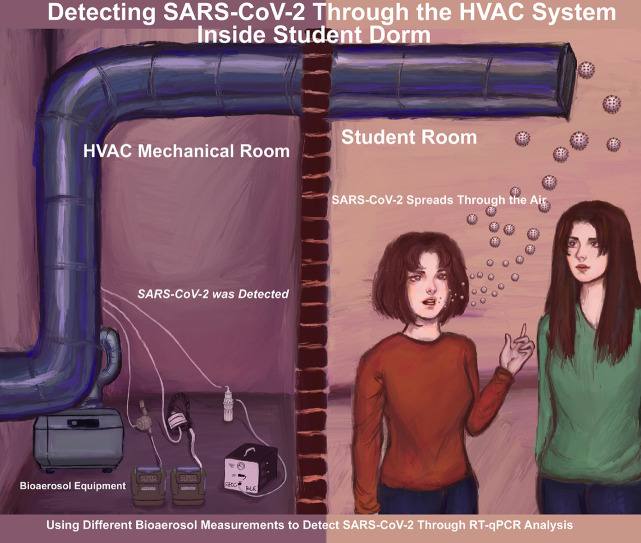
Graphical Abstract
Key words: Bioaerosol, Virus detection, AerosolSense sampler, SKC samplers, qRT-PCR analysis, Coronavirus
Abstract
Background
The COVID-19 pandemic affected universities and institutions and caused campus shutdowns with a transition to online teaching models. To detect infections that might spread on campus, we pursued research towards detecting SARS-CoV-2 in air samples inside student dorms.
Methods
We sampled air in 2 large dormitories for 3.5 months and a separate isolation suite containing a student who had tested positive for COVID-19. We developed novel techniques employing 4 methods to collect air samples: Filter Cassettes, Button Sampler, BioSampler, and AerosolSense sampler combined with direct qRT-PCR SARS-CoV-2 analysis.
Results
For the 2 large dorms with the normal student population, we detected SARS-CoV-2 in 11 samples. When compared with student nasal swab qRT-PCR testing, we detected SARS-CoV-2 in air samples when a PCR positive COVID-19 student was living on the same floor of the sampling location with a detection rate of 75%. For the isolation dorm, we had a 100% SARS-CoV-2 detection rate with AerosolSense sampler.
Conclusions
Our data suggest air sampling may be an important SARS-CoV-2 surveillance technique, especially for buildings with congregant living settings (dorms, correctional facilities, barracks). Future building designs and public health policies should consider implementation of Heating, Ventilation, and Air Conditioning surveillance.
Background
The COVID-19 pandemic caused institutions and universities worldwide to shut down and move to online models during Spring.1 , 2 However, online models established due to emerging circumstances are different from traditional online teaching models and in-person interactions.3 Pandemic distance work and online teaching presented educational challenges and effects on mental health due to isolation and lack of social interaction.1 , 4 , 5 Businesses and college campuses put mitigation methods into practice, including wearing masks, social distancing, isolation and contact tracing, and hand and surface sanitizing.6, 7, 8 In addition, various surveillance methods were developed, including quantitative detection of SARS-CoV-2 in wastewater from student dorms.9 For example, the University of Arizona10 implemented a surveillance program that successfully contained campus outbreaks using wastewater sampling and nasopharyngeal swab sample testing. In order to keep the campus open and detect any virus before widespread transmission could occur, we investigated SARS-CoV-2 detection in air samples from the Heating, Ventilation, and Air Conditioning (HVAC) systems inside student dorms, since the virus is known to spread in the air. 11
SARS-CoV-2 airborne detection inside buildings is a challenge, and researchers are actively pursuing different air sampling methods.12 Air samples may be collected using several filters and devices, and the virus can be detected by reverse transcriptase quantitative polymerase chain reaction (qRT-PCR). Borges, et al.13 reviewed some SARS-CoV-2 air sampling methods in indoor settings and highlighted the efficacy of each method based on positive detected samples. Chia, et al.,14 Kenarkoohi, et al.,15 and Zhu, et al.16 performed air sampling in COVID-19 infection isolation rooms with solid and liquid collections. These studies used primers to detect different gene sequences using different amplification kits and showed that detecting COVID-19 in air samples in clinical settings may provide random and unreliable results. Further research should be conducted in this area. In addition, there are no reports of sampling air directly from the HVAC system to perform surveillance for COVID inside a large building.
We report here the successful detection of SARS-CoV-2 in student dorm HVAC air samples by qRT-PCR. The study was conducted in 2 parts (1) a 3.5-month period that represented air sampling inside HVAC of 2 dorms without specific knowledge of a COVID-19 case; and (2) air sampling from HVAC associated with a suite containing one student who tested positive for COVID-19 by nasal swab RT-PCR.
Material and methods
Site description and study period
Two student dorms were selected for the current study, designated here as Dorm1 and Dorm2. The HVAC floor plan for Dorm1, where sampling was performed, is shown in Figure S1 in the Supplemental Material, where Dorm2 has a similar layout. The buildings have 5 floors and contain 225 rooms each, including student units, offices, common bathrooms, professional staff apartments, and storage units. In addition, Dorm1 and Dorm2 are connected on the second floor with an indoor hallway bridge. These 2 buildings were chosen for monitoring where the return air inside the HVAC system for all the rooms is connected at one location inside locked mechanical rooms. We chose sampling locations in the mechanical room before the air passes through multiple filters and outside makeup air is added. Sampling inside Dorm1 and Dorm2 was conducted during the Spring semester from January 19 to April 29, 2021, except for the week between March 1 and March 5, because we had a COVID case in the lab and were required to quarantine.
By early March, there were few COVID-19 cases in the surrounding county (see Supplemental Material Figure S2), and on campus, so we selected the isolation dorm to collect additional air samples. The dorm contained suites, and each suite contains a common room, a kitchen, and 2 bedrooms. The HVAC system for each suite was located adjacent to the suite inside a locked mechanical room. The HVAC floor plan for the IsolationDorm, where sampling was performed, is shown in Figure S3 in the Supplemental Material. Therefore, we used different sampling methods for one suite, while a COVID-19 RT-PCR positive student occupied one of the rooms inside the suite. Sampling inside the IsolationDorm was conducted for 2 weeks, from April 19 to April 29.
Air sampling methods
Four bioaerosol sampling methods were used for this study, 3 SKC samplers, and one ThermoFisher Scientific sampler. The specifications for the 4 methods are shown in Table 1 . Methods 1-3 were used at Dorm1, Dorm2, and the IsolationDorm. In late April, we began using an additional collection device, Method 4, which was only used at the IsolationDorm. Methods 1-3 collected air samples from the HVAC system using isokinetic sampling probes inserted inside the duct, and Method 4 used a 3″ tube to sample directly from the HVAC system. Different filter types and PBS solutions (dilutions of normal physiologic saline solution, which is defined as 1x) were tested for Method 3, as shown in Table 2 . Sampling was performed on Monday, Tuesday, and Thursday of each week, beginning at 8 am, and samples were retrieved after 30-minutes, 90-minutes, or 24-hours, depending on the method and filter used. Weekly samples were placed inside test tubes, stored at -20°C, and analyzed by qRT-PCR within 10 days.
Table 1.
Air sampling methods
| Sampling method | Name | Company | Part number | Collection media |
|---|---|---|---|---|
| 1 | Filter Cassettes | SKC | 225-1723 | 37-mm Filters |
| 2 | Button Sampler | SKC | 225-360 | 25-mm Filters |
| 3 | BioSampler | SKC | 225-9595 | PBS Solution |
| 4 | AerosolSense sampler | ThermoFisher Scientific | NA | Sample Cartridge |
Table 2.
Different collection media and sampling times
| Collection media | Filter size (mm)/ Solution |
Pore size (µm)/ PBS concentration |
Sampling time |
|---|---|---|---|
| Filter | 37 | 5 | 24-h |
| Filter | 37 | 0.3 | 24-h |
| Filter | 25 | 5 | 24-h |
| Filter | 25 | 3 | 24-h |
| Filter | 25 | 1.2 | 24-h |
| Filter | 25 (Gelatin) | 3 | 30-min |
| Solution | NA | 0.5 x | 30-min |
| Solution | NA | 0.25 x | 30-min |
| Solution | NA | 0.25 x | 90-min |
| Solution | NA | 0.15 x | 90-min |
| Solution | NA | 0.15 x | 30-min |
| Sample Cartridge | NA | NA | 24-h |
Filter cassettes
For Method 1, 37-mm filters (SKC Inc., Eighty Four, PA, USA) were loaded inside a cassette and on top of a support pad, inserted into a cassette holder. An AIRCHEK TOUCH PUMP (SKC Inc.), operating at 5 LPM, was attached to the cassette outlet. The inlet of the cassette was attached to the HVAC system. Method 1 was stopped on April 15 for Dorm1 and Dorm2.
For Methods 1-3, the pump was calibrated before sampling, and the flow was checked after sampling for deviation.
Button sampler
For Method 2, 25-mm filters (SKC Inc.) were loaded inside the Button sampler. An AIRCHEK TOUCH PUMP, operating at 4 LPM, was attached to the outlet of the button. The calibration adaptor of the Button sampler was attached to the inlet of the button, and the inlet of the calibration adaptor was attached to the HVAC system.
BioSampler
For Method 3, the BioSampler (SKC Inc.) is a liquid impingement collection method, where buffer solution was filled inside the collection vessel. First, the impinger was attached on top of the collection vessel, followed by an inlet section which was attached to the HVAC system. Next, a BioLite+ pump (SKC Inc.), operating at 12.5 LPM, was attached to the outlet of the impinger. We wanted to collect the virus into a solution that would preserve its integrity before RT-PCR analysis. Therefore, we hypothesized that a physiologic phosphate buffered saline solution (eg, 1x PBS) would preserve virus integrity. However, significant water evaporation occurs during the sample collection time, thus reducing the volume approximately 5-6 times depending on ambient humidity, so we decided to employ dilutions down to a 0.15 x PBS solution in the impinger (Table 2).
AerosolSense sampler
Method 4 was the AerosolSense sampler (Thermo Fisher Scientific, Waltham, MA, USA), a standalone device with a built-in pump that operates at 200 LPM and a proprietary Sample Cartridge. The AerosolSense sampler did not require flow calibration and was less time-consuming to operate compared to Methods 1-3. The AerosolSense sampler flow rate was at least 50 times higher than Methods 1 and 2 within the same collection time (24-hours). The AerosolSense sampler is a new device released in Spring 2021, therefore, the sampling duration was only performed in April.
Experimental setup
The collection media for Methods 1-4 are listed in Table S1 in the Supplemental Material. For Dorm1 and Dorm2, we used one device for Methods 1-3, with different collection media and sampling times. We performed measurements from the return air of the HVAC system in Dorm1 and Dorm2, as shown in Figure S4 in the Supplemental Material.
SARS COV-2 detection in air samples by qRT-PCR
The qRT-PCR analysis was carried out using Go Script RT Mix for 1-Step RT-qPCR and Go Taq Probe qPCR Master Mix with dUTP and standard techniques. At least 2 positive and negative controls were run each week with samples. Positive control purified SARS-CoV-2 RNA was obtained from BEI Resources ATCC (5.5×10^7 genome Equiv/mL). We were able to detect SARS-CoV-2 RNA down to 3 genome equivalent copies. A dilution series was run, resulting in increasing Cycle quantification values as expected. Primers used were SARS-CoV-2 Nucleocapsid 1, 2, and RNase P for human cell detection from Integrated DNA Technologies (cat# 10006770). The reactions were run on the Quant Studio 3 Applied Biosystems instrument. The viral genomes were not sequenced, because the purpose of the study was to determine SARS-CoV-2 detection in air, not to link it to a particular individual.
We drew upon the background of previously developed protocols for detecting virus genomes from various preparations and samples,17, 18, 19, 20, 21 and we have successfully amplified viral genomes without purification of the nucleic acids. Given the expected paucity of virus in the sample, we opted to attempt viral detection without any nucleic acid purification step to maximize efficiency and sensitivity for sample detection.
All filters, impingers, and cartridges were tested for inhibitors of the PCR reaction. The blank filters were treated in the standard way and the samples were spiked with known SARS-CoV-2 RNA. Different volumes and salt solution concentrations were tested for the impinger detection method considering the time of detection and volume loss due to evaporation. None of the filter materials or impinger solutions used here inhibited the PCR.
Liquid from impingers and dry filters were collected in microfuge tubes and stored at -20°C prior to PCR analysis (up to 10 days). On the day of analysis, we added 1 mL of water to the filters 1 hour before the PCR to liberate the samples from the filter. Tubes were vortexed vigorously for 5 seconds, and 16.2 uL of samples were taken from the dirty side of the filter while scraping the filters with the pipette tip gently to use in a 40 uL PCR reaction. Because these samples are dilute and low copy number, viral genome was often detected around 28-30 cycles, but sometimes up to 39 cycles.
Campus track tracing
The campus conducted random and planned COVID-19 testing for students living inside dorms using saliva and nasopharyngeal PCR testing. If the student tested positive, the student was then moved to the IsolationDorm.
Results
Air sampling
A total of 41 sampling days were completed at Dorm1 and Dorm2 and 6 days at the IsolationDorm. The number of samples collected for each collection method are shown in Table 3 . The complete list of locations, dates, filter types, PBS concentrations, and sampling time of Methods 1-4 are shown in Table S2, S3, S4, and S5, respectively, in the Supplement Material. Problems experienced during the sampling period include random pump shut down during the night, pump failure, and inconsistent flow due to the calibration adapter on the Button sampler. Sample collection interruptions were noted for 8 samples each for Methods 1 and 2, and 1 sample for Method 3, however, the downtime for each method was minimal. The total numbers of samples collected for each method were 66 for Method 1, 94 for Method 2, 85 for Method 3, and 3 for Method 4. Therefore, the total number of samples collected for the project was 248.
Table 3.
The number of samples for each collection media, filter type or PBS concentration, used at each dorm
| Dorm | Collection media | Filter size (mm)/ Solution |
Pore size (µm)/ PBS concentration |
Number of samples |
|---|---|---|---|---|
| Dorm1 | Filter | 37 | 5 | 8 |
| Dorm2 | Filter | 37 | 5 | 8 |
| Dorm1 | Filter | 37 | 0.3 | 25 |
| Dorm2 | Filter | 37 | 0.3 | 19 |
| IsolationDorm | Filter | 37 | 0.3 | 6 |
| Dorm1 | Filter | 25 | 5 | 27 |
| Dorm2 | Filter | 25 | 5 | 24 |
| IsolationDorm | Filter | 25 | 5 | 6 |
| Dorm1 | Filter | 25 | 3 | 3 |
| Dorm2 | Filter | 25 | 3 | 4 |
| Dorm1 | Filter | 25 | 1.2 | 10 |
| Dorm2 | Filter | 25 | 1.2 | 10 |
| IsolationDorm | Filter | 25 | 1.2 | 6 |
| IsolationDorm | Filter | 25 (Gelatin) | 3 | 4 |
| Dorm1 | Solution | NA | 0.5 x | 8 |
| Dorm2 | Solution | NA | 0.5 x | 8 |
| Dorm1 | Solution | NA | 0.25 x | 4 |
| Dorm2 | Solution | NA | 0.25 x | 4 |
| Dorm1 | Solution | NA | 0.15 x | 28 |
| Dorm2 | Solution | NA | 0.15 x | 27 |
| IsolationDorm | Solution | NA | 0.15 x | 6 |
| IsolationDorm | Sample Cartridge | collection substrate | NA | 3 |
SARS-CoV-2 detection in air samples by qRT-PCR
SARS-CoV-2 RNA was detected in 14 samples by qRT-PCR in air samples from dorms, as shown in Table 4 . However, human testing was not performed on each person in the dorms each day. This table identifies the date and number of individuals who tested positive by PCR analysis in scheduled clinical testing and the distance from the sampling location, for example, the same floor or the number of floors distant from the sampling locations. In addition, the table provides the air sampling unit, date, filter type, method, sampling time, the PCR results, and the number of COVID-positive students detected by the human clinical testing.
Table 4.
qRT-PCR analysis positive tests and campus COVID-19 confirmed cases during the study period
| Dorm name | Number of floors from collection unit | Date | Filter size (mm)-pore size (µm) / PBS concen-tration | Method number | Sampling time (hours) | Presence of COVID-19 in air samples | Number of COVID-19 positive students |
|---|---|---|---|---|---|---|---|
| January | |||||||
| Dorm1 | 1 | 17 | 1 | ||||
| Dorm2 | 19 | 25-5 | 2 | 24 | Positive | ||
| Dorm1 | 19 | 0.5x | 3 | 1 | Positive (Inconclusive) | ||
| Dorm2 | 21 | 25-5 | 2 | 24 | Positive | ||
| Dorm1 | 25 | 25-5 | 2 | 24 | Positive | ||
| Dorm2 | 25 | 25-5 | 2 | 24 | Positive | ||
| Dorm2 | 25 | 0.5x | 3 | 0.5 | Positive (Inconclusive) | ||
| Dorm1 | 26 | 25-5 | 2 | 24 | Positive | ||
| Dorm2 | 26 | 25-5 | 2 | 24 | Positive | ||
| Dorm2 | same floor | 26 | 1 | ||||
| Dorm1 | 28 | 0.5x | 3 | 0.5 | Positive (Inconclusive) | ||
| Dorm2 | 2 | 29 | 1 | ||||
| February | |||||||
| Dorm2 | 2 | 3 | 1 | ||||
| Dorm2 | 2 | 4 | 1 | ||||
| Dorm2 | 3 | 5 | 1 | ||||
| Dorm1 | 3 | 13 | 1 | ||||
| Dorm1 | 2 | 13 | 1 | ||||
| Dorm1 | 3 | 25 | 1 | ||||
| March | |||||||
| Dorm1 | same floor | 16 | 1 | ||||
| Dorm2 | 1 | 18 | 1 | ||||
| Dorm2 | 22 | 25-5 | 2 | 24 | Positive (Inconclusive) | ||
| Dorm1 | 23 | 25-5 | 2 | 24 | Positive | ||
| Dorm1 | same floor | 25 | 2 | ||||
| Dorm2 | same floor | 25 | 1 | ||||
| Dorm1 | same floor | 26 | 2 | ||||
| Dorm2 | 1 | 29 | 1 | ||||
| Dorm1 | 2 | 29 | 1 | ||||
| April | |||||||
| Dorm1 | 3 | 1 | 1 | ||||
| Dorm1 | same floor | 14 | 1 | ||||
| Dorm2 | 3 | 20 | 1 | ||||
| Dorm2 | 3 | 22 | 1 | ||||
| Dorm2 | 4 | 26 | 1 | ||||
| Dorm2 | 3 | 27 | 1 | ||||
| Dorm1 | 2 | 28 | 1 | ||||
| IsolationDorm | 21 | 1⁎ | |||||
| IsolationDorm | 22 | 1⁎ | |||||
| IsolationDorm | 23 | 1⁎ | |||||
| IsolationDorm | 24 | 1⁎ | |||||
| IsolationDorm | 25 | 1⁎ | |||||
| IsolationDorm | 26 | - | 4 | 24 | Positive | 1⁎ | |
| IsolationDorm | 27 | 1⁎ | |||||
| IsolationDorm | 28 | - | 4 | 24 | Positive | 1⁎ | |
| IsolationDorm | 29 | - | 4 | 24 | Positive | 1⁎ |
The number of floors separating the location where the identified COVID positive student was living, and the sample collection unit are shown. The dates when SARS-CoV-2 was detected both by air sampling and by human clinical testing (saliva or nasopharyngeal PCR testing) are shown in bold in italic.
The student in the isolation dorm was only tested once, and the same student (only one) was in the IsolationDorm from April 21 through 29.
We detected SARS-CoV-2 RNA by RT-PCR in air January 19-25 using Methods 2 and 3 in both dorms at a time when no student testing was scheduled. We detected SARS-CoV-2 in air samples on January 25 and 26 for Dorm2, where we also detected SARS-CoV-2 in a student by scheduled testing (January 26, shown in italic bold). These results show the timely ability of the HVAC detection method in identifying previously unrecognized cases, which can alert staff to implement testing and quarantine to prevent spread. In the period between January 29 and March 18, cases were detected in students, but our air sampling did not detect SARS-CoV-2. It is possible that the distance of the infected person from the air sampling unit caused dilution of the sample such that it was below the level of detection by our methods. For the qRT-PCR air analysis, the results reported as positive were identified as positive by the software. Results reported as “positive inconclusive” were cases where the viral RNA was detected but at such low levels that the software identified the results as “inconclusive”. Since our multiple negative controls run each week never return an inconclusive result or any amplification, we believe the “inconclusive” cases are actually detecting quantities of SARS-CoV-2 at the limit of detection.
In the week of March 22, we detected SARS-CoV-2 on March 22 and 23 using Method 2 for Dorm1 and Dorm2, where 5 students tested positive in both Dorm1 and Dorm2 on March 25 and 26. However, we did not detect SARS-CoV-2 on March 16 and April 14, when 2 students tested COVID-19 positive on the same floor of the sampling location. Therefore, from the 8 patients living on the same floor of the sampling location, in Dorm1 and Dorm2, we detected 6 out of 8 (75%). Of course, it is possible that some of the students who tested positive were not actively infected since others have reported that people may remain COVID RNA positive by RT-PCR even if they are not infectious or releasing virus into the air.22
For the IsolationDorm, with a known positive case, SARS-CoV-2 was detected in the HVAC air samples from the suite with Method 4 in 3 consecutive samples (from April 26 to 29), showing reproducibility. We did not detect SARS-CoV-2 using Methods 1-3 inside the IsolationDorm on April 22-25 when the same student was present in the room. This suggests the superiority of sampling method 4 with a 24 hours collection time over other filter and impinger methods and collection times.
Discussion
We successfully detected COVID in 4 of the 14 weeks we collected samples. Compared to the clinical sampling of humans (nasopharyngeal swabs or saliva), we had a 75% detection rate when a PCR positive COVID-19 student lived on the same floor as the sampling location. The HVAC qRT-PCR analysis detected SARS-CoV-2 on January 19 and 21 in Dorm2 and January 28 in Dorm1, indicating the likely presence of asymptomatic infected students. We also detected SARS-CoV-2 in the air on March 22 and 23 with subsequent detection of COVID cases in students when testing was performed on March 25 and 26. These data indicate that the HVAC sampling is a useful COVID surveillance method. In our sampling, Method 1 did not detect SARS-CoV-2 with any of the filters used for this study. Method 2 detected SARS-CoV-2 only with the 5 µm pore size filters. Method 3 only detected SARS-CoV-2 with the 0.5x PBS solution with sampling times of 30 and 60 minutes. In addition, on many days, there was no COVID-19 infection in students in the dorms, and our HVAC RT-PCR results were negative. This suggests that we did not have a high false positive rate in our sampling.
Our data suggest the HVAC surveillance is more sensitive when the infected individual is closer to the air sampling location as the HVAC detection correlates with cases detected on the same floor of the sampling location. Presumably, the virus concentration is diluted as it travels through the duct and is combined with clean air from different locations. In addition, it is possible that virus may be deposited on the duct walls.
For the 2 cases identified by nasopharyngeal swab testing on March 16 and April 14, we did not detect SARS-CoV-2 in air samples. There is a possibility that these 2 COVID-19 PCR positive students were not spreading the virus in the air, or that it was a false positive PCR test. A previous study has reported that individuals may be PCR positive when they are not infectious22 but have enough residual viral RNA in the respiratory system to be detectable by a clinical PCR COVID-19 laboratory test. The PCR analysis developed for this study only detects COVID-19 if a person was shedding virus into the air, the salient feature that is important for controlling viral spread.
In the IsolationDorm we detected SARS-CoV-2 in all 3 consecutive samples, showing reproducible and reliable results. We did not detect SARS-CoV-2 by sampling Methods 2 and 3 in that suite; however, Method 4 has 50 times the flow rate of Method 2, and Method 3 was only performed for 30 minutes compared to 24 hours for Method 4. These results suggest that increased sampling flow rates or longer collection times may be needed for detection of dilute virus particles.
In this study, we were able to detect the SARS-CoV-2 viral RNA without any purification of the RNA prior to the RT-PCR analysis. Our method takes advantage of the fact that PCR does not require purified genome samples, it simply requires accessible genome sequences and the absence of specific RT-PCR inhibitors, which is similar to previous studies.17, 18, 19, 20, 21 We did not attempt to assess viral “viability” on the filters in tissue culture, because the air sampling techniques in most cases would desiccate the virus over time making its ability to infect and replicate unlikely. We tested the materials used for sampling (filters and cartridges) and found no PCR inhibitors. The absence of RNA purification step allows for more rapid and efficient PCR testing of samples and likely increases the sensitivity of the testing since genome materials are not lost or damaged during purification steps.
To our knowledge, this is the first study successfully detecting SARS-CoV-2 by PCR detection on air samples from HVAC systems. However, we present literature for detecting SARS-CoV-2 PCR compared to the methods we used. Ong et al.23 performed air sampling using our Method 1 for 2 days in a COVID-19 hospital infection isolation, for a sampling duration of 2 hours with a total of 32 samples. The researchers used qRT-PCR with TaqPath Combo Kit to target the N, S, and ORF1ab (RdRP) gene regions to detect SARS-CoV-2 with a positive rate of 53.8%. Lednicky et al.11 performed air sampling inside 2 hospital rooms designated for COVID-19 patients, using in-house aerosol samplers with a flow of 8 LPM, at 3 different locations in the rooms. The researchers used qRT-PCR with 5 mL of purified viral RNA, primers, and probe to detect SARS-CoV-2 with a positive rate of 100%. Similarly, we could detect COVID-19 with Methods 2, 3, and 4, but not with Method 1. For the first environment, Dorm1 and Dorm2, we had a 75% detection rate when PCR positive COVID-19 students lived on the same floor of the sampling location. Compared to Lednicky, et al.11 and Ong et al.23, we did not achieve 100% detection, and Method 1 did not detect COVID-19. However, both studies sampled directly inside the hospital rooms, compared to our studies where we detected virus in the HVAC system for the buildings. In addition, Lednicky et al.11 used an aerosol sampler with a higher flow rate and a water-vapor condensation method for collection. For the second environment, IsolationDorm, we had a 100% detection rate for Method 4, on 3 consecutive testing days. Our sampling time was 24 hours, compared to 2 hours maximum used by Dietz, et al.24
We conducted the current study as a cleaner, easier and safer alternative method to the wastewater track tracing method10 and we have shown that it is possible to detect viruses in HVAC air samples collected at one location from the dorm's return air. It is possible that performing air sampling for SARS-CoV2 on each floor of a large building may yield greater sensitivity.
Conclusion
We developed methods for successful detection of SARS-CoV-2 in HVAC air samples. Compared to confirmed cases by human clinical testing, we had a success rate of 75%-100%. HVAC air sampling can be an important surveillance technique for the control of viral spread in large buildings, especially congregant living settings. Such monitoring of air may allow early intervention to stop the spread of SARS-CoV-2 inside buildings. Future building designs should include HVAC access for such sampling, and public health policies should consider implementation of HVAC surveillance testing either routinely or during times of contagion. Our data suggest air sampling at each floor would be beneficial, and that more distant sampling is less sensitive. More frequent sampling would detect the virus more quickly, but this must be balanced against cost and the level of concern for viral transmission at the time.
Acknowledgments
We want to thank the East Carolina University Division of Research, Economic Development and Engagement, Brody School of Medicine, and the North Carolina Coronavirus Disease 2019 (COVID-19) Crisis House Bill 1043 for the support to conduct this research. In addition, we would like to thank Jim Menke, Lanika Wright, Griffin Avin, Will Bullock, and John Fallon from ECU for their help during the study. Finally, we want to thank Matthew Nonnenmann and Ralph Altmaier from the University of Iowa for their recommendations and help before the study started.
Footnotes
Conflicts of interest: The authors declare no conflict of interest.
Ethics approval: This research did not meet federal definition of Human Subjects research because the research team had no intervention or interaction with human subjects and had no identifiable private information on the specimens, which were collected by housing staff acting as an honest broker.
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.ajic.2021.10.009.
Appendix. SUPPLEMENTARY MATERIALS
References
- 1.Adedoyin OB, Soykan E. Covid-19 pandemic and online learning: the challenges and opportunities. Interactive Learning Environments. 2020:1–13. [Google Scholar]
- 2.Johnson N, Veletsianos G, Seaman J. US faculty and administrators' experiences and approaches in the early weeks of the COVID-19 pandemic. Online Learning. 2020;24:6–21. [Google Scholar]
- 3.Hodges C, Moore S, Lockee B, Trust T, Bond A. The difference between emergency remote teaching and online learning. Educause rev. 2020;27:1–12. [Google Scholar]
- 4.Kar N, Kar B, Kar S. Stress and coping during COVID-19 pandemic: result of an online survey. Psychiatry Res. 2021;295 doi: 10.1016/j.psychres.2020.113598. /01/01/2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahu P. Closure of Universities Due to Coronavirus Disease 2019 (COVID-19): Impact on Education and Mental Health of Students and Academic Staff. Cureus. Apr 4 2020;12:e7541. doi: 10.7759/cureus.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherr TF, Hardcastle AN, Moore CP, DeSousa JM, Wright DW. Understanding on-campus interactions with a semiautomated, barcode-based platform to augment COVID-19 contact tracing: app development and usage. JMIR Mhealth Uhealth. 2021;9:e24275. doi: 10.2196/24275. 2021/3/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losina E, Leifer V, Millham L, et al. College campuses and COVID-19 mitigation: clinical and economic value. Ann Internal Med. 2020;174:472–483. doi: 10.7326/M20-6558. 2021/04/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillam TB, Cole J, Gharbi K, et al. Norwich COVID-19 testing initiative pilot: evaluating the feasibility of asymptomatic testing on a university campus. J Public Health. 2020;43:82–88. doi: 10.1093/pubmed/fdaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen DA, Wigginton KR. Tracking COVID-19 with wastewater. Nature Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. 2020/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betancourt WQ, Schmitz BW, Innes GK, et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Science of The Total Environment. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. 2021/07/20/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lednicky JA, Lauzardo M, Fan ZH, et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int J Infectious Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. 2020/11/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robie ER, Abdelgadir A, Binder RA, Gray GC. Live SARS-CoV-2 is difficult to detect in patient aerosols. Influenza and other respiratory viruses. 2021;15:554–557. doi: 10.1111/irv.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borges JT, Nakada LYK, Maniero MG, Guimarães JR. SARS-CoV-2: a systematic review of indoor air sampling for virus detection. Environ Sci Pollut Res Int. 2021;28:40460–40473. doi: 10.1007/s11356-021-13001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nature Communications. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. 2020/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenarkoohi A, Noorimotlagh Z, Falahi S, et al. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Sci Total Environ. 2020;748 doi: 10.1016/j.scitotenv.2020.141324. Dec 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. New England J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roper RL. Simple, Rapid preparation of poxvirus DNA for PCR cloning and analysis. Methods Mol Biol. 2019;2023:63–71. doi: 10.1007/978-1-4939-9593-6_3. [DOI] [PubMed] [Google Scholar]
- 18.Roper RL. Rapid preparation of vaccinia virus DNA template for analysis and cloning by PCR. Methods Mol Biol. 2004;269:113–118. doi: 10.1385/1-59259-789-0:113. [DOI] [PubMed] [Google Scholar]
- 19.White M, Freistaedter A, Jones GJB, Zervos E, Roper RL. Development of improved therapeutic mesothelin-based vaccines for pancreatic cancer. PloS one. 2018;13:1–17. doi: 10.1371/journal.pone.0193131. e0193131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.See RH, Petric M, Lawrence DJ, et al. Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. J Gen Virol. Sep 2008;89(Pt 9):2136–2146. doi: 10.1099/vir.0.2008/001891-0. [DOI] [PubMed] [Google Scholar]
- 21.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. 2003;77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiwani RA, Mao Y, Pona A, et al. Discontinuation of transmission precautions for COVID-19 patients: polymerase chain reaction diagnostics, patient delays, and cycle threshold values. Infectious Dis in Clin Practice, ahead of print [DOI] [PMC free article] [PubMed]
- 23.Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietz L, Constant DA, Fretz M, et al. Exploring integrated environmental viral surveillance of indoor environments: a comparison of surface and bioaerosol environmental sampling in hospital rooms with COVID-19 patients. medRxiv. 2021:1–28. 2021.03.26.21254416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.