Abstract
Ferroptosis is a lipid peroxidation-dependent cell death caused by metabolic dysfunction. Ferroptosis-associated enzymes are promising therapeutic targets for cancer treatment. However, such therapeutic strategies show limited efficacy due to drug resistance and other largely unknown underlying mechanisms. Here we report that cystine transporter SLC7A11 is upregulated in lung cancer stem-like cells (CSLC) and can be activated by stem cell transcriptional factor SOX2. Mutation of SOX2 binding site in SLC7A11 promoter reduced SLC7A11 expression and increased sensitivity to ferroptosis in cancer cells. Oxidation at Cys265 of SOX2 inhibited its activity and decreased the self-renewal capacity of CSLCs. Moreover, tumors with high SOX2 expression were more resistant to ferroptosis, and SLC7A11 expression was positively correlated with SOX2 in both mouse and human lung cancer tissue. Together, our study provides a mechanism by which cancer cells evade ferroptosis and suggests that oxidation of SOX2 can be a potential therapeutic target for cancer treatment.
STATEMENT OF SIGNIFICANCE
This study uncovers a SOX2-SLC7A11 regulatory axis that confers resistance to ferroptosis in lung cancer stem-like cells.
Keywords: ferroptosis, SLC7A11, cancer stem-like cells, SOX2, oxidation
INTRODUCTION
Ferroptosis is a recently characterized cell death caused by iron-dependent aberrant accumulation of lipid hydroperoxides and membrane damage [1-3]. Ferroptosis has drawn great interest due to its promising therapeutic functions in multiple human diseases, such as tumorigenesis, neurodegenerative diseases and renal function failure [4, 5]. Targeting ferroptosis has great potential in developing novel therapeutic strategy for various human disorders [6, 7].
Dysregulations of iron, amino acids and lipid metabolism are involved in the induction of ferroptosis [8]. Among them, homeostasis of intracellular cysteine is essential for protecting cells from ferroptosis [9]. SLC7A11, a key component of the cystine/glutamate antiporter system xc−, is critical to cystine uptake, glutathione (GSH) synthesis and ferroptosis resistance [10]. Pharmacological inhibition of SLC7A11 by Erastin or Imidazole ketone erastin (IKE) reduces intracellular GSH level and induces ferroptosis [11]. The expression and activity of SLC7A11 are precisely regulated at multiple levels, including transcription, epigenetical regulation and post-translational modification. For example, NRF2 and p53 directly bind to the promoter region of SLC7A11 and regulate its transcription [12]. Epigenetically, BAP1 inhibits SLC7A11 expression through de-ubiquitinating H2A [13], while chromatin remodeling factor ARID1A increases SLC7A11 expression [14]. In protein level, SLC7A11 is regulated by BECN1, mTORC2, OTUB1 and CD44v [15-18]. Yet, the specific regulatory mechanism of SLC7A11 under different pathological conditions remains to be fully elucidated.
Accumulated evidence demonstrates that inducing ferroptosis has great potential to inhibit tumor growth and may be an efficient strategy for cancer treatment. For instance, treatment with SLC7A11 inhibitor IKE to block cysteine import or cyst(e)inase to deplete extracellular cysteine significantly limits tumor growth [19-21]. Moreover, ferroptosis induction in combination with immune checkpoint inhibitors or radiation are promising in cancer therapy [22-24]. However, as the complex and highly heterogeneous diseases, different types of cancers are resistant to ferroptosis via multiple mechanisms, such as reducing polyunsaturated lipid levels [25, 26] or increasing the antioxidant capacity [12, 13]. Because evading from ferroptosis may greatly attenuate the efficacy of ferroptosis-dependent cancer therapy and probably involve in drug resistance and cancer relapse, it is important to uncover the mechanism of ferroptosis resistance in various cancer types as well as at different stages of tumor progression.
Cancer stem-like cells (CSLCs), a small portion of tumor cells with the self-renewal and clonal tumor initiation capacity, play important roles in therapy resistance [27]. Targeting CSLCs can greatly augment the efficacy in cancer treatment [28]. Accumulating evidence indicates that cell metabolism is reprogrammed in CSLCs and differs dramatically from that of bulk tumor cells [29]. For example, compared to the bulk tumor cells, CSLCs contain higher levels of labile iron due to increased iron importing but decreased iron exporting [28, 30, 31]. Theoretically, excess of labile iron should increase the sensitivity to ferroptosis in CSLCs, which is mediated by Fenton reaction [32]. However, whether and how CSLCs survive from ferroptosis remain largely elusive. In this study, we report that CSLCs are more resistant to ferroptosis than bulk tumor cells. Mechanistically, we found that stem cell transcriptional factor SOX2, which played pivotal roles in stemness maintenance of CSLCs [33], promoted the expression of cystine transporter SLC7A11 and prevented lung cancer from ferroptosis.
MATERIALS AND METHODS
Cell culture
All cell lines were obtained from Cell Bank, Chinese Academy of Sciences (Shanghai, China) unless otherwise specified. H5889, L78, H1299 and SW620 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. HEK293T and HCT116 cells were cultured in DMEM medium supplemented with 10% FBS. E14 mESC (mouse embryonic stem cells) and R1 mESC cells (from Dr. Ke Wei lab) were maintained feeder free on gelatin-coated plastics in mES media (DMEM with 15% FBS, 2 mM GlutaMAX, 0.1 mM NEAAs, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin and 100 μg/ml streptomycin supplemented with 1,000 U/ml LIF and passaged every 3 days. HTS cells (from Dr. Yufeng Shi lab) were cultured in serum free defined medium. The cells were actively passaged for less than one month and were routinely tested for Mycoplasma contamination using the Myco-Blue® Mycoplasma Detector Kit (Vazyme, D101-01). Cells were cultured at 37°C supplied with 5% CO2. Transfection was performed by using calcium phosphate-DNA coprecipitation for HEK293T cells and Lipofectamine 2000 for H1299 cells.
Antibodies and Drugs
Anti-HA (sc-7392, RRID:AB_627809) antibody was obtained from Santa Cruz, anti-Flag (F3165, RRID:AB_259529) antibody was obtained from Sigma, anti-SOX2 (23064, RRID:AB_2714146), anti-Nanog (8822, RRID:AB_11217637), anti-AKT (9272, RRID:AB_329827) and anti-SLC7A11 (12691, RRID:AB_2687474) antibodies were obtained from Cell Signaling Technology, anti-SLC7A11 (26864-1-AP, RRID:AB_2880661) for mouse, anti-Tubulin (11224-1-AP, RRID:AB_2210206), anti-Actin (60008-1-Ig, RRID:AB_2289225) and anti-GAPDH (60004-1-Ig, RRID: AB_2107436) antibodies were obtained from Proteintech, anti-Oct4 (ab18976, RRID:AB_444714) antibody was obtained from Abcam. Erastin (S7242), Imidazole ketone erastin (S8877), Fer-1 (S7243) and Cisplatin (S1166) were obtained from Selleck. MG132 (C2211), cycloheximide (CHX, C7698), PD146176 (P4620) and DFO (S1712) were obtained from Sigma.
Sphere formation assay
Oncospheres were enriched from H1299, SW620 or HCT116 cells. Single-cell suspension cells (2000 cells per well) were plated on 96-well Ultra-Low Attachment Plates (Corning Incorporated, catalog number: 3474) and cultured in DMEM or Cysteine-free DMEM supplemented with 20 ng/ml EGF, 5 μg/ml insulin, 10 ng/ml bFGF, 1:50 B27 and 0.4% BSA for 14 days. Oncospheres were visualized under phase contrast microscope, photographed, counted and represented graphically. Tumor spheres larger than 100 μm were counted.
Gene interference
siRNAs were purchased from GenePharma. siRNA transfection was performed according to the manufacturer’s instructions. Briefly, siRNAs and Lipofectamine® 2000 reagent were mixed and incubated in serum-free medium for 20 mins at room temperature, respectively, and then added to cells at 50% confluence.
Lentiviral infection
For shRNA, sgRNA and PCDH vectors, the virus was produced from transfected HEK 293T cells, by harvesting the viral supernatant 48-72 hours after transfection and used to infect the target cells at 70% confluence. PCDH lentivirus was used to generate SOX2 wild-type (WT) and C265S mutant overexpression cell lines. SOX2 knockout (KO) cell lines were generated using lentiCRISPR methods. Briefly, the lentiCRISPR vector was linearized using BsmBI, guide RNA (sgRNA) was constructed into the lentiviral expression vector for Cas9 and sgRNA.
Generation of SLC7A11 promoter mutant cell lines
For SLC7A11 promoter mutation H1299 cell lines, we selected the gRNA around SOX2 binding site in SLC7A11 promoter (sgRNA F: CACCGCGAATTTACTACTTCTGGAT; R: AAACATCCAGAAGTAGTAAATTCGC) and cloned it to PX458 vector, which expressing Cas9 and GFP. After transfected for 48 hours, GFP positive cells were FACS sorted into 96 well plates to obtain single-cell derived colonies and extracted genomic DNA. SLC7A11 promoter mutation was confirmed by DNA Sanger sequencing.
Cell viability assay
Cells were seeded in 96 well plates (2000 cells per well) and incubated with indicated treatments. Cell viability was determined by using CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7572).
Lipid peroxidation assay
Lipid peroxidation levels were measured according to the manufacturer’s instructions. Briefly, cells were incubated in a 6 well plates containing 5 μM BODIPY 581/591 C11 dye (Invitrogen, D3861). After incubation at 37°C for 30 mins, cells were washed twice with PBS and trypsinized. Cells were then subjected to flow cytometry analysis. The FL1 channel signal was plotted as shown in the figures.
Chromatin immunoprecipitation (ChIP) analysis
ChIP assay was performed according to the manufacturer’s instructions (CST, 9004). Briefly, indicated H1299 cells were cross-linked with 1% formaldehyde for 10 mins and terminated by glycine. The extracted chromatin was digested and fragmented to 0 to 90 bp and then immunoprecipitated using SOX2 antibody or normal IgG with protein A/G agarose beads. The complexes were then uncrosslinked to obtain pure DNA fragments. The primers for the SOX2 binding site of SLC7A11 promoter were then used for PCR.
GSH and Cysteine assay
The relative GSH concentration was assessed using a Glutathione Assay Kit (Beyotime, S0053) according to the manufacturer’s instructions. Briefly, the measurement of GSH used a kinetic assay in which catalytic amounts (nmoles) of GSH caused a continuous reduction of 5,5’-dithiobis (2-nitrobenzoic acid) to 5-thio-2-nitrobenzoic acid; the formed GSSG was recycled by GSR/glutathione reductase and NADPH. The reaction rate was proportional to the concentration of glutathione up to 2 mM. The yellow product (5-thio-2-nitrobenzoic acid) was measured spectrophotometrically at 412 nm. The relative cysteine concentration was assessed using Cysteine Assay Kit (Sigma, MAK255) according to the manufacturer’s instructions. All samples were normalized to cell number.
Protein oxidation analysis
The cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.0, 120 mM NaCl, 5 mM EDTA, and 0.5% NP-40) containing protease inhibitors. After centrifuged, the supernatant was incubated with 0.1 mM biotin-maleimide (Sigma, 116919-18-7) for 4 hours to label reduced thiols. Afterwards, the lysate was incubated with 10 μl NeutrAvidin agarose (Thermo Fisher Scientific, 29201) and incubated for 2 hours at 4°C. The beads were then washed 3 times with lysis buffer and boiled in SDS-PAGE loading buffer to perform SDS-PAGE analysis. For mass spectrometry of SOX2 oxidation, H1299 cells were transfected with HA-SOX2 plasmid for 24 hours and cultured in cysteine-free medium for additional 6 hours. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol and 0.5% NP-40) supplemented with protease inhibitors and incubated with anti-HA beads at 4°C for 3 hours. The immunoprecipitate was washed three times. The sample was separated on the 10% SDS-PAGE gel and stained with Coomassie blue. The candidate gel was digested for 16 hours with trypsin. Peptides were extracted and analyzed using a liquid chromatography system (Thermo Fisher Easy-nLC 1000) coupled with a mass spectrometer (Thermo Fisher LTQ Obitrap ETD).
Luciferase reporter assay
HEK293T cells were transfected with Luciferase reporter gene, Renilla and other indicated plasmids. After 24 hours, cells were lysed with the lysis buffer (25 mM Tris-Cl (pH 7.8), 2 mM 1,2-diaminocyclo-hoxane N,N,N,N-tetracetic acid, 25 mM dithiothreitol (DTT), 1% Triton X-100 and 10% glycerol), and Luciferase assays were performed using the dual-Luciferase reporter assay system (Promega, E1960).
Immunoprecipitation (IP) and Western blot (WB)
Transfected HEK293T cells were lysed in CoIP lysis buffer (50 mM Tris-cl, pH 7.4, 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, 10% glycerophosphate). After lysis for 30 mins, the cell lysates were centrifuged at 12,000 rpm for 15 mins at 4°C to remove the cell debris and were incubated in HA-conjugated beads (Abmart, M20013) or M2 beads (Sigma, M8823) for 2 hours. The beads were boiled after three times washing and analyzed via immunoblotting.
Immunofluorescence (IF)
Cells were seeded on fibronectin coated glass coverslips in 24 well culture plates. After 24 hours, the cells were washed once with PBS and fixed in 4% paraformaldehyde for 10 mins at room temperature. The fixed cells were permeabilized by using 0.1% Triton X-100 and washed twice with PBS. The coverslips were blocked for 1 hour (0.1% BSA) and incubated in primary antibody overnight at 4°C. Next, the coverslips were washed twice and incubated in secondary antibodies for 1 hour at room temperature. The glass coverslips were examined using a Zeiss LSM 510 Meta confocal system.
Mice and Immunohistochemistry (IHC)
KrasLSL-G12D/+;Lkb1flox/flox (KL) mice were originally generously provided by T. Jacks, and R. Depinho, respectively [34]. All the animal experimental procedures were approved by the Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Chinese Academy of Sciences, with a project license number of IBCB0011. Mice were housed in the SIBCB animal facility under SPF conditions with a 12 hours light/dark cycle at room temperature in accordance with the institutional guidelines and ethical regulations, and fed with regular chow and water by the facility staff. Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (2 X 106 p.f.u for Ad-Cre) as described [35, 36]. For Erastin treatment, we sequentially dissolved Erastin in 5%DMSO, 30%PEG300, 5%Tween80 and ddH2O according to the manufacturer’s instructions, it should be noted that the solvent needed to be added from left to right, after the dissolution was completely clear, added the next reagent, 40 mg/kg Erastin was intraperitoneal injected, every other day for 2 weeks. For IKE treatment, 50 mg/kg IKE was intraperitoneal injected, once a day for 2 weeks. Mice were then killed at indicated times for gross inspection and histopathological examination. Mice lungs were inflated with 4% paraformaldehyde, fixed overnight, dehydrated in ethanol, embedded in paraffin, sectioned at 5 mm followed by staining with haematoxylin and eosin (H&E). Immunohistochemistry was performed as previously described [37]. Oil Red O staining was carried out using standard protocols. The following antibodies were used: anti-SOX2 (ab92494, Abcam, 1:500, RRID:AB_10585428), 4HN (ab46545, Abcam, 1:200, RRID:AB_722490) and anti-SLC7A11 antibodies (ab175186, Abcam, 1:500, RRID:AB_2722749).
TCGA and GEO data analysis
The mRNA expression data from TCGA and GEO database was downloaded and processed using standard methods. Indicated mRNA expression in normal and cancer samples was analyzed, and the data was represented as box-and-whisker plots or dot plots. Gene correlation was assessed by the Spearman rank correlation test. Statistical significance was assessed by the Wilcoxon test.
Correlation of cancer stemness and ferroptosis genes
The stem cell signature is the stemness index based on the epigenetic features (enhancer elements, DNA methylation, histone markers, e.g., H3K4me3, H3K27ac, H3K4me1, H3K36me3, H3K9me3, and H3K27me3) and gene expression (e.g., NANOG, OCT4, SOX2, and c-MYC) according to previous study [38]. For correlation between cancer stemness and ferroptosis genes expression, the mRNAsi indices of pan-cancer cases in TCGA were obtained from previous study[38]. The TCGA pan-cancer data consist of 33 types of cancers. A total of 11,057 tissues samples were useful for the study. The RNA sequencing (RNA-Seq) expression data of all samples was downloaded from the TCGA database (https://portal.gdc.cancer.gov/). We used the Perl language (http://www.perl.org/) to combine the RNA-Seq results of each sample and the Ensemble database (http://asia.ensembl.org/index.html) to convert gene IDs to gene symbols in a matrix profile. The Kruskal test in R was used to test the correlation between mRNAsi scores and gene expression. P value < 0.05 was considered statistically significant.
Data Availability
A complete software environment and all necessary data to reproduce the analysis and figure described in this manuscript are available on Code Ocean (https://codeocean.com/capsule/8756276/tree/v1).
Statistical analysis
Statistical analyses were performed with a two-tailed unpaired Student’s t-test or one/two-way ANOVA as indicated. The data was presented as the means ± SEM. P value < 0.05 was considered statistically significant. Gene expression correlation was assessed by the Spearman test between the indicated two groups.
RESULTS
Cystine transporter SLC7A11 is upregulated in CSLCs
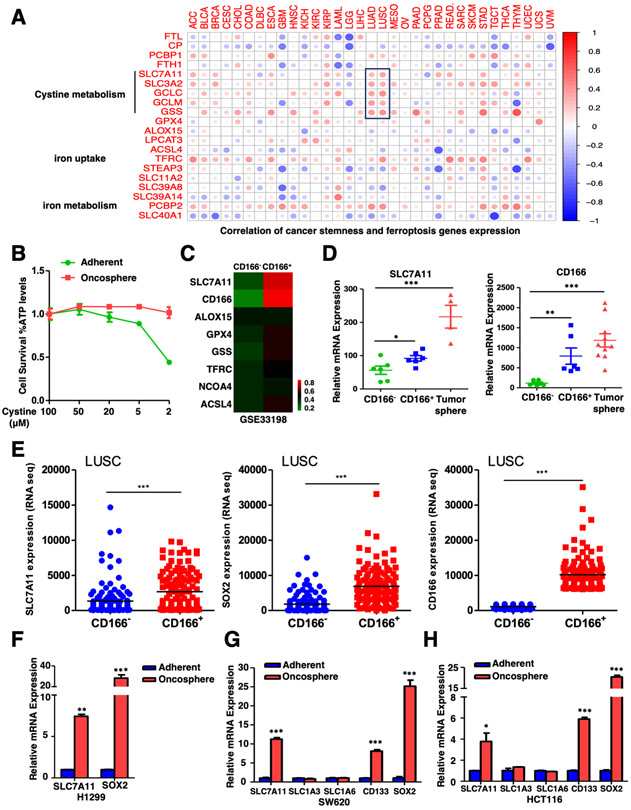
To characterize the role of ferroptosis in CSLCs, we analyzed the correlation between quantified stem cell signatures and ferroptosis-related gene expression in nearly 12,000 samples of 33 tumor types, by using a previously established one-class logistic regression (OCLR) machine learning algorithm [38]. Our results showed that, in addition to two iron metabolism-related genes, expression levels of cystine metabolism genes were most positively correlated with cancer stemness, especially in lung cancer (LUAD & LUSC) (Figure 1A). This led us to propose that cystine metabolism, probably ferroptosis, might play vital roles in CSLCs. To test our hypothesis, we collected CSLCs by culturing cells under 3-D oncosphere condition, which was a model system for studying the CSLCs functions, and examined its sensitivity to cysteine-deprivation induced ferroptosis. Interestingly, we found that 3-D oncosphere cells were more resistant to cysteine-deprivation induced ferroptosis than parallel adherent cells (Figures 1B, S1A).
Figure.1. Cystine transporter SLC7A11 is upregulated in CSLCs.
A. Correlation between cancer stemness and ferroptosis genes expression in 33 tumor types was shown. The Kruskal test in R was used to test the correlation, P value < 0.05 was considered statistically significant. ACC (Adrenocortical Carcinoma), BLCA (Bladder Urothelial Carcinoma), BRCA (Breast Invasive Carcinoma), CESC (Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma), CHOL (Cholangiocarcinoma), COAD (Colon Adenocarcinoma), DLBC (Lymphoid Neoplasm Diffuse Large B-cell Lymphoma), ESCA (Esophageal Carcinoma), GBM (Glioblastoma Multiforme), HNSC (Head and Neck Squamous Cell Carcinoma), KICH (Kidney Chromophobe), KIRC (Kidney Renal Clear Cell Carcinoma), KIRP (Kidney Renal Papillary Cell Carcinoma), LAML (Acute Myeloid Leukemia), LGG (Brain Lower Grade Glioma), LIHC (Liver Hepatocellular Carcinoma), LUAD (Lung Adenocarcinoma), LUSC (Lung Squamous Cell Carcinoma), MESO (Mesothelioma), OV (Ovarian Serous Cystadenocarcinoma), PAAD (Pancreatic Adenocarcinoma), PCPG (Pheochromocytoma and Paraganglioma), PRAD (Prostate Adenocarcinoma), READ (Rectum Adenocarcinoma), SARC (Sarcoma), SKCM (Skin Cutaneous Melanoma), STAD (Stomach Adenocarcinoma), TGCT (Testicular Germ Cell Tumors), THCA (Thyroid Carcinoma), THYM (Thymoma), UCEC (Uterine Corpus Endometrial Carcinoma), UCS (Uterine Carcinosarcoma) and UVM (Uveal Melanoma).
B. H1299 adherent and oncosphere cells were cultured in different concentrations of cysteine for 24 hours and cell viability of indicated cells was measured. The data was representative of three independent experiments.
C. Gene expression data of CD166− control and CD166+ lung CSLCs was obtained from GEO (GSE33198) and heatmap of ferroptosis related gene expression was shown.
D. Expression levels of SLC7A11 and CD166 in CD166− control cells, CD166+ lung CSLCs and lung tumor sphere were showed, *P<0.05, **P<0.01, ***P<0.001 (Student's t-test).
E. Expression levels of SLC7A11, SOX2 and CD166 in CD166− versus CD166+ lung tumor cells from TCGA database were showed as dot plots, The data represented as scatter dot plot and the line showed mean. Significance was assessed by the Wilcoxon test compared between the indicated two groups, ***P<0.001 (Student's t-test).
F. SLC7A11 was upregulated in H1299 tumor oncospheres when compared with adherent cells, **P<0.01, ***P<0.001 (Student's t-test).
G. SLC7A11 but not other transporters was upregulated in SW620 tumor oncospheres when compared with adherent cells, ***P<0.001 (Student's t-test).
H. SLC7A11 but not other transporters was upregulated in HCT116 tumor oncospheres when compared with adherent cells, *P<0.05, ***P<0.001 (Student's t-test).
To explore the potential mechanism of ferroptosis resistance in CSLCs, we compared the expression of ferroptosis-related genes by analyzing the GEO dataset (GSE33198), which was derived from human lung tumor xenograft. Interestingly, we found that SLC7A11, but not other genes related to ferroptosis such as GPX4, ALOX15 or NCOA4, was significantly upregulated in lung CD166+ CSLCs when compared to those CD166− cells (Figures 1C-D, S1B). The upregulation of SLC7A11 in CSLCs was also confirmed in TCGA (The Cancer Genome Atlas) database (Figure 1E). Besides, our data further revealed that SLC7A11 was indeed upregulated in oncospheres (Figures 1F-H), as well as glioblastoma stem-like HTS cells [39] (Figure S1C). In addition, the expression level of SLC7A11 was significantly reduced upon differentiation in embryonic stem cells (ESCs) (Figures S1D-G). Thus, our data together indicate that SLC7A11 is upregulated in multiple stem cells and suggest that CSLCs may have strong capacity to uptake cystine and are more resistant to ferroptosis.
SOX2 transcriptionally up-regulates SLC7A11 expression
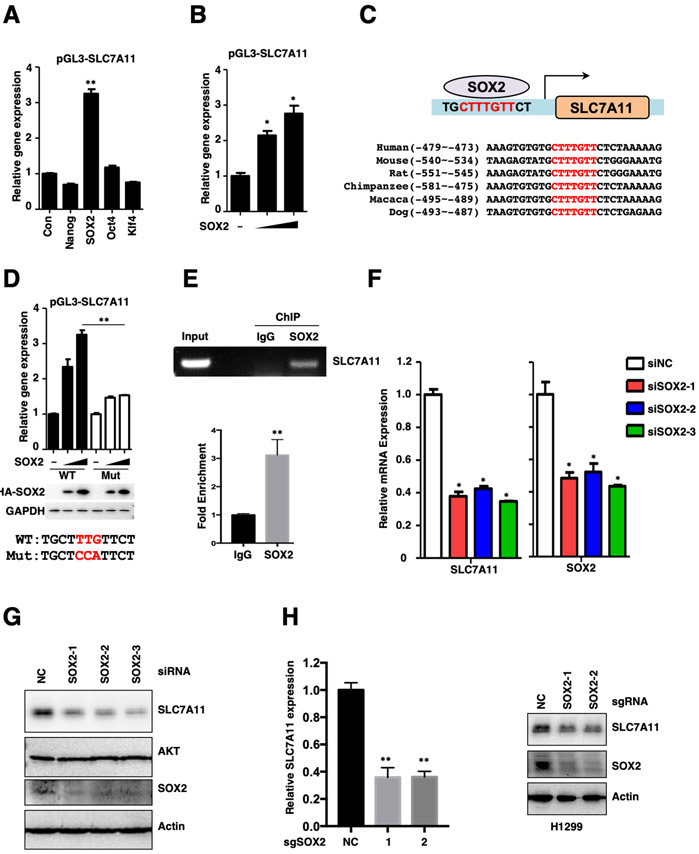
The increased expression of SLC7A11 in various stem cells led us to examine whether SLC7A11 was regulated by those stem cell transcriptional factors. To this end, we constructed a pGL3-SLC7A11 promoter reporter gene system (Figure S2A). Our data showed that SOX2, but not other stem cell factors including Nanog, Oct4 or Klf4, efficiently enhanced the promoter-driven expression of Luciferase in a dose-dependent manner (Figures 2A-B). Transcription factor SOX2 usually exhibits higher binding affinity to the consensus CTTTGTT sequence localized at the promoter of its target genes. Our analysis identified a potential SOX2 binding site on SLC7A11 promoter, which was highly conserved across multiple species (Figure 2C). Mutation of this site dramatically reduced the SOX2-mediated Luciferase expression (Figure 2D). Furthermore, chromatin immunoprecipitation (ChIP) assay revealed that endogenous SOX2 readily bound to the SLC7A11 promoter (Figure 2E). Consistently, depletion of endogenous SOX2 reduced the expression of SLC7A11 in multiple lung cancer cell lines (Figures 2F-H, S2B). Besides, SOX2 overexpression was sufficient to activate endogenous SLC7A11 expression in both mRNA and protein levels, and also largely promote ferroptosis resistance (Figures S2C-F). These data collectively suggest that SLC7A11 is transcriptionally regulated by SOX2.
Figure.2. SOX2 transcriptionally activates the expression of SLC7A11.
A. pGL3-SLC7A11 promoter reporter gene was co-expressed with Nanog, SOX2, Oct4 and Klf4 in HEK293T cells for 24 hours, and the expression of Luciferase was measured and normalized to Renilla, **P<0.01 (Student's t-test).
B. pGL3-SLC7A11 promoter reporter gene was co-expressed with different dose of SOX2 in HEK293T cells for 24 hours, and the expression of Luciferase was measured and normalized to Renilla, *P<0.05 (Student's t-test).
C. Schematic diagram of SOX2 binding site on SLC7A11 promoter and sequence homology across different species.
D. WT and mutated pGL3-SLC7A11 promoter reporter genes were co-expressed with different dose of SOX2 in HEK293T cells for 24 hours, and the expression of Luciferase was measured and normalized to Renilla, **P<0.01 (two-way ANOVA test), the protein level of SOX2 was analyzed by WB.
E. ChIP results of SOX2 binding on SLC7A11 promoter in indicated H1299 cells.
F. siRNA mediated SOX2 knock down resulted in decreased expression of SLC7A11 in H1299 cells, mRNA expression level was analyzed by qRT-PCR, *P<0.05 (Student's t-test).
G. siRNA mediated SOX2 knock down resulted in decreased expression of SLC7A11 in H1299 cells, protein levels of SLC7A11 and SOX2 were analyzed by WB (related to Fig. 2F).
H. Both the mRNA and protein expression levels of SLC7A11 were decreased upon CRISPR mediated SOX2 KO in H1299 cells, mRNA expression level was analyzed by qRT-PCR, **P<0.01 (Student's t-test) and the protein level was analyzed by WB.
SOX2 protects cancer cells from ferroptosis through SLC7A11
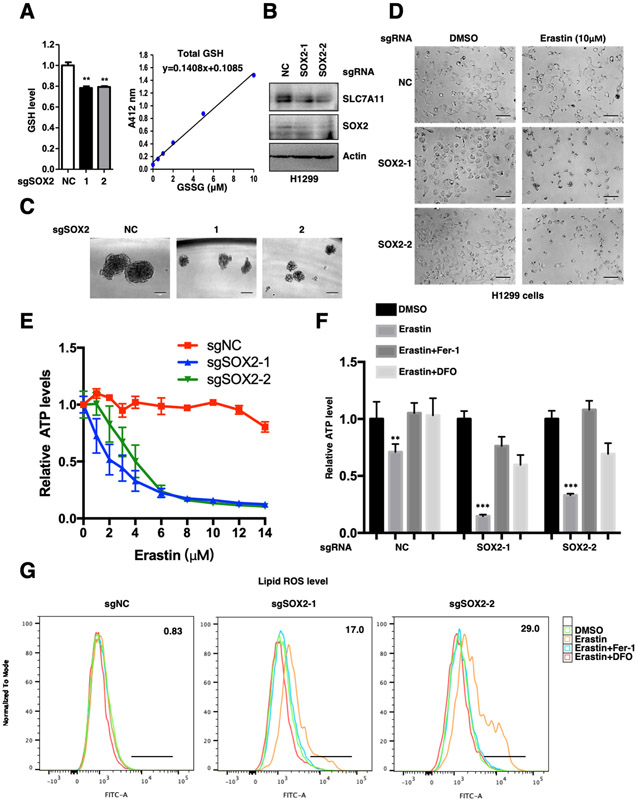
Since SLC7A11 is one of the indispensable components of glutamate/cysteine antiporter xc− system, we intended to investigate whether SOX2 could regulate cysteine metabolism and GSH synthesis in cancer cells. Our data indeed showed that the intracellular GSH levels were significantly decreased upon SOX2 depletion (Figures 3A-C). Moreover, we analyzed the Cancer Cell Line Encyclopedia (CCLE) metabolism profiling database, which including 913 human cancer cell lines [40] and found that the intracellular GSH levels were positively correlated with the expression of both SOX2 and SLC7A11 (Figures S2G-H). In line with this, SOX2 depletion resulted in increased lipid peroxidation and ferroptosis sensitivity in H1299, H5889 and L78 cells (Figures 3D-G) (Figures S3A-J). Notably, Erastin-induced cell death could be rescued by ferroptosis inhibitors ferrostatin-1 and DFO, further supporting the occurring of ferroptosis (Figure 3F) (Figures S3E-F). Moreover, the increased ferroptosis caused by SOX2 ablation was significantly rescued by SLC7A11 overexpression (Figures S3K-M). Besides, our data suggested that SOX2 depletion had little effect on apoptosis (Figure S3N). These data together indicate that SOX2 protects lung tumor cells against ferroptosis by maintaining a fit SLC7A11 level.
Figure.3. SOX2 protects lung cancer cells from ferroptosis.
A. Relative GSH levels in SOX2 WT and KO H1299 cell lines were showed, all samples were normalized to cell number. **P<0.01 (Student's t-test). GSH assay standard curve line was also showed.
B. Protein levels of SLC7A11 and SOX2 were analyzed by WB.
C. Representative sphere images from each condition of H1299 cells. Scale bar: 100 μm. SOX2 KO severely impaired the self-renewal of tumor spheres.
D. SOX2 WT and KO H1299 cell lines were treated with 10 μM Erastin for 20 hours and representative images from each condition were shown. Scale bar: 100 μm.
E. Cell viability of indicated H1299 cells was measured. The data was representative of three independent experiments.
F. Indicated H1299 cells were treated with 10 μM Erastin for 20 hours, 1 μM Fer-1 and 50 μM DFO treatment could rescue Erastin-induced cell death. Cell viability of indicated cells was measured, **P<0.01, ***P<0.001 (two-way ANOVA test).
G. Lipid ROS levels of indicated H1299 cells treated with 5 μM Erastin for 10 hours. 1 μM Fer-1 and 50 μM DFO treatment could rescue Erastin-induced lipid peroxidation.
Loss of SOX2 binding motif in SLC7A11 promoter renders lung cancer cells more vulnerable to ferroptosis
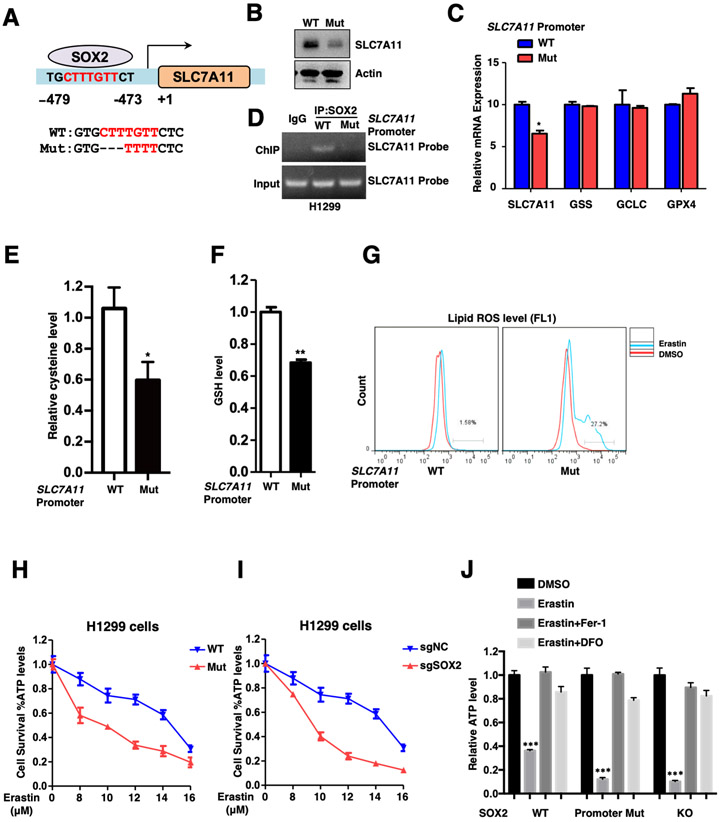
To further specify the role of SOX2-SLC7A11 axis in ferroptosis, we utilized CRISPR-Cas9 technology to establish SLC7A11 promoter mutant cancer cell lines, in which SOX2 binding sequence CTTTGTT localized on SLC7A11 promoter was mutated (Figures 4A, S4A-B). As expected, the mutant cells showed dramatically decreased SLC7A11 expression at both mRNA and protein levels compared to control cells (Figures 4B-C), and such mutation also largely abolished binding of SOX2 to the SLC7A11 promoter (Figure 4D). Consistently, intracellular cysteine and GSH levels were significantly reduced in mutant cells (Figures 4E-F). Importantly, the mutant cells exhibited increased lipid ROS levels (Figure 4G) and a higher susceptibility to ferroptosis (Figures 4H-I), which could be rescued by ferrostatin-1 and DFO (Figure 4J). To strengthen our conclusion, we generated SOX2 overexpressed WT and mutant cell lines and found that the mutation largely attenuated the effect of SOX2 on SLC7A11 expression and ferroptosis resistance (Figures S4C-F). These data demonstrate that SOX2 binds to SLC7A11 promoter to maintain a proper level of intracellular cysteine and confers resistance to ferroptosis in lung cancer cells.
Figure.4. SLC7A11 promoter mutation cells are susceptible to ferroptosis.
A. Schematic diagram of SLC7A11 promoter WT and mutant H1299 cells.
B. The protein levels of SLC7A11 in SLC7A11 promoter WT and mutant H1299 cells were analyzed by WB.
C. Relative mRNA expression levels of SLC7A11, GSS, GCLC and GPX4 in SLC7A11 promoter WT and mutant H1299 cells were analyzed by qRT-PCR, *P<0.05 (Student's t-test). SLC7A11 but not other ferroptosis related genes was decreased in SLC7A11 promoter mutation cells.
D. ChIP results of SOX2 binding on SLC7A11 promoter in WT and mutant H1299 cells. The mutation largely abolished SOX2’s binding on the SLC7A11 promoter.
E. Relative cysteine levels in SLC7A11 promoter WT and mutant H1299 cells were showed, *P<0.05 (Student's t-test).
F. Relative GSH levels in SLC7A11 promoter WT and mutant H1299 cells were showed, **P<0.01 (Student's t-test).
G. Lipid ROS levels of SLC7A11 promoter WT and mutation H1299 cells treated with 5 μM Erastin for 10 hours.
H. SLC7A11 promoter WT and mutant H1299 cells were treated with different dose of Erastin for 20 hours and cell viability of indicated cells was measured. The data was representative of three independent experiments.
I. SOX2 WT and KO H1299 cells were treated with different dose of Erastin for 20 hours and cell viability of indicated cells was measured. The data was representative of three independent experiments.
J. Indicated H1299 cells were treated with 10 μM Erastin for 20 hours, 1 μM Fer-1 and 50 μM DFO treatment could rescue Erastin-induced cell death. Cell viability of indicated cells was measured, ***P<0.001 (two-way ANOVA test).
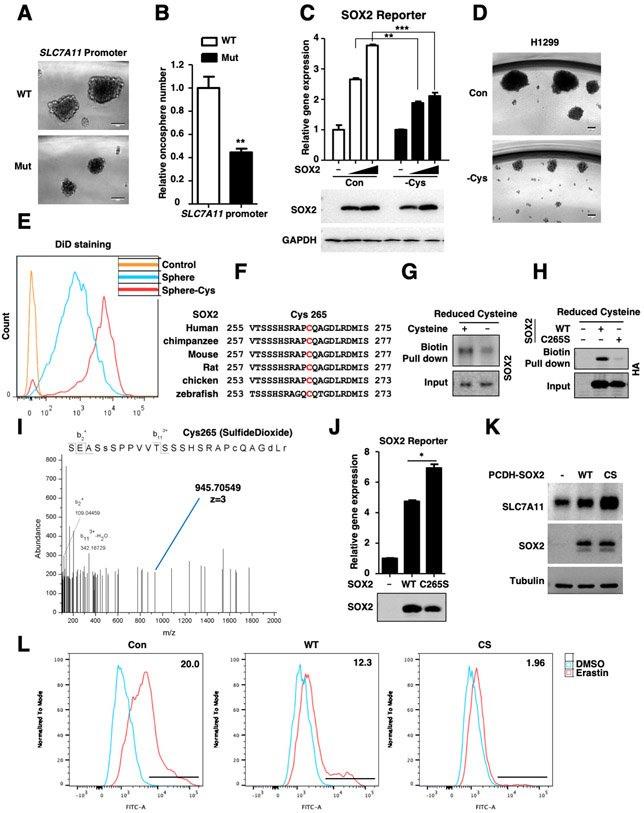
Cysteine-deprivation induces SOX2-Cys265 oxidative modification
Notably, in vitro oncosphere formation assay showed that both the size and number of oncospheres were decreased in mutant cells compared with WT cells (Figures 5A-B), indicating that the ability to form tumor sphere was reduced in SLC7A11 promoter mutant cells. Given to the essential role of SLC7A11 in cystine uptake, we hypothesized that the SOX2 activity might be modulated by cysteine levels. Our data showed that cysteine-deprivation significantly inhibited the SOX2 transcriptional activity without affecting its protein stability (Figure 5C) and weakened the self-renewal capability of CSLCs (Figures 5D-E). In contrast, cysteine-deprivation did not affect the transcriptional activity of stem cell factor Nanog (Figure S5A). These data collectively suggest that homeostasis of intracellular cysteine is required for maintaining SOX2 activity and cancer cell stemness in a feedback regulatory manner.
Figure.5. Cysteine deprivation induces SOX2-Cys265 oxidative modification.
A. Representative sphere images derived from SLC7A11 promoter WT and mutant H1299 cells. Scale bar: 100 μm.
B. Relative tumor spheres number derived from SLC7A11 promoter WT and mutant cells, **P < 0.01 versus WT (Student’s t test).
C. SOX2 activity reporter gene was co-expressed with different dose of SOX2 in HEK293T cells for 24 hours, then the cells were treated with cysteine deprivation for 6 hours or not, and the expression of Luciferase was measured and normalized to Renilla, **P<0.01, ***P<0.001 (two-way ANOVA test). The protein levels were analyzed by WB. Cysteine deprivation had little effect on expression level of SOX2.
D. Representative sphere images derived from H1299 cells treated with cysteine deprivation for 14 days or not, Scale bar: 100 μm.
E. DiD staining assay was performed to test the self-renewal of H1299 CSLCs. DiD, the cell labeling dye which is quite stable in cells, will decrease upon cell division. CSLCs treated with cysteine deprivation showed slower cell renewal rate compared to that cultured in normal medium.
F. Sequence alignment of SOX2 Cys265 across different species.
G. H1299 cells were treated with cysteine deprivation for 6 hours or not, the cell lysate was incubated with maleimide-biotin to label reduced cysteine groups. Pull-down of biotin-labeled proteins was followed by WB.
H. HA-SOX2 and C265S mutant were transfected in HEK293T cells for 24 hours, the cell lysate was incubated with maleimide-biotin to label reduced cysteine groups. Pull-down of biotin-labeled proteins was followed by WB.
I. Tandem mass spectrometry spectrum of Glu-C-digested SOX2 fragment containing the oxidized cysteine residues (blue line) was shown.
J. SOX2 activity reporter gene was co-expressed with SOX2 WT or C265S mutant plasmids in HEK293T cells for 24 hours, the expression of Luciferase was measured and normalized to Renilla, *P<0.05 (Student's t-test). The protein levels were analyzed by WB.
K. The protein levels of SLC7A11 and SOX2 in indicated H1299 cells were analyzed by WB.
L. Lipid ROS levels of indicated H1299 cells treated with 5 μM Erastin for 10 hours.
Cysteine-deprivation can induce the oxidation of intracellular proteins on cysteine residue, which contains a highly reactive sulfhydryl group [41]. Interestingly, we found that SOX2 protein contained only one cysteine residue-Cys265, which localized at its C-terminal transactivation domain (TAD) (Figure 5F). Thereby, we examined whether SOX2 was oxidized at Cys265 by using biotin-maleimide to label reduced cysteine groups and pull-down the biotin-labeled proteins followed by an immunoblot for SOX2. Our data showed that cysteine-deprivation significantly increased SOX2 oxidation (Figure 5G). Moreover, mutation of Cys265 to Ser largely impaired the binding (Figure 5H), suggesting that Cys265 was essential for SOX2 oxidation. This result was further confirmed by mass spectrometry, which showed that SOX2 was indeed oxidatively modified at Cys265 upon cysteine-deprivation (Figure 5I). Moreover, the C265S mutant, which could not undergo oxidation modification, exhibited increased transcriptional activity (Figure 5J), while had little effect on the promoter binding ability, subcellular location, dimer formation and protein stability of SOX2 (Figures S5B-G). Given that Cys265 localizes in transcriptional activation domain of SOX2, it is possible that the oxidation at Cys265 may inhibit the binding of SOX2 to its co-activators, thus decreasing the transcriptional activity. Consistently, SOX2-C265S overexpression induced stronger SLC7A11 expression and conferred more ferroptosis resistance when compared to WT (Figures 5K-L, S5H-I). These data indicate that SOX2 activity is inhibited by oxidation at Cys265 and intracellular cysteine level is essential for keeping SOX2 transcriptional activity, which is critical for stemness maintenance of CSLCs.
To further examine whether SOX2 was a critical regulator of SLC7A11 expression during occurring of ferroptosis, we measured SLC7A11 expression upon ferroptosis induction. Our data showed that Erastin, IKE and cysteine-deprivation upregulated the mRNA and protein expression of SLC7A11 in both sgNC and sgSOX2 cells (Figures S6A-B). This was consistent with the previous studies that other mechanisms beyond SOX2 were involved in SLC7A11 expression under ferroptotic conditions [1, 42].
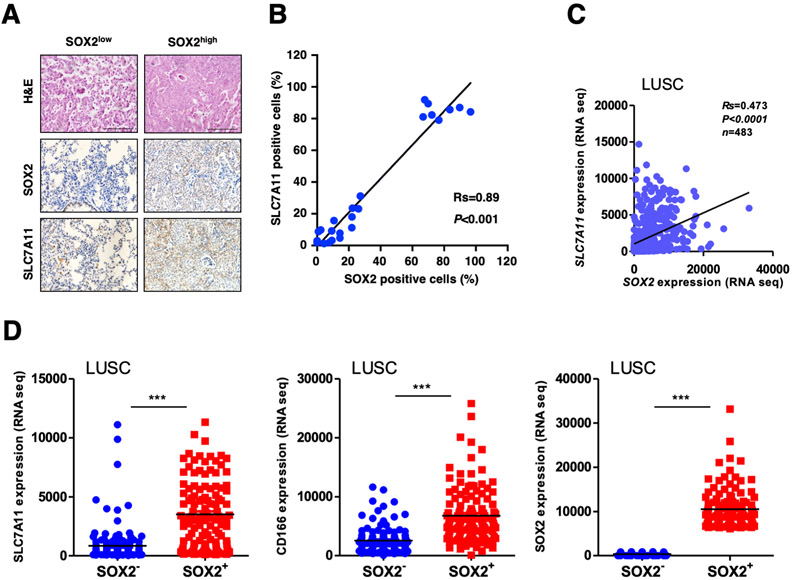
SLC7A11 expression is positively correlated with SOX2 in lung cancer tissues
Given that SOX2 is closely related with lung cancer progression [43], we examined whether the expression levels of SOX2 and SLC7A11 were correlated in mouse and human lung cancers. We found that SLC7A11 protein expression was significantly higher in SOX2high lung tumors than SOX2low tumors in KrasLSL-G12D/+;Lkb1flox/flox (KL) mouse model (Figure 6A). Consistently, the SLC7A11 protein levels were positively correlated with that of SOX2 (Figure 6B). We further analyzed and found that expression levels of SLC7A11 were positively correlated with SOX2 in human lung squamous cell carcinoma (n=483) (Figures 6C, S6C). The expression level of SLC7A11 was also higher in SOX2high than SOX2low human lung tumors (Figure 6D). Importantly, the levels of SLC7A11 were reversely correlated with overall survival in human lung cancer patients, indicating that higher SLC7A11 expression was associated with poorer outcome in lung cancer (Figure S6D). These data collectively suggest that SLC7A11 expression is positively correlated with SOX2 in both mouse and human lung cancers.
Figure.6. SLC7A11 expression positively correlates with SOX2 in lung cancer.
A. Representative images of SLC7A11 and SOX2 immunohistochemical staining in SOX2 high and low mouse lung tumor specimens. Scale bar: 100 μm.
B. The correlation of SLC7A11 and SOX2 level was showed in dot plots in lung tumor lesions from KL mice. Gene expression correlation was assessed by the Spearman test between the indicated two groups. We counted SOX2 and SLC7A11 positive cells per high-power fields (HPF), respectively.
C. The gene expression correlation of SLC7A11 and SOX2 was showed in dot plots in lung squamous cell carcinoma (LUSC). Gene expression correlation was assessed by the Spearman test between the indicated two groups.
D. The mRNA expression levels of SLC7A11, SOX2 and CD166 were compared using scatter dot plots in SOX2low versus SOX2high lung squamous cell carcinoma. The data represented as scatter dot plots and the line showed mean. Significance was assessed by the Wilcoxon test compared between the indicated two groups, ***P<0.001.
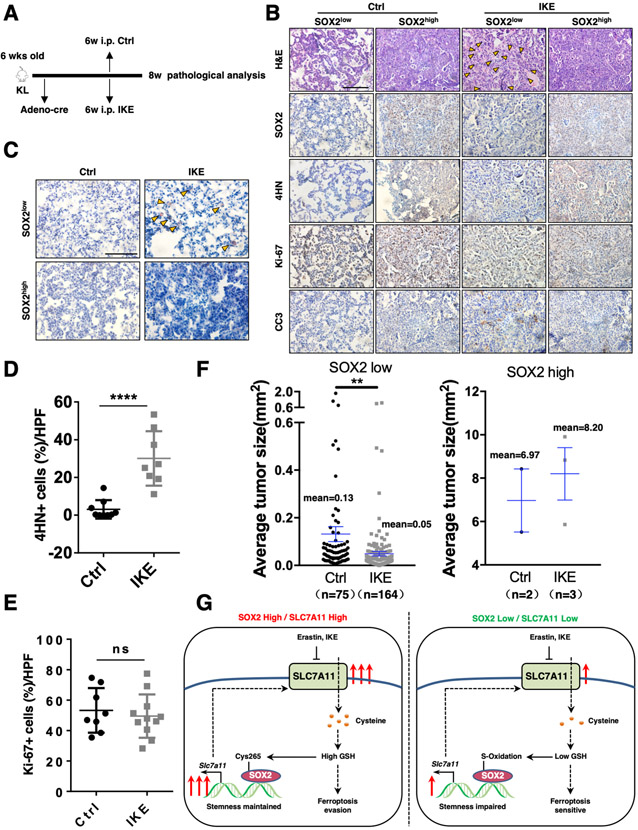
SOX2high lung tumors are more resistant to IKE treatment versus SOX2low tumors
As our data showed that SOX2-SLC7A11 axis protected lung cancer cells from ferroptosis, we further investigated whether this regulation was involved in tumor ferroptosis in vivo by using a KL spontaneous lung cancer mouse model. Because it was reported that Erastin was not suitable for direct use in in vivo experiment due to its poor solubility [21, 44], we optimized the formulation for in vivo study by sequentially dissolving Erastin in 5%DMSO, 30%PEG300, 5%Tween80 and ddH2O, which largely improved its solubility. We also repeated the experiment using a metabolically stable IKE, which had been proved to efficiently induce ferroptosis in vivo. Our data showed that both Erastin and IKE treatment induced more lipid droplet-like structures in SOX2low than SOX2high lung tumors (Figures 7A-B, S7D-E), which had been demonstrated as an in vivo ferroptosis marker [19]. Oil Red O staining confirmed the presence of large lipid droplets in SOX2low lung tumors (Figures 7C, S7F). Furthermore, Erastin and IKE treated tumors showed higher levels of 4-Hydroxynonenal (4HN) than control group while they displayed similar Ki-67 staining intensity (Figures 7D-E, S7G-H). Moreover, both Erastin and IKE treatment dramatically decreased the tumor size of SOX2low but had less effect on SOX2high tumors (Figures 7F, S7A-B, S7I-K), without affecting the mice body weight (Figures S7C, S7L). These results together indicate that SLC7A11 inhibition is selectively to restrict the growth of SOX2low but not SOX2high tumors in vivo.
Figure.7. SOX2 high lung tumors are more resistant to IKE treatment versus SOX2low tumors.
A. The scheme of IKE treatment. The KL mice at 6 weeks post Ad-Cre infection were treated with IKE for 2 weeks, followed by tumor analysis.
B. Representative H&E and IHC staining of SOX2, 4HN, Ki-67 and CC3 of Ad-Cre-infected KL mice from control and IKE groups, yellow arrowheads indicated lipid droplets. Scale bar: 100 μm.
C. Oil Red O staining of IKE treated tumors showing lipid droplets (in red) of large size. Scale bar: 100 μm.
D. Statistical analysis of the percentage of 4HN+ cells in the lung tumors control and IKE groups in Ad-Cre-infected KL mice. Data was shown as mean ± SEM. Significance was assessed by the Wilcoxon test compared between the indicated two groups, ****P<0.0001. IKE treatment induced higher levels of 4HN.
E. Statistical analysis of the percentage of Ki-67+ cells in the lung tumors control and IKE groups in Ad-Cre-infected KL mice. Data was shown as mean ± SEM. Significance was assessed by the Wilcoxon test, ns, not significant. IKE treatment had little effect on Ki-67 levels.
F. Quantification of individual tumor size of Ad-Cre-infected KL mice from control and IKE groups. Data was shown as mean ± SEM. Significance was assessed by the Wilcoxon test compared between the indicated two groups, **P<0.01. IKE treatment decreased the tumor size of SOX2 low but not SOX2 high tumors.
G. Model of SOX2-SLC7A11 regulation axis.
DISCUSSION
In the current study, we demonstrated that cystine transporter SLC7A11 was regulated by stem cell transcriptional factor SOX2. Based on this, we proposed a model that SOX2high lung tumor cells transcriptionally activated the expression of SLC7A11 to increase cystine uptake and GSH synthesis, which protected lung cancer from ferroptosis. On the other hand, upregulated GSH consequently blocked the oxidative modification of SOX2-Cys265, thus enhancing SOX2 activity and maintaining cancer cells stemness (Figure 7G). Notably, it is reported that SOX2 could also upregulate the expression of glucose transporter to control intracellular NADPH and GSH levels [45, 46], together with our study, these data indicate that SOX2 is a key redox regulator to confer ferroptosis resistance.
Activation of cystine transporter SLC7A11 is crucial for promoting tumor growth and drug resistance. The expression level and cystine uptake activity of SLC7A11 are precisely regulated at transcriptional, epigenetical and post-translational levels. Our results revealed that SLC7A11 was transcriptionally upregulated by stem cell factor SOX2 in CSLCs. Although the study mainly focused on lung cancer cells, our data also indicated that SOX2 promoted SLC7A11 expression in ESCs and neuron stem-like cells, suggesting that the SOX2-SLC7A11 regulatory axis might participate in embryonic development and differentiation process. Yet, the physiological functions of ferroptosis and SLC7A11 regulation in embryonic and adult stem cells remain to be further studied. Besides, although oncosphere formation assay is widely used for studying CSLCs and depletion of SOX2 indeed largely impairs the oncosphere formation ability (Figure 3C), it needs further validating whether these 3D structures are bona fide stem-like cells. Notably, in our KL lung cancer model, the number of SOX2high tumors is much less than SOX2low tumors, which may largely affect statistical analysis, additional study is needed to further strengthen our findings. Future studies are also needed to fully explore the physiological role of SOX2-SLC7A11 axis in cancer stem cells and its direct relevance to ferroptosis in vivo.
SOX2 is an essential pluripotent factor for stem cell self-renewal capacity and its upregulated expression is closely related to the occurrence and development of various cancers [47]. Yet, SOX2 function is precisely regulated post-translationally by phosphorylation, methylation, ubiquitination and acetylation [48], in this study, we uncovered a novel post-translational mechanism of SOX2 and demonstrated that cysteine oxidation inhibited its transcriptionally activity. Thus, our data together suggest that SOX2 may function as a potential redox sensor in cancer cells.
As an iron-dependent oxidative stress and cell death, ferroptosis has gained a lot of interests, especially in the field of cancer. Inducing ferroptosis in cancer cells is regarded as a promising way to kill therapy-resistant cancers [7, 49, 50]. Our study indicates that SOX2 may be a potential biomarker to predict the sensitivity of cancer cells to ferroptosis. Notably, tumors expressed high level of SOX2 are more resistant to ferroptosis, which may largely attenuate the potential therapeutic efficacy by inducing ferroptosis. Interestingly, SOX2-Cys265 oxidation could probably act as an Achilles' Heel in such cancers. ROS inducer such as chemotherapeutic drugs may be used to induce the oxidation of SOX2, thus reducing SOX2 activity and decreasing SLC7A11 expression, which could largely sensitize SOX2high cancer cells to ferroptosis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Yufeng Shi and Dr. Ke Wei for kindly providing cells and reagents. This study was supported by Grants from the National Natural Science Foundation of China (31920103007, 81625019, 31830053, 31900525, 31800757, 81872312, 82011540007, 31621003, 82030083), National Basic Research Program of China (2017YFA0505501, 2020YFA0803300), Shanghai International Science and Technology Cooperation Fund Project (18410722000), Shanghai Sailing Program (18YF1419300), Shanghai Chenguang Program (19CG20), Shanghai Municipal Health Commission (20204Y0029), Chinese Academy of Sciences (XDB19020201, ZDBS-LY-SM006), China Postdoctoral Science Foundation (2019M650090, 2019T120353, 2019M651586), Shanghai Post-doctoral Excellence Program, NIH R01 CA204232 and NIH P30 CA008748.
Footnotes
CONFLICTS OF INTEREST STATEMENT:
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Dixon Scott J., Lemberg Kathryn M., Lamprecht Michael R., Skouta R, Zaitsev Eleina M., Gleason Caroline E., et al. , Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell, 2012. 149(5): p. 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J and Conrad M, The Metabolic Underpinnings of Ferroptosis. Cell Metab, 2020. 32(6): p. 920–937. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, Stockwell BR, and Conrad M, Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. , Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell, 2017. 171(2): p. 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DR, The Coming Decade of Cell Death Research: Five Riddles. Cell, 2019. 177(5): p. 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedmann Angeli JP, Krysko DV, and Conrad M, Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer, 2019. 19(7): p. 405–414. [DOI] [PubMed] [Google Scholar]
- 7.Hassannia B, Vandenabeele P, and Vanden Berghe T, Targeting Ferroptosis to Iron Out Cancer. Cancer Cell, 2019. 35(6): p. 830–849. [DOI] [PubMed] [Google Scholar]
- 8.Kang YP, Mockabee-Macias A, Jiang C, Falzone A, Prieto-Farigua N, Stone E, et al. , Non-canonical Glutamate-Cysteine Ligase Activity Protects against Ferroptosis. Cell Metab, 2021. 33(1): p. 174–189.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren E and Fuchs Y, Modes of Regulated Cell Death in Cancer. Cancer Discov, 2021. [DOI] [PubMed] [Google Scholar]
- 10.Koppula P, Zhuang L, and Gan B, Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppula P, Zhang Y, Zhuang L, and Gan B, Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun (Lond), 2018. 38(1): p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Kon N, Li T, Wang S-J, Su T, Hibshoosh H, et al. , Ferroptosis as a p53-mediated activity during tumour suppression. Nature, 2015. 520(7545): p. 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. , BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol, 2018. 20(10): p. 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, et al. , Targeting the Vulnerability of Glutathione Metabolism in ARID1A-Deficient Cancers. Cancer Cell, 2019. [DOI] [PubMed] [Google Scholar]
- 15.Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, et al. , AMPK-Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System Xc(−) Activity. Curr Biol, 2018. 28(15): p. 2388–2399 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y, Albuquerque CP, Braas D, Zhang W, Villa GR, Bi J, et al. , mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT. Mol Cell, 2017. 67(1): p. 128–138 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, et al. , CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(−) and thereby promotes tumor growth. Cancer Cell, 2011. 19(3): p. 387–400. [DOI] [PubMed] [Google Scholar]
- 18.Liu T, Jiang L, Tavana O, and Gu W, The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee HJ, Purohit V, et al. , Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science, 2020. 368(6486): p. 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, et al. , Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature, 2019. 572(7769): p. 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Tan H, Daniels JD, Zandkarimi F, Liu H, Brown LM, et al. , Imidazole Ketone Erastin Induces Ferroptosis and Slows Tumor Growth in a Mouse Lymphoma Model. Cell Chem Biol, 2019. 26(5): p. 623–633.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang X, Green MD, Wang W, Yu J, Choi JE, Jiang L, et al. , Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov, 2019. 9(12): p. 1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, et al. , CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature, 2019. 569(7755): p. 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei G, Zhang Y, Koppula P, Liu X, Zhang J, Lin SH, et al. , The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res, 2020. 30(2): p. 146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou Y, Henry WS, Ricq EL, Graham ET, Phadnis VV, Maretich P, et al. , Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature, 2020. 585(7826): p. 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubellacker JM, Tasdogan A, Ramesh V, Shen B, Mitchell EC, Martin-Sandoval MS, et al. , Lymph protects metastasizing melanoma cells from ferroptosis. Nature, 2020. 585(7823): p. 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder S, Crabtree JS, Golde TE, Minter LM, Osborne BA, and Miele L, Targeting Notch in oncology: the path forward. Nat Rev Drug Discov, 2020. [DOI] [PubMed] [Google Scholar]
- 28.Mai TT, Hamai A, Hienzsch A, Caneque T, Muller S, Wicinski J, et al. , Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem, 2017. 9(10): p. 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. , Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell, 2018. 34(5): p. 724–740.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Recalcati S, Gammella E, and Cairo G, Dysregulation of iron metabolism in cancer stem cells. Free Radic Biol Med, 2019. 133: p. 216–220. [DOI] [PubMed] [Google Scholar]
- 31.Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS, et al. , Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell, 2015. 28(4): p. 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torti SV, Manz DH, Paul BT, Blanchette-Farra N, and Torti FM, Iron and Cancer. Annu Rev Nutr, 2018. 38: p. 97–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. , Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther, 2020. 5(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang MC, Ma J, Chen L, Kozlowski P, Qin W, Li D, et al. , TSC1 loss synergizes with KRAS activation in lung cancer development in the mouse and confers rapamycin sensitivity. Oncogene, 2010. 29(11): p. 1588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han X, Li F, Fang Z, Gao Y, Li F, Fang R, et al. , Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun, 2014. 5: p. 3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, and Berns A, Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell, 2011. 19(6): p. 754–64. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. , YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun, 2014. 5: p. 4629. [DOI] [PubMed] [Google Scholar]
- 38.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. , Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell, 2018. 173(2): p. 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, Lim SK, Liang Q, Iyer SV, Wang HY, Wang Z, et al. , Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature, 2019. 567(7748): p. 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Ning S, Ghandi M, Kryukov GV, Gopal S, Deik A, et al. , The landscape of cancer cell line metabolism. Nat Med, 2019. 25(5): p. 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul BD, Sbodio JI, and Snyder SH, Cysteine Metabolism in Neuronal Redox Homeostasis. Trends Pharmacol Sci, 2018. 39(5): p. 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo M, Ling V, Wang YZ, and Gout PW, The xc- cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer, 2008. 99(3): p. 464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, et al. , SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell, 2016. 30(4): p. 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi J, Minikes AM, and Jiang X, Aiming at Cancer In Vivo: Ferroptosis-Inducer Delivered by Nanoparticles. Cell Chem Biol, 2019. 26(5): p. 621–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh MH, Choe JH, Gadhvi J, Kim YJ, Arguez MA, Palmer M, et al. , p63 and SOX2 Dictate Glucose Reliance and Metabolic Vulnerabilities in Squamous Cell Carcinomas. Cell Rep, 2019. 28(7): p. 1860–1878.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreucci E, Pietrobono S, Peppicelli S, Ruzzolini J, Bianchini F, Biagioni A, et al. , SOX2 as a novel contributor of oxidative metabolism in melanoma cells. Cell Commun Signal, 2018. 16(1): p. 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, Xiong X, and Sun Y, Functional characterization of SOX2 as an anticancer target. Signal Transduct Target Ther, 2020. 5(1): p. 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Kang L, Zhang H, Huang Y, Fang L, Li M, et al. , AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene, 2019. 38(26): p. 5250–5264. [DOI] [PubMed] [Google Scholar]
- 49.Crielaard BJ, Lammers T, and Rivella S, Targeting iron metabolism in drug discovery and delivery. Nat Rev Drug Discov, 2017. 16(6): p. 400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, et al. , Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell, 2018. 33(5): p. 890–904.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A complete software environment and all necessary data to reproduce the analysis and figure described in this manuscript are available on Code Ocean (https://codeocean.com/capsule/8756276/tree/v1).