This study identifies an LPAR1-axis critical for melanoma invasion and intrinsic/acquired therapy resistance.
Abstract
Metastatic melanoma is challenging to clinically address. Although standard-of-care targeted therapy has high response rates in patients with BRAF-mutant melanoma, therapy relapse occurs in most cases. Intrinsically resistant melanoma cells drive therapy resistance and display molecular and biologic properties akin to neural crest-like stem cells (NCLSC) including high invasiveness, plasticity, and self-renewal capacity. The shared transcriptional programs and vulnerabilities between NCLSCs and cancer cells remains poorly understood. Here, we identify a developmental LPAR1-axis critical for NCLSC viability and melanoma cell survival. LPAR1 activity increased during progression and following acquisition of therapeutic resistance. Notably, genetic inhibition of LPAR1 potentiated BRAFi ± MEKi efficacy and ablated melanoma migration and invasion. Our data define LPAR1 as a new therapeutic target in melanoma and highlights the promise of dissecting stem cell–like pathways hijacked by tumor cells.
Significance:
This study identifies an LPAR1-axis critical for melanoma invasion and intrinsic/acquired therapy resistance.
Introduction
Immense progress has been made for patients with advanced melanoma with 13 new FDA-approved therapies since 2011 (1, 2). The majority (76%) of patients whose melanomas harbor activating BRAFV600E/K mutations (∼50% of patients) respond dramatically to dual BRAF/MEK inhibitor therapy (BRAFi/MEKi; ref. 3). However, resistance arises within 2 years in near all cases, and 4 of 5 of these patients show no long-term benefit with immunotherapy due to toxicity and/or nonresponsiveness. This expanding BRAFi/MEKi-resistant patient cohort is the greatest challenge of the melanoma field as no alternative effective targeted therapies exist.
We and others have identified melanoma cells that dedifferentiate to a neural crest-like stem cell (NCLSC) state capable of surviving challenging environmental contexts including metastasis and therapy (4). Molecularly, these NCLSC-like melanoma cells express NCLSC markers including JARID1B (5), nerve growth factor receptor (NGFR; ref. 6), EGFR (7), SerpinE2 (8), and/or AXL, as well as a NCLSC transcriptional signature (9). Biologically, NCLSC-like melanoma cells exhibit high invasiveness and plasticity. Critically, NCLSC-like melanoma cells are intrinsically resistant to chemotherapy and targeted therapy (4, 9). These findings underscore the need to develop novel therapeutic strategies that potently eliminate NCLSC-like melanoma cells and define the molecular mechanisms responsible for the therapy resistance and aggressive phenotypes of NCLSC-like melanoma cells.
Toward this goal, we executed a transcriptome juxtaposition study to compare gene expression profiles of human skin-derived NCLSCs, melanocytes, and melanoma cultures. During embryonic development, multipotent trunk NCLSCs migrate from the neural plate to the epidermis/dermis and undergo lineage specification to generate differentiated melanocytes (10, 11). In turn, melanoma cells arise following the malignant transformation of melanocytes. We focused on genes highly expressed in both melanoma cells and NCLSCs, but not in melanocytes to identify genes associated with a dedifferentiated NCLSC state. This approach led to the delineation of a cluster of stem cell–like genes enriched in melanoma. Using a targeted RNAi strategy coupled with functional in vitro and in vivo studies, we have identified lysophosphatidic acid receptor 1 (LPAR1) as critical for targeted therapy resistance in melanoma cells. LPAR1 is elevated in patient tumor tissue and drives melanoma proliferation, mobility, invasiveness, and resistance to therapy by promoting mTOR and YAP activity. Our findings reveal an unanticipated role of LPAR1 in regulating both mTOR and YAP activity, and define LPAR1 as a novel target for the treatment of patients with BRAFi/MEKi-resistant melanoma.
Materials and Methods
Additional molecular and cell biology techniques, list of oligonucleotides, high-throughput methods, and detailed computational analyses are described in Supplementary Data.
Cell culture of NCLSC, melanocytes, and fibroblasts
NCLSC, melanocytes, and fibroblasts were isolated from human foreskins and were cultured as described previously (12). Following the investigation of LPAR1 levels in a panel of 23 melanoma cell lines, four melanocyte cultures, and four NCLSC cultures, we selected the top highest three LPAR1-expressing cell lines in the BRAF-MT cohort, with similar expression to that found in NCLSCs, as LPAR1 high. Conversely, we selected the lowest three LPAR1-expressing cell lines in the BRAF-MT cohort, with similar expression to melanocytes, as LPAR1 low cells. We also selected the top three highest LPAR1-expressing cell lines in the NRAF-MT cohort as LPAR1 high and the lowest three LPAR1-expressing cell lines in the NRAS-MT cohort as the LPAR1 low. All of the BRAF/NRAS-WT/WT cell lines had low LPAR1 expression akin to melanocytes.
Melanoma patient single-cell RNA-sequencing data
Invasive/NCSC/Nutrient depleted and Pigmented gene sets were derived from Rambow and colleagues Cell 2018 (9), and the Stress-like gene set was derived from Baron and colleagues Cell Systems 2020 (13). Normalized single-cell expression data from Tirosh and colleagues Science 2016 article [Gene Expression Omnibus (GEO) accession number GSE72056; ref. 14] was used to calculate cell expression signatures for 1,257 tumor cells as an average value among genes from the set. Expression signatures were compared using t test between cells with no detected LPAR1 expression versus LPAR1-positive cells. Fold changes of mean LPAR1+/LPAR1− values were calculated.
Cell lines established from human melanomas and patient-derived xenografts
All normal skin epidermal melanocytes, keratinocytes, dermal stem cells, and human metastatic melanoma cell lines that were established at The Wistar Institute (Philadelphia, PA) have been documented (https://www.wistar.org/lab/meenhard-herlyn-dvm-dsc/page/resources). UACC-62 and UACC-903 cells were kind gifts from Marianne B. Powell (Stanford University, Stanford, CA). A375 cells were purchased from the ATCC. All resistant cell lines that acquired drug resistance to PLX4720 (hereafter referred to as “BR” cell lines) or the combination of PLX4720 and PD0325901 (hereafter referred to as “CR” cell lines) were established after continuous exposure to PLX4720 at 10 μmol/L or the combination of PLX4720 at 10 μmol/L and PD0325901 at 1 μmol/L. All cell lines were maintained in Tumor media (MCDB 153 and L-15) supplemented with 2% FBS (Tissue Culture Biologicals) or DMEM (Mediatech, Inc.) supplemented with 10% FCS. Cells were cultured in a 37°C humidified incubator supplied with 5% CO2. All cell lines were authenticated by DNA fingerprinting.
Accession number
The GEO database accession number for the gene expression microarray and RNA sequencing (RNA-seq) data is GSE92765.
Drug sensitivity IC50 data
IC50 data for human melanoma cell lines with BRAFV600 mutations treated with selective BRAFis and MEKis were curated from the Cancer Cell Line Encyclopedia.
Reverse phase protein array and analysis
The reverse phase protein array (RPPA) assay was performed by the MD Anderson Cancer Center RPPA core facility (Houston, TX) using the method described on the MD Anderson Cancer Center website. The same differential expression analysis method used for the microarray data was applied to RPPA data. Proteins that were most significantly downregulated were clustered by their functional pathways and were used to generate the heatmaps.
Chemicals
The BRAFis PLX4032, PLX4720, and GSK2118436 and the MEKi PD0325901 were purchased from Selleckchem. For in vivo oral gavage, PLX4032 was suspended in Klucel EF 2% (w/v), and the pH was adjusted to 4 using HCl. The compound was dosed using feeding tubes (Instech Laboratories, Inc).
Cell growth/viability and assessment of cell clonogenicity
Cell viability was measured by the MTT assay. For the assessment of cell clonogenicity, cells were seeded in 6-well tissue culture plates at a density of 4,000 cells/well as biological triplicates in drug-free medium. Medium was refreshed every 2 or 3 days for 15–21 days. Colonies were then stained overnight with methanol containing 0.05% crystal violet. After extensive washing with distilled H2O, cells were air dried and subjected to image acquisition using an Epson scanner (Epson perfection V700 photo).
GST pulldown assay
The GST pulldown assay was carried out using an Active Rap1 Detection Kit (Cell Signaling Technology) according to the manufacturer's instructions. Briefly, 50% glutathione resin slurry was bound to the column tube, followed by the addition of 20 μG GST-RalGDS-RBD. Cell lysates (1 mg total protein) from melanoma cells with different treatments were added to the column tube and incubated at 4°C for 1 hour. After three cycles of washing, the reducing sample buffer was added and the eluted samples were collected and subjected to SDS electrophoresis and Western blotting.
Results
Transcriptome juxtaposition and targeted screens identify LPAR1
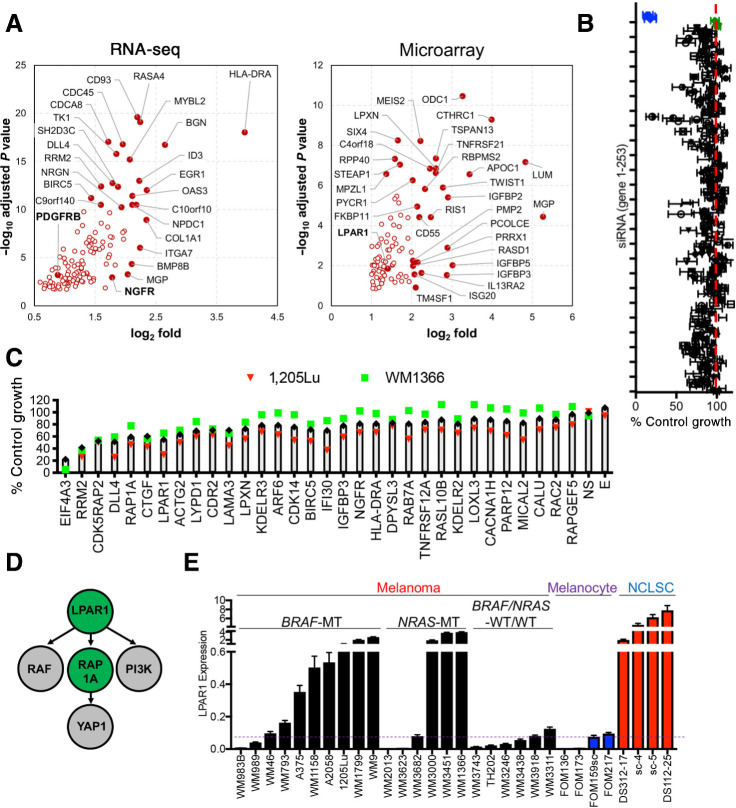
In an effort to identify shared stem cell–like programs between melanoma cells and NCLSC, gene expression profiles of human skin-derived NCLSC, melanocyte and melanoma cultures were juxtaposed to distinguish gene candidates enriched in NCLSC and melanoma cultures, but not melanocytes (Fig. 1A). Analysis of gene expression microarray and RNA-seq data identified 98 (Padjusted < 0.05, fold>2) and 122 (Padjusted < 0.05) gene candidates, respectively, including novel and known (i.e., NGFR, PDGFRβ) drivers of melanoma aggressiveness (Fig. 1A; Supplementary Table S1; Supplementary Fig. S1A; refs. 6, 15, 16), resulting in a set of 208 unique genes. To experimentally validate the functional relevance of these gene candidates on melanoma cell proliferation, a targeted RNAi screen was executed (188 of the 208 genes were included in the siRNA library), which in addition included 52 genes that serve functional roles in stem cell biology, resulting in a set of 240 unique genes. Four distinct siRNA sequences were incorporated against each gene (eight genes IGFBP2, MEIS2, ODC1, PALLD, RASD1, TM4SF1, TNFRSF12A, TNFRSF21 were tested twice) along with the housekeeping gene EIF4A3 (K), five nonspecific (NS) siRNA and a media only well (E). Two melanoma cell lines (one harboring a BRAFV600E mutation, one harboring an NRASQ61R mutation) were included in the screen (Fig. 1B; Supplementary Table S2). Approximately 19% of the gene candidates could inhibit melanoma cell proliferation by ≥ 25% across the melanoma cell lines. The top 31 gene candidates were subjected to a second independent validation screen (Fig. 1C; Supplementary Table S3). Genes previously reported to contribute to melanoma aggressiveness and therapy resistance emerged both in our shared NCLSC and melanoma transcriptome analysis and second validation screen, further supporting our hypothesis that novel therapeutic targets could be defined among genes associated with a dedifferentiated NCLSC state. NGFR emerged in our screen, which is a canonical marker of dedifferentiated melanoma cells that display adaptive resistance to BRAF inhibition (6), and melanoma cells resistant to anti-PD-1 blockade (17). The Notch signaling ligand, delta like canonical notch ligand 4 (DLL4), emerged in our screen, which agrees with the literature showing a role for notch signaling in promoting cancer stem cell viability through tissue remodeling (18). Connective tissue growth factor was detected in our screen and has been reported to increase in acute myeloid leukemia–derived mesenchymal stem cells (19) and serve a critical role in the metastatic capacity of melanoma cells (20). Ribonucleotide reductase regulatory subunit M2 (RRM2), Baculoviral IAP repeat containing 5 (BIRC5) and KDEL endoplasmic reticulum protein retention receptor 3 (KDELR3) are additional genes that were detected in our screen that have been previously shown to play a role in melanoma metastasis and therapy resistance (21–23). The top gene candidates were next evaluated by Ingenuity Pathway Analysis software, which identified a lysophosphatidic acid receptor 1 (LPAR1)-axis as a critical signaling network (Fig. 1D; Supplementary Fig. S1B). LPAR1 is a G12/13-coupled receptor that drives the proliferation of neuronal progenitor cells (24), and promotes the aggressiveness of breast (25), lung (26), and ovarian cancer (27). Interrogation of LPAR1 expression in an extended panel of NCLSC, melanocyte, and melanoma cultures confirmed melanoma cells exhibit higher expression levels of LPAR1 relative to melanocytes (Fig. 1E). A subset of five BRAF-mutant and three NRAS-mutant melanoma cell lines display higher levels of LPAR1 relative to melanocytes and equivalent to those in NCLSCs (Fig. 1E). Conversely, a subset of four BRAF-mutant and three NRAS-mutant melanoma cell lines display relatively low LPAR1 expression levels akin to those observed in melanocytes. LPAR1 expression within BRAF/NRAS-wildtype/wildtype melanoma cells is lower overall relative to other genotypes (Fig. 1E). Collectively, these data identify LPAR1 as a stem cell gene enriched in subsets of BRAF-MT and NRAS-MT melanoma cells.
Figure 1.
Transcriptome juxtaposition and targeted screens identify LPAR1. A, Volcano plot for genes selected from RNA-seq and microarray data. Highlighted are best novel genes enriched in melanoma and NCLSC cultures relative to melanocytes by fold change and Padjusted (for RNA-seq: P < 10–10 or fold < 4, for microarrays: P < 10–6 or fold < 4). B, Scatter plot showing the averaged results of the targeted siRNA screen in 1205Lu and WM1366 cells. The x-axis represents the normalized growth and the y-axis represents each individual gene from 1–253. Four individual siRNAs against each gene were used. The blue dots represent the siEIF4A3 and the green dots represent nonspecific siRNAs as negative control. C, The top 30 genes identified in the primary screen were used for the secondary screen. D, Ingenuity Pathway Analysis of the top 30 genes in the screen identifies the LPAR1-RAP1A axis as the most significant network among the gene candidates. E, Relative gene expression levels of LPAR1 in melanoma, NCLSC, and melanocytes assayed by qRT-PCR. Biological replicates (n = 3) for each condition are included.
LPAR1 sustains melanoma and NCLSC survival
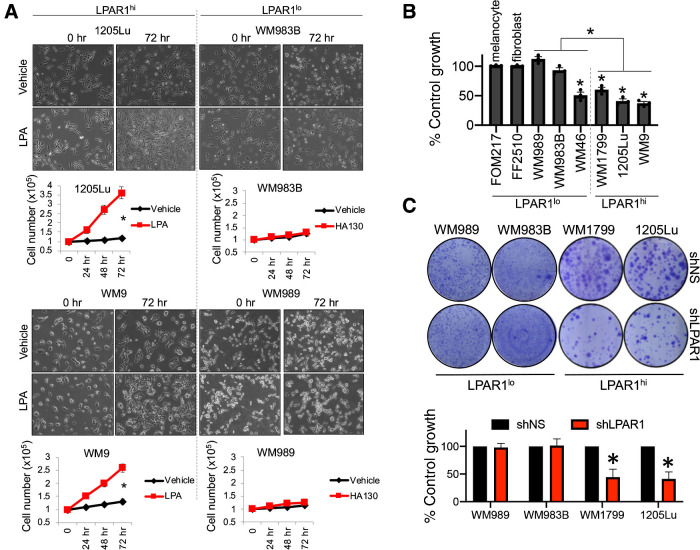
A subset of three BRAF-mutant and three NRAS-mutant melanoma cell lines with the highest levels of LPAR1 (hereafter called LPAR1hi cells) and a subset of three BRAF-mutant and three NRAS-mutant melanoma cell lines with the lowest levels of LPAR1 (hereafter called LPAR1lo cells) were chosen to perform additional analyses to understand the functional role of LPAR1 in melanoma. We found that ectopic addition of lysophosphatidic acid (LPA), the activating ligand of LPAR1, increases the proliferation of a subset of melanoma cell lines across different genotypes (Fig. 2A; Supplementary Fig. S2A), which was positively correlated to LPAR1 expression (Fig. 1E; ref. 28). Notably, LPAR1 expression correlated with sensitivity to genetic silencing of LPAR1, whereby BRAF-mutant melanoma cell lines with high LPAR1 expression (LPAR1hi) display significant inhibition of proliferation following siLPAR1 (Fig. 2B; Supplementary Fig. S2B). In contrast, BRAF-mutant melanoma cells or normal cells (i.e., melanocytes, fibroblasts) with low LPAR1 expression (LPAR1lo) experience reduced or no cytotoxicity, with the exception of the WM46 cell line. Loss of LPAR1 with short hairpin (shRNA) also blunted the long-term colony forming capacity of LPAR1hi melanoma cells, in contrast to LPAR1lo melanoma cells (Fig. 2C; Supplementary Fig. S2D). The positive relationship between LPAR1 expression and genetic silencing of LPAR1-induced inhibition of proliferation was also observed in NRAS-mutant melanoma cells (Supplementary Fig. S2C). In accordance, genetic silencing of LPAR1 induces NCLSC cytotoxicity (Supplementary Fig. S2E). Collectively, these data demonstrate the importance of LPAR1 for NCLSCs and LPAR1hi melanoma cells independent of genotype.
Figure 2.
LPAR1 promotes melanoma and NCLSC survival. A, 1205Lu, WM9, WM989, and WM983B cells were seeded in 6-well plates, serum starved for 24 hours, and then treated with LPA (10 μmol/L, 0–72 hours). Cell numbers were quantified for LPA or vehicle-treated cells (n = 3). Student t test; *, P < 0.05. Error bars, SD. B, Melanoma cells, melanocytes, and fibroblasts were transfected with control siNS or siLPAR1 and incubated for 96 hours, followed by the cell proliferation MTT assay. n = 3; *, P < 0.05, all statistics compared proliferation between siNS or siLPAR1 for each respective cell line. C, Melanoma cells were infected with lentiviral constructs expressing control shNS or shLPAR1 and grown for 3 weeks. Cells were stained with crystal violet; n = 3.
LPAR1 drives melanoma migration and invasion
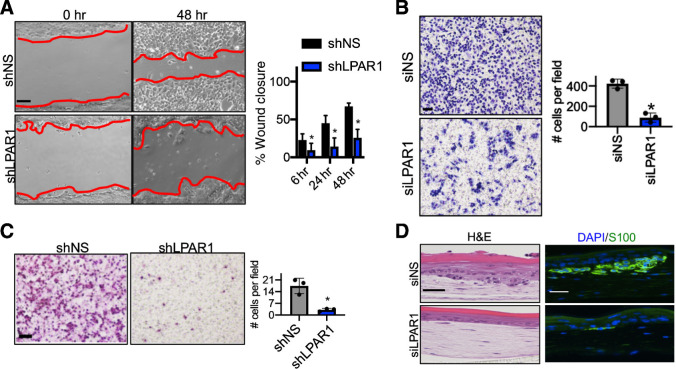
We next interrogated the role of LPAR1 in melanoma invasiveness. LPAR1 has been reported to promote metastasis of breast and ovarian cancer (29, 30). In wound healing and transwell migration assays, genetic inhibition of LPAR1 reduced wound closure and migratory capacity of LPAR1hi melanoma cells grown in complete media, respectively (Fig. 3A and B). In Boyden chamber assays, genetic (shLPAR1) inhibition of LPAR1 also reduced melanoma invasiveness (Fig. 3C). To better recapitulate the normal physiology of the skin, we established human skin reconstructs including LPAR1hi melanoma cells transfected with siLPAR1, which decreased melanoma proliferation and invasion (Fig. 3D; Supplementary Fig. S3B; ref. 31). To ensure our observations on melanoma invasion were independent of the impact of LPAR1 inhibition on melanoma viability, we performed extracellular matrix degradation assays demonstrating viable melanoma cells expressing shLPAR1 have reduced ability to degrade gelatin (Supplementary Fig. S3A). Taken together, these results suggest that LPAR1 signaling plays an important role in promoting melanoma migration and invasion.
Figure 3.
LPAR1 drives melanoma motility and invasiveness. A, A wound healing assay was performed in WM9 cells with the knockdown of LPAR1. Red lines indicate the leading edges of migrating cells. Scale bar, 50 μm. The percentage of wound closure after different time periods of migration was calculated and is shown. Error bars, SD; n = 3; *, P < 0.05. B, Transwell migration assay (without Matrigel coating) was performed for WM9 cells infected with luciferase shRNA (shLuc) or shLPAR1. Scale bar, 100 μm. Right, the number of migrated cells were quantified. Error bars, SD; n = 3; *, P < 0.05. C,Transwell invasion assay (with Matrigel coating) was performed in 1205Lu cells infected with shLuc or shLPAR1. Scale bar, 100 μm. The numbers of invaded cells were quantified. Error bars, SD; n = 3; *, P < 0.05. D, 1205Lu cells transfected with siNS or siLPAR1 were used to make skin reconstructs. S100, green; DAPI, blue. Scale bar for hematoxylin and eosin (H&E) staining, 50 μm. Scale bar for immunofluorescence staining, 20 μm.
LPA regulates LPAR1 signaling through a RAPGEF5-RAP1A-axis
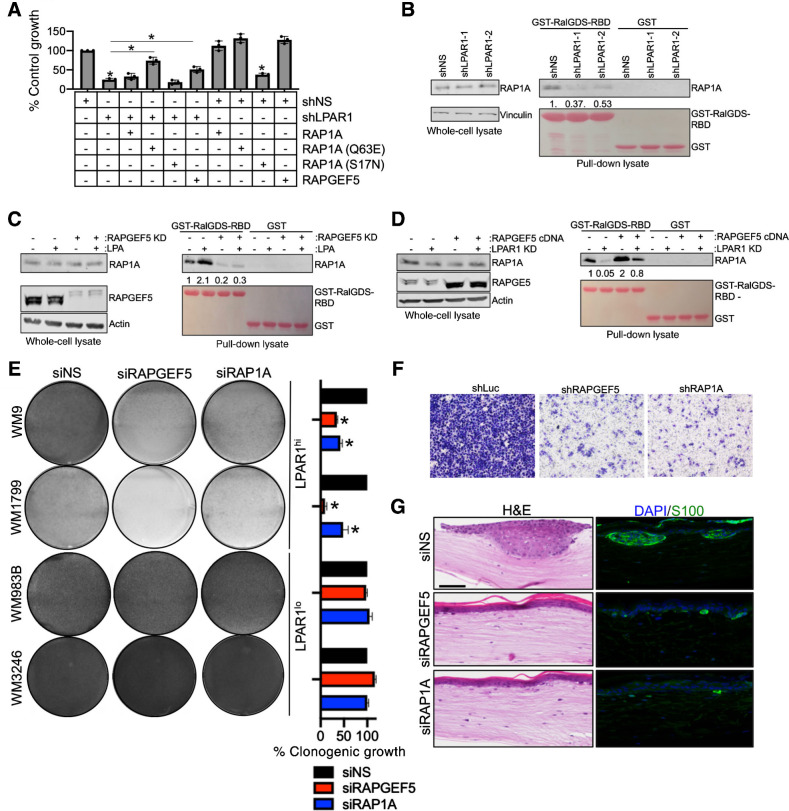
We next dissected the requisite molecules for LPAR1 signal transduction using BRAF-MT melanoma cells. RAP-GTPases have been reported to be regulated by LPAR receptors (32, 33). Two members of the RAS subfamily of GTPases (RAP1A and RAPGEF5) emerged from our targeted siRNA screen (Fig. 1). In accordance, overexpression of RAPGEF5 could rescue proliferation in melanoma cells expressing shLPAR1 (Fig. 4A; Supplementary Fig. S4A). Furthermore, expression of a constitutively active RAP1A (Q63E) mutant rescues proliferation in shLPAR1 melanoma cells, whereas expression of the constitutively inactive RAP1A (S17N) mutant does not rescue proliferation. To more directly examine the role of LPAR1 on RAP1A activity, a GST pulldown assay was performed using GST-tagged RAP binding domain (RBD) of RalGDS (GST-RalGDS-RBD), a direct downstream effector of RAP1A that specifically binds to active RAP1A (34). RAP1A activity is suppressed in shLPAR1 melanoma cells, as evidenced by a decrease in GST-RalGDS-GBD levels in pulldown lysate (Fig. 4B). Activation of LPAR1 following ectopic LPA hyperactivates RAP1A, which is ablated by genetic silencing of RAPGEF5 (Fig. 4C). In concordance, overexpression of RAPGEF5 recovers RAP1A activity in shLPAR1 melanoma cells (Fig. 4D). Although acute genetic silencing of RAPGEF5 did not cause significant inhibition of cell growth (Fig. 1C), long-term genetic silencing of RAPGEF5 and RAP1A reduces colony formation only in LPAR1hi melanoma cells (Fig. 4E; Supplementary Fig. S4B). Genetic silencing of RAPGEF5 and RAP1A also ablates melanoma invasiveness (Fig. 4F) and decreases proliferation in human skin reconstructs (Fig. 4G; Supplementary Fig. S4C, S4D, S4E, and S4F). As LPAR1 is a G12/13-coupled receptor (35), we confirmed the importance of G12/13 in the transduction of LPAR1 survival signals in LPAR1hi melanoma cells (Supplementary Fig. S4G, S4H, and S4I). Indeed, silencing of G12/13 phenocopies the effect of genetic silencing of LPAR1 on proliferation. Proliferation could not be rescued following siG12/13 by ectopic LPA treatment demonstrating its role downstream of LPAR1. Collectively, these data demonstrate LPAR1 signal transduction locally transmits through a RAPGEF5-RAP1A-axis.
Figure 4.
LPA regulates LPAR1 signaling through a RAPGEF5-RAP1A-axis. A, MTT assay was performed in WM9 LPAR1 knockdown cells as well as in LPAR1 knockdown cells with the expression of RAP1A (Q63E, constitutively active mutant), RAP1A (S17N, inactive mutant), overexpression of RAP1A (wt) or RAPGEF5. n = 3; ANOVA with post hoc Holm-Sidak multiple comparisons test were applied. *, Holm-Sidak multiple comparisons Padjusted < 0.05; error bars, SD. B, GST-tagged RalGDS-RBD was conjugated on Glutathione Sepharose 4B and incubated with WM9 cells infected with shLuc, shLPAR1-1, or shLPAR1-2. C and D, GSTtagged RalGDS-RBD was conjugated on Glutathione Sepharose 4B and incubated with WM9 cells with the addition of LPA followed by the knockdown of RAPGEF5 (C) or the overexpression of RAPGEF5 followed by the knockdown of LPAR1 (D). E, Melanoma cell lines were transfected with siNS, siRAPGEF5, or siRAP1A and grown for 3 weeks in long-term colony formation assays. Cells were subsequently stained with crystal violet and imaged, n = 3. ANOVA with post hoc Holm-Sidak multiple comparisons test was applied. *, Holm-Sidak multiple comparisons Padjusted < 0.05; error bars, SD. F, Transwell invasion assay (with Matrigel coating) was performed in WM9 cell lines infected with shNS, shRAPGEF5, or shRAP1A. G, WM9 cells transfected with siNS, siRAPGEF5, or siRAP1A were used to make skin reconstructs. S100, green; DAPI, blue. Scale bar for hematoxylin and eosin (H&E) staining, 50 μm. Scale bar for immunofluorescence staining, 20 μm.
mTOR and YAP are downstream effectors of LPAR1-axis
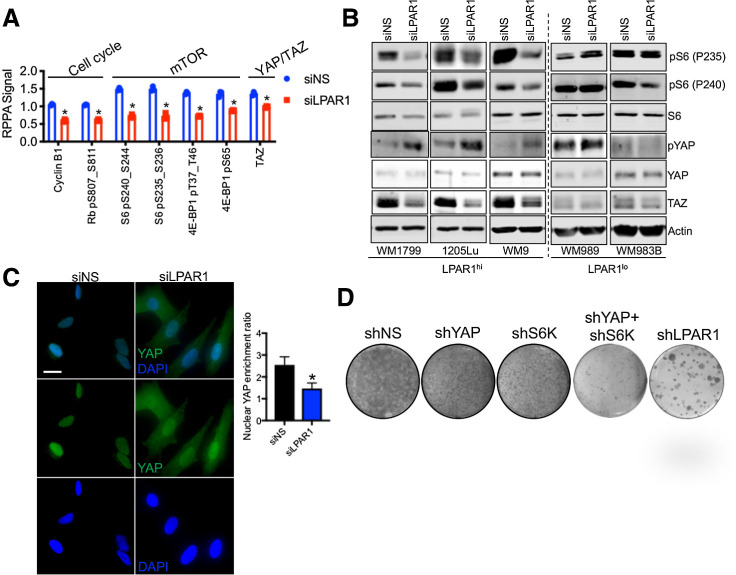
LPAR1 stimulation by LPA has been previously shown to induce downstream YAP1 activation (35) and activation of the PI3K-mTOR pathway (36). To gain further mechanistic insight into LPAR1 signal transduction in melanoma, we exploited a systems biology approach to investigate functional proteomic alterations following LPAR1 inhibition. In agreement, RPPA revealed a signature associated with inactivation of YAP (TAZ), as well as inhibition of mTOR (pS6, p4E-BP1) and cell-cycle progression (cyclin B1, pRb) following LPAR1 inhibition (Fig. 5A; Supplementary Table S4). Notably, genetic (siLPAR1) inhibition of LPAR1 inactivates mTOR and inhibits YAP/TAZ as validated by Western blotting specifically in LPAR1hi melanoma cells across genotypes (Fig. 5B; Supplementary Fig. S5A). Phosphorylation of YAP at S127 inactivates YAP by promoting its translocation from the nucleus to the cytoplasm (37). Genetic silencing of LPAR1 triggers cytoplasmic expression of YAP (Fig. 5C). Notably, genetic silencing of neither YAP or S6K could achieve equivalent suppression of long-term clonogenic growth to silencing of LPAR1 (Fig. 5D). However, concurrent silencing of S6K and YAP more completely blunted clonogenic growth only in LPAR1hi melanoma cells, suggesting LPAR1 promotes melanoma cell-cycle progression via both mTOR and YAP signaling (Supplementary Fig. S5B and S5C).
Figure 5.
mTOR and YAP are downstream effectors of the LPAR1-axis. A, WM9 cells were transfected with siNS or siLPAR1 for 48 hours. Protein lysate was analyzed by RPPA. Shown are the proteins most significantly altered. B, A panel of LPAR1lo and LPAR1hi melanoma cell lines was transfected with siNS or siLPAR1 for 48 hours. Protein lysate was immunoblotted to validate findings in A. C, Immunostaining of YAP in WM9 cells transfected with siNS or siLPAR1 using an anti-YAP antibody (green); nuclei were stained with DAPI (blue). Right, ratio of YAP localized to the nucleus was quantified. D, WM9 cells expressing shNS, shYAP, shS6K, shYAP+shS6K, or shLPAR1 were grown for 3 weeks in long-term colony formation assays and subsequently stained with crystal violet. An unpaired two-tailed t test was used for all studies.
LPAR1 promotes intrinsic and acquired resistance to targeted therapy
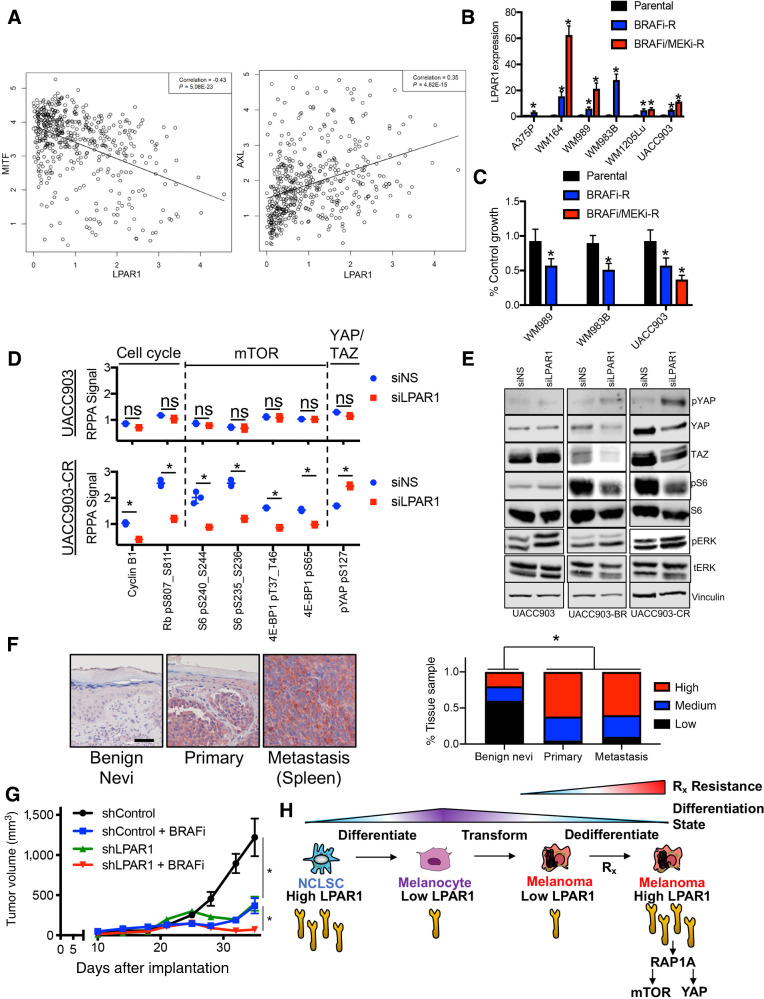
We next interrogated the role LPAR1 serves in the context of targeted therapy. Publicly available genome-wide expression data in The Cancer Genome Atlas (TCGA) revealed LPAR1hi melanoma cells express a negative correlation with a master lineage regulator in melanocytes, MITF, and a positive correlation with a dedifferentiated melanoma marker, AXL (Fig. 6A). To place LPAR1hi cells into context with other recently characterized therapy resistant melanoma subpopulations (i.e., NCSC, invasive, pigmented, stress-like cancer cell; refs. 9, 13, 38), we analyzed publicly available single-cell RNA-sequencing (scRNA-seq) datasets that revealed LPAR1hi cells significantly correlate with a recently reported neural crest stem cell melanoma subpopulation that plays a role during minimal residual disease (Supplementary Fig. S6A; refs. 9, 14). In agreement, LPAR1hi cells possess intrinsic resistance to MAPK pathway inhibitors (MAPKi; Supplementary Fig. S6B, S6C, and S6D; ref. 39). LPAR1hi melanoma cell lines are less sensitive to BRAFi (PLX4720) and MEKi (PD0325901, AZD6244) therapy relative to LPAR1lo melanoma cells. No difference in β-catenin levels are observable between LPAR1hi and LPAR1lo cells (Supplementary Fig. S6E). Notably, LPAR1 expression increases in LPAR1lo melanoma cell lines (Fig. 6B) and patient-derived xenograft models (Supplementary Fig. S6F) once resistance to BRAFi or BRAFi/MEKi is acquired. LPAR1 is functional in acquired therapy resistance, as genetic (siLPAR1) targeting of LPAR1 impairs proliferation (Fig. 6C; Supplementary Fig. S6G) and blunts long-term colony formation Supplementary Fig. S6H) exclusively in melanoma cells with acquired BRAFi- or BRAFi/MEKi-resistance, whereas paired parental cells displayed little to no sensitivity. Furthermore, genetic silencing of RAGPEF or RAP1A phenocopied these effects in therapy resistant melanoma cells (Supplementary Fig. S6H). Genetic silencing of LPAR1 also increased the efficacy of BRAFi (Supplementary Fig. S6I). RPPA analyses revealed that melanoma cells with acquired BRAFi/MEKi-resistance display higher phosphorylation of Rb at S807 (associated with elevated cell-cycle progression) and higher mTOR activity (increased phosphorylation of S6 and 4E-BP1) relative to therapy-naïve parental cells, which has been reported to denote therapy refractory melanomas (ref. 40; Fig. 6D; Supplementary Table S5). Genetic silencing of LPAR1 significantly inhibited cell-cycle progression (decreased cyclin B1 and pRb S807), decreased mTOR activity (decreased pS6 and p4E-BP1) and blunted YAP activity (increased pYAP S127), which was validated by Western blotting (Fig. 6D and E). Notably, genetic (siLPAR1) inhibition of LPAR1 does not inhibit the MAPK pathway in melanoma cells. Taken together, these data reveal LPAR1 promotes intrinsic and acquired resistance to BRAFi and BRAFi/MEKi by sustaining mTOR and YAP activity.
Figure 6.
LPAR1 promotes intrinsic and acquired resistance to targeted therapy. A, A scatterplot was generated to examine the correlation between LPAR1 and MITF, as well as the correlation between LPAR1 and AXL using gene expression data in TCGA. A line of best fit is shown. B, Relative gene expression levels of LPAR1 were assessed by quantitative PCR in a panel of paired melanoma cell lines that were therapy naïve, or developed acquired resistance to BRAF inhibition or combination BRAF/MEK inhibition. n = 3; ANOVA with post hoc Holm-Sidak multiple comparisons test was applied. *, Holm-Sidak multiple comparisons Padjusted < 0.05. C, Melanoma cells was transfected with siNS or siLPAR1 for 96 hours, followed by the cell proliferation MTT assay, n = 3. ANOVA with post hoc Holm-Sidak multiple comparisons test was applied. *, Holm-Sidak multiple comparisons Padjusted < 0.05. D, UACC903 and UACC903CR cells were transfected with siNS or siLPAR1 for 48 hours. Lysate was analyzed by RPPA and shown are the proteins most significantly altered in UACC903CR (LPAR1hi) versus parental UACC903 (LPAR1lo) cells. E, Western blotting validation of lysate from UACC903, UACC903BR, and UACC903CR cells treated as in D. F, IHC for LPAR1 staining in patient-derived tissue. Quantification of 61 tissues is shown to the right. Red, LPAR1 staining. Fisher exact test was used to compare percentage of tissue samples by the LPAR1 staining level. G, Tumor volumes from WM9 xenografts infected with lentiviral constructs expressing shCon or shLPAR1 were treated with vehicle control or PLX4032 (25 mg/kg/day, chow). *, P < 0.05. n = 10 mice/arm. The tumors in the shLPAR1 + BRAFi arm are statistically smaller than all other arms. The tumors in the shControl + BRAFi arm and the shLPAR1 arm are both statistically smaller than the tumors in the shControl arm. H, Schematic illustration of LPAR1 expression fluctuation with differentiation status of melanoma cells. Under targeted therapy, LPAR1lo cells undergo dedifferentiation to a LPAR1hi, NCSC-like state that is resistant to targeted therapy via elevated LPAR1→RAPGEF5→RAP1A→mTOR/YAP activity. An unpaired two-tailed t test was used for two group comparisons, unless otherwise stated.
We next investigated the in vivo relevance of LPAR1 in tumorigenesis. Staining of a large set patient tissue derived from benign nevi, primary melanoma and melanoma metastases revealed a striking increase in LPAR1 expression in primary and metastatic melanoma tumor tissue from patients versus benign nevi (Fig. 6F). To characterize the in vivo antitumor activity of targeting LPAR1, melanoma xenografts were established in the flanks of NSG mice using the WM9 cell line expressing shLuc or shLPAR1 (Fig. 6G; Supplementary Fig. S6J). Genetic inhibition of LPAR1 had a significant effect on tumor growth, mTOR signaling and YAP activity relative to control tumors (Supplementary Fig. S6K). Notably, the antitumor BRAFi efficacy is significantly increased in tumor cells expressing shLPAR1 relative to control tumors (Fig. 6G). Altogether, these data suggest the stem cell–like LPAR1-axis is reduced in NCSLCs as they differentiate into melanocytes and increased in a subset of melanoma cultures that possess intrinsic resistance to therapy (Fig. 6H). Notably, LPAR1 expression is elevated in therapy-naïve cells following the acquisition of resistance to BRAFi and/or MEKi, representing a novel target to increase therapy efficacy.
Discussion
Tumor cells often hijack stem cell–like pathways active in their normal progenitor cells favoring tumor growth and survival (41, 42). Identifying and dissecting the stem cell–like pathways that are responsible for the maintenance of normal stem cells thus has the potential to help us better define tumor-specific changes and to discover novel therapeutic approaches. There are an increasing number of studies that show correlations between genes that regulate neural crest/melanocyte development and their contribution to melanoma tumorigenesis (4, 7–9, 43, 44). By integrating combinatorial gene knockout approaches, cell-based assays and immunohistochemical observations, recent studies have illustrated several genes and pathways including Wnt (45) and Sox (46) proteins that serve important roles in melanocyte specification and melanoma progression (47). The evidence that high WNT5A expression also correlates with a MITFlo/AXLhi signature and that YAP signaling serves as a downstream effector of WNT5A would suggest that LPAR1 and WNT5A may coregulate downstream YAP signaling through distinct yet parallel pathways (48). Furthermore, YAP has been shown to rescue RAS-mutant cancer cell viability following suppression of KRAS in KRAS-mutant colon cancer cells (49) and MEK inhibition in NRAS-mutant melanoma cells (50), with evidence that KRAS and YAP converge to regulate epithelial–mesenchymal transition and tumor survival.
Using zebrafish, a chemical screen recently identified small-molecule suppressors of the neural crest lineage that may have inhibitory effects on melanoma (51). However, a systematic investigation of human cells is still lacking to directly explore shared gene signatures between NCLSCs and melanoma cells for translational studies. In this study, we took advantage of our extensive collection of melanoma cell lines and normal human skin-derived melanocytes to compare them with human skin-derived NCLSCs that behave similarly to embryonic neural crest cells (12). Using a systems biology approach in which computational analyses were coupled with targeted siRNA screens, we identified a panel of genes upregulated both in NCLSC and melanoma cells, but not in melanocytes. To our knowledge, this is the first study where the common gene signature shared between NCLSCs and melanoma cells was subjected to computational prediction, biological assays and functional characterization.
We here identify that LPAR1 is upregulated in NCLSC and melanoma cells relative to melanocytes. LPAR1 is highly expressed in regions of the central nervous system during embryonic neurogenesis and serves a role in stem cell processes (52). LPA signaling functions in nervous system processes, such as the mobilization of intracellular calcium, changes in neuroblast and neuron conductance, and survival and migration of Schwann cells (53). LPA gradients have previously been shown to be a major driver of melanoma chemotaxis (54, 55). To our knowledge, no previous connection between LPA or LPAR1 and targeted therapy (i.e., BRAFi/MEKi resistance) has been made in the melanoma field. In this study, we observe inhibition of LPAR1 signaling disrupts the acquisition of resistance. Furthermore, LPAR1 drives the formation of stem cell spheres. Collectively, these data suggest that LPA signaling plays a vital role in cells with stem cell–like properties.
RAP small GTPases are one group of downstream effectors of LPA signaling that can be activated via G proteins or specific GDP/GTP exchange factors (GEF; ref. 56). We identified RAPGEF5 and RAP1A as critical for the local signal transduction of LPAR1. Through functional rescue experiments and GST pulldown assays, we discovered that RAP1A is a direct downstream effector of LPAR1 signaling that is regulated through RAPGEF5 by LPAR1. Collectively, these results underscore a core-signaling network comprised of LPAR1, RAPGEF5, and RAP1A in melanoma. LPAR1 is a widely expressed LPAR receptor that is upregulated in advanced cancers including ovarian and breast cancer, and has been implicated in promoting aggressiveness and metastasis in these tumors (29, 36). Our work investigates the functions of LPAR1 in melanoma and demonstrates that LPAR1-RAPGEF5-RAP1A signaling is required for the growth, viability, and invasiveness of LPAR1hi melanoma cells, independent of genotype.
LPA signaling has been previously reported to regulate cell survival signaling through two main pathways; the PI3K/AKT (57) and the RAS/MAPK pathways (58). Interestingly, we did not observe any significant alterations of pERK or pAKT levels upon genetic inhibition of LPAR1 signaling. These findings indicate that LPAR1 signaling does not contribute to human melanoma cell growth through the canonical MAPK or PI3K/AKT pathways, but rather via alternative downstream effectors. Strikingly, we find LPAR1 inhibition results in the potent and concurrent inactivation of YAP and mTOR signaling in melanoma, in agreement with previous reports (35). LPA can stimulate the nuclear accumulation of YAP, resulting in YAP activation (35); however, its role in mTOR is novel in melanoma. mTOR (59) and YAP (60) have both been implicated as resistance mechanisms against MAPKis in melanoma. YAP downregulation has been shown to sensitize BRAFi-resistant melanoma cells to MAPK pathway inhibitors, and YAP and Wnt signaling (via β-catenin-LEF1) coregulate apoptosis of acquired MAPKi-resistant melanoma (61). LPAR1 inhibition inhibits melanoma growth by loss of both mTOR and YAP activity as shown by cells expressing either shS6K or shYAP not able to achieve the equivalent level of growth suppression relative to shLPAR1. Concurrently silencing both S6K and YAP could achieve equivalent growth suppression to shLPAR1, notably only in LPAR1hi melanoma cells. These data suggest LPAR1 expression could serve as a biomarker for melanoma cells that heavily rely on mTOR and YAP activity.
Collectively, our studies establish the molecular basis of how the LPAR1-RAPGEF5-RAP1A signaling axis underlies both intrinsic and acquired drug resistance to targeted therapies for BRAFV600E/K melanoma cells. We also provide a rationale for combining inhibitors of LPAR1 signaling with FDA-approved MAPKis to trigger synthetic lethality both in innate resistant and in acquired resistant cells. Our data warrant further investigation of prognostic biomarkers of LPAR1 signaling that could predict success in combining LPAR1 inhibitors and targeted therapies to overcome both intrinsic and acquired therapy resistance by expanding in vitro and in vivo models. To date, there have been a number of melanoma subpopulations characterized to drive therapy resistance including NCSC-like (6), invasive (9), pigmented and a recently identified stress subpopulation that all exhibit transient yet detectable levels of differentiation/dedifferentiation (13). We believe our LPAR1 studies yield an actionable vulnerability necessary for dedifferentiated NCSC-like melanoma cells to persist and drive therapy resistance. In summary, these data provide the scientific rationale to clinically explore LPAR1 as a novel therapeutic target for therapy-naïve patients as well as in patients with melanomas that are intrinsically resistant or have developed acquired resistant to targeted therapies. As LPAR1 is important for the maintenance and growth of stem cells, it will be interesting to further examine the link between LPAR1 signaling and melanoma stem-like cells. Future studies of the functions of LPAR1 signaling in other types of cancers and its potential role in response to immunotherapy are needed.
Authors' Disclosures
M. Fukunaga-Kalabis reports grants from NIH, other support from Pfizer, and other support from Merck outside the submitted work. C. Krepler reports other support from Merck&Co outside the submitted work. D.M. Hristova reports grants from NIH, grants from Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and grants from Melanoma Research Foundation during the conduct of the study. G.B. Mills reports grants, personal fees, nonfinancial support, and other support from Amphista, AstraZeneca, Chrysallis Biotechnology, GlaxoSmithKline, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, Zentalis Pharmaceuticals; HRD assay to Myriad Genetics DSP to Nanostring; Adelson Medical Research Foundation, Breast Cancer Research Foundation, Komen Research Foundation, Ovarian Cancer Research Foundation, Prospect Creek Foundation, Nanostring Center of Excellence, Ionis (provision of tool compounds), and Genentech during the conduct of the study. X. Xu reports grants from NIH during the conduct of the study; grants, personal fees, and other support from CureBiotech Inc. and other support from Exio Biosciences outside the submitted work. T.C. Mitchell reports grants and personal fees from Merck, grants and personal fees from Bristol Myers Squibb, and personal fees from OncoSec outside the submitted work. R.K. Amaravadi reports other support from Pinpoint Therapeutics, personal fees from Deciphera, personal fees from Sprint Biosciences, other support from Immunaccel, personal fees from Merck, grants from Novartis, and grants from Bristol Myers Squibb outside the submitted work. G.M. Boland reports grants from Takeda Oncology, grants from Olink Proteomics, grants from Palleon Pharmaceuticals, grants from InterVenn Biosciences, personal fees from Nektar Therapeutics, personal fees from Merck, and personal fees from Novartis outside the submitted work. D.W. Speicher reports being on the science advisory board MOBILion Inc. K.T. Flaherty reports personal fees from Loxo Oncology, personal fees from Clovis Oncology, personal fees from Strata Oncology, personal fees from Vivid Biosciences, personal fees from Checkmate Pharmaceuticals, personal fees from Kinnate Biopharma, personal fees from Scorpion Therapeutics, personal fees from X4 Pharmaceuticals, personal fees from PIC Therapeutics, personal fees from Apricity, personal fees from Oncoceutics, personal fees from Fog Pharma, personal fees from Tvardi, personal fees from xCures, personal fees from Monopteros, personal fees from Vibliome, grants and personal fees from Sanofi, personal fees from Amgen, personal fees from Asana Biosciences, personal fees from Adaptimmune, personal fees from Aeglea Biosciences, personal fees from Shattuck Labs, personal fees from Tolero Pharmacetuicals, personal fees from Neon Therapeutics, personal fees from Lilly, grants and personal fees from Novartis, personal fees from Genentech, personal fees from Bristol Myers Squibb, personal fees from Merck, personal fees from Takeda, personal fees from Verastem, personal fees from Boston Biomedical, personal fees from Pierre Fabre, personal fees from Debiopharm, and personal fees from ALX Oncology during the conduct of the study. Z.A. Ronai reports being the founder and scientific advisor of Pangea Therapeutics. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures
Supplementary Table
Supplementary Table
Supplementary Table
Supplementary Table
Supplementary Table
Acknowledgments
The authors thank all former and current lab members for comments and helpful discussions; J. Hayden and F. Keeney (Wistar Microscopy Facility), C. Chang, S. Billouin, and T. Nguyen (Wistar Genomics Facility), and J.S. Faust and D. Ambrose (Wistar Flow Cytometry Facility) for technical support. The authors apologize to those whose work was not cited or mentioned here due to space constraints. The research was supported by NIH grants R01 CA 182890, U54 CA224070, P01 CA114046, P01 CA025874, P30 CA010815, R01 CA047159, K01 CA245124, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Melanoma Research Foundation. The support for Shared Resources utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 and S10 OD023586 to the Wistar Institute.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
J. Liu: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. V.W. Rebecca: Conceptualization, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. A.V. Kossenkov: Investigation. T. Connelly: Investigation. Q. Liu: Formal analysis, investigation. A. Gutierrez: Investigation. M. Xiao: Investigation. L. Li: Investigation. G. Zhang: Investigation. A. Samarkina: Investigation. D. Zayasbazan: Investigation. J. Zhang: Investigation. C. Cheng: Investigation. Z. Wei: Investigation. G.M. Alicea: Investigation. M. Fukunaga-Kalabis: Investigation. C. Krepler: Investigation. P. Aza-Blanc: Investigation. C.-C. Yang: Investigation. B. Delvadia: Investigation. C. Tong: Investigation. Y. Huang: Investigation. M. Delvadia: Investigation. A.S. Morias: Investigation. K. Sproesser: Investigation. P. Brafford: Investigation. J.X. Wang: Investigation. M. Beqiri: Investigation. R. Somasundaram: Investigation. A. Vultur: Investigation. D.M. Hristova: Investigation. L.W. Wu: Investigation. Y. Lu: Investigation. G.B. Mills: Investigation. W. Xu: Investigation. G.C. Karakousis: Investigation. X. Xu: Investigation. L.M. Schuchter: Investigation. T.C. Mitchell: Investigation. R.K. Amaravadi: Investigation. L.N. Kwong: Investigation. D.T. Frederick: Investigation. G.M. Boland: Investigation. J.M. Salvino: Investigation. D.W. Speicher: Investigation. K.T. Flaherty: Investigation. Z.A. Ronai: Supervision, funding acquisition, investigation, project administration. M. Herlyn: Formal analysis, supervision, funding acquisition, investigation, project administration.
References
- 1. Carlino MS, Long GV, Schadendorf D, Robert C, Ribas A, Richtig E, et al. Outcomes by line of therapy and programmed death ligand 1 expression in patients with advanced melanoma treated with pembrolizumab or ipilimumab in KEYNOTE-006: a randomised clinical trial. Eur J Cancer 2018;101:236–43. [DOI] [PubMed] [Google Scholar]
- 2. Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2018;19:1315–27. [DOI] [PubMed] [Google Scholar]
- 3. Long GV, Eroglu Z, Infante J, Patel S, Daud A, Johnson DB, et al. Long-term outcomes in patients with BRAF V600-mutant metastatic melanoma who received dabrafenib combined with trametinib. J Clin Oncol 2018;36:667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013;23:811–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010;141:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin JR, Boswell SA, et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol 2017;13:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017;546:431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perego M, Maurer M, Wang JX, Shaffer S, Muller AC, Parapatics K, et al. A slow-cycling subpopulation of melanoma cells with highly invasive properties. Oncogene 2018;37:302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, et al. Toward minimal residual disease-directed therapy in melanoma. Cell 2018;174:843–55. [DOI] [PubMed] [Google Scholar]
- 10. Dupin E, Calloni G, Real C, Goncalves-Trentin A, Le Douarin NM. Neural crest progenitors and stem cells. C R Biol 2007;330:521–9. [DOI] [PubMed] [Google Scholar]
- 11. Cichorek M, Wachulska M, Stasiewicz A, Tyminska A. Skin melanocytes: biology and development. Postepy Dermatol Alergol 2013;30:30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li L, Fukunaga-Kalabis M, Yu H, Xu X, Kong J, Lee JT, et al. Human dermal stem cells differentiate into functional epidermal melanocytes. J Cell Sci 2010;123:853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baron M, Tagore M, Hunter MV, Kim IS, Moncada R, Yan Y, et al. The stress-like cancer cell state is a consistent component of tumorigenesis. Cell Syst 2020;11:536–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd TD, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alonso-Curbelo D, Riveiro-Falkenbach E, Perez-Guijarro E, Cifdaloz M, Karras P, Osterloh L, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell 2014;26:61–76. [DOI] [PubMed] [Google Scholar]
- 17. Boshuizen J, Vredevoogd DW, Krijgsman O, Ligtenberg MA, Blankenstein S, de Bruijn B, et al. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat Commun 2020;11:3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, et al. Targeting notch signalling pathway of cancer stem cells. Stem Cell Investig 2018;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H, Li J, Cheng J, Chen X, Zhou L, Li Z. AML-derived mesenchymal stem cells upregulate CTGF expression through the BMP pathway and induce K562-ADM fusiform transformation and chemoresistance. Oncol Rep 2019;42:1035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finger EC, Cheng CF, Williams TR, Rankin EB, Bedogni B, Tachiki L, et al. CTGF is a therapeutic target for metastatic melanoma. Oncogene 2014;33:1093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep 2013;3:1252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKenzie JA, Liu T, Jung JY, Jones BB, Ekiz HA, Welm AL, et al. Survivin promotion of melanoma metastasis requires upregulation of alpha5 integrin. Carcinogenesis 2013;34:2137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marie KL, Sassano A, Yang HH, Michalowski AM, Michael HT, Guo T, et al. Melanoblast transcriptome analysis reveals pathways promoting melanoma metastasis. Nat Commun 2020;11:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park H, Kim S, Rhee J, Kim HJ, Han JS, Nah SY, et al. Synaptic enhancement induced by gintonin via lysophosphatidic acid receptor activation in central synapses. J Neurophysiol 2015;113:1493–500. [DOI] [PubMed] [Google Scholar]
- 25. Marshall JC, Collins JW, Nakayama J, Horak CE, Liewehr DJ, Steinberg SM, et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. J Natl Cancer Inst 2012;104:1306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magkrioti C, Oikonomou N, Kaffe E, Mouratis MA, Xylourgidis N, Barbayianni I, et al. The autotaxin-lysophosphatidic acid axis promotes lung carcinogenesis. Cancer Res 2018;78:3634–44. [DOI] [PubMed] [Google Scholar]
- 27. Zhang G, Cheng Y, Zhang Q, Li X, Zhou J, Wang J, et al. ATXLPA axis facilitates estrogeninduced endometrial cancer cell proliferation via MAPK/ERK signaling pathway. Mol Med Rep 2018;17:4245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat 2010;91:130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sahay D, Leblanc R, Grunewald TG, Ambatipudi S, Ribeiro J, Clezardin P, et al. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget 2015;6:20604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo EJ, Kwon YW, Jang IH, Kim DK, Lee SI, Choi EJ, et al. Autotaxin regulates maintenance of ovarian cancer stem cells through lysophosphatidic acid-mediated autocrine mechanism. Stem Cells 2016;34:551–64. [DOI] [PubMed] [Google Scholar]
- 31. Li L, Fukunaga-Kalabis M, Herlyn M. The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J Vis Exp 2011;(54):2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelley GG, Reks SE, Smrcka AV. Hormonal regulation of phospholipase Cepsilon through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem J 2004;378:129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y, Kim JG, Kim HJ, Moon MY, Lee JY, Kim J, et al. Small GTPases Rap1 and RhoA regulate superoxide formation by Rac1 GTPases activation during the phagocytosis of IgG-opsonized zymosans in macrophages. Free Radic Biol Med 2012;52:1796–805. [DOI] [PubMed] [Google Scholar]
- 34. Albright CF, Giddings BW, Liu J, Vito M, Weinberg RA. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J 1993;12:339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012;150:780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cui R, Cao G, Bai H, Zhang Z. LPAR1 regulates the development of intratumoral heterogeneity in ovarian serous cystadenocarcinoma by activating the PI3K/AKT signaling pathway. Cancer Cell Int 2019;19:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee Y, Kim NH, Cho ES, Yang JH, Cha YH, Kang HE, et al. Dishevelled has a YAP nuclear export function in a tumor suppressor context-dependent manner. Nat Commun 2018;9:2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun 2015;6:6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 2015;520:368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X, Han H, Liu GP, Ma YX, Pan RL, Sang LJ, et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J 2017;36:3325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nicolas FJ, Hill CS. Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene 2003;22:3698–711. [DOI] [PubMed] [Google Scholar]
- 43. Liu X, Zhang SM, McGeary MK, Krykbaeva I, Lai L, Jansen DJ, et al. KDM5B promotes drug resistance by regulating melanoma-propagating cell subpopulations. Mol Cancer Ther 2019;18:706–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 2018;362:1055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ndoye A, Budina-Kolomets A, Kugel CH 3rd, Webster MR, Kaur A, Behera R, et al. ATG5 mediates a positive feedback loop between wnt signaling and autophagy in melanoma. Cancer Res 2017;77:5873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim Y, Yeon M, Jeoung D. DDX53 regulates cancer stem cell-like properties by binding to SOX-2. Mol Cells 2017;40:322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell 2008;3:242–52. [DOI] [PubMed] [Google Scholar]
- 48. Webster MR, Fane ME, Alicea GM, Basu S, Kossenkov AV, Marino GE, et al. Paradoxical role for wild-type p53 in driving therapy resistance in melanoma. Mol Cell 2020;77:633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 2014;158:171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 2015;47:250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 2011;471:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hecht JH, Weiner JA, Post SR, Chun J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol 1996;135:1071–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci U S A 2000;97:13384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muinonen-Martin AJ, Susanto O, Zhang Q, Smethurst E, Faller WJ, Veltman DM, et al. Melanoma cells break down LPA to establish local gradients that drive chemotactic dispersal. PLoS Biol 2014;12:e1001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Susanto O, Koh YWH, Morrice N, Tumanov S, Thomason PA, Nielson M, et al. LPP3 mediates self-generation of chemotactic LPA gradients by melanoma cells. J Cell Sci 2017;130:3455–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol 2011;21:615–23. [DOI] [PubMed] [Google Scholar]
- 57. Hawes BE, Luttrell LM, van Biesen T, Lefkowitz RJ. Phosphatidylinositol 3-kinase is an early intermediate in the G beta gamma-mediated mitogen-activated protein kinase signaling pathway. J Biol Chem 1996;271:12133–6. [DOI] [PubMed] [Google Scholar]
- 58. van Biesen T, Hawes BE, Luttrell DK, Krueger KM, Touhara K, Porfiri E, et al. Receptor-tyrosine-kinase- and G beta gamma-mediated MAP kinase activation by a common signalling pathway. Nature 1995;376:781–4. [DOI] [PubMed] [Google Scholar]
- 59. Gopal YN, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, et al. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1alpha and oxidative phosphorylation in melanoma. Cancer Res 2014;74:7037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fisher ML, Grun D, Adhikary G, Xu W, Eckert RL. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget 2017;8:110257–72. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 61. Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 2015;162:1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table
Supplementary Table
Supplementary Table
Supplementary Table
Supplementary Table