Author Roles
(1) Research project: A. Conception, B. Organization, C. Execution; (2) Statistical Analysis: A. Design, B. Execution, C. Review and Critique; (3) Manuscript Preparation: A. Writing of the first draft, B. Review and Critique.
L.C.‐M.: 1A, 1B, 1C, 2A, 2B, 2C, 3A, 3B
C.M.F.: 1B, 1C, 2C, 3B
B.H.: 1C, 3B
Y.M.: 1C, 3B
J.S.: 1C, 3B
M.J.R.: 2A, 2B, 2C, 3B
U.J.K.: 2A, 2B, 2C, 3B
S.H.: 1B, 3B
K.M.: 3B
M.S.: 3B
C.S.: 1A, 2C, 3B
Financial Disclosure
Dr. Soto, Dr. Concha, Ms. Farris, Mr. Ma, and Mr. Holguin are inventors on several patents related to the SAA (PMCA) technology and are affiliated to Amprion Inc., a biotech company focusing on the commercial utilization of SAA (PMCA) for diagnosis. Dr. Shahnawaz is also an inventor on several patents related to SAA (PMCA) technology but he is not associated with Amprion. Dr. Kang is on the advisory board of Amprion.
Parkinson's disease (PD) diagnosis relies primarily on clinical evaluation due to lack of validated tests and biomarkers. DaTscan imaging has been used to distinguish psychogenic and drug‐induced parkinsonism from idiopathic PD. Some clinically diagnosed PD patients show scans without evidence of dopaminergic deficit (SWEDD), including some that respond to dopaminergic treatment. Some SWEDD patients present abnormal scans consistent with PD many years later and it is unknown if these patients developed PD in between scans or presented PD with low dopaminergic degeneration.
α‐Synuclein seed amplification assays (αS‐SAAs) detect α‐synuclein (αSyn) aggregates in the cerebrospinal fluid (CSF) of PD, dementia with Lewy bodies (DLB), and isolated rapid eye movement (REM) sleep behavior disorder (iRBD) patients with high sensitivity and specificity.1, 2, 3, 4 We used an optimized high‐throughput αS‐SAA (based on a previously described α‐Syn protein misfolding cyclic amplification (PMCA) assay)2, 5, 6 that detects αSyn aggregates in CSF, to evaluate 140 blinded samples from the Parkinson's Progression Markers Initiative (PPMI). Samples included baseline (BL) and 3‐year follow‐up (V08) from 30 PD and 30 healthy controls (HC), and BL samples from 20 SWEDD patients. PD‐BL samples were collected within 2 years from diagnosis and presented abnormal DaTscans, while SWEDD patients presented normal DaTscans. PPMI classified enrollees as PD or SWEDD based on visual inspection of their baseline DaTscans.
Figure 1A shows the assay results. The assay performed with 96.2% sensitivity (95% CI: 80.4%–99.9%) and 96.7% specificity (95% CI: 82.8%–99.9%) for PD versus HC at BL, and 96.4% sensitivity (95% CI: 81.7%–99.9%) and 93.8% specificity (95% CI: 79.2%–99.2%) at V08. After αS‐SAA analysis, PPMI reclassified two of the three αS‐SAA‐negative subjects in the PD cohort as non‐PD, therefore they were excluded from the above calculation. There were three false‐positive samples: #3112‐V08 and both samples from patient #3264. The latter was found to be a probable RBD case based on their RBD questionnaire score. Unfortunately, confirmatory polysomnography is not available and both samples from this patient were considered to be false‐positives. Two BL‐PD samples were inconclusive. Retest was not possible due to lack of sample and they were excluded from analysis.
FIG. 1.
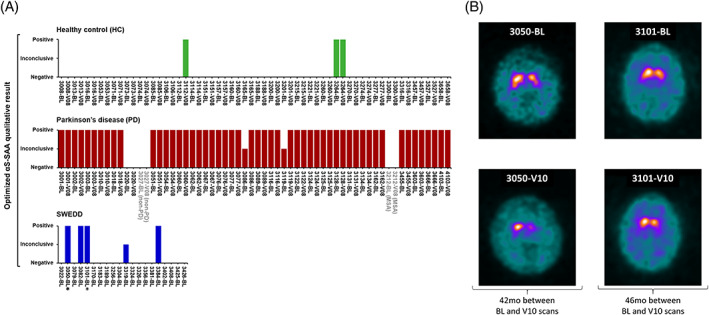
α‐Synuclein seed amplification assay (αS‐SAA) results of the 140 cerebrospinal fluid (CSF) samples and DaTscan images of αS‐SAA‐positive scans without evidence of dopaminergic deficit (SWEDD) patients. (A) The data analysis algorithm produced a trinary qualitative outcome (positive, inconclusive, and negative), which was graphed for all 60 healthy control (HC), 60 Parkinson's disease (PD) and 20 SWEDD CSF samples. Each bar represents the αS‐SAA result for a sample from a patient collected at a given time point (BL for baseline and V08 for visit 08). Reclassified PD patients are shown in grey; one was reclassified as multiple system atrophy (MSA) confirmed by postmortem pathological examination and the other one as unknown non‐PD clinically. SWEDD patients whose abnormal brain scans were predicted by the optimized αS‐SAA 42 and 46 months (mo) before the abnormal DaTscan are indicated with an asterisk (*). (B) Transverse DaTscan from patient #3050 at BL was considered borderline normal by visual inspection and abnormal 42 months later at visit 10 (V10). BL scan (2010/09/23) showed slightly asymmetric uptake with heterogenous appearance left striatum with specific binding ratio (SBR) lowest putamen equal to 1.04. V10 scan (2014/03/05) shows significant bilateral reduction, with the greatest changes in the left striatum. Within 4 years, there was a significant 25% SBR reduction (SBR‐V10 = 0.78). Transverse DaTscan from patient #3101 at BL was considered borderline normal by visual inspection and abnormal 46 months later. BL scan (2010/10/21) was read borderline normal with rotation effect creating a faux reduction in the left putamen (BL‐SBR lowest putamen = 1.02). V10 scan (2014/08/22) shows decided asymmetric signal loss, bilateral, with greatest involvement of signal loss on the left, with relatively preserved right caudate. Within 4 years, there was a significant 47% SBR reduction (SBR‐V10 = 0.54). [Color figure can be viewed at wileyonlinelibrary.com]
Of the 20 BL‐SWEDD samples, we found 4 positive, 15 negative, and 1 inconclusive. Second DaTscans available at V10 (4 years from BL) of the negatives were normal. Interestingly, two of the four αS‐SAA‐positive SWEDD subjects (#3050 and #3101) showed a substantial increase in dopaminergic degeneration by DaTscans at 42 and 46 months after enrollment, with substantial putamen deficits consistent with a PD diagnosis (Fig. 1B). After αS‐SAA analysis and after reviewing all longitudinal, clinical, and imaging data, the PPMI analytic cohort consensus committee decided to change the enrollment diagnosis of #3050 and #3101 from SWEDD to PD.
Our results indicate that the optimized αS‐SAA is highly accurate compared to the gold standard (longitudinal, clinical, and imaging data). The potential value of unbiased αS‐SAA results in a clinical setting can be appreciated in cases with disputable diagnosis, such as the two αS‐SAA‐negative clinical PD patients (reclassified as non‐PD) and the two αS‐SAA‐positive SWEDD cases (reclassified as PD). The αS‐SAA‐positive HC with probable RBD is in agreement with recent reports showing prodromal PD diagnosis.4, 7 Detailed introduction, results, methods, comparison to the original assay,2, 5 and discussion are included as Appendix S1.
Supporting information
AppendixS1. Supporting information.
Acknowledgments
We thank the sample donors and their families for supporting this research by participating in the PPMI study. We also want to thank The Michael J. Fox Foundation for their assistance in accessing CSF samples and online information of the cohort.
Relevant conflicts of interest/financial disclosures: Dr. Soto, Dr. Concha, Ms. Farris, Mr. Ma, and Mr. Holguin are inventors on several patents related to the SAA (PMCA) technology and are affiliated to Amprion Inc., a biotech company focusing on the commercial utilization of SAA (PMCA) for diagnosis. Dr. Shahnawaz is also an inventor on several patents related to SAA (PMCA) technology but he is not associated with Amprion. Dr. Kang is on the advisory board of Amprion.
Funding agency: This work was funded by grant #16712 from The Michael J. Fox Foundation for Parkinson's Research to Amprion. The results are available online in the PPMI website under project #155.
Data Availability Statement
All data presented in this letter is available at the PPMI database (https://ida.loni.usc.edu/login.jsp).
References
- 1. Fairfoul G, McGuire LI, Pal S, et al. Alpha‐synuclein RT‐QuIC in the CSF of patients with alpha‐synucleinopathies. Ann Clin Transl Neurol 2016;3(10):812–818. 10.1002/acn3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shahnawaz M, Tokuda T, Waragai M, et al. Development of a biochemical diagnosis of Parkinson disease by detection of α‐synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol 2017;74(2):163. 10.1001/jamaneurol.2016.4547. [DOI] [PubMed] [Google Scholar]
- 3. Groveman BR, Orrù CD, Hughson AG, et al. Rapid and ultra‐sensitive quantitation of disease‐associated α‐synuclein seeds in brain and cerebrospinal fluid by αSyn RT‐QuIC. Acta Neuropathol Commun 2018;6(1):7. 10.1186/s40478-018-0508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iranzo A, Fairfoul G, Ayudhaya ACN, et al. Detection of α‐synuclein in CSF by RT‐QuIC in patients with isolated rapid‐eye‐movement sleep behaviour disorder: a longitudinal observational study. Lancet Neurol 2021;20(3):203–212. 10.1016/S1474-4422(20)30449-X. [DOI] [PubMed] [Google Scholar]
- 5. Concha‐Marambio L, Shahnawaz M, Soto C. Detection of misfolded α‐synuclein aggregates in cerebrospinal fluid by the protein misfolding cyclic amplification platform. Methods Mol Biol 1948;2019:35–44. 10.1007/978-1-4939-9124-2_4. [DOI] [PubMed] [Google Scholar]
- 6. Shahnawaz M, Mukherjee A, Pritzkow S, et al. Discriminating α‐synuclein strains in Parkinson's disease and multiple system atrophy. Nature 2020;578(7794):273–277. 10.1038/s41586-020-1984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stefani A, Iranzo A, Holzknecht E, et al. Alpha‐synuclein seeds in olfactory mucosa of patients with isolated REM sleep behaviour disorder. Brain 2021;144(4):1–9. 10.1093/brain/awab005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AppendixS1. Supporting information.
Data Availability Statement
All data presented in this letter is available at the PPMI database (https://ida.loni.usc.edu/login.jsp).


