prism software, version 6 (GraphPad, Inc, San Diego, CA). Differences were considered statistically significant at P < 0.05.
RESULTS
AOAH-Deficient Mice Have Enlarged Ceca and Increased Gut Permeability
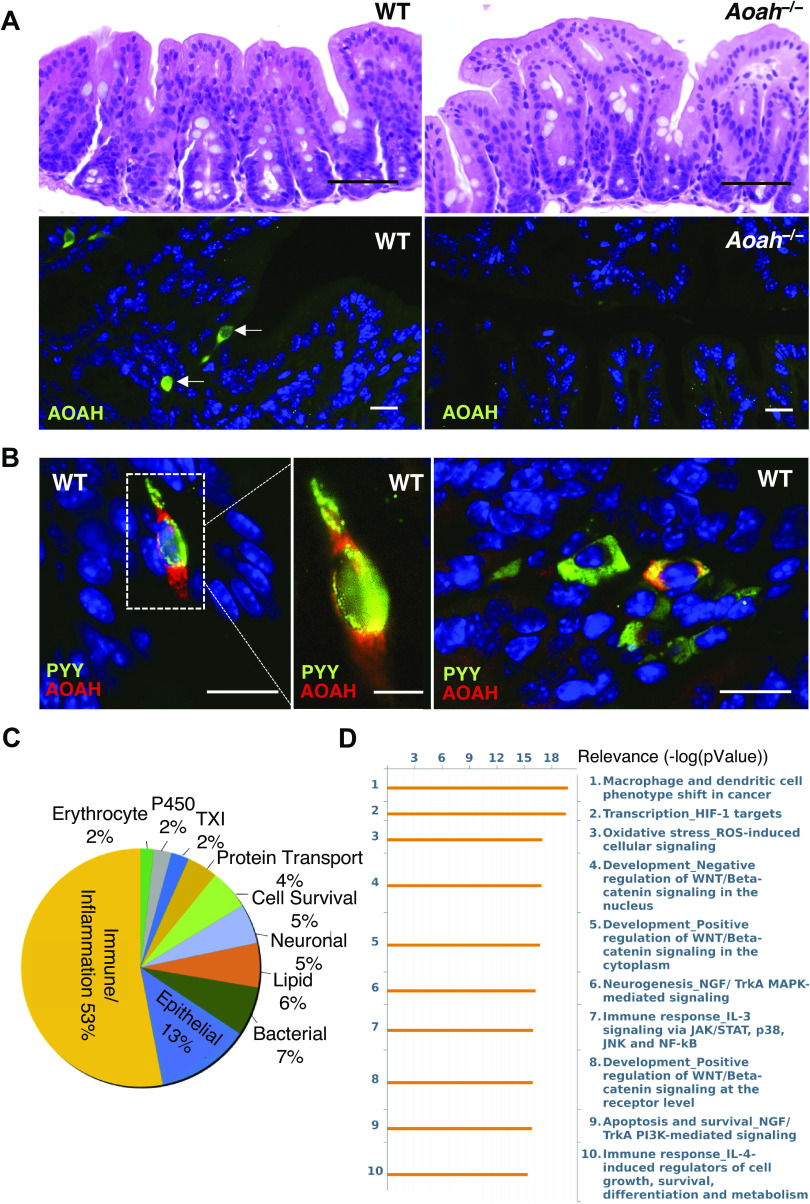
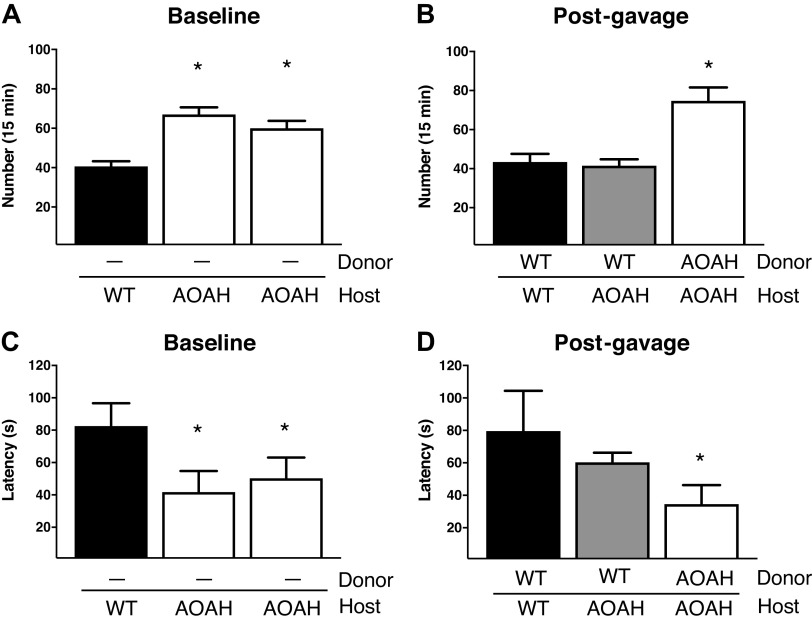
Dissection of AOAH-deficient mice suggested a gastrointestinal (GI) phenotype relative to wild-type mice where AOAH-deficient mice ceca appeared larger and distended in both females (Fig. 1A) and males (Fig. 1D). This difference in appearance was at least partially borne out in mass because the cecum and cecal stool as a function of body mass were significantly heavier in male AOAH-deficient mice than in wild-type mice (Fig. 1E). In contrast, despite the enlarged morphology of AOAH-deficient ceca, neither cecal tissue nor cecal stool were significantly more massive in AOAH-deficient females relative to wild-type mice (Fig. 1B). We further characterized the cecum phenotype by assessing cecum barrier function in female and male wild-type and AOAH-deficient mice by quantifying cecum transepithelial electrical resistance (TEER) ex vivo via an Ussing chamber (Fig. 1, C and F, respectively). Cecum TEER was significantly lower in AOAH-deficient mice than in wild-type mice in both females (33.58 ± 4.77 Ω·cm2 for wild type and 19.94 ± 6.66 Ω·cm2 for Aoah—/—; Fig. 1C) and males (27.66 ± 6.63 Ω·cm2 for wild type and 17.11 ± 6.11 Ω·cm2 for Aoah—/—; Fig. 1F), indicating that AOAH-deficient mice have increased cecum permeability or “leakiness” in both sexes. Although number of cases in males may exceed previous reports (25), IC/BPS is more common in women, and therefore, we focused further characterization on female mice.
Figure 1.
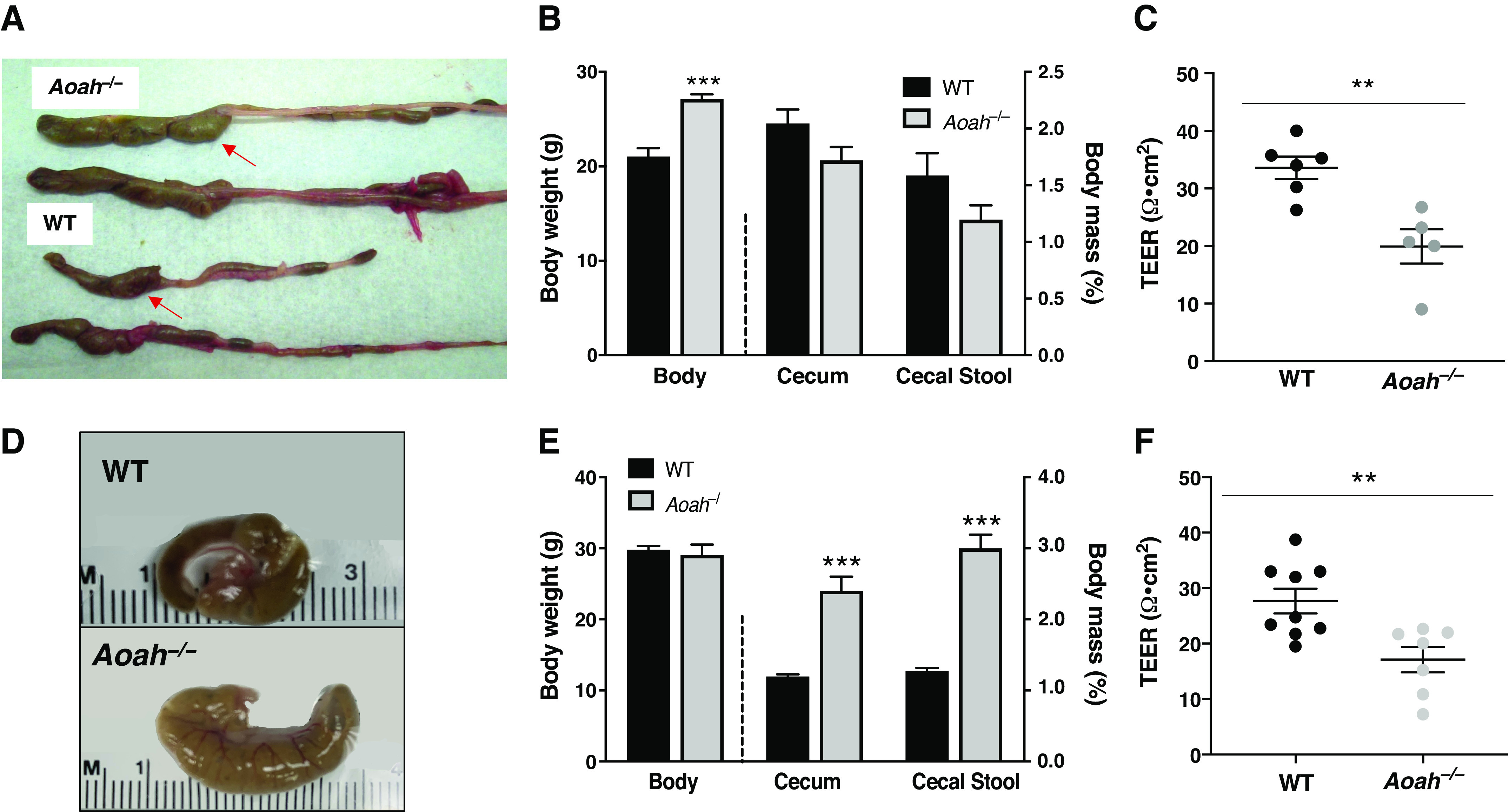
Female acyloxyacyl hydrolase-deficient (Aoah−/−) mice exhibit enlarged ceca and compromised gut permeability. A: comparison of representative cecum sizes (arrows) of female wild-type (WT) C57BL/6 (bottom) and Aoah−/− (top) mice. B: female Aoah−/− mice showed increased body weight vs. WT mice (WT mice: n = 4 and Aoah–/– mice: n = 7, ***P < 0.0001, Student’s t test, two-tailed). No differences were observed in the mass of the cecum and cecal stool (WT mice: n = 4 and Aoah−/− mice: n = 6, P > 0.05, Student’s t test, two-tailed). Bars represented as average ± SE. C: ceca of female Aoah−/− mice showed a decrease in transepithelial electrical resistance (TEER) compared with WT (WT mice: n = 6 and Aoah−/− mice: n = 5, **P < 0.01, Student’s t test, two-tailed). Data are represented as individual values and average (represented by horizontal line) ± SE. Male Aoah−/− mice exhibit enlarged ceca, greater mass of cecal contents, and compromised gut permeability. D: comparison of representative cecum sizes of male WT (top) and Aoah−/− (bottom) mice. E: male Aoah−/− mice showed increased cecal weight vs. WT mice (WT mice: n = 5 and Aoah−/− mice: n = 4, P < 0.001, Student’s t test, two-tailed) and increased mass of cecal contents (WT mice: n = 3 and Aoah−/− mice: n = 4, ***P < 0.0001, Student’s t test, two-tailed). No differences were observed in body weights (n = 4 for both groups, P > 0.05, Student’s t test, two-tailed). Bars are represented as average ± SE. F: male Aoah−/− mice showed a decrease in TEER compared with WT (WT mice: n = 9 and Aoah−/− mice: n = 7, P < 0.01, Student’s t test, two-tailed). Data are represented as individual values and average (represented by horizontal line) ± SE.
AOAH-Deficient Mice Have Altered Gut Signaling Pathways
The compromised cecal TEER of AOAH-deficient mice suggests functionally altered epithelium, yet histological examination uncovered no obvious differences in cecal epithelia of wild-type and AOAH-deficient mice (Fig. 2A, top). Immunostaining of AOAH protein in wild-type mouse cecum revealed sparse expression of AOAH in the epithelium, which was mainly absent in AOAH-deficient cecum (Fig. 2A, bottom). AOAH-positive cells in wild-type cecum were characteristic of enteroendocrine cells (EECs) containing neuropods (Fig. 2B, left and middle), which function as GI sensory cells that can signal directly to the CNS (26). Immunostaining of AOAH and an EEC marker peptide YY (PYY) revealed colocalization of AOAH in a small subset of EECs within the cecum (Fig. 2B, right). Transcriptome profiling by microarray analyses revealed significant differences in gene expression consistent with altered signaling pathways and disease risk in the AOAH-deficient ceca (Fig. 2, C and D, and Supplemental Fig. S1; all Supplemental material is available at https://doi.org/10.6084/m9.figshare.14428700, respectively). Examination of genes increased/decreased at least twofold indicated altered mRNA abundance in AOAH-deficient mice of genes associated with immune response/inflammation (53%), epithelia (13%), bacteria (7%), lipids (6%), neurons (5%), cell survival (5%), protein transport (4%), thioredoxin (2%), P450 (2%), and erythrocytes (2%) (Fig. 2C). Consistent with this, pathway analyses also revealed increased immune response signaling in AOAH-deficient mice compared with wild type (Fig. 2D). Together, these findings suggest that an inflammatory milieu and alterations in mRNA associated with epithelia contribute to increased cecal epithelial permeability underlying a leaky gut phenotype of AOAH-deficient mice.
Figure 2.
AOAH-deficient mice have normal gut morphology but altered gut signaling pathways. A: comparison of representative cecum H&E staining of female WT (top left) and Aoah−/− (top right) mice (n = 3 mice for both conditions, scale bar: 50 μm). Immunostaining of AOAH (green) in WT (bottom left) and Aoah−/− (bottom right) cecum. DAPI staining nuclei shown in blue (n = 3 mice for both conditions, scale bar: 17 μm). B: immunostaining of AOAH (red) and PYY (green) in WT cecum. DAPI staining nuclei shown in blue (n = 3 mice, scale bar: 17 μm for left and right, 7 μm for middle). C: pie chart of transcriptome profiling by microarray analysis showing levels of mRNA with 2-fold difference in AOAH-deficient cecum compared with wild-type (n = 5 mice for both conditions). D: pathway analysis indicating top 10 signaling pathways altered in AOAH-deficient cecum compared with wild-type (n = 5 mice for both conditions). AOAH, acyloxyacyl hydrolase; H&E, hematoxylin and eosin; HIF-1, hypoxia-inducible factor 1; WT, wild type.
AOAH-Deficient Mice Have Altered Gut Microbiota
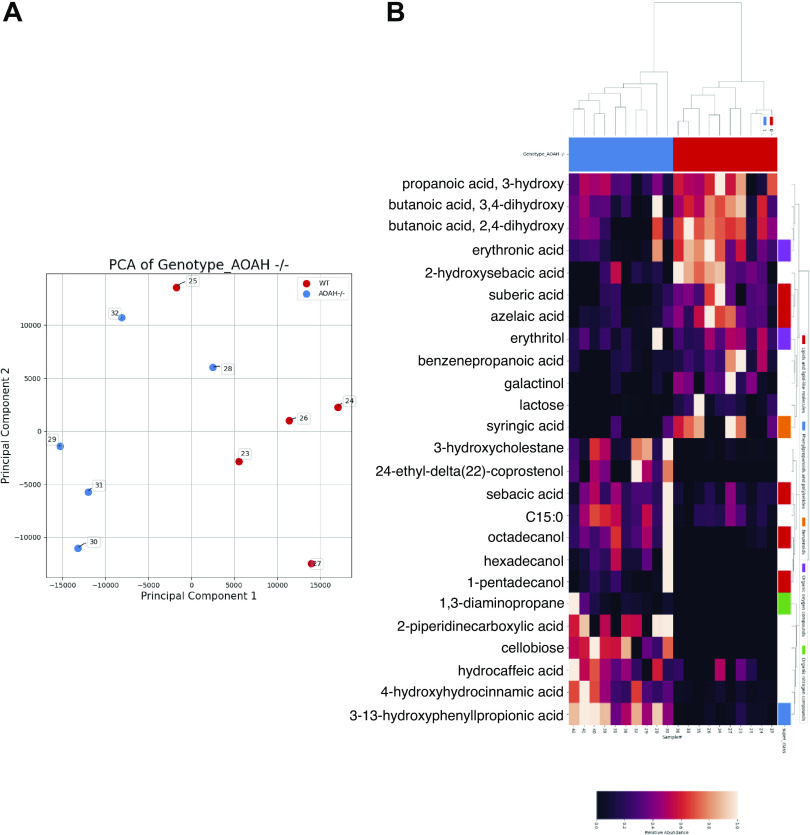
Previous studies have linked increased gut permeability to gut dysbiosis, and cecal enlargement is a long-known phenotype of the altered microbiome of germ-free rodents, both suggesting potential gut dysbiosis in AOAH-deficient mice (27–29). We used 16S rRNA amplicon sequencing data to assess large compositional differences in fecal (Fig. 3) and cecal (Supplemental Fig. S2) microbiota between wild-type and Aoah−/− mice. As shown by principal component analysis (PCA) and the corresponding heat maps, AOAH-deficient mice demonstrate an altered gut microbiome compared with wild-type control in both fecal and cecal stool (Fig. 3, A and C, and Supplemental Fig. S2, A and D, respectively). AOAH-deficient mice show group variance along the PC1 axis in bacterial composition of gut flora compared with wild-type mice in both fecal and cecal stool (Fig. 3A and Supplemental Fig. S2A, respectively). In addition, α-diversity calculations revealed a greater number of operational taxonomic units (OTUs) in AOAH-deficient fecal stool (444.4 ± 30.67 OTUs) compared with WT fecal stool (366.6 ± 43.93 OTUs, Fig. 3B). We did not observe a significantly different number of observed OTUs between groups in cecal stool (Supplemental Fig. S2B). However, PC2 axes reveal larger variances among individual AOAH-deficient mice compared with wild type, suggesting greater bacterial diversity in mice deficient for AOAH (Fig. 3A and Supplemental Fig. S2A).
Figure 3.
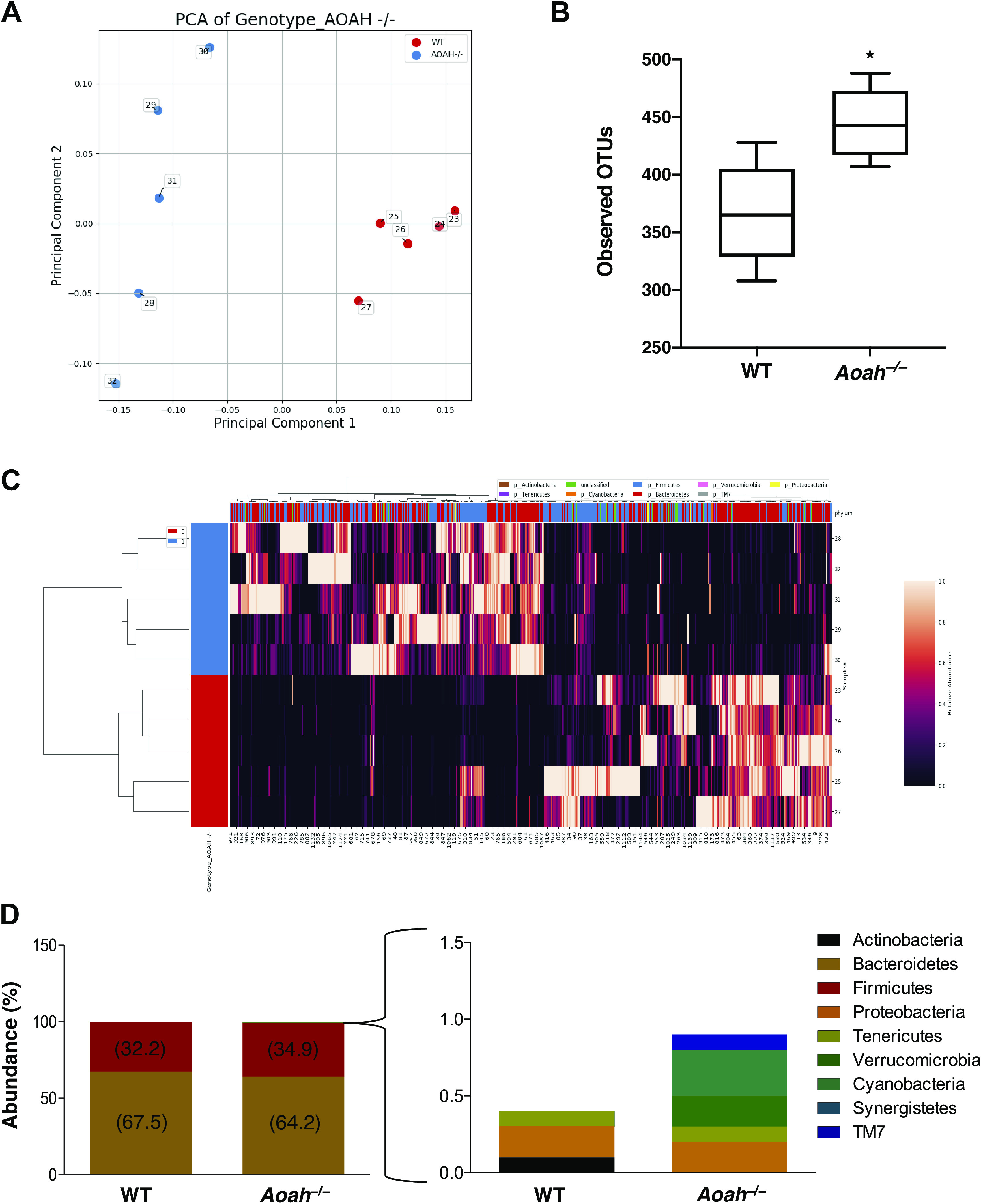
Aoah−/− mice exhibit gut dysbiosis. A: PCA plot of 16S rRNA analyses of fecal stool. Dots represent individual mice; AOAH-deficient mice are shown in blue (n = 5) and wild-type mice are shown in red (n = 5). B: α-diversity as measured by observed operational taxonomic units (OTUs), n = 5 for both groups, *P < 0.05. C: heat map of 16S rRNA analyses of AOAH-deficient (blue) and wild-type (red) fecal stool (n = 5). D: quantification of bacterial phyla using 16S rRNA sequencing in wild-type and AOAH-deficient fecal stool (n = 5). AOAH, acyloxyacyl hydrolase; PCA, principal component analysis.
To determine the specificity of AOAH modulation of gut microbiota, we examined the relative abundance of bacterial phyla (Fig. 3D). Over 99% of bacteria identified in both mouse groups belonged to Bacteroidetes and Firmicutes phyla, with no significant differences in abundance of these groups between wild-type and AOAH-deficient stool. However, there were differences in less common phyla such as Verrucomicrobia (0% expression for WT and 0.2% expression for Aoah−/−), Cyanobacteria (0% expression for wild type and 0.3% expression for Aoah−/−), and TM7 (0% expression for wild type and 0.1% expression for Aoah−/−), where AOAH-deficient mice showed more abundant expression of these phyla compared with wild type. A decrease in abundance was observed for Actinobacteria in AOAH-deficient mice compared with wild type (0.10% expression for wild type and 0% expression for Aoah−/−). These findings suggest that a gut microbiome is richer in diversity in AOAH-deficient mice than in wild type.
To identify changes in the presence and absence of bacterial species between wild-type and AOAH-deficient mice, the top 500 bacterial OTUs were compared in fecal (Table 1) and cecal (Supplemental Table S1) samples. We observed several bacterial species that were present in AOAH-deficient stool but absent in wild-type stool (Table 1). In addition, several bacterial species were absent in AOAH-deficient stool but present in wild-type stool. Cecal samples exhibited a modest overlap with our findings in fecal samples but were identified to have several more bacterial species unique to AOAH-deficient mice compared with wild-type mice (see Supplemental Table S1 for full list). On further analyses of bacterial functionality, many of the bacterial species with altered abundance were associated with carbohydrate metabolism (Table 1 and Supplemental Table S1).
Table 1.
Characterization of fecal bacteria in Aoah−/− stool compared with WT
| Fecal Bacterial Species | Absent or Present | Functionality |
|---|---|---|
| Alistipes onderdonkii | P | Carbohydrate fermentation; end product: succinic acid (30) |
| Butyrivibrio crossotus | P | Th1/Th2 inflammation balance; fermentation of maltose, starch, glycogen, and dextrin (31,32) |
| Cellulosibacter alkalithermophilus | P | Cellulose and xylan hydrolysis (33) |
| Clostridium colinum | P | Fatty acid metabolism; associated with enteric disease (34) |
| Lactonifactor longoviformis | P | Dehydrogenation of enterodiol to enterolactone (35) |
| Lachnoanaerobaculum orale | P | No active metabolism (36) |
| Olivibacter sitiensis | P | Diphenol metabolism (37) |
| Pediococcus parvulus | P | Carbohydrate metabolism (38) |
| Siphonobacter aquaeclarae | P | Production of catalase and gelatinase (39) |
| Anaerorhabdus furcosa | A | Esculin hydrolysis; end products: acetic and lactic acids (40) |
| Bacillus samanii | A | Glucose, lactose, maltose, sucrose, and trehalose fermentation (41) |
| Clostridium methylpentosum | A | Fermentation of methylpentoses and pentoses (42) |
| Lachnospira pectinoschiza | A | Carbohydrate metabolism (43) |
| Melissococcus plutonius | A | Glucose, fructose, and mannose metabolism (44) |
| Parabacteroides gordonii | A | l-arabinose production (45) |
Aoah, acyloxyacyl hydrolase; WT, wild type.
Cohousing Alters the Functional Characterization of Bacterial Communities
We used in silico metagenome prediction of function with PICRUSt to gain functional insights into the effects of AOAH deficiency and the impacts of bacterial communities by microbiome manipulation (22). PICRUSt revealed that AOAH-deficient mice have several significant differences relative to controls including genes that mediate cell growth and death, signaling molecules and interaction, DNA replication and repair, transcription, human diseases, lipid metabolism, the circulatory system, the digestive system, and the immune system (Fig. 4 and Supplemental Fig. S1). On further analyses, we observed that cohousing AOAH-deficient mice with wild-type mice to allow for coprophagia-mediated microbiome manipulation, altered the predictive metagenomes in both mouse strains (Fig. 4). Wild-type and AOAH-deficient mice showed differences in genes associated with several different taxa, including the retinoic acid inducible gene 1 (RIG-I)-like receptor signaling pathway (Fig. 4A), which plays a role in the innate immune system’s ability to recognize pathogens (46). The differences observed in a subset of functional classes were rescued in cohoused AOAH-deficient mice (Fig. 4, A–D and H). Interestingly, for other functional classes, cohousing resulted in no changes in AOAH-deficient mice but altered the predictive values in cohoused wild-type to similar values observed in AOAH-deficient mice (Fig. 4, E–G). These data suggest that both wild-type and AOAH-deficient microbiota can influence changes in the metagenome.
Figure 4.
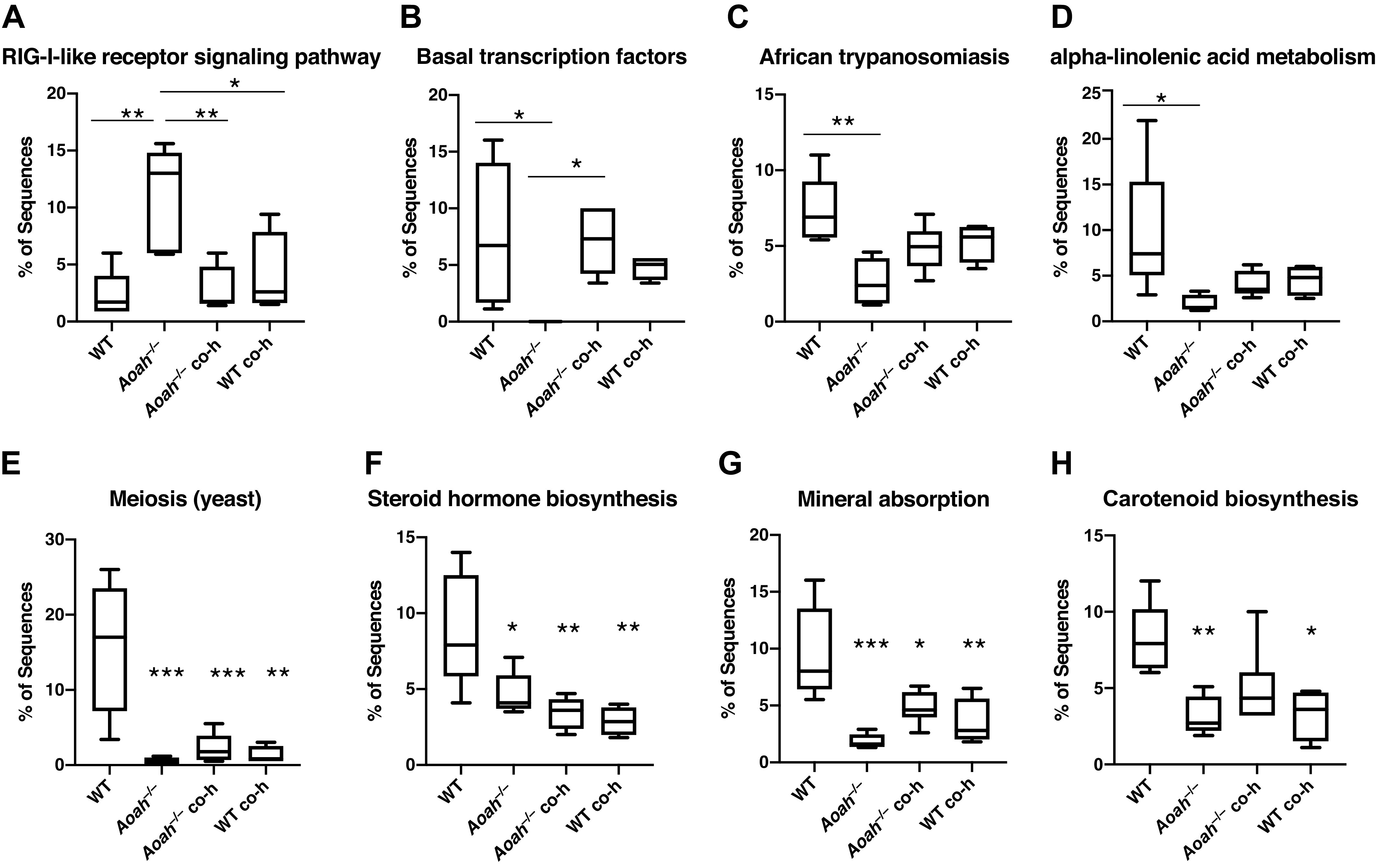
Microbiome-dependent changes in the functional characterization of bacterial communities. Biological functions of bacterial communities were predicted by implementing Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analyses using 16S rRNA sequence data from wild-type and AOAH-deficient fecal samples. Changes in the gut microbiome by genetic manipulation (Aoah−/−) or by cohousing (Aoah−/− co-h and B6 co-h) resulted in altered predictions including functions related to the retinoic acid-inducible gene I (RIG-I)-like receptor signaling pathway (A), basal transcription factors (B), African trypanosomiasis (C), α-linolenic acid metabolism (D), meiosis (yeast, E), steroid hormone biosynthesis (F), mineral absorption (G), and carotenoid biosynthesis (H). n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA followed by post hoc Tukey’s HSD. Data represented as average ± SE. AOAH, acyloxyacyl hydrolase; co-h, cohoused; HSD, honestly significant difference.
AOAH-Deficient Mice Have Altered Gut Metabolites
To identify the biochemical consequences of dysbiosis associated with AOAH deficiency, we performed untargeted metabolomics. In both fecal and cecal stool, AOAH-deficient mice show altered gut metabolomes compared with controls (Fig. 5 and Supplemental Fig. S2C, respectively). Among the top 500 feature metabolites, we identified numerous metabolites either uniquely present or absent from fecal or cecal AOAH-deficient mice (Table 2 and Supplemental Tables S2 and S3). We observed that 24-ethyl-delta(22)-coprostenol, cellobiose, and hexadecanol were present in AOAH-deficient fecal samples but absent in wild type, whereas methionine was absent in AOAH-deficient fecal samples (Table 2). Similarly, cecal samples also expressed the same unique metabolites as fecal samples in AOAH-deficient mice (Supplemental Tables S2 and S3). In addition to overlap with fecal metabolites, we identified 23 newly present metabolites and 8 absent metabolites in AOAH-deficient cecal stool compared with controls (Supplemental Tables S2 and S3). Similar to our observations of bacterial species, functionality of the metabolites with aberrant presence/absence were primarily associated with carbohydrate metabolism (compare Table 1 and Supplemental Table S1 with Table 2 and Supplemental Tables S2 and S3). In addition, we observed the presence/absence of gut metabolites associated with lipid and amino acid metabolism (Tables 2 and Supplemental Tables S2 and S3). Taken together, our findings show that AOAH-deficient mice have an altered gut microbiome that corresponds with altered accumulation of gut metabolites.
Figure 5.
Aoah−/− mice exhibit altered fecal metabolomics. A: PCA plot of metabolomics of fecal stool. Dots represent individual mice; AOAH-deficient mice are shown in blue (n = 5) and wild-type mice are shown in red (n = 5). B: heat map of metabolomics of AOAH-deficient (blue) and wild-type (red) fecal stool (n = 5). AOAH, acyloxyacyl hydrolase; PCA, principal component analysis.
Table 2.
Characterization of fecal metabolites in Aoah−/− stool compared with WT
| Fecal Metabolites | Absent or Present | Functionality |
|---|---|---|
| 24-Ethyl-delta(22)-coprostenol | P | Derived from cholesterol metabolism (47) |
| Cellobiose | P | Reducing sugar; glucose hydrolysis (48) |
| Hexadecanol | P | Palmitic acid reduction (49) |
| Methionine | A | Polyamine and creatine synthesis; conversion to S-adenosylmethionine; goblet cell development in small intestine (50,51) |
Aoah, acyloxyacyl hydrolase; WT, wild type.
Cohousing Converges Microbiota and Alters Gut Phenotype in AOAH-Deficient Mice
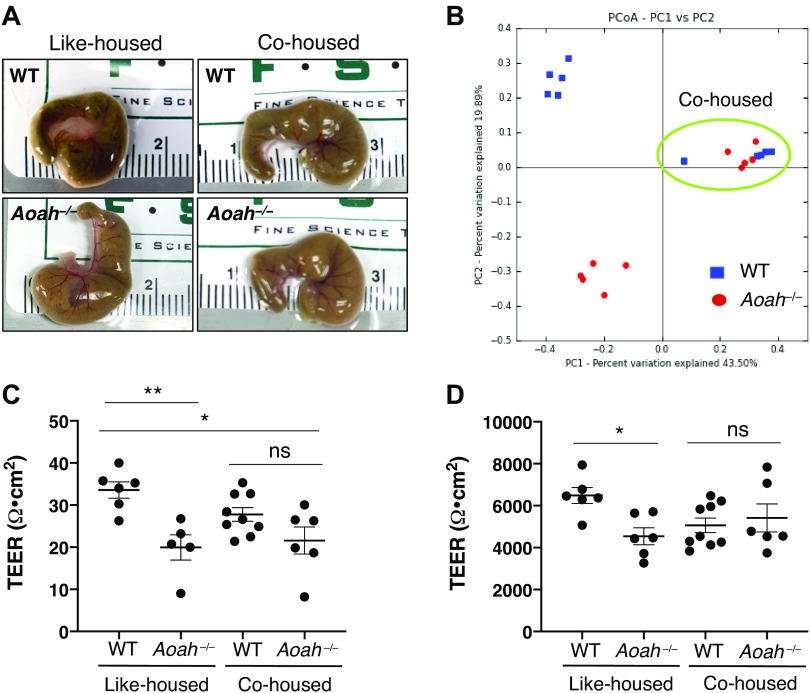
To determine whether gut dysbiosis could be rescued in AOAH-deficient mice, we cohoused AOAH-deficient and wild-type mice for 7 days to allow for coprophagia-mediated microbiome manipulation. Cohousing resulted in a convergence of cecum size between wild-type and AOAH-deficient mice, in contrast with the distinct morphologies in like-housed mice (Fig. 6A). We next determined whether our phenotypic observations in cohoused mice would match in genotype. Similar to our observations of converged gut morphology (Fig. 6A), 16S rRNA amplicon sequencing revealed that cohousing resulted in a microbiota genotype that was converged in both mouse strains and distinct from both wild-type and AOAH-deficient mice that were like-housed (Fig. 6B).
Figure 6.
Converging microbiota by cohousing results in altered gut morphology and TEER. A: comparison of representative cecum sizes of female WT (top left), Aoah−/− (bottom left), cohoused WT (top right), and cohoused Aoah−/− (bottom right) mice. B: principle component analysis (PCoA) plot of 16S rRNA analyses of fecal stool in like-housed and cohoused animals. Dots represent individual mice; AOAH-deficient mice are shown in red and wild-type mice are shown in blue. n = 5 for like-housed conditions and n = 10 for cohoused condition (circled in green). C: Aoah−/− mice showed an improvement in cecum TEER after cohousing (like-housed WT mice: n = 6, like-housed Aoah−/− mice: n = 5, cohoused WT mice: n = 8, cohoused Aoah−/− mice: n = 6; *P < 0.05, **P < 0.01, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as individual values and average (represented by horizontal line) ± SE. D: Aoah−/− mice showed an improvement in bladder TEER after cohousing (like-housed WT mice: n = 6, like-housed Aoah−/− mice: n = 6, cohoused WT mice: n = 9, cohoused Aoah−/− mice: n = 6; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as individual values and average (represented by horizontal line) ± SE. AOAH, acyloxyacyl hydrolase; HSD, honestly significant difference; ns, not significant; PC1, principal component 1; PC2, principal component 2; TEER, transepithelial electrical resistance; WT, wild type.
We next addressed whether cohousing altered the leaky gut phenotype of AOAH-deficient mice (Fig. 6C). Similar to cecum morphology, quantifying TEER showed a convergence of cecum barrier function (4.85 Ω·cm2 for cohoused wild-type mice and 21.58 ± 7.85 Ω·cm2 for cohoused Aoah−/− mice; Fig. 6C). However, cohoused AOAH-deficient mice still exhibited significantly increased cecum permeability compared with like-housed wild-type mice, suggesting that cohousing does not fully resolve barrier dysfunction in AOAH-deficient mice (Fig. 6C).
Bladder biopsies of patients with IC/BPS suggest defects in bladder barrier function based on histopathological features (52–54). Also, clinical studies using the potassium sensitivity test have revealed increased sensitivity in patients with IC/BPS to intravesical potassium consistent with functionally diminished barrier function (55–57). Similarly, we reported that AOAH-deficient mice have bladder barrier dysfunction as measured by increased bladder permeability (11). Because previous studies in pelvic organ cross talk have shown that the gut can modulate bladder inflammation and pelvic pain (58–62), we examined whether microbiome manipulation altered the bladder permeability phenotype of AOAH-deficient mice. Cohoused AOAH-deficient mice showed bladder TEER similar to cohoused wild-type mice (5,062 ± 1,037 Ω·cm2 for co-housed WT mice and 5,416 ± 1,634 Ω·cm2 for Aoah−/− mice, Fig. 6D). In addition, bladder permeability was not significantly different between cohoused mice and like-housed wild-type mice (Fig. 6D), suggesting that convergence of microbiota can alleviate the leaky bladder phenotype of AOAH-deficient mice.
Microbiota Manipulation Alters Pelvic Pain
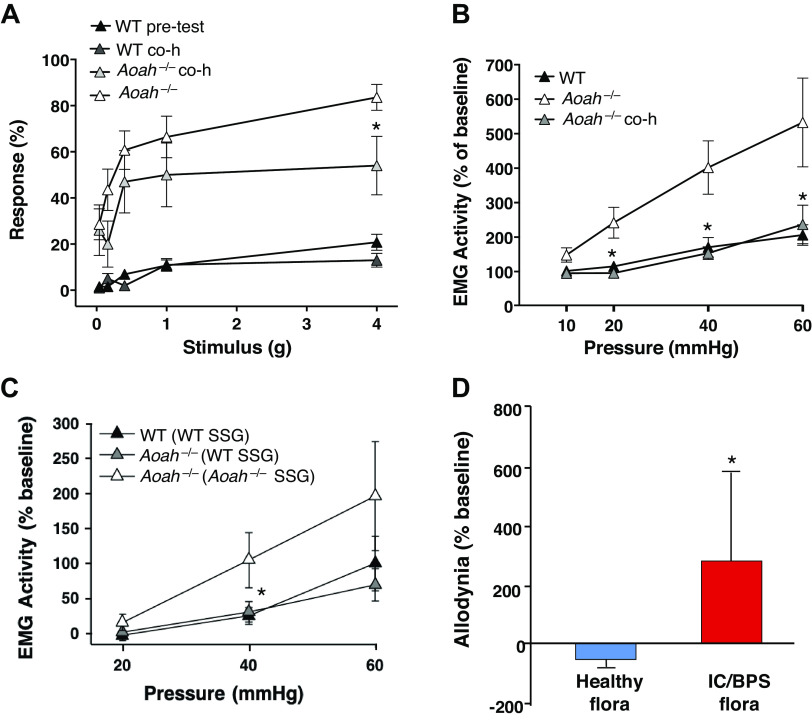
AOAH-deficient mice exhibit pelvic pain behaviors that include pelvic allodynia and increased visceromotor response (VMR) (6). To determine whether altered gut flora affected the pelvic pain phenotype of AOAH-deficient mice, we cohoused wild-type and AOAH-deficient mice and quantified allodynia in response to von Frey filaments applied to the pelvic region (Fig. 7A). Significant differences in pelvic allodynia were observed after 28 days of cohousing. AOAH-deficient mice showed a 35% reduction in pelvic allodynia from baseline in response to the highest stimulus compared with AOAH-deficient mice before being cohoused (Fig. 7A, 83.57% ± 20.98% response at baseline vs. 54.00% ± 40.06% response after cohousing). These data suggest that changes in the gut microbiome, in part, can decrease pelvic allodynia in AOAH-deficient mice but do not completely alleviate the pelvic pain phenotype.
Figure 7.
Pelvic pain phenotype is microbiome dependent. A: AOAH-deficient mice exhibited increased pelvic allodynia compared with WT, which was partially alleviated by cohousing for 28 days as shown through response to von Frey filaments stimulating the pelvic region (n = 10; *P < 0.05, Student’s t test, two-tailed). Data are represented as average response (%) ± SE. B: visceromotor response (VMR) was quantified in mice as electromyography (EMG) activity in response to bladder distension under anesthesia. EMG was elevated in AOAH-deficient mice, which was alleviated by cohousing for 7 days [WT mice: n = 15, Aoah−/− mice: n = 9, cohoused (co-h) Aoah−/− mice: n = 9; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD]. Data are represented as average EMG activity (%) ± SE. C: measurement of VMR following stool slurry gavage (SSG) from WT and Aoah−/− donors. EMG was elevated in AOAH-deficient mice that received SSG from other AOAH-deficient mice compared with WT mice that received SSG from other WT mice. SSG in AOAH-deficient mice from WT donors resulted in reduced EMG activity (WT mice: n = 5 and Aoah−/− mice conditions: n = 4; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as average EMG activity (%) ± SE. D: male AOAH-deficient mice exhibited increased pelvic allodynia following SSG of anaerobic cultures from patients with IC/BPS compared with AOAH-deficient mice that received SSG from healthy patients, as shown through response to von Frey filaments stimulating the pelvic region (n = 10 for healthy flora, n = 12 for IC/BPS flora; *P < 0.05, Student’s t test, two-tailed). Data are represented as average response (%) ± SE. AOAH, acyloxyacyl hydrolase; HSD, honestly significant difference; IC/BPS, interstitial cystitis/bladder pain syndrome; WT, wild type.
To confirm the findings of altered allodynia, we quantified the effects of cohousing on VMR measured by superior oblique abdominal muscle electromyography (EMG) activity following urinary bladder distension (Fig. 7, B and C). Cohousing significantly reduced bladder distension-induced VMR in AOAH-deficient mice to levels similar to wild type (Fig. 7B), suggesting decreased pelvic pain following microbiome manipulation. These findings were further corroborated through FMT by stool slurry gavage. AOAH-deficient mice that received gavage of wild-type stool slurry showed reduced EMG activity on bladder distension compared with AOAH-deficient mice that received gavage of stool from other AOAH-deficient donors (Fig. 7C).
Previous reports have shown gut dysbiosis in patients with IC/BPS (16, 17), but whether this dysbiosis modulates pelvic pain remains speculative. To address this question, AOAH-deficient mice were gavaged with anaerobic cultures of human stool from healthy controls or patients with IC/BPS. After 1 wk of microbiota gavage, mice were assessed for pelvic allodynia. AOAH-deficient mice that were exposed to IC/BPS microbiota showed significantly increased pelvic allodynia from baseline compared with AOAH-deficient mice gavaged with control microbiota (Fig. 7D), suggesting that the gut microbiota of patients with IC/BPS can influence pelvic pain. Taken together, our data from manipulating the microbiome of AOAH-deficient mice suggest that gut microbiota modulate pelvic pain.
Fecal Microbiota Transplantation Reduces Anxiety-like Behavior in AOAH-Deficient Mice
We have previously reported that AOAH-deficient mice exhibit comorbid anxious/depressive behaviors, similar to patients with IC/BPS (3, 5, 12). To assess whether the gut microbiome contributes to the anxious/depressive phenotype of AOAH-deficient mice, we measured defensive burying behavior after placing mice into a novel holding cage with deep, clean bedding. Anxious behaviors were quantified as latency to the first burying event and the number of burying behaviors. Corroborating our prior findings, AOAH-deficient mice exhibited higher levels of anxious behaviors as observed by increased number of burying events and decreased latency to burying (Fig. 8, A and C, respectively). Anxiety-like behavior was rescued in AOAH-deficient mice that received gavage of wild-type stool, showing fewer burying events and longer latency to burying compared with AOAH-deficient mice that received gavage of AOAH-deficient stool (Fig. 8, B and D, respectively). Our findings show that the gut microbiome plays a significant role in the anxiety phenotype of AOAH-deficient mice.
Figure 8.
Anxiety-like behavior in AOAH-deficient mice is microbiome dependent. A: analyses of defensive burying behavior revealed AOAH-deficient mice exhibited increased anxiety-like behavior as measured through number of burying attempts compared with WT mice (WT mice: n = 5 and Aoah−/− mice conditions: n = 4; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as average number of burying attempts (for 15 min) ± SE. B: AOAH-deficient mice showed reduced burying attempts 7 days following SSG from WT donors compared with AOAH-deficient mice that received SSG from AOAH-deficient donors (WT mice: n = 5 and Aoah−/− mice conditions: n = 4; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data represented as average number of burying attempts (for 15 min) ± SE. C: defensive burying behavior revealed AOAH-deficient mice exhibited increased anxiety-like behavior as observed through decreased latency of their first burying attempt compared with WT mice (WT mice: n = 5 and Aoah−/− mice conditions: n = 4; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as latency of burying attempt (in seconds) ± SE. D: AOAH-deficient mice showed increased latency in their first burying attempt 7 days following SSG from WT donors compared with AOAH-deficient mice that received SSG from AOAH-deficient donors (WT mice: n = 5 and Aoah−/− mice conditions: n = 4; *P < 0.05, one-way ANOVA followed by post hoc Tukey’s HSD). Data are represented as latency of burying attempt (in seconds) ± SE. AOAH, acyloxyacyl hydrolase; HSD, honestly significant difference; SSG, stool slurry gavage; WT, wild type.
DISCUSSION
AOAH deficiency mimics many aspects of IC/BPS, and here, we identified Aoah as a novel genetic modulator of gut microbiota. Prior studies have implicated host genetic factors in microbiota, including twin studies that showed increased microbiota heritability among monozygotic twins and identified specific host loci in interacting with specific taxa and mediating metabolic processes (63, 64). Additional human genome-wide association studies (GWAS) and murine quantitative trait loci studies have furthered this understanding of interactions between microbiota and host genetics and provided new mechanistic insights, yet few loci implicated by such studies have been interrogated functionally. For example, 42 human loci were identified that were associated with bacterial diversity, including the vitamin D receptor, and vitamin D receptor-deficient mice had increased Parabacteroides abundance (65). Although loci on chromosome 7, where AOAH resides, were significantly associated with altered microbiota, none of these lay nearer than 2MB from AOAH, so GWAS studies have not yet directly implicated human AOAH in microbiome modulation. However, PLA2G3 was significantly associated with microbiota diversity, suggesting a role for arachidonic acid metabolism as a modulator of the human microbiome (65). That finding dovetails with in silico metagenome analyses that identified arachidonic acid metabolism as significantly associated with microbiota of patients with IC/BPS (16). We recently reported that AOAH-deficient mice accumulate elevated levels of arachidonic acid and PGE2 in the CNS and found evidence that AOAH is a transacylase-mediating arachidonic acid homeostasis via sequestration in phospholipid pools (13). Thus, together these studies are consistent with an emerging model where loss of AOAH-dependent arachidonic acid homeostasis contributes to gut dysbiosis associated with IC/BPS symptoms.
AOAH-deficient mice exhibited enlarged ceca with decreased barrier function, and transcriptome analyses indicated association with Wnt signaling and inflammatory/immune pathways (Figs. 1 and 2). Altered Wnt signaling in AOAH deficiency may be due to elevated PGE2 in these mice and the known role of PGE2 as a modulator of Wnt expression (13). Moreover, Wnt expression has been previously implicated in pelvic pain, where a putative urinary marker of IC/BPS was identified as a glycosylated fragment of Frizzled-8 (66). Alternatively, in the absence of AOAH that would otherwise detoxify LPS, upregulation of inflammatory and immune responses may be due to excess toll-like receptor (TLR)-mediated signals evoked by LPS of the gut microbiota. Increased inflammatory responses, in turn, may contribute to loss of barrier function and consequent “leaky gut” (67, 68). Consistent with this possibility, in silico metagenome analyses by PICRUSt revealed changes in AOAH-deficient mice associated increased RIG-I-like receptor signaling pathway, a regulator of innate immune activation in various cell types (46). These data were particularly interesting as at least a subset of patients with IC/BPS exhibit bladder inflammation associated with activated mast cells (69), and we previously reported that AOAH-deficient mice exhibit significantly more bladder mast cells (6). In addition, mast cells in the gastrointestinal (GI) tract show increased activation in patients with GI disorders such as irritable bowel syndrome (IBS) (70, 71). As dysfunctional bowel-bladder cross talk may also influence GI mast cells in AOAH-deficient mice and gut “leakiness,” activation of these cells should be analyzed.
Immunostaining did not reveal widespread expression of AOAH in gut epithelium or underlying tissue layers (Fig. 2), suggesting that AOAH effects on gut gene expression lie elsewhere. Indeed, we previously reported that AOAH is a genetic regulator of the locus encoding corticotropin-releasing factor (CRF) in brain sites mediating bladder and stress responses where AOAH deficiency leads to increased Crf expression and stress responses (11, 12). CRF alters gut function at multiple levels (reviewed in Refs. 72 and 73). At the enteric epithelium, elevated CRF engages CRF receptor 1 receptors on underlying mast cells, resulting in increased epithelial permeability (74). In addition, centrally administered CRF suppresses gut motility. This raises the possibility that elevated CRF alters cecal motility, contributing to cecal enlargement in AOAH-deficient mice while also altering the lumenal environment and driving dysbiosis. The resulting combination of dysbiosis and leaky gut may then contribute to AOAH-deficient phenotypes of pain and anxiety (15).
Although AOAH expression is not widespread in enteric mucosa, we observed occasional cells within the epithelium that label brightly (Fig. 2). The appearance and prevalence of these AOAH-positive cells in enteric epithelium is reminiscent of neuropods that mediate gut sensory responses (75). Indeed, we observed that AOAH was expressed in a subset of EECs in wild-type gut possessing neuropod morphology (Fig. 2). Neuropods are neuroendocrine cells of the enteric epithelium that synapse with vagal neurons, hence providing enteric sensory information to the brainstem through a single synapse and thus poised to convey information about microbiota (26). Centrally, AOAH deficiency leads to arachidonic acid accumulation, a known mediator of neuronal excitability and nociception (13, 76). If arachidonic acid metabolism is similarly dysregulated in putative neuropods, this may lead to increased excitability and enhanced sensory responses to microbiota in AOAH-deficient mice. Alternatively, increased intestinal permeability can lead to altered CNS responses when microbial constituents cross the blood brain barrier (68, 77), and future studies will dissect the relative contributions of neuropods and gut leakage to Aoah phenotypes.
We observed increased mass of the cecum and cecal stool in male AOAH-deficient mice compared with male WT mice, but not in female AOAH-deficient mice compared with female WT mice (Fig. 1). These findings suggest potential sex differences in cecum phenotype and the differential roles that AOAH may play in cecum development and maintenance. Several studies in mice and humans have shown sex-specific differences in gut microbiota (78). Although levels of testosterone and estrogen are directly linked to altered gut flora, other factors such as metabolism, body mass index, and colonic transit time differ among males and females and have been linked to changes in the gut microbiome (78), so future studies will examine sex differences in AOAH-mediated microbiota. Nonetheless, we observed significant differences in female microbiota at the levels of phyla and individual taxa, where AOAH-deficient mice showed more bacterial diversity in both fecal and cecal stool and increased verrucomicrobia, cyanobacteria, and TM7 in AOAH-deficient feces (Fig. 3). Cyanobacteria have previously been linked to gastrointestinal symptoms in humans, including abdominal pain, and mucosa of patients with Crohn’s disease and ulcerative colitis show significant differences in TM7 bacteria composition (79, 80). Together, these studies suggest a role for cyanobacteria and TM7 phyla in microbiota-associated pelvic pain.
We also observed several OTUs that were present/absent in AOAH-deficient fecal and cecal stool compared with wild type (Table 1 and Supplemental Table S1, respectively). Based on the known functions of these OTUs, our findings suggest that AOAH-deficient mice may have altered carbohydrate metabolism. This possibility is corroborated by the increased abundance of metabolites that play a role in carbohydrate metabolism (Table 2 and Supplemental Tables S2 and S3). For example, cecal stool shows an absence of Bacteroides acidifaciens and Parabacteroides distasonis in AOAH-deficient mice (Supplemental Table S1), bacterial species that regulate glucose metabolism and homeostasis as well as play a role in obesity (81–83). Previous studies have shown that B. acidifaciens and P. distasonis prevent obesity or weight gain in mice (82, 83). Here, we also observed that female AOAH-deficient mice have increased body mass compared with controls, consistent with previous studies that associated reduced Aoah expression with increased poultry size (84). Thus, it is possible that AOAH modulation of microbiota, resulting in altered prevalence of species including B. acidifaciens and P. distasonis and consequent altered carbohydrate metabolism may explain the increase in body mass observed in diverse species.
Manipulating microbiota demonstrated that gut flora underlie phenotypes of AOAH-deficient mice (Figs. 6, 7, and 8). Cohousing AOAH-deficient and wild-type mice to manipulate microbiota by coprophagia resulted in a merged gut flora composition that was distinct from like-housed mice and corresponded to converged cecum morphology and epithelial barrier function (Fig. 6). In addition, we observed that cohousing or direct FMT by stool slurry gavage resulted in reduced pelvic pain in AOAH-deficient mice (Fig. 7). These findings are similar to the beneficial effects of cohousing on symptomatology and epithelial barrier function of the colon in ulcerative colitis models (85, 86). However, cohousing colitis mice with controls also increased histologic colitis scores in control mice, suggesting that microbiota transmission may result in a diseased phenotype in previously healthy mice (86). Although we observed improvement in the pelvic pain phenotype of AOAH-deficient mice after cohousing without adverse pain effects on cohoused control mice, we nonetheless found that cohousing impacted several PICRUSt functional classes in wild-type mice, including steroid hormone biosynthesis, mineral absorption, and carotenoid biosynthesis (Figs. 4 and 7). Thus, microbiota alterations associated with AOAH deficiency are likely sufficient to impact biological processes and phenotypes.
Our previous studies show that AOAH-deficient mice mimic symptoms and comorbidities of IC/BPS (6, 11, 12). Here, we show that the pelvic pain phenotype and anxiety/depressive-like behavior can be regulated by microbiota (Figs. 7 and 8). We identified 2-quinolinecarboxylic acid, 4,8-dihydroxy (also known as xanthurenic acid) as a gut metabolite of AOAH-deficient cecal stool of mice (Supplemental Table S3). Xanthurenic acid is a tryptophan metabolite and an agonist of metabotropic glutamate receptor mGluRII that is elevated in urine of patients with depression, drawing another clinical parallel between patients and AOAH-deficient mice and suggesting this metabolite in AOAH-deficient mice ceca may contribute to the depressive-like phenotype (87–89). Finally, we found IC/BPS microbiota exacerbate the pelvic pain phenotype of AOAH-deficient mice (Fig. 7). In light of our prior report of altered microbiota among patients with IC/BPS (16), this suggests dysbiosis in IC/BPS is functionally significant and sufficient to impact the defining clinical parameter of IC/BPS, pelvic pain. Thus, addressing gut dysbiosis is a therapeutic target for IC/BPS, and novel IC/BPS therapies should be developed that modify the gut microbiome. Such therapies may include prebiotic diets, probiotic supplements to complement deficiencies in specific OTUs or the functions thereof, and even transplant of stool or complex communities derived from stool.
Perspectives and Significance
The data presented here show that AOAH-deficient mice exhibit gut dysbiosis and an altered gut phenotype that mediates pelvic pain and anxiety-like behavior. These findings demonstrate that the gut microbiome is a promising therapeutic target for treating IC/BPS.
SUPPLEMENTAL DATA
Supplemental Tables S1–S3 and Supplemental Figs. S1 and S2: https://doi.org/10.6084/m9.figshare.14428700.
GRANTS
This work was supported by NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Award R01 DK103769 (B.A.W., A.J.S., and D.J.K.) and by NIH/NIDDK T32 DK062716 postdoctoral fellowship to A.R.-E. Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory which is supported by National Cancer Institute (NCI) P30-CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.R.-E., W.Y., R.E.Y., M.B., J.M.R., A.J.S., and D.J.K. conceived and designed research; A.R.-E., W.Y., R.E.Y., B.A.W., M.W., L.A., M.B., C.B., J.M.R., C.N.R., A.J.S., and D.J.K. performed experiments; A.R.-E., W.Y., R.E.Y., B.A.W., M.B., and C.B. analyzed data; A.R.-E., W.Y., R.E.Y., M.B., J.M.R., A.J.S., and D.J.K. interpreted results of experiments; A.R.-E., W.Y., R.E.Y., B.A.W., M.B., J.M.R., and D.J.K. prepared figures; A.R.-E., W.Y., R.E.Y., M.B., J.M.R., and , D.J.K. drafted manuscript; A.R.-E., W.Y., M.B., J.M.R., C.N.R., A.J.S., and D.J.K. edited and revised manuscript; A.R.-E., W.Y., R.E.Y., B.A.W., M.W., L.A., M.B., C.B., J.M.R., C.N.R., A.J.S., and D.J.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Robert Munford for generously providing AOAH-deficient mice.
REFERENCES
- 1.Akiyama Y, Homma Y, Maeda D. Pathology and terminology of interstitial cystitis/bladder pain syndrome: a review. Histol Histopathol 34: 25–32, 2019. doi: 10.14670/HH-18-028. [DOI] [PubMed] [Google Scholar]
- 2.Kim H-J. Update on the pathology and diagnosis of interstitial cystitis/bladder pain syndrome: a review. Int Neurourol J 20: 13–17, 2016. doi: 10.5213/inj.1632522.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuang Y-C, Weng S-F, Hsu Y-W, Huang CL-C, Wu M-P. Increased risks of healthcare-seeking behaviors of anxiety, depression and insomnia among patients with bladder pain syndrome/interstitial cystitis: a nationwide population-based study. Int Urol Nephrol 47: 275–281, 2015. doi: 10.1007/s11255-014-0908-6. [DOI] [PubMed] [Google Scholar]
- 4.Hanno PM, Erickson D, Moldwin R, Faraday MM; American Urological Association. Diagnosis and treatment of interstitial cystitis/bladder pain syndrome: AUA guideline amendment. J Urol 193: 1545–1553, 2015. doi: 10.1016/j.juro.2015.01.086. [DOI] [PubMed] [Google Scholar]
- 5.McKernan LC, Walsh CG, Reynolds WS, Crofford LJ, Dmochowski RR, Williams DA. Psychosocial co-morbidities in interstitial cystitis/bladder pain syndrome (IC/BPS): a systematic review. Neurourol Urodyn 37: 926–941, 2018. doi: 10.1002/nau.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Yaggie RE, Jiang MC, Rudick CN, Done J, Heckman CJ, Rosen JM, Schaeffer AJ, Klumpp DJ. Acyloxyacyl hydrolase modulates pelvic pain severity. Am J Physiol Regul Integr Comp Physiol 314: R353–R365, 2018. doi: 10.1152/ajpregu.00239.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erwin AL, Munford RS. Deacylation of structurally diverse lipopolysaccharides by human acyloxyacyl hydrolase. J Biol Chem 265: 16444–16449, 1990. doi: 10.1016/S0021-9258(17)46242-9. [DOI] [PubMed] [Google Scholar]
- 8.Hagen FS, Grant FJ, Kuijper JL, Slaughter CA, Moomaw CR, Orth K, O’Hara PJ, Munford RS. Expression and characterization of recombinant human acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides. Biochemistry 30: 8415–8423, 1991. doi: 10.1021/bi00098a020. [DOI] [PubMed] [Google Scholar]
- 9.Lu M, Varley AW, Ohta S, Hardwick J, Munford RS. Host inactivation of bacterial lipopolysaccharide prevents prolonged tolerance following gram-negative bacterial infection. Cell Host Microbe 4: 293–302, 2008. doi: 10.1016/j.chom.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munford RS, Hunter JP. Acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides, has phospholipase, lysophospholipase, diacylglycerollipase, and acyltransferase activities in vitro. J Biol Chem 267: 10116–10121, 1992. doi: 10.1016/S0021-9258(19)50207-1. [DOI] [PubMed] [Google Scholar]
- 11.Aguiniga LM, Searl TJ, Rahman-Enyart A, Yaggie RE, Yang W, Schaeffer AJ, Klumpp DJ. Acyloxyacyl hydrolase regulates voiding activity. Am J Physiol Renal Physiol 318: F1006–F1016, 2020. doi: 10.1152/ajprenal.00442.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguiniga LM, Yang W, Yaggie RE, Schaeffer AJ, Klumpp DJ; MAPP Research Network Study Group. Acyloxyacyl hydrolase modulates depressive-like behaviors through aryl hydrocarbon receptor. Am J Physiol Regul Integr Comp Physiol 317: R289–R300, 2019. doi: 10.1152/ajpregu.00029.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Yaggie RE, Schaeffer AJ, Klumpp DJ. AOAH remodels arachidonic acid-containing phospholipid pools in a model of interstitial cystitis pain: A MAPP Network study. PLoS One 15: e0235384, 2020. doi: 10.1371/journal.pone.0235384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutgendorf SK, Kreder KJ, Rothrock NE, Hoffman A, Kirschbaum C, Sternberg EM, Zimmerman MB, Ratliff TL. Diurnal cortisol variations and symptoms in patients with interstitial cystitis. J Urol 167: 1338–1343, 2002. doi: 10.1016/S0022-5347(05)65295-0. [DOI] [PubMed] [Google Scholar]
- 15.Pusceddu MM, Gareau MG. Visceral pain: gut microbiota, a new hope? J Biomed Sci 25: 73, 2018. doi: 10.1186/s12929-018-0476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braundmeier-Fleming A, Russell NT, Yang W, Nas MY, Yaggie RE, Berry M, Bachrach L, Flury SC, Marko DS, Bushell CB, Welge ME, White BA, Schaeffer AJ, Klumpp DJ. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci Rep 6: 26083, 2016. doi: 10.1038/srep26083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KW, Song HY, Kim YH. The microbiome in urological diseases. Investig Clin Urol 61: 338–348, 2020. doi: 10.4111/icu.2020.61.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du H-X, Liu Y, Zhang L-G, Zhan C-S, Chen J, Zhang M, Chen X-G, Zhang L, Liang C-Z. Abnormal gut microbiota composition is associated with experimental autoimmune prostatitis-induced depressive-like behaviors in mice. Prostate 80: 663–673, 2020. doi: 10.1002/pros.23978. [DOI] [PubMed] [Google Scholar]
- 19.Codagnone MG, Spichak S, O'Mahony SM, O'Leary OF, Clarke G, Stanton C, Dinan TG, Cryan JF. Programming bugs: microbiota and the developmental origins of brain health and disease. Biol Psychiatry 85: 150–163, 2019. doi: 10.1016/j.biopsych.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Moloney RD, Johnson AC, O’Mahony SM, Dinan TG, Greenwood-Van Meerveld B, Cryan JF. Stress and the microbiota-gut-brain axis in visceral pain: relevance to irritable bowel syndrome. CNS Neurosci Ther 22: 102–117, 2016. doi: 10.1111/cns.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malykhina AP, Wyndaele J-J, Andersson K-E, De Wachter S, Dmochowski RR. Do the urinary bladder and large bowel interact, in sickness or in health? ICI-RS 2011. Neurourol Urodyn 31: 352–358, 2012. doi: 10.1002/nau.21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821, 2013. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142, 2000. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 24.Rudick CN, Jiang M, Yaggie RE, Pavlov VI, Done J, Heckman CJ, Whitfield C, Schaeffer AJ, Klumpp DJ. O-antigen modulates infection-induced pain states. PLoS One 7: e41273, 2012. doi: 10.1371/journal.pone.0041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai HH, Krieger JN, Pontari MA, Buchwald D, Hou X, Landis JR; MAPP Research Network. Painful bladder filling and painful urgency are distinct characteristics in men and women with urological chronic pelvic pain syndromes: a MAPP research network study. J Urol 194: 1634–1641, 2015. doi: 10.1016/j.juro.2015.05.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liddle RA. Neuropods. Cell Mol Gastroenterol Hepatol 7: 739–747, 2019. doi: 10.1016/j.jcmgh.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 375: 2369–2379, 2016. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 28.Dupont HL, Jiang Z-D, Dupont AW, Utay NS. The intestinal microbiome in human health and disease. Trans Am Clin Climatol Assoc 131: 178–197, 2020. [PMC free article] [PubMed] [Google Scholar]
- 29.Viganò D, Zara F, Usai P. Irritable bowel syndrome and endometriosis: New insights for old diseases. Dig Liver Dis 50: 213–219, 2018. doi: 10.1016/j.dld.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Song Y, Könönen E, Rautio M, Liu C, Bryk A, Eerola E, Finegold SM. Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin. Int J Syst Evol Microbiol 56: 1985–1990, 2006. doi: 10.1099/ijs.0.64318-0. [DOI] [PubMed] [Google Scholar]
- 31.Renson A, Mullan Harris K, Dowd JB, Gaydosh L, McQueen MB, Krauter KS, Shannahan M, Aiello AE. Early signs of gut microbiome aging: biomarkers of inflammation, metabolism, and macromolecular damage in young adulthood. J Gerontol A Biol Sci Med Sci 75: 1258–1266, 2020. doi: 10.1093/gerona/glaa122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotta M, Forster R, The family lachnospiraceae, including the genera butyrivibrio, lachnospira and roseburia. In: The Prokaryotes, edited by Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. New York, NY: Springer US, 2006, p. 1002–1021. [Google Scholar]
- 33.Watthanalamloet A, Tachaapaikoon C, Lee YS, Kosugi A, Mori Y, Tanasupawat S, Kyu KL, Ratanakhanokchai K. Cellulosibacter alkalithermophilus gen. nov., sp. nov., an anaerobic alkalithermophilic, cellulolytic-xylanolytic bacterium isolated from soil of a coconut garden. Int J Syst Evol Microbiol 62: 2330–2335, 2012. doi: 10.1099/ijs.0.027854-0. [DOI] [PubMed] [Google Scholar]
- 34.Kondo F, Tottori J, Soki K. Ulcerative enteritis in broiler chickens caused by Clostridium colinum and in vitro activity of 19 antimicrobial agents in tests on isolates. Poult Sci 67: 1424–1430, 1988. doi: 10.3382/ps.0671424. [DOI] [PubMed] [Google Scholar]
- 35.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 57: 1–24, 2018. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedberg ME, Moore ERB, Svensson-Stadler L, Hörstedt P, Baranov V, Hernell O, Wai SN, Hammarström S, Hammarström M-L. Lachnoanaerobaculum gen. nov., a new genus in the Lachnospiraceae: characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from the human small intestine, and Lachnoanaerobaculum orale sp. nov., isolated from saliva, and reclassification of Eubacterium saburreum (Prevot 1966) Holdeman and Moore 1970 as Lachnoanaerobaculum saburreum comb. nov. Int J Syst Evol Microbiol 62: 2685–2690, 2012. doi: 10.1099/ijs.0.033613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ntougias S, Lapidus A, Han J, Mavromatis K, Pati A, Chen A, Klenk H-P, Woyke T, Fasseas C, Kyrpides NC, Zervakis GI. High quality draft genome sequence of Olivibacter sitiensis type strain (AW-6T), a diphenol degrader with genes involved in the catechol pathway. Stand Genomic Sci 9: 783–793, 2014. doi: 10.4056/sigs.5088950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velasco SE, Yebra MJ, Monedero V, Ibarburu I, Dueñas MT, Irastorza A. Influence of the carbohydrate source on β-glucan production and enzyme activities involved in sugar metabolism in Pediococcus parvulus 2.6. Int J Food Microbiol 115: 325–334, 2007. doi: 10.1016/j.ijfoodmicro.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 39.Táncsics A, Kéki Z, Márialigeti K, Schumann P, Tóth EM. Siphonobacter aquaeclarae gen. nov., sp. nov., a novel member of the family ’Flexibacteraceae’, phylum Bacteroidetes. Int J Syst Evol Microbiol 60: 2567–2571, 2010. doi: 10.1099/ijs.0.019398-0. [DOI] [PubMed] [Google Scholar]
- 40.Shah Hn A. Anaerorhabdus. In: Bergey’s Manual of Systematics of Archaea and Bacteria, edited by Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S.. Hoboken, NJ: Wiley, 2015, p. 1–2.doi: 10.1002/9781118960608. [DOI] [Google Scholar]
- 41.Saman S, Sarman S, Slattery P. Isolation of a potential new member of the Bacillus cereus group from snow covered soil. Life Sciences and Medicine Research, Gale OneFile: Health and Medicine, 2010.
- 42.Himelbloom BH, Canale-Parola E. Clostridium methylpentosum sp. nov.: a ring-shaped intestinal bacterium that ferments only methylpentoses and pentoses. Arch Microbiol 151: 287–293, 1989. doi: 10.1007/BF00406553. [DOI] [PubMed] [Google Scholar]
- 43.Cornick NA, Jensen NS, Stahl DA, Hartman PA, Allison MJ. Lachnospira pectinoschiza sp. nov., an anaerobic pectinophile from the pig intestine. Int J Syst Bacteriol 44: 87–93, 1994. doi: 10.1099/00207713-44-1-87. [DOI] [PubMed] [Google Scholar]
- 44.Arai R, Tominaga K, Wu M, Okura M, Ito K, Okamura N, Onishi H, Osaki M, Sugimura Y, Yoshiyama M, Takamatsu D. Diversity of Melissococcus plutonius from honeybee larvae in Japan and experimental reproduction of European foulbrood with cultured atypical isolates. PLoS One 7: e33708, 2012. doi: 10.1371/journal.pone.0033708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakamoto M, Suzuki N, Matsunaga N, Koshihara K, Seki M, Komiya H, Benno Y. Parabacteroides gordonii sp. nov., isolated from human blood cultures. Int J Syst Evol Microbiol 59: 2843–2847, 2009. doi: 10.1099/ijs.0.010611-0. [DOI] [PubMed] [Google Scholar]
- 46.Loo Y-M, Gale M Jr.. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692, 2011. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 3: 14–24, 2013. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrić P, Meyer AS, Jensen PA, Dam-Johansen K. Reactor design for minimizing product inhibition during enzymatic lignocellulose hydrolysis: I. Significance and mechanism of cellobiose and glucose inhibition on cellulolytic enzymes. Biotechnol Adv 28: 308–324, 2010. doi: 10.1016/j.biotechadv.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Peng B, Zhao C, Kasakov S, Foraita S, Lercher JA. Manipulating catalytic pathways: deoxygenation of palmitic acid on multifunctional catalysts. Chemistry 19: 4732–4741, 2013. doi: 10.1002/chem.201203110. [DOI] [PubMed] [Google Scholar]
- 50.Seyyedin S, Nazem MN. Histomorphometric study of the effect of methionine on small intestine parameters in rat: an applied histologic study. Folia Morphol (Warsz) 76: 620–629, 2017. doi: 10.5603/FM.a2017.0044. [DOI] [PubMed] [Google Scholar]
- 51.Brosnan JT, Brosnan ME, Bertolo RFP, Brunton JA. Methionine: a metabolically unique amino acid. Livest Sci 112: 2–7, 2007. doi: 10.1016/j.livsci.2007.07.005. [DOI] [Google Scholar]
- 52.Tomaszewski JE, Landis JR, Russack V, Williams TM, Wang LP, Hardy C, Brensinger C, Matthews YL, Abele ST, Kusek JW, Nyberg LM; Interstitial Cystitis Database Study Group . Biopsy features are associated with primary symptoms in interstitial cystitis: results from the interstitial cystitis database study. Urology 57, Suppl 1: 67–81, 2001. doi: 10.1016/S0090-4295(01)01166-9. [DOI] [PubMed] [Google Scholar]
- 53.Ratliff TL, Klutke CG, McDougall EM. The etiology of interstitial cystitis. Urol Clin North Am 21: 21–30, 1994. doi: 10.1016/S0094-0143(21)00588-7. [DOI] [PubMed] [Google Scholar]
- 54.Johansson SL, Fall M. Clinical features and spectrum of light microscopic changes in interstitial cystitis. J Urol 143: 1118–1124, 1990. doi: 10.1016/s0022-5347(17)40201-1. [DOI] [PubMed] [Google Scholar]
- 55.Parsons CL, Zupkas P, Parsons JK. Intravesical potassium sensitivity in patients with interstitial cystitis and urethral syndrome. Urology 57: 428–432; discussion 432, 2001. doi: 10.1016/S0090-4295(00)01110-9. [DOI] [PubMed] [Google Scholar]
- 56.Parsons CL, Greenberger M, Gabal L, Bidair M, Barme G. The role of urinary potassium in the pathogenesis and diagnosis of interstitial cystitis. J Urol 159: 1862–1866; discussion 1866–1867, 1998. [Erratum in J Urol 191: 1936, 2014]. doi: 10.1016/S0022-5347(01)63178-1. [DOI] [PubMed] [Google Scholar]
- 57.Parsons CL. The role of a leaky epithelium and potassium in the generation of bladder symptoms in interstitial cystitis/overactive bladder, urethral syndrome, prostatitis and gynaecological chronic pelvic pain. BJU Int 107: 370–375, 2011. doi: 10.1111/j.1464-410X.2010.09843.x. [DOI] [PubMed] [Google Scholar]
- 58.Kaplan SA, Dmochowski R, Cash BD, Kopp ZS, Berriman SJ, Khullar V. Systematic review of the relationship between bladder and bowel function: implications for patient management. Int J Clin Pract 67: 205–216, 2013. doi: 10.1111/ijcp.12028. [DOI] [PubMed] [Google Scholar]
- 59.Rudick CN, Chen MC, Mongiu AK, Klumpp DJ. Organ cross talk modulates pelvic pain. Am J Physiol Regul Integr Comp Physiol 293: R1191–R1198, 2007. doi: 10.1152/ajpregu.00411.2007. [DOI] [PubMed] [Google Scholar]
- 60.Malykhina AP. Neural mechanisms of pelvic organ cross-sensitization. Neuroscience 149: 660–672, 2007. doi: 10.1016/j.neuroscience.2007.07.053. [DOI] [PubMed] [Google Scholar]
- 61.Pezzone MA, Liang R, Fraser MO. A model of neural cross-talk and irritation in the pelvis: implications for the overlap of chronic pelvic pain disorders. Gastroenterology 128: 1953–1964, 2005. doi: 10.1053/j.gastro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Qin C, Malykhina AP, Akbarali HI, Foreman RD. Cross-organ sensitization of lumbosacral spinal neurons receiving urinary bladder input in rats with inflamed colon. Gastroenterology 129: 1967–1978, 2005. doi: 10.1053/j.gastro.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. Human genetics shape the gut microbiome. Cell 159: 789–799, 2014. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19: 731–743, 2016. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 48: 1396–1406, 2016. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keay SK, Szekely Z, Conrads TP, Veenstra TD, Barchi JJ Jr, Zhang C-O, Koch KR, Michejda CJ. An antiproliferative factor from interstitial cystitis patients is a frizzled 8 protein-related sialoglycopeptide. Proc Natl Acad Sci USA 101: 11803–11808, 2004. doi: 10.1073/pnas.0404509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68: 1516–1526, 2019. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obrenovich MEM. Leaky gut, leaky brain? Microorganisms 6: 107, 2018. doi: 10.3390/microorganisms6040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grover S, Srivastava A, Lee R, Tewari AK, Te AE. Role of inflammation in bladder function and interstitial cystitis. Ther Adv Urol 3: 19–33, 2011. doi: 10.1177/1756287211398255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wouters MM, Vicario M, Santos J. The role of mast cells in functional GI disorders. Gut 65: 155–168, 2016. doi: 10.1136/gutjnl-2015-309151. [DOI] [PubMed] [Google Scholar]
- 71.Ramsay DB, Stephen S, Borum M, Voltaggio L, Doman DB. Mast cells in gastrointestinal disease. Gastroenterol Hepatol (NY) 6: 772–777, 2010. [PMC free article] [PubMed] [Google Scholar]
- 72.Taché Y, Perdue MH. Role of peripheral CRF signalling pathways in stress-related alterations of gut motility and mucosal function. Neurogastroenterol Motil 16, Suppl 1: 137–142, 2004. doi: 10.1111/j.1743-3150.2004.00490.x. [DOI] [PubMed] [Google Scholar]
- 73.Tache Y, Larauche M, Yuan P-Q, Million M. Brain and gut CRF signaling: biological actions and role in the gastrointestinal tract. Curr Mol Pharmacol 11: 51–71, 2018. doi: 10.2174/1874467210666170224095741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodiño-Janeiro BK, Alonso-Cotoner C, Pigrau M, Lobo B, Vicario M, Santos J. Role of corticotropin-releasing factor in gastrointestinal permeability. J Neurogastroenterol Motil 21: 33–50, 2015. doi: 10.5056/jnm14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X, Bohórquez DV. A gut-brain neural circuit for nutrient sensory transduction. Science 361: eaat5236, 2018. doi: 10.1126/science.aat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jang Y, Kim M, Hwang SW. Molecular mechanisms underlying the actions of arachidonic acid-derived prostaglandins on peripheral nociception. J Neuroinflammation 17: 30, 2020. doi: 10.1186/s12974-020-1703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Logsdon AF, Erickson MA, Rhea EM, Salameh TS, Banks WA. Gut reactions: How the blood-brain barrier connects the microbiome and the brain. Exp Biol Med (Maywood) 243: 159–165, 2018. doi: 10.1177/1535370217743766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim YS, Unno T, Kim BY, Park MS. Sex differences in gut microbiota. World J Mens Health 38: 48–60, 2020. doi: 10.5534/wjmh.190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lévesque B, Gervais M-C, Chevalier P, Gauvin D, Anassour-Laouan-Sidi E, Gingras S, Fortin N, Brisson G, Greer C, Bird D. Prospective study of acute health effects in relation to exposure to cyanobacteria. Sci Total Environ 466–467: 397–403, 2014. doi: 10.1016/j.scitotenv.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 80.Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S, Ott SJ. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 57: 1569–1576, 2008. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 81.Miyamoto Y, Itoh K. Bacteroides acidifaciens sp. nov., isolated from the caecum of mice. Int J Syst Evol Microbiol 50: 145–148, 2000. doi: 10.1099/00207713-50-1-145. [DOI] [PubMed] [Google Scholar]
- 82.Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, Wang Y, Liu C, Wang W, Wang J, Liu S-J, Liu H. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep 26: 222–235.E5, 2019. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 83.Yang J-Y, Lee Y-S, Kim Y, Lee S-H, Ryu S, Fukuda S, Hase K, Yang C-S, Lim HS, Kim MS, Kim H-M, Ahn S-H, Kwon B-E, Ko H-J, Kweon M-N. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol 10: 104–116, 2017. doi: 10.1038/mi.2016.42. [DOI] [PubMed] [Google Scholar]
- 84.Claire D'Andre H, Paul W, Shen X, Jia X, Zhang R, Sun L, Zhang X. Identification and characterization of genes that control fat deposition in chickens. J Anim Sci Biotechnol 4: 43, 2013. doi: 10.1186/2049-1891-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu M, Li P, An Y, Ren J, Yan D, Cui J, Li D, Li M, Wang M, Zhong G. Phloretin ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating the gut microbiota. Pharmacol Res 150: 104489, 2019. doi: 10.1016/j.phrs.2019.104489. [DOI] [PubMed] [Google Scholar]
- 86.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131: 33–45, 2007. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maes M, De Ruyter M, Suy E. Xanthurenic acid flow in 24-hour urine following L-tryptophan loading in depressive patients. Acta Psychiatr Belg 86: 120–130, 1986. [PubMed] [Google Scholar]
- 88.Hoes MJ. The clinical significance of an elevated excretion of xanthurenic acid in psychiatric patients. Acta Psychiatr Belg 79: 638–646, 1979. [PubMed] [Google Scholar]
- 89.Hoes MJAJ, Sijben N. Xanthurenic acid in depression. J Orthomol Med 2: 129–136, 1987. [Google Scholar]