Abstract
Given a large number of SARS-CoV-2 infected individuals, clinical detection has proved challenging. The wastewater-based epidemiological paradigm would cover the clinically escaped asymptomatic individuals owing to the faecal shedding of the virus. We hypothesised using wastewater as a valuable resource for analysing SARS-CoV-2 mutations circulating in the wastewater of Pune region (Maharashtra; India), one of the most affected during the covid-19 pandemic. We conducted study in open wastewater drains from December 2020–March 2021 to assess the presence of SARS-CoV-2 nucleic acid and further detect mutations using ARTIC protocol of MinION sequencing. The analysis revealed 108 mutations across six samples categorised into 39 types of mutations. We report the occurrence of mutations associated with Delta variant lineage in March-2021 samples, simultaneously also reported as a Variant of Concern (VoC) responsible for the rapid increase in infections. The study also revealed four mutations; S:N801, S:C480R, NSP14:C279F and NSP3:L550del not currently reported from wastewater or clinical data in India but reported worldwide. Further, a novel mutation NSP13:G206F mapping to NSP13 region was observed from wastewater. Notably, S:P1140del mutation was detected in December 2020 samples while it was reported in February 2021 from clinical data, indicating the instrumentality of wastewater data in early detection. This is the first study in India to demonstrate utility of sequencing in wastewater-based epidemiology to identify mutations associated with SARS-CoV-2 virus fragments from wastewater as an early warning indicator system.
Keywords: Wastewater, Epidemiology, Nanopore sequencing, Metagenomics, SARS-CoV-2, ARTIC protocol
Graphical abstract
1. Introduction
The respiratory distress virus, Severe Acute Respiratory Syndrome – Corona Virus – 2 (SARS-CoV-2), has unprecedented effects on human life and the healthcare system worldwide. The findings of Tang et al. (2020) revealed the high viral load in the faecal matter of infected individual, irrespective of the individuals showing any symptoms. The wastewater containing viral load from infected individuals would enter the wastewater system of Sewage Treatment Plant in well-planned regions or directly into the river system as untreated wastewater, raising concerns worldwide (Mohapatra et al., 2020). The diagnostics limited to clinical context was eventually hypothesised for wastewater system to gain insight into the infection dynamics of the infected population. The Wastewater-based Epidemiological Study (WBE) provided a comprehensive depiction of infection dynamics in the population by enabling asymptomatic individuals to be included, who would otherwise escape the clinical settings. The WBE approach was previously employed to identify illicit drug use and specific infective agents like SARS and Polio (Zuccato et al., 2005, Heijnen and Medema, 2011, Lago et al., 2003). The foundation of the WBE tool was laid quickly for a better understanding of SARS-CoV-2 spread worldwide.
It was crucial to evaluate the current wastewater viral concentration protocols to optimise novel viral detection and the work was started promptly. Ahmed et al. (2020) provided an evidence-based protocol for concentrating Murine Hepatitis Virus, a positive sense single-stranded enveloped virus, as a surrogate for SARS-CoV-2, from wastewater using seven methods and MCE electronegative membrane filter protocol showed highest recovery. The work for isolating and concentrating SARS-CoV-2 from wastewater was also started worldwide. The statistical model-based evidence suggested by Peccia et al. (2020) provided insight into the correlation of the fluctuations observed in the number of infected individuals and viral load present in the wastewater. Wastewater is the metagenomic landscape with various organisms; hence, detection of specific viral nucleic acids posed a challenge (Che et al., 2019). The MinION sequencer from Oxford Nanopore Technologies can be very useful in such scenarios as the total genomic material from the sample can be sequenced to identify the potential candidate (Che et al., 2019; Pantha et al., 2021). The development of ARTIC protocol facilitated the study of the metagenomic landscape of SARS-CoV-2 from wastewater utilizing the amplicon sequencing to obtain whole-genome sequences (Tyson et al., 2020, Josh Quick, 2021).
The numerous mutations of SARS-CoV-2 were observed worldwide and raised concerns about the effectiveness of treatment and vaccines. The mutants were studied, and particular mutants were speculated for the higher spreading of infection, such as the rapid spread of B.1.617.1 variant by mediating increment of viral entry into certain cell lines by Hoffmann et al., 2021. Studies were also performed for the effectiveness of vaccine candidates among the Variants of Concern (VOC), such as the effectiveness of BBV152 (Covaxin) being able to generate neutralising serological response against B.1.617.1 (Yadav et al., 2021). The tracking of genomic variants from wastewater was assumed essential to understand the spread. The phylogenetic assessment of SARS-CoV-2 from wastewater was carried out by Nemudryi et al., 2020 using a long-read sequencing platform. Genomic variants were studied using Next Generation Sequencing (NGS) platforms by Agrawal et al., 2021 in Germany, Landgraff et al., 2021 in Canada, Wilton et al., 2021 in London, Crits-Christoph et al., 2021 in California, Jahn et al., 2021 in Switzerland and others. The studies were essential to analyse the regionally prevalent strains in circulation, aiding the assumption of Variants of Concerns as causal elements in rising cases in the region.
Presently India is one of the worst affected countries in the world, and Pune in the state of Maharashtra recording one of the highest CoViD19 infections (Pune-Maharashtra covid cases). It was necessary to evaluate the wastewater from Pune city to understand the infection dynamics and focus on the variants circulating in the population. However, no studies are currently being recorded in India, allowing the variant analysis of SARS-CoV-2 from wastewater. To emphasise the importance of the variant study, we present the first study in India for the amplicon-based metagenomic landscape of SARS-CoV-2 in the wastewater of the Pune region.
We hypothesised that wastewater in Pune, being a highly affected region, would demonstrate SARS-CoV-2 RNA presence, which eventually could be employed to analyse the genomic variants. Our goal was to examine the presence of SARS-CoV-2 RNA in the wastewater streams and employ the NGS platform of the MinION sequencer to identify mutations. The study could provide essential information regarding the variants circulating in the community while also examining the potential source of wastewater as an early warning system.
2. Material and methods
2.1. Sample collection and processing
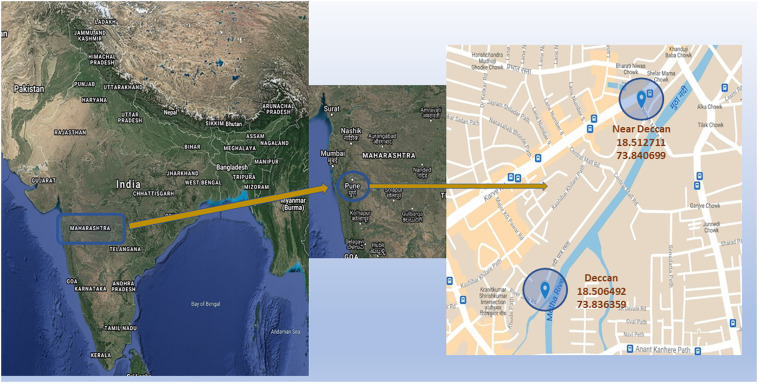
Samples WW9, WW10 and WWP (WWP is SARS-CoV-2 RT-qPCR positive RNA sample collected on 23/12/2020, 26/12/2020, 19/02/2021 and 22/02/2021, pooled for sequencing) from Deccan (18.506492,73.836359; Kothrud Basin) and samples WW8, WW10 and WW12 from Near Deccan (18.512711,73.840699; Prabhat Road Basin) are open wastewater drains entering the Mutha river near the sample collection site (Fig. 1 ). The samples were collected as 1 l- 1 h grab sample between morning 09:00 am to 10:00 am in a sterile plastic container (Himedia Solution Bottles - TCP040-1x12NO) throughout December 2020 to April 2021 (Rimoldi et al., 2020). The sample bottles were thoroughly cleaned from outside by 70% alcohol and 1% hypochlorite solution and transported to the laboratory at 4 °C. The wastewater sample container was kept in the water bath for 60 min at 60 °C for heat inactivation (Wang et al., 2020). After the heat treatment, the bottles were allowed to cool down to room temperature and were immediately processed (Fig. 2 ). The permissions regarding sample collection and processing were obtained from Pune Municipal Corporation.
Fig. 1.
Sampling was done in open drains containing wastewater that was directly entering the Mutha river in Pune, one of India's worst SARS-CoV-2 affected cities.
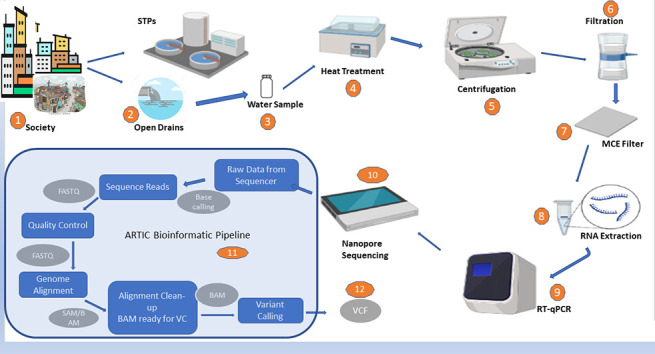
Fig. 2.
The collected sample was transported to the laboratory in a cold chain, and then heat treated. A 200 ml aliquot was centrifuged, and the virus was concentrated using electronegative membrane filtration. The filter was used to extract RNA, and positive RT-qPCR samples were subjected to Nanopore amplicon sequencing to identify genomic sequences.
(Image: BioRender.com).
2.2. Virus concentration
An aliquot of 200 ml was transferred into a sterile Fluorinated HDPE Bottle (Tarson; 584,230) and centrifuged at 4500g for 10 min to settle down larger debris. The sample was then filtered through Whatman filter paper (Whatman; 1001-047) using the vacuum filtration assembly (Tarson; 050030A) and Vacuum pump. The filtrate was transferred into a sterile glass flask, and 25 mM of MgCl2 was added. The sample was then filtered through a 0.45um Mixed Cellulose Ester filter (Millipore; HAWP04700). The MCE filter was immediately transferred to a bead beating tube from the RNA extraction kit. Contaminated glassware and plastic wares were decontaminated or disposed of according to the rules of the institute.
2.3. RNA extraction and Realtime-qPCR
RNeasy Power Water Kit (Qiagen; 14700-50-NF) was used for RNA extraction following the instruction by the manufacturer. The Real-time quantitative Polymerase Chain Reaction (RT-qPCR) was performed for the detection of SARS-CoV-2 using ICMR validated kit TRUPCR® SARS-CoV-2 RT qPCR kit (V-3.2) (3B BlackBio Biotech India Limited; 3B306) on Applied Biosystem ™ 7500 plus (Applied Biosystems). The threshold for cycle cut-off was set manually, and positive samples were detected (Supplementary Table 1). The SARS-CoV-2 positive RNA was employed further for the Oxford Nanopore Sequencing platform using ARTIC protocol (Tyson et al., 2020, Josh Quick, 2021).
2.4. cDNA and nanopore library preparation
According to manufacturer instructions, the Real-time-qPCR positive RNA from six samples was subjected to cDNA preparation using Maxima H minus Reverse Transcriptase Enzyme (Thermofisher; EP0752). The cDNA prepared was then purified using Agencourt Ampure XP beads (Beckman Coulter; A63881). The purified cDNA was further subjected to nCoV-2019 sequencing protocol v3 (LoCost) V.3, which uses two primer pool to amplify the whole genome of SARS-CoV-2 present in sample (Tyson et al., 2020, Josh Quick, 2021). The reverse-transcribed cDNA was amplified using Q5 High-Fidelity DNA Polymerase (New England Biolabs; M0491S), 5× Q5 Reaction Buffer (New England Biolabs; M0491S), dNTPs mix (New England Biolabs; N0447S) and primer pools 40 U/ul SARS-CoV-2 primers (Pool A & B) 100uM (ARCTIC) (New England Biolabs; GTR_066_COVID25). The amplified pools were mixed and purified using Agencourt Ampure XP beads (Beckman Coulter; A63881). End preparation and Barcoding was performed using Blunt/TA Ligase MasterMix (New England Biolabs; M0367L), NEBNext Ultra II End Repair/dA-Tailing Module (New England Biolabs; E7546L) and Native Barcoding Expansion 1–12 (PCR-free) (Oxford Nanopore Technologies; EXP-NBD104). The quantification was performed with Qubit Fluorometer (Invitrogen), and the 24 ng library was loaded onto the flow cell. The barcoded samples were pooled together, and the run was set up on the MinION device (Oxford Nanopore Technologies). The sequencing was allowed to run for 24 h, and data was collected. The raw reads from the Nanopore sequencer were base-called using Guppy High Accuracy - dna_r9.4.1_450bps_hac.cfg, and further analysis was carried out using ARTIC Bioinformatic Pipeline with few required modifications (Tyson et al., 2020, Josh Quick, 2021). All the sequences obtained were analysed with reference to the SARS-CoV-2 reference genome Wuhan-Hu-1 (NCBI Accession: MN908947). The GSAID database was utilised to obtain information regarding the reported mutation and was last accessed on 01st May 2021.
3. Results
The Ct values for the sample are provided in the Supplementary Table 1. All the wastewater samples collected between December 2020 through April 2021 consistently were positive for the presence of SARS-CoV-2 nucleic acids. The cycle threshold values obtained from all the samples have shown variability attributed to the changing infection dynamics. The amplicon sequenced Sequence Read Archive data is submitted to the National Center for Biotechnology Information database with accession number SRA:PRJNA728440. The analysis revealed several mutations in multiple genomic regions of SARS-CoV-2, including 3′UTR, ORF1a, ORF1b, Spike, ORF3a, ORF7a, M, ORF6, N, ORF8 and 3′UTR. In total, 108 mutations, categorised into 39 types based on nucleotide position, were detected in all the samples (Table 1 , Supplementary Table 2). We detected 15 mutations from WW8, 19 mutations from WW9, 17 mutations from WW10, 20 mutations from WW11, 23 mutations from WW12 and 13 mutations from WW-P (Table 1, Supplementary Table 2). Notably, nine mutations in the Spike region (S: L452R, S:C480R, S: E484Q, S: D614G, S: P681R, S: N801, S: D950N, S: Q1071H, S: P1140) were observed in this study (Table 1, Supplementary Table 2). The March-2021 samples showed L452R and E484Q mutations, while these mutations were absent in the sample collected from December-2020 to February-2021 (WWP). We detected five novel mutations not reported from Indian clinical sequence data on Global Initiative on Sharing Avian flu Data (GISAID)21. These mutations are as follows: 23964 AT>A (S: N801del), 4369 TG > T (NSP3:L550), 18,875C > T (NSP14:C279F), 16,852/16853 GG > TT (NSP15:C206F), 23,000 T > C (S:C480R).
Table 1.
Mutations identified in the six samples collected from December 2020 throughout April 2021. ‘-’ indicates- no mutation detected.
The table displays the detailed mutations detected in the samples using Oxford Nanopore amplicon sequencing. Notably, samples collected in March (WW8, WW9, WW10, WW11, and WW12) contain the SARS-CoV-2 Delta variant lineage mutations S:L452R and S:E484Q, which are not present in WWP. The six samples revealed nine mutations associated with the spike region of the SARS-CoV-2 genome.
| Samples | SARS-CoV-2 genomic regions |
||||||
|---|---|---|---|---|---|---|---|
| 5′UTR | ORF | NSP | N | S | M | 3′UTR | |
| WW8, WW9, WW10, WW11, WW12 WWP | 5′-UTR:210 5′-UTR:241 |
ORF3a:S26L | NSP3:Y246Y NSP12b:P314L |
– | S:P1140del | – | – |
| WW8, WW9, WW10, WW11, WW12 | – | – | NSP13:M429I | – | S:L452R S:E484Q |
– | 3′UTR:28270 |
| WWP, WW8, WW9, WW10, WW11 | – | – | – | – | S:D614G | – | – |
| WW8, WW9, WW10, WW11 | – | – | NSP3:T749I NSP6:T77A |
– | S:Q1071H | – | – |
| WW9, WW10, WW11 | – | ORF6:I33T ORF7a:V82A |
– | – | – | – | – |
| WW8, WW9, WW11 | – | – | – | – | – | – | 3′UTR:29742 |
| WWP | – | ORF3a:E261* ORF8:S97I |
NSP3:H1630H NSP10:H80H |
– | – | – | – |
| WW12 | 5′-UTR:75 | – | NSP3:L550del NSP13:P77L NSP13:G206F NSP13:V484F NSP14:C279F |
– | S:C480R S:D950N |
M:V10A | 3′-UTR:21555 3′UTR:26493 |
| WWP, WW12 | – | – | NSP14:C279C | – | – | – | – |
| WWP, WW10 | – | – | – | – | S:N801 | – | – |
| WW11, WW12 | – | – | NSP3:P822L | – | – | – | – |
| WW9 | – | – | – | N:R203M | – | – | 3′-UTR:29700 |
| WW11 | – | – | – | N:D63G | S:P681R | – | – |
4. Discussion
The wastewater based epidemiological approach can be used for predicting the infection dynamics of the population (Peccia et al., 2020). However, an exact estimation of the infected individuals is currently unattainable. Currently, exact estimation of viral load using recovery and stability of virus from the faecal source to sewage treatment plant has not been carried out. However, WBE can be applied to obtain comparative infection dynamics of a particular region to obtain information regarding the severity of affected regions (Peccia et al., 2020). Since the water has shown consistent viral presence, as seen in current study, it is essential to bring the public and government attention to the constant viral presence to take actions accordingly. The study has also provided us instances where the mutations obtained from the wastewater sequenced data are either not reported in GISAID from India or, in case of novel mutation, not across the world.
The WBE study can provide us information regarding genomic variants in the population as mentioned earlier (Hoffmann et al., 2021; Agrawal et al., 2021; Landgraff et al., 2021; Wilton et al., 2021; Crits-Christoph et al., 2021; Jahn et al., 2021). The clinical evaluation of variants in circulation is an arduous exercise, where asymptomatic can be overlooked, along with the time-consuming protocols for viral culture and whole-genome sequencing. The state of Maharashtra recorded very high cases of infection, and it has been raised concern as B.1.617.1 variant to be a causal factor (Maharashtra double mutant). The clinical evaluation raised concern over rising cases from B.1.617.1 variant from March 2021, and the data analysis also revealed the presence of mutations L452R and E484Q associated with B.1.617.1 variant lineage in samples collected during March 2021. While the mentioned mutations were absent in samples collected from December 2020 to February 2021, the mutations concerning the variant were also observed in the wastewater sample from a similar period. When clinical genome sequencing data became available, the speculation was confirmed, as the percentage of SARS-CoV-2 infections associated with the B.617.2 lineage began to rise in March 2021 (https://clingen.igib.res.in/covid19genomes/). Here, it can be concluded that wastewater can act as early warning system and regular wastewater monitoring can be a critical resource. Hence, regular monitoring of wastewater is an essential criterion for the study of variants in circulation as the required results can be obtained from a smaller sample volume of wastewater, as compared to the larger number of individuals.
There are mutations found in wastewater such as S:P1140del which was reported in late February from India and earlier only from Africa-Egypt and North America (GISAID - Shu and McCauley, 2017). However, the mutation, which is in the Spike region, was present in all the samples collected from December 2020 to March 2021. This identification provides conclusive evidence of how clinical data can lag behind wastewater sequencing data to identify the presence of the mutations and a possible variant in circulation. WW12 was the most recent sample with maximum mutations recorded and has 12 mutations unique to the sample, in which NSP3:L550del, NSP14:C279F and S:C480R are not yet reported from India (GISAID - Shu and McCauley, 2017). We also report a novel mutation NSP13:G206F (NSP13 region). This identification of a novel mutation can be an instance where novel mutations are identified across the wastewater sample before they are identified clinically. The WWP is the pooled sample from WW1, WW2, WW3 and WW4 from Deccan and Near Deccan site showing 13 reported mutations, and we report the mutation S:N801 (Spike region), not yet reported from India (GISAID - Shu and McCauley, 2017). WW9 has shown mutation N:R203M and 3′-UTR:29700, while WW11 has shown mutation S:P618R and N:D63G, present in samples collected in March 2021, and absent in sample WWP taken before March. It can be predicted that the mentioned mutations prevailed in wastewater from March 2021 and might have been absent before. These mutations have not yet been reported from India, while reported in other countries (GISAID - Shu and McCauley, 2017). This detection of mutation from wastewater sample provides an instance where mutations that are clinically neglected can be recorded in wastewater sample, providing thorough information for mutations of the virus in circulation.
It has been observed in studies around the world; how variants of the SARS-CoV-2 virus has shown distinct infectivity (Hoffmann et al., 2021). Hence, it is crucial to constantly examine the evolving mutations and classify the variants in circulation. In order to understand the genomic variations in SARS-CoV-2, an open drain system can be used to obtain information regarding the mutations and strain analysis. The methods used in the experiment followed the protocols of Ahmed et al. (2020) for viral concentration. The virus concentration was carried out using MCE electronegative filtration, which provides a mean recovery of 65.7% ± 23.8 (Mean ± SD of % recovery) of the viral particles from sample (Ahmed et al., 2020). The regular monitoring and repetitive extraction of such samples can increase the possibility of concentrating virus that can be identified for novel mutations. The studies conducted in India have reported the viral presence across the STPs or wastewater. However, no studies have yet revealed NGS platform sequencing to identify the variants in circulation from wastewater (Srivastava et al., 2021; Chakraborty et al., 2021; Hemalatha et al., 2021; Arora et al., 2020; Kumar et al., 2020). Our study is first to report the approach of variant analysis in the WBE and provides an insight into the variants in circulation, along with reporting novel mutations.
5. Conclusion
Wastewater can be considered a vital source to understand the mutations in circulation and understand the infection dynamics. The prevalence of clinically unreported mutations, such as S:P1140del, NSP3:L550del, NSP14:C279F and S:C480R, in circulation from wastewater data, provide conclusive evidence for the potential utilization of wastewater as early warning system. We also report novel mutations such as NSP13:G206F (NSP13), which conclude the capability of wastewater sequencing data to provide mutations in circulation before they are observed clinically. The observation of mutations L452R and E484Q associated with B.1.617.1 variant lineage in similar period of declaration as variant of concern provides a conclusive evidence of WBE as early warning system. Regular monitoring of wastewater system for analysing mutations can act as an early warning system to understand the community infection dynamics.
Funding
This project is funded by Science Engineering and Research Board (SERB), India under special covid grant (CVD/2020/001002).
Author contributions
All authors have read and review the manuscript for submission. TD: performing experiments, writing original draft and art design; RKY: performing experiments, editing and review; VR: bioinformatic analysis, review and editing; RB: sampling permissions, coordination with authorities; DP: sampling plan, review; SPK: study plan and logistics; SD: review and funding; MD: editing, review, funding and corresponding author.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgements
Authors are indebted to the Directors of CSIR-NCL and Ecosan Services Foundation for providing facilities, infrastructure and support for this study. Sincere thanks to Pune Municipal Corporation for giving the required permissions. Authors also extend acknowledgement to BioRender.com for creation of images. Authors thank Mr. Nitya Jacob, India Coordinator, Sustainable Sanitation Alliance (SuSanA) India Chapter for providing technical guidance. Authors also thank Mr. Sandeep More (Ecosan Services Foundation) for his able assistance in sampling. TD, VR and RKY acknowledge SERB and University Grant Commission (UGC), India respectively for fellowship support.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.151038.
Appendix A. Supplementary data
Supplementary tables
References
- Agrawal S., Orschler L., Lackner S. Metatranscriptomic analysis reveals SARS-CoV-2 mutations in wastewater of the Frankfurt metropolitan area in southern Germany. 2021;10(15) doi: 10.1128/MRA.00280-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., Gupta A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020;82(12):2823–2836. doi: 10.2166/wst.2020.540. [DOI] [PubMed] [Google Scholar]
- Chakraborty P., Pasupuleti M., Shankar M.J., Bharat G.K., Krishnasamy S., Dasgupta S.C., Sarkar S.K., Jones K.C. First surveillance of SARS-CoV-2 and organic tracers in community wastewater during post lockdown in Chennai, South India: methods, occurrence and concurrence. Sci. Total Environ. 2021;15(778) doi: 10.1016/j.scitotenv.2021.146252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y., Xia Y., Liu L., Li A.D., Yang Y., Zhang T. Mobile antibiotic resistome in wastewater treatment plants revealed by nanopore metagenomic sequencing. Microbiome. 2019;7(1):1–3. doi: 10.1186/s40168-019-0663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A., Kantor R.S., Olm M.R., Whitney O.N., Al-Shayeb B., Lou Y.C., Flamholz A., Kennedy L.C., Greenwald H., Hinkle A., Hetzel J. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. MBio. 2021;12(1) doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of influenza a and the pandemic influenza a (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health. 2011;9(3):434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Hemalatha M., Kiran U., Kuncha S.K., Kopperi H., Gokulan C.G., Mohan S.V., Mishra R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: comprehensive study. Sci. Total Environ. 2021;10(768) doi: 10.1016/j.scitotenv.2020.144704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Sidarovich A., Moldenhauer A.S., Winkler M.S., Schulz S., Jäck H.M. bioRxiv; 2021. SARS-CoV-2 variant B. 1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination.https://clingen.igib.res.in/covid19genomes/ Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., Bänziger C., Stachler E., Fuhrmann L., Jablonski K.P., Chen C. medRxiv; 2021. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. Jan 1. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;1(746) doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago P.M., Gary H.E., Jr., Pérez L.S., Cáceres V., Olivera J.B., Puentes R.P., Corredor M.B., Jímenez P., Pallansch M.A., Cruz R.G. Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. 2003;32(5):772–777. doi: 10.1093/ije/dyg185. [DOI] [PubMed] [Google Scholar]
- Landgraff C., Wang L.Y., Buchanan C., Wells M., Schonfeld J., Bessonov K., Ali J., Robert E., Nadon C. medRxiv; 2021. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B. 1.1. 7 variant of concern from a Canadian municipality concurrent with an outbreak. Jan 1. [DOI] [Google Scholar]
- Maharashtra double mutant. https://indianexpress.com/article/explained/maharashtra-double-mutant-found-b-1-617-variant-and-the-surge-7274080/. (last accessed on 29 May 2020)
- Mohapatra S., Menon N.G., Mohapatra G., Pisharody L., Pattnaik A., Menon N.G., Bhukya P.L., Srivastava M., Singh M., Barman M.K., Gin K.Y. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2020;6 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantha K., Acharya K., Mohapatra S., Khanal S., Amatya N., Ospina-Betancourth C., Butte G., Shrestha S.D., Rajbhandari P., Werner D. Faecal pollution source tracking in the holy Bagmati River by portable 16S rRNA gene sequencing. NPJ CleanWater. 2021;4(1):1. [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pune-Maharashtra covid cases. https://timesofindia.indiatimes.com/city/mumbai/maharashtra-reports-highest-single-day-spike-of-63729-covid-19-cases/articleshow/82106311.cms (last accessed on 29 May 2020.)
- Quick J. nCoV-2019 sequencing protocol v3 (LoCost) V.3. 2021. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (Last accessed on 22 October 2021)
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;20(744) doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. Mar 30. (GISAID last accessed on 01 May 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V., Gupta S., Patel A.K., Joshi M., Kumar M. 2021. Comparative analysis of SARS-CoV-2 RNA load in wastewater from three different cities of Gujarat, India. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A.N., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26(6):1337. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., Choi J.H., Lapointe H., Kamelian K., Smith A.D., Prystajecky N. bioRxiv; 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. Sep 4. [DOI] [Google Scholar]
- Wang T., Lien C., Liu S., Selveraj P. Medrxiv; 2020. Effective heat inactivation of SARS-CoV-2. Jan 1. [DOI] [Google Scholar]
- Wilton T., Bujaki E., Klapsa D., Fritzsche M., Mate R., Martin J. medRxiv; 2021. Rapid increase of SARS-CoV-2 variant B. 1.1. 7 detected in sewage samples from England between October 2020 and January 2021. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P., Sapkal G.N., Abraham P., Ella R., Deshpande G., Patil D.Y., Nyayanit D., Gupta N., Sahay R.R., Shete A.M., Panda S. bioRxiv; 2021. Neutralization of variant under investigation B. 1.617 with sera of BBV152 vaccinees. Jan 1. [DOI] [PubMed] [Google Scholar]
- Zuccato E., Chiabrando C., Castiglioni S., Calamari D., Bagnati R., Schiarea S., Fanelli R. Cocaine in surface waters: a new evidence-based tool to monitor community drug abuse. Environ. Health. 2005;4(1):1–7. doi: 10.1186/1476-069X-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables