Abstract
Staphylococcus epidermidis and Staphylococcus aureus are the most common causes of medical device-associated infections, including septicemic loosenings of orthopedic implants. Frequently, the microbiological diagnosis of these infections remains ambiguous, since at least some staphylococci have the capacity to reduce their growth rate considerably. These strains exhibit a small-colony phenotype, and often they are not detectable by conventional microbiological techniques. Moreover, clinical isolates of S. aureus and S. epidermidis adhere to polymer and metal surfaces by the generation of thick, multilayered biofilms consisting of bacteria and extracellular polysaccharides. This study reports improved detection and identification of S. aureus and S. epidermidis by an in situ hybridization method with fluorescence-labeled oligonucleotide probes specific for staphylococcal 16S rRNA. The technique has proven to be suitable for the in situ detection of staphylococci, which is illustrated by the identification of S. epidermidis in a connective tissue sample obtained from a patient with septicemic loosening of a hip arthroplasty. We also show that this technique allows the detection of intracellularly persisting bacteria, including small-colony variants of S. aureus, and the differentiation of S. epidermidis from other clinically relevant staphylococci even when they are embedded in biofilms. These results suggest that the 16S rRNA in situ hybridization technique could represent a powerful diagnostic tool for the detection and differentiation of many other fastidious microorganisms.
Staphylococci are the most important pathogens in polymer-associated infections. They cause a wide range of polymer-related infections as well as infections associated with prosthetic joints and other implanted biomaterials (13, 21, 30). Although staphylococci are easy to identify by classical microbiological techniques, in some cases the correct species diagnosis is difficult. This might be due to the fact that at least some clinical staphylococcal isolates can differ in a wide range of phenotypic properties, leading to misinterpretations of biochemical tests (10, 11, 23, 29). In Staphylococcus aureus, the appearance of small-colony variants (SCVs) has been associated with recurrent and persistent infections (29). SCVs are characterized by a reduced growth rate, diminished exoprotein production, and increased resistance to aminoglycosides. Moreover, these bacteria were found to persist intracellularly. It was shown that this phenotype is due to defects in the hemin and menadione metabolism (5, 29, 33). The microbiological identification of SCVs is often complicated by the reduced growth rate and altered cell wall and exoprotein production. In S. epidermidis, variation of phenotypic features, including variation of enzyme expression, has also been described (8–11, 27, 35).
In the pathogenesis of foreign body-related infections caused by S. epidermidis, adhesion of the bacteria to the biomaterials is an essential step (12, 28). Scanning electron microscope investigations of polymer devices revealed that multilayered cell clusters of staphylococci are embedded in a thick matrix of slime substance (14, 28). This process is suggested to occur in two stages: (i) a rapid initial attachment of the bacteria to the polymer surfaces is followed by (ii) a cell proliferation process and the production of an extracellular polysaccharide substance which mediates the intercellular adherence of the bacteria and the accumulation of a multilayered biofilm (17–19, 24, 25, 31). The production of this substance, termed the polysaccharide intercellular adhesin, was found to be associated with the presence and expression of the ica operon, which is widespread in clinical S. epidermidis isolates (19, 35).
This study was aimed at the development of an improved identification method for clinically important staphylococcal species by using a 16S rRNA-based oligonucleotide in situ hybridization technique which also detects biofilm-forming staphylococci and small-colony phenotypes in tissue samples. The study demonstrates the potential of the technique for the detection of other fastidious microorganisms which often remain unidentified by classical microbiological approaches.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used are listed in Table 1.
TABLE 1.
Bacterial strains
| Straina | Sourceb or reference |
|---|---|
| 1. S. epidermidis RP62A (ATCC 35984) | ATCC |
| 2. S. epidermidis 163 | Clinical isolate |
| 3. S. epidermidis 165 | Clinical isolate |
| 4. S. epidermidis 161 | Clinical isolate |
| 5. S. epidermidis 183 | Clinical isolate |
| 6. S. epidermidis 195 | Clinical isolate |
| 7. S. epidermidis 9 | Clinical isolate |
| 8. S. warneri DSM 20316 | DSM |
| 9. S. warneri 193 | Clinical isolate |
| 10. S. aureus 8325/4 (NCTC 8325) | NCTC |
| 11. S. aureus 173 | Clinical isolate |
| 12. S. aureus 175 | Clinical isolate |
| 13. S. aureus 3 | Clinical isolate |
| 14. S. saprophyticus DSM 20229 | DSM |
| 14. S. haemolyticus DSM 20263 | DSM |
| 15. S. cohnii 194 | Clinical isolate |
| 16. S. cohnii DSM 20260 | DSM |
| 17. S. carnosus TM300 | 28 |
| 18. S. lugdunensis DSM 4804 | DSM |
| 19. Enterococcus faecalis DSM 2570 | DSM |
| 21. 20. E. coli K-12 strain C600 | |
| 22. Klebsiella pneumoniae IA565 | Clinical isolate |
| 23. Proteus mirabilis 102 | Clinical isolate |
| 24. S. typhimurium C17 | Clinical isolate |
| 25. Citrobacter freundii 3009 | Clinical isolate |
| 26. S. capitis DSM 20326 | DSM |
| 27. S. aureus 453 | Clinical isolate |
| 28. C. albicans ATCC 44808 | ATCC |
| 29. S. aureus hemB mutant | 28 |
| 30. E. coli K-12 strain HB101 |
Numbers correspond to those in Fig. 3a.
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; NCTC, National Collection of Type Cultures.
16S rRNA-targeted DNA probes.
DNA oligonucleotide probes were synthesized and labeled with Cy3 or fluorescein by MWG Biotech GmbH, Ebersberg, Germany. The probe SEP-1, which is specific for S. epidermidis, S. saccharolyticus, S. caprae, and S. capitis, was described by Zakrzewska-Czerwinska et al. (34). The DNA probe EUB338, which is specific for all eubacteria, was reported by Amann et al. (2). The S. aureus-specific oligonucleotide probe SA-P1 was described by Bentley et al. (7). All oligonucleotide sequences are shown in Fig. 1.
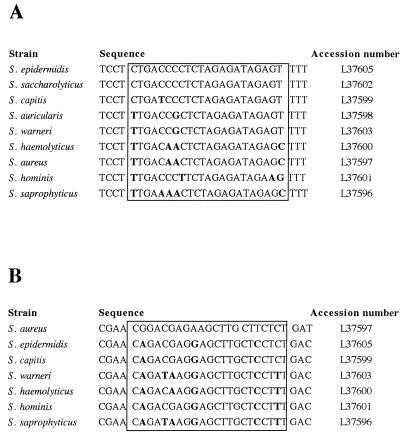
FIG. 1.
(A) Sequence alignment of the region in the 16S rRNA genes of staphylococci corresponding to nucleotides 1014 to 1043 of the E. coli 16S rRNA. The target region of the oligonucleotide probe SEP-1 is boxed. Nucleotides different from those in the S. epidermidis sequence are in boldface. (B) Sequence alignment of the region in the 16S rRNA genes of staphylococci corresponding to nucleotides 66 to 95 of the E. coli 16S rRNA. The target region of the oligonucleotide SA-P1 is boxed. Nucleotides different from those in the S. aureus sequence are in boldface.
RNA preparation for 16S rRNA hybridization.
Overnight cultures were diluted 1:40 in 10 ml of brain heart infusion broth (Difco) and grown at 37°C in a shaking water bath to the mid-log phase. Bacterial cells were harvested by centrifugation for 5 min at 4,000 rpm. The pellet was resuspended in 500 μl of protoplast buffer containing 15 mM Tris-HCl (pH 7.5), 0.45 M sucrose, 8 mM EDTA (pH 8.0), and 100 μl of lysostaphin (500-μg/ml stock) (Sigma, Deisenhofen, Germany). The mixture was chilled for 20 minutes on ice and then incubated for 5 to 20 min at 37°C. Following the addition of 500 μl of lysis buffer (30 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate [SDS], 100 μg of proteinase K per ml), the suspension was twice frozen at −80°C and thawed at room temperature. Following the last thawing, the suspension was incubated for 1 h at 37°C, and phenolchloroform-isoamyl alcohol extraction was performed five times. The RNA was precipitated with 5 M sodium chloride and 2 volumes of chilled 96% ethanol. Following centrifugation, the RNA pellet was rinsed with 80% ethanol and air dried.
DIG labeling of oligonucleotides for dot blot hybridization.
For digoxigenin (DIG) labeling, the DIG Oligonucleotide 3′-End Labeling Kit (no. 1362372; Boehringer, Mannheim, Germany) was used. The labeling procedure was performed according to the instructions of the manufacturer.
Dot blot hybridization with DIG-labeled oligonucleotides.
Ten micrograms of RNA was blotted onto a nylon membrane (PALL Biodyne B membrane) by using a dot blot apparatus (Schleicher & Schuell SRC 96 D Minifold I Dot Blotter) and UV fixed. For the hybridization procedure high-SDS buffer containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 7% SDS, 50 mM sodium phosphate (pH 7.0), 2% blocking reagent (Boehringer), and 0.1% N-lauroylsarcosine was used. All buffers and solutions were used according to the instructions of the supplier. The hybridization temperature was 42°C. Washing of the membrane was performed by using a high-stringency washing buffer containing 0.1% SDS and 0.1× SSC at 50°C. Detection of the DIG-labeled oligonucleotide was performed by using alkaline phosphatase-labeled anti-DIG antibody Fab fragments (Boehringer) and the chemiluminescent substrate CSPD (Boehringer) by the procedure recommended by the supplier. The membrane was exposed to Hyperfilm-ECL (Amersham) for 1 to 60 min.
Bacterial cell fixation and permeabilization for whole-cell hybridization.
Lysis of the bacterial cells was performed as described previously by Matsuhisa et al. (26) with some modifications. Briefly, bacteria were grown to exponential phase and harvested by centrifugation. The pellet was resuspended and washed in phosphate-buffered saline (PBS) (130 mM sodium chloride, 10 mM sodium phosphate buffer, pH 7.4). Cells were fixed in 4% paraformaldehyde–PBS for 1 h, fixed again in 70% ethanol for 30 min, and stored in a 1:1 mixture of ethanol and PBS as described previously (2). For the hybridization, 1 μl of bacterial cells was dropped onto a microscope slide and air dried. For permeabilization, the cells were treated with an enzyme mix (750 μg of lysostaphin per ml, 5 mg of lysozyme per ml, 50 mM sodium phosphate, 0.05% saponin) for 1 h at 37°C.
Whole-cell hybridization.
Fifteen microliters of hybridization solution (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 0.001% SDS, 43% formamide, 50 ng of Cy3- or fluorescein-labeled probe) was applied to microscope slides and incubated for 3 h at 43°C in an isotonically equilibrated humid chamber (6). Removal of the unbound probe and washing were performed at 43°C as described by Amann et al. (3). The slides were rinsed briefly with distilled water, air dried, and mounted with Citifluor solution (Citifluor Ltd., London, United Kingdom). Fluorescence was detected by using an Axioplan microscope (Zeiss, Oberkochen, Germany) equipped with an epifluorescence unit.
Detection of S. epidermidis in biofilms.
The biofilm-producing strains S. epidermidis RP62A and S. aureus 453 were used. Bacterial strains were grown overnight in chamber slides (Lab-Tek II chamberslide no. 154534; Nalge Nunc International, Naperville, Ill.) in Tripticase soy broth at 37°C. After decanting, the supernatant slides were washed three times with PBS to remove all nonadhering bacteria. Bacterial biofilms were fixed by air drying and treated for hybridization as described above for the whole-cell hybridization assay.
Detection of an S. aureus hemB mutant in a human endothelial cell line by in situ hybridization.
For the intracellular persistence assay, the following bacterial strains were used: S. aureus 8325/4 (noninvasive), an invasive S. aureus 8325/4 hemB mutant (33), Salmonella typhimurium C17 (invasive positive control) (15), and Escherichia coli HB101 (noninvasive negative control).
The human endothelial cell line E.Ahy 926 was grown to a confluent monolayer in RPMI medium with 10% fetal calf serum and 2 mM glutamine in chamber slides (Lab-Tek II chamberslide no. 154534). Washed bacteria were adjusted to nearly equal numbers for each strain and added to the washed monolayers. The infected monolayers were incubated for 3.5 h (2.5 h for the hemB mutant) at 37°C in 5% CO2 to allow adhesion and invasion of the bacteria. The monolayers were washed three times with PBS. In order to remove all extracellular bacteria, 1 ml of medium containing 10 μg of lysostaphin (Sigma) per ml was added in case of the S. aureus strains. Extracellular S. typhimurium and E. coli were removed by adding 100 μg of gentamicin/ml. Incubation in both cases was for 30 min at 37°C. The monolayers were washed three times with PBS and fixed for 20 min in 4% freshly prepared paraformaldehyde at room temperature. Dehydration was performed stepwise for 5 min in 50, 70, and 100% ethanol. For the detection by the fluorescent oligonucleotide probes, the fixed and dehydrated monolayers were treated as described above for the permeabilization for whole-cell hybridization. The conditions for the in situ hybridization were exactly the same as for the whole-cell hybridization assay.
In situ hybridization of paraffin-embedded tissue samples.
To remove paraffin, the tissue sections were treated twice with Rotihistol (Roth, Karlsruhe, Germany) for 10 min and then with Rotihistol-ethanol (1:1) for 10 min. For the detection by the fluorescent oligonucleotide probes, the tissue sections were treated as described above for the permeabilization for whole-cell hybridization. The conditions for the in situ hybridization were exactly the same as for the whole-cell hybridization assay.
RESULTS AND DISCUSSION
Specificities of the oligonucleotide probes.
The detection and identification of microorganisms based on their 16S rRNAs have many advantages. First, each bacterial cell contains multiple copies of the 16S rRNA in its ribosomes. Hence, the technique is sensitive enough to detect single bacterial cells. Second, 16S rRNA genes are highly conserved throughout bacterial evolution. They consist of regions which are common to all eubacteria and other regions which are extremely species specific. By using the appropriate gene probes, it is possible either to detect any bacterial pathogen or, when highly specific probes are used, to identify single bacterial species. Also, this technique allows for the identification of microorganisms independently of bacterial growth rates and metabolic activities. This is a special advantage for the detection of dormant and metabolically inactive bacteria, since the number of ribosomes is not significantly affected in such organisms. This article reports the development of improved diagnostic tools for both S. aureus and S. epidermidis, which are the most common causative agents of infections in orthopedic implants (30).
For the detection of S. aureus and S. epidermidis, two oligonucleotide probes which are directed against specific regions in the 16S rRNA were used. The nucleotide sequence of the S. epidermidis-specific probe SEP-1 was described by Zakrzewska-Czerwinska et al. (34). For the S. aureus-specific probe SA-P1, the sequence published by Bentley et al. (7) was used. Figure 1 shows an alignment of the 16S rRNA genes of different Staphylococcus species and the positions of the SEP-1 and SA-P1 probes.
In a first set of experiments, the specificities of the gene probes used in this study were analyzed by dot blot hybridizations (Fig. 2 and 3). To this end, RNAs of (i) different staphylococcal species, (ii) a range of nonrelated bacteria, and (iii) Candida albicans were isolated, dotted onto nylon membranes, and hybridized with the appropriate DIG-labeled gene probes. In a control experiment, the stability and the amount of the filter-immobilized RNA were determined by using the oligonucleotide probe EUB338 (2), which binds to the 16S rRNAs of all eubacteria (Fig. 3b) but not to the rRNA of C. albicans (Fig. 2b).
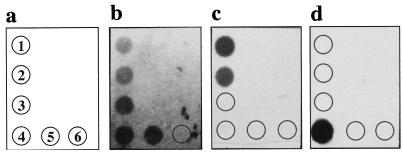
FIG. 2.
RNA dot blot hybridization with S. epidermidis 163 (1), S. capitis DSM 20326 (2), S. warneri DSM 20316 (3), S. aureus 8325/4 (4), S. saprophyticus DSM 20229 (5), and C. albicans ATCC 44808 (6). The oligonucleotide probes were labeled with DIG, and hybridizations were performed as described in Materials and Methods at 42°C in a buffer containing 50% formamide. (a) Strain numbers; (b) hybridization with probe EUB338; (c) hybridization with probe SEP-1; (d) hybridization with probe SA-P1.
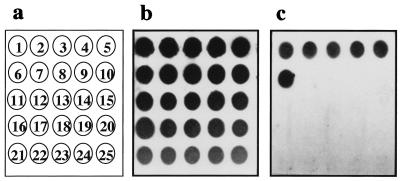
FIG. 3.
RNA dot blot hybridizations with bacterial strains listed in Table 1. The oligonucleotide probes were labeled with DIG, and hybridizations were performed as described in Materials and Methods at 42°C in a buffer containing 50% formamide. (a) Strain numbers, corresponding to the first 25 strains, respectively, in Table 1; (b) hybridization with probe EUB338; (c) hybridization with probe SEP-1.
Figure 1A shows that the sequence of the SEP-1 probe differs from those of most staphylococcal species by more than two nucleotides. In the cases of S. capitis and S. warneri, however, the SEP-1 probe contains only one and two nucleotide mismatches, respectively. As expected, the S. epidermidis-specific probe, SEP-1, hybridized to the RNAs from the S. epidermidis isolates but not to RNAs isolated from all of the other bacterial species analyzed in this experiment (Fig. 3c). The data also show that it was not possible to distinguish between S. epidermidis and the very closely related species S. capitis (Fig. 2c). Since the S. epidermidis-specific gene probe contains only one base exchange in comparison with the corresponding S. capitis 16S rRNA region, S. capitis strains were misidentified as S. epidermidis. This cross-hybridization of the SEP-1 gene probe with S. capitis, S. saccharolyticus, and S. caprae has also been described recently by Zakrzewska-Czerwinska et al. (34). S. capitis, S. saccharolyticus, and S. caprae have been shown to have a certain pathogenic potential (e.g., S. caprae can be associated with bone and joint infections in humans [20, 32]), but clearly, they are rarely isolated human pathogens (1, 22), whereas the vast majority of this type of infections are caused by S. epidermidis. Therefore, it is unlikely that the cross-reactivity of the SEP-1 probe will pose a real problem, particularly as the treatment strategy would be the same as that for an S. epidermidis infection. The S. aureus-specific oligonucleotide probe SA-P1 revealed positive signals only with the RNA isolated from S. aureus and not with that from any other species tested in this study (Fig. 2d). Thus, both oligonucleotide probes proved to be suitable for the identification of S. aureus and a group of four coagulase-negative staphylococci, including S. epidermidis, and for their discrimination from a range of other bacterial species. However, it should be noted that an extensive study involving a broad range of related and nonrelated bacterial species is required to completely exclude possible unspecific reactions of the oligonucleotide probes described here.
Fluorescent hybridization of whole bacterial cells.
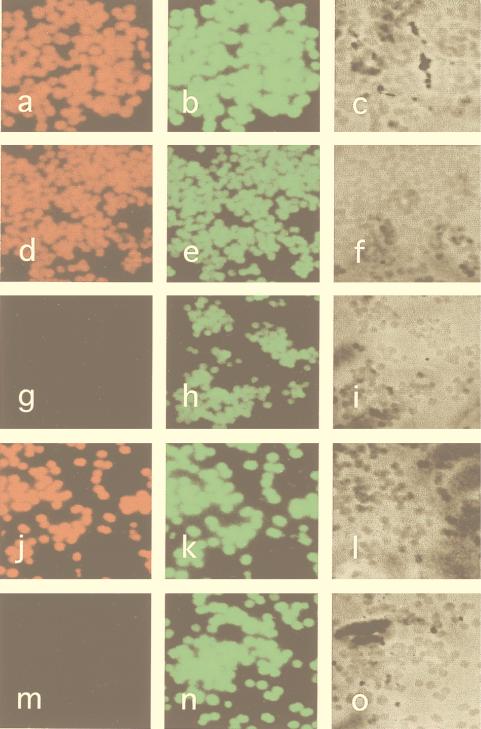
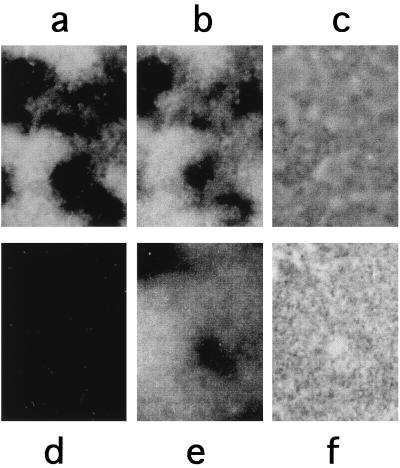
The goal of the experiments described next was to establish a 16S rRNA-based in situ hybridization method that would allow for an improved diagnosis of staphylococcal infections. First, it was important to show that staphylococcal cells can be sufficiently permeabilized to allow for intracellular hybridization with fluorescence-labeled oligonucleotide probes. Next, we attempted to answer the question of whether staphylococci can be reliably differentiated by this technique. To address these issues, different staphylococcal species were analyzed in a whole-cell hybridization assay. The microorganisms were grown in liquid cultures to the exponential phase, harvested, and treated as described in Materials and Methods. For the detection of S. aureus and S. epidermidis, the Cy3-labeled oligonucleotide probes SA-P1 and SEP-1 were used. The FLUOS-labeled probe EUB338 was used as a positive control specific for all eubacterial species. The results obtained by fluorescence microscopy can be summarized as follows: (i) hybridization with the positive control (EUB338) revealed that all eubacterial cells are visualized by this technique (Fig. 4b, e, h, k, and n), (ii) the SEP-1 probe specifically stained all S. epidermidis and S. capitis cells (Fig. 4a and d), and (iii) with SEP-1, no specific fluorescence signals were obtained for S. warneri (Fig. 4g) and S. aureus, S. haemolyticus, and S. saprophyticus (data not shown). We concluded from these data that the two-nucleotide mismatch of the SEP-1 probe with the S. warneri target sequence appeared to be sufficient for a reliable discrimination between S. epidermidis and other staphylococcal species (with the exception of S. capitis). Finally, the S. aureus-specific probe SA-P1 was found to specifically label S. aureus cells, whereas S. epidermidis and other staphylococci remained unstained (Fig. 4j and m).
FIG. 4.
Epifluorescence micrographs of S. epidermidis RP62A (a to c), S. capitis DSM 20326 (d to f), S. warneri DSM 20316 (g to i), S. aureus 8325/4 (j to l), and S. epidermidis RP62A (m to o) under high-stringency conditions (43°C, 43% formamide). The SEP-1 probe (a, d, and g), the SA-P1 probe (j and m) the EUB338 probe (b, e, h, k, and n), or phase contrast (c, f, i, l, and o) was used.
Obviously, a gene probe capable of identifying all staphylococcal species at the genus level is highly desirable. In the absence of such a reagent, we are restricted to the identification of bacterial infections with the eubacterial gene probe EUB338 and the subsequent discrimination of S. aureus and S. epidermidis with specific gene probes. There is a clear need for the development of additional gene probes which could be used in combination for the identification of any bacterial pathogen.
Detection of S. epidermidis in bacterial biofilms.
The adherence and formation of biofilms on smooth surfaces is an essential step in the pathogenesis of staphylococcal infections. Therefore, we decided to extend the approach described above to address the question of whether it is possible to identify staphylococci in biofilms. In these experiments, the biofilm-positive strain S. epidermidis RP62A, which produces the polysaccharide intercellular adhesin, was used. In parallel, an S. aureus clinical isolate which had been found to form biofilms on polystyrene surfaces was analyzed and served as a control. The fluorescent hybridization technique with the SEP-1 probe proved to be suitable for the detection of S. epidermidis cells, even when they were embedded in a thick matrix of extracellular polysaccharides (Fig. 5a). In the control experiment with the biofilm-forming S. aureus isolate, no specific staining was obtained. This finding confirms the specificity of SEP-1 for S. epidermidis and also rules out the possibility that the positive signal was generated by an unspecific binding to extracellular proteins or polysaccharides. We concluded from these data that SEP-1 is an appropriate diagnostic tool for the detection and differentiation of S. epidermidis isolates even when they form biofilms.
FIG. 5.
Detection of staphylococci in biofilms. Phase-contrast (c and f) and epifluorescence (a to e) micrographs of S. epidermidis RP62A (a, b, and c) and S. aureus 8325/4 (d, e, and f) in biofilms following hybridization with the Cy3-labeled probe SEP-1 (a and d) and the FLUOS-labeled probe EUB338 (b and e) under high-stringency conditions (43°C, 43% formamide) are shown.
Detection of an S. aureus SCV phenotype by the fluorescent hybridization technique.
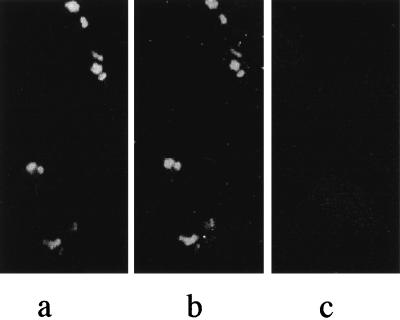
The abilities of clinically relevant staphylococci both to generate biofilms and to vary their phenotypic properties (e.g., growth rate, antibiotic susceptibility, and adherence properties) have been suggested to contribute considerably to staphylococcal virulence. The occurrence of S. aureus SCVs has been associated with recurrent and persistent infections, especially in patients with osteomyelitis and in patients with orthopedic implants (29). SCVs are characterized by a reduced growth rate, an increased resistance to aminoglycosides, and intracellular persistence. In order to investigate whether SCVs can be detected by the direct hybridization technique, we did an invasion assay by using a site-directed S. aureus hemB mutant which exhibits all properties of a small-colony phenotype, including the invasion of endothelial cells (33). The human endothelial cell line E.Ahy 926 was incubated with the S. aureus hemB mutant as described in Materials and Methods. Thereafter, the fixed cells were hybridized with the oligonucleotide probes EUB338 and SA-P1. Fluorescence microscopy analysis revealed that the S. aureus SCVs can be detected by both the EUB338 and SA-P1 gene probes, even when the bacteria occur intracellularly (Fig. 6a and b). An S. typhimurium isolate which is invasive in epithelial cells was used as a positive control for the invasion assay (data not shown), and the noninvasive strain E. coli HB101 was used as a negative control (Fig. 6c). We conclude from this experiment that the 16S rRNA technique is indeed suitable for the detection of metabolically inactive bacteria even when they occur intracellularly. Therefore, the method gives the opportunity to improve the diagnosis of infections caused by staphylococcal SCVs.
FIG. 6.
Detection of intracellularly persisting bacterial cells of an S. aureus hemB mutant in the human endothelial cell line E.Ahy 926. (a) Hybridization with the FLUOS-labeled probe EUB338. (b) Hybridization with the Cy3-labeled probe SA-P1. (c) Negative control with the noninvasive strain E. coli HB101 probed with EUB338.
In situ hybridization of a tissue section sample from a patient with hip arthroplasty loosening.
A further important problem in the diagnosis of orthopedic staphylococcal infections is the evaluation of the microbiological results, especially when coagulase-negative staphylococci have been identified. Often it is difficult to decide whether an isolate represents the causative agent of an infection or an unspecific contamination from the skin or the environment. One possible way to overcome these problems is to use diagnostic tools which allow the direct detection and differentiation of the bacteria in situ (e.g., in tissues, smears, or biopsy material, etc.).
To conclude our study, we addressed the question of whether the 16S rRNA hybridization technique is also appropriate for the direct detection of staphylococci in tissues. We therefore used the method in a clinical case of hip arthroplasty loosening that was thought to be caused by S. epidermidis.
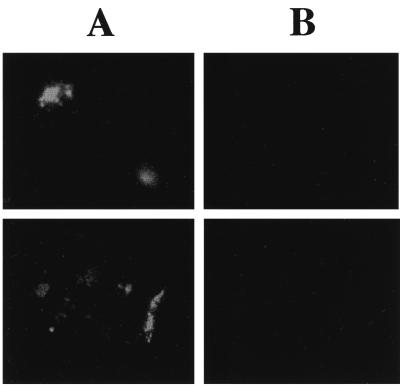
In a 65-year old female suffering from progressive loosening of an artificial hip arthroplasty, a replacement of the implant had to be performed. Since the clinical course was typical for an infection of the implant, specimens for microbiological investigations were obtained during surgery. Initially, microbiological cultivation failed to identify any causative agent. However, after prolonged cultivation (1 week) in Trypticase soy broth, S. epidermidis was identified in several specimens of the material. The S. epidermidis isolate was found to be resistant to oxacillin, gentamicin, and erythromycin. To decide whether the isolate represented the cause of the infection or a contamination, the 16S rRNA hybridization technique was used again. A tissue sample which had been obtained during the joint replacement surgery was fixed with paraformaldehyde and embedded in paraffin for thin-layer dissection. For the in situ hybridization with the fluorescence-labeled oligonucleotide probes, the tissue sections were treated as described in Materials and Methods. Both the EUB338 and SEP-1 probes were used in this experiment. In all tissue sections analyzed, several foci which gave signals with both oligonucleotide probes were identified. The foci were localized in the vicinity of blood vessels and resembled bacterial structures in size and shape. To ensure that the observed structures indeed represented bacteria, any unspecific reaction was excluded by probing another section with the S. epidermidis-specific SEP-1 probe and a Legionella pneumophila-specific probe, LEGPNE1 (16), as a negative control. No specific signal of the foci was obtained with the Legionella-specific probe in this control experiment (Fig. 7B). In contrast, the structures were strongly stained with the S. epidermidis-specific SEP-1 probe (Fig. 7A). In a prospective study using a mathematical model, it was shown recently that the investigation of at least three independent specimens gives results that are highly predictive of an infection (4). Although we had only one specimen at our disposal, we feel that the high number of the S. epidermidis-specific bacterial foci in the sample and their localization, combined with the microbiological cultivation results from several independent specimens, strongly suggest an involvement of an S. epidermidis infection in the loosening of the implant. The direct detection and identification of S. epidermidis in the infected tissue by this method support the clinical diagnosis that the loosening of the arthroplasty was caused by an infection.
FIG. 7.
In situ hybridization of connective tissue from a patient suffering from progressive loosening of an artificial hip arthroplasty with the S. epidermidis specific gene probe SEP-1 (A) and the Legionella-specific gene probe LEGPNE1 (16) as a negative control (B). Two different sectors of a sample that was obtained during replacement surgery are shown.
In summary, this initial report shows that the in situ hybridization technique with 16S rRNA-directed oligonucleotide probes is suitable for the diagnosis of staphylococcal infections associated with orthopedic implants. It should be noted that, for several reasons, this method cannot be expected to replace conventional culture techniques. In those cases, however, where the standard cultivation fails to identify a causative infectious agent, the 16S rRNA in situ hybridization could represent a useful additional diagnostic tool. We have applied this method to the diagnosis of one clinical case of hip arthroplasty loosening. However, in order to explore its general usefulness in the diagnostic laboratory (in comparison with currently available culture techniques), a broad clinical study would be extremely desirable.
Encouraged by the results of this study, we believe that the method can be extended to allow for the identification of a wide range of other fastidious microorganisms (e.g., Legionella, Burkholderia, or streptococci). For this purpose, however, the method has to be optimized to make it practicable for use in the routine microbiological laboratory.
ACKNOWLEDGMENTS
We thank Rudolf I. Amann of the Max-Planck-Institut für Marine Biologie, Bremen, Germany, for helpful discussions and technical support with whole-cell hybridizations, and we thank Bernd Schmaußer of the Institut für Pathologie, Universität Würzburg, Würzburg, Germany, for carrying out the sectioning of paraffin-embedded tissue.
This work was supported by a grant from the BMBF (no. 01K19608), by the Graduiertenkolleg Infektiologie, and by the Fond der Chemischen Industrie.
REFERENCES
- 1.al-Rashdan A, Bashir R, Khan F A. Staphylococcus capitis causing aortic valve endocarditis. J Heart Valve Dis. 1998;7:518–520. [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins B L, Athanasou N, Deeks J J, Crook D W M, Simpson H, Peto T E A, McLardy-Smith P, Berendt A R the OSIRIS Collaborative Study Group. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. J Clin Microbiol. 1998;36:2932–2939. doi: 10.1128/jcm.36.10.2932-2939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balwit J M, van Langevelde P, Vann J M, Proctor R A. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within endothelial cells. J Infect Dis. 1994;170:1033–1037. doi: 10.1093/infdis/170.4.1033. [DOI] [PubMed] [Google Scholar]
- 6.Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 7.Bentley R W, Harland N M, Leigh J A, Collins M D. A Staphylococcus aureus-specific oligonucleotide probe derived from 16S rRNA gene sequences. Lett Appl Microbiol. 1993;16:203–206. doi: 10.1111/j.1472-765x.1993.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 8.Christensen G D, Baddour L M, Simpson W A. Phenotypic variation of Staphylococcus epidermidis slime production in vitro and in vivo. Infect Immun. 1987;55:2870–2877. doi: 10.1128/iai.55.12.2870-2877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen G D, Baddour L M, Madison B M, Parisi J T, Abraham S N, Hasty D L, Lowrence J H, Josephs J A, Simpson W A. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1169. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- 10.Deighton M, Pearson S, Capstick J, Spelman D, Borland R. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J Clin Microbiol. 1992;30:2385–2390. doi: 10.1128/jcm.30.9.2385-2390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deighton M A, Capstick J, Borland R. A study of phenotypic variation of Staphylococcus epidermidis using Congo red agar. Epidemiol Infect. 1992;109:423–432. doi: 10.1017/s095026880005041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Mitoma F, Harding G K, Hoban D J, Roberts R S, Low D E. Clinical significance of a test for slime production in ventriculoperitoneal shunt infections caused by coagulase-negative staphylococci. J Infect Dis. 1987;156:555–560. doi: 10.1093/infdis/156.4.555. [DOI] [PubMed] [Google Scholar]
- 13.Emori T G, Gaynes R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franson T R, Sheth N K, Rose H D, Sohnle P G. Scanning electron microscopy of bacteria adherent to intravascular catheters. J Clin Microbiol. 1984;20:500–505. doi: 10.1128/jcm.20.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fumagalli O, Tall B D, Schipper C, Oelschläger T. N-glycosylated proteins are involved in efficient internalization of Klebsiella pneumoniae by cultured human epithelial cells. Infect Immun. 1997;65:4445–4451. doi: 10.1128/iai.65.11.4445-4451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm D, Merkert H, Ludwig W, Schleifer K H, Hacker J, Brand B C. Specific detection of Legionella pneumophila: construction of a new 16S rRNA-targeted oligonucleotide probe. Appl Environ Microbiol. 1998;64:2686–2690. doi: 10.1128/aem.64.7.2686-2690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann C, Gerke C, Perdreau-Remington F, Götz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1023. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 19.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura Y, Hou X G, Sultana F, Hirose K, Miyake M, Shu S E, Ezaki T. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J Clin Microbiol. 1998;36:2038–2042. doi: 10.1128/jcm.36.7.2038-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloos W E, Bannermann T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan S, Haglund L, Ashfaq A, Leist P, Roat T. Prosthetic valve endocarditis due to Staphylococcus saccharolyticus. Clin Infect Dis. 1996;22:722–723. doi: 10.1093/clinids/22.4.722. [DOI] [PubMed] [Google Scholar]
- 23.Lewis L A, Li K, Bharosay M, Cannella M, Jorgenson V, Thomas R, Pena D, Velez M, Pereira B, Sassine A. Characterization of gentamicin-resistant respiratory-deficient (Res−) variant strains of Staphylococcus aureus. Microbiol Immunol. 1990;34:587–605. doi: 10.1111/j.1348-0421.1990.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 24.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuhisa A, Saito Y, Ueyama H, Aikawa Y, Ohono T. Detection of staphylococci in mouse phagocytic cells by in situ hybridization using biotinylated DNA probes. Biotech Histochem. 1994;69:31–37. doi: 10.3109/10520299409106258. [DOI] [PubMed] [Google Scholar]
- 27.Mempel M, Feucht H, Ziebuhr W, Endres M, Laufs R, Grüter L. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother. 1994;38:1251–1255. doi: 10.1128/aac.38.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters G, Locci R, Pulverer G. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J Infect Dis. 1982;146:479–482. doi: 10.1093/infdis/146.4.479. [DOI] [PubMed] [Google Scholar]
- 29.Proctor R A, van Langevelde P, Kristjansson M, Maslow J N, Arbeit R D. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin Infect Dis. 1995;20:95–102. doi: 10.1093/clinids/20.1.95. [DOI] [PubMed] [Google Scholar]
- 30.Rupp M E, Archer G D. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–245. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher-Perdreau F, Heilmann C, Peters G, Götz F, Pulverer G. Comparative analysis of a biofilm-forming Staphylococcus epidermidis strain and its adhesion-positive, accumulation-negative mutant M7. FEMS Microbiol Lett. 1994;117:71–78. doi: 10.1111/j.1574-6968.1994.tb06744.x. [DOI] [PubMed] [Google Scholar]
- 32.Shuttleworth R, Behme R J, McNabb A, Colby W D. Human isolates of Staphylococcus caprae: association with bone and joint infections. J Clin Microbiol. 1997;35:2537–2541. doi: 10.1128/jcm.35.10.2537-2541.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Eiff C, Heilmann C, Proctor R A, Woltz C, Peters G, Götz F. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol. 1997;179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakrzewska-Czerwinska J, Gaszewska-Mastalarz A, Pulverer G, Mordarski M. Identification of Staphylococcus epidermidis using a 16S rRNA-directed oligonucleotide probe. FEMS Microbiol Lett. 1992;79:51–58. doi: 10.1111/j.1574-6968.1992.tb14018.x. [DOI] [PubMed] [Google Scholar]
- 35.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of an intercellular adhesin gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]