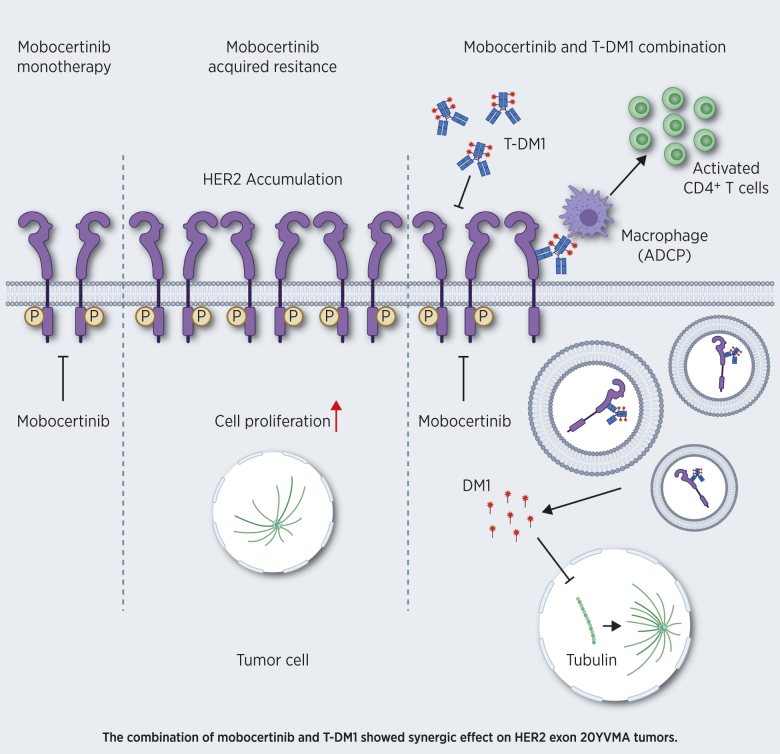
This study elucidates the potent inhibitory activity of mobocertinib against HER2 exon 20 insertion–mutant lung cancer and the synergic effect of combined mobocertinib and T-DM1, providing a strong rationale for clinical investigation.
Abstract
No targeted treatments are currently approved for HER2 exon 20 insertion–mutant lung adenocarcinoma patients. Mobocertinib (TAK-788) is a potent irreversible tyrosine kinase inhibitor (TKI) designed to target human epidermal growth factor receptor 2 (HER2/ERBB2) exon 20 insertion mutations. However, the function of mobocertinib on HER2 exon 20 insertion–mutant lung cancer is still unclear. Here we conducted systematic characterization of preclinical models to understand the activity profile of mobocertinib against HER2 exon 20 insertions. In HER2 exon 20 insertion–mutant cell lines, the IC50 of mobocertinib was higher than poziotinib and comparable with or slightly lower than afatinib, neratinib, and pyrotinib. Mobocertinib had the lowest HER2 exon 20 insertion IC50/wild-type (WT) EGFR IC50 ratio, indicating that mobocertinib displayed the best selectivity profile in these models. Also, mobocertinib showed strong inhibitory activity in HER2 exon 20YVMA allograft and patient-derived xenograft models. In genetically engineered mouse models, HER2 exon 20G776>VC lung tumors exhibited a sustained complete response to mobocertinib, whereas HER2 exon 20YVMA tumors showed only partial and transient response. Combined treatment with a second antibody–drug conjugate (ADC) against HER2, ado-trastuzumab emtansine (T-DM1), synergized with mobocertinib in HER2 exon 20YVMA tumors. In addition to the tumor cell autonomous effect, sustained tumor growth control derived from M1 macrophage infiltration and CD4+ T-cell activation. These findings support the ongoing clinical development of mobocertinib (NCT02716116) and provide a rationale for future clinical evaluation of T-DM1 combinational therapy in HER2 exon 20YVMA insertion–mutant lung adenocarcinoma patients.
Significance:
This study elucidates the potent inhibitory activity of mobocertinib against HER2 exon 20 insertion–mutant lung cancer and the synergic effect of combined mobocertinib and T-DM1, providing a strong rationale for clinical investigation.
Graphical Abstract
Introduction
Lung cancer remains a major threat to public health, with an estimated 230,000 new cases and 140,000 deaths in the United States in 2020 (1). Non–small cell lung cancer (NSCLC) represents the major subtype of lung cancer, with adenocarcinoma the predominant histology. In the past few decades, several oncogenes have been characterized as driver mutations in NSCLC (2). Subsequent development of pharmacologic agents targeting these mutations, such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), has led to tremendous clinical benefits for patients (3).
Despite these advances, many driver mutations still do not have targeted agents, indicating the need for novel therapeutic strategies. Among these are alterations in human epidermal growth factor receptor 2 (HER2/ERBB2), an essential gene harbored by 2% to 3% of lung adenocarcinoma (4–8). Most of these mutations occur in the exon 20 as in-frame insertion of with the duplication of amino acids YVMA being the most prevalent (77%–100%; refs. 4–7). Considering the large number of lung cancer patients (estimated 230,000 new cases in 2020), it is extremely important to investigate potential treatment strategies for HER2-mutant lung adenocarcinoma (1). Although several clinical trials have tested HER2-targeting tyrosine kinase inhibitors (TKI), such as poziotinib, afatinib, neratinib, and pyrotinib, only poziotinib have shown some clinical benefits in lung cancer and currently there are no approved therapies (9–12). Therefore, HER2 exon 20 insertion–mutant NSCLC represents an urgent unmet clinical need.
Mobocertinib is a potent EGFR and HER2 TKI currently under investigation against exon 20 insertions. In a phase I/II clinical trial, NSCLC patients with EGFR exon 20 insertions showed promising responses to mobocertinib with an objective response rate of 43% (13, 14). However, the activity of mobocertinib on HER2 exon 20 insertions is still unclear and requires further investigation.
We utilized systemic preclinical models to study how tumors harboring HER2 exon 20 insertions respond to mobocertinib. We also investigated acquired resistance mechanisms to mobocertinib and utilized combination treatments to overcome these mechanisms. Importantly, HER2 exon 20 YVMA insertion tumors showed a potent and sustained response to the combination of mobocertinib and ado-trastuzumab emtansine (T-DM1). In addition to tumor cell autonomous effect, the infiltration of M1 macrophages and the activation of CD4+ T cells are potential mechanisms involved in the combinational therapy.
Materials and Methods
Cell culture
H1781 and A431 cells were obtained from American Type Culture Collection, and HER2 exon 20 insertion–mutant Ba/F3 cells were generated using lentiviral vectors. All cell lines were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) and incubated in a humidified chamber with 5% CO2.
Cell proliferation assay
Cells were plated in 96-well dishes at a certain density (3–5 × 103/well). Compound was added to the cells in serial dilution in culture medium to achieve indicated concentration after 24 hours' incubation. Compounds at each dilution were evaluated in triplicate and cells treated with DMSO (1% final concentration) served as the untreated control. MTS-based cholecystokinin-8 (CCK-8; cat. #CK04, Dojindo) assay was used for the measurement and absorption at 450 nm was detected 2–4 hours after adding CCK-8 into the culture medium.
Western blot
Cells were resuspended with cold phosphate-buffered saline (PBS) to wash out the medium and then collected by centrifuging at 3,000 rpm, 4°C. Protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific; cat. #78440) was added into RIPA lysis buffer (Thermo Fisher Scientific; cat. #89900) used for the protein extraction and the concentration was measured by a BCA protein assay kit (Thermo Fisher Scientific; cat. #23225). After normalizing the protein concentration, loading buffer was added, and the samples were heated at 95°C for 7 minutes. All the samples were loaded to 4%–20% gradient gel SDS-PAGE with the same volume and the gel was transferred to a nitrocellulose membrane (Bio-Rad; cat. #1704271), which would be blocked in blocking buffer [LICOR; cat. #927-50003) for 1 hour at room temperature. Then, antibodies were added and incubated overnight at 4°C (Cell Signaling, HER2; cat. #2165S; phospho-HER2; cat. #2243S; AKT; cat. #4691L; phospho-AKT (Ser473); cat. #4060; Erk1/2; cat. #9102; phospho-ERK1/2; cat. #9101]. Antibodies were diluted into the indicated concentration according to the instructions with blocking buffer supplemented with 0.01% Tween 80. On the following day, TBST (TBS with 0.1% Tween 80) was used to wash the membrane 4 times (7 minutes/wash) to wipe out the antibodies and the membrane was incubated with the second antibody (LICOR, anti-Rabbit; cat. #925-32213; anti-Mouse; cat #925-68072) for 1 hour at room temperature. Finally, we used TBST to wash the membrane 3 times (7 minutes/time) and TBS 1 time (7 minutes/time) and imaged the membrane using the Odyssey Classic Infrared Imaging System.
Colony formation assay
Single-cell suspension was produced and plated in a 6-well cell culture dish (5,000 cells/well). Medium containing DMSO or mobocertinib was changed every 2 days. Cells were cultured for 11 days and fixed with 100% ethanol for 7 minutes. Crystal violet 0.5% (dissolved in 20% methanol) was used for staining and the cells were stained for 5 minutes. After washing, images were taken, and ImageJ was used for quantification.
Ba/F3 allograft experiment
Ba/F3 cells expressing HER2 exon 20YVMA insertion mutation were cultured in RPMI-1640 medium supplemented with 10% FBS and incubated at 37°C with 5% CO2. On the day of implantation, cells were harvested, resuspended in serum-free RPMI-1640, and a 100-μL cell suspension (107 cells) was implanted subcutaneously in the right flank of female severe combined immunodeficiency (SCID) mice. All mice were weighed prior to dosing and throughout the study once daily. The tumors were measured in 2 dimensions (length and width) at least twice per week with a caliper in millimeters. Tumor volume (mm3) was calculated with the following formula: tumor volume = L × W2 × 0.5.
Patient-derived xenografts experiment
The patient-derived xenograft (PDX) ST3107 (START) was derived from a primary NSCLC tumor bearing the HER2 exon20 insertion YVMA. ST3107 tumor fragments (5 × 5 × 5 mm) were implanted subcutaneously in the right flank of 7-week-old female athymic Nude, Outbred Homozygous mice (Jackson Laboratory); all experiments were conducted at START. When the mean tumor volume (MTV) MTV reached approximately 150–250 mm3, the animals were randomized into treatment groups and dosing was initiated on day 0 with mobocertinib or vehicle orally administered daily. Tumor size and body weight were measured twice weekly, and the MTV was calculated using the formula (0.5 × [length × width2]).
Mouse generation
The chicken β-actin (pGK) promoter, a loxP flanked STOP cassette, and human HER2 with exon 20 insertion sequences of G776>VC were inserted into the mouse collagen A1 locus. Sequence-verified targeting vectors were coelectroporated with an FLPe recombinase plasmid into C10 C57BL/6J embryonic stem cells (Mirimus). Then, transgene-positive embryonic stem clones were injected into C57BL/6 blastocysts, and the resulting chimaeras were mated with wild-type mice to determine germline transmission of G776>VC transgene. Upon Cre-mediated recombination, the STOP cassette was excised promoting expression of the mutant HER2 protein. The mouse gDNA was used as PCR template, and the hHER2ex20ins GVC sequence was confirmed with Sanger sequencing. The genotyping primers used are: HER2-forward: CAGATGCGGATCCTGAAAGAG and HER2-reverse: CCAGCCCGAAGTCTGTAATTT. The detailed strategy was previously described (15). All animal experiments, including breeding and treatment studies, were performed with approval of the NYU Langone Medical Center Institutional Animal Care and Use Committee.
Genetically engineered mouse model treatment studies
HER2 exon 20G776>VC mice were monitored by magnetic resonance imaging (MRI) for tumor development after intranasal induction with adeno-Cre (5 × 107 pfu). Tumor-bearing mice were dosed with mobocertinib [30 mg/kg, orally (p.o.), daily] and monitored by MRI every 2 weeks.
HER2 exon 20YVMA mice were fed a continuous doxycycline diet from 6 weeks of age. Mice were evaluated by MRI to quantify lung tumor burden before and after drug treatment. Mice with equal initial tumor volume were nonblindly randomized to the following groups: vehicle control, mobocertinib (30 mg/kg, p.o., daily), T-DM1 (10 mg/kg, tail vein, once every week), combination of mobocertinib (30 mg/kg, p.o., daily), and T-DM1 (10 mg/kg, tail vein, once every week), alisertib (20 mg/kg, p.o., daily), combination of mobocertinib (30 mg/kg, p.o., daily) and alisertib (20 mg/kg, p.o., daily), sapanisertib (0.3 mg/kg, p.o., daily), combination of mobocertinib (30 mg/kg, p.o., daily) and sapanisertib (0.3 mg/kg, p.o., daily). For macrophage-depletion experiments, Clodrosome was administered to mice via tail vein (intravenously) at 50 mg/kg. The first dosage was conducted 2 days before treatments with 200 μL, followed by 100 μL per mouse twice a week.
Progression-free survival (PFS) was analyzed based on the standard criteria in clinical trials. Briefly, PFS was the duration between treatment start and progression, which was defined by increase of tumor size compared with the previous scan of MRI or the appearance of new lesions.
MRI quantification
Mice were anesthetized with isoflurane to perform lung MRI using the BioSpec USR70/30 horizontal bore system (Bruker) to scan 24 consecutive sections. Tumor volume within the whole lung was quantified using 3-D slicer software to reconstruct MRI volumetric measurements, as described previously (16). Acquisition of the MRI signal was adapted according to cardiac and respiratory cycles to minimize motion effects during imaging.
IHC staining and multi-immunofluorescence
Mice were euthanized and lung tissues were collected and fixed with 10% formalin. IHC staining and multi-immunofluorescence (IF) were performed in iHisto Inc., using the following antibodies: TTF1 (Epitomics, 5883-1), p63 (Abcam, ab53039), phospho-HER2 (Tyr1221/1222; Cell Signaling Technology, #2243), HER2 (Cell Signaling Technology, #2165), phospho-ERK1/2 (Thr202/Tyr204; Cell Signaling Technology, #4370), and F4/80 (Cell Signaling Technology, #70076 for IHC and #30325 for IF). ImageJ was used for the quantifications of all IHC staining.
RNA-seq and data analyses
RNA-seq on vehicle and treated nodules was performed in NYU Langone Health Genome Technology Core. Reads were aligned to the reference genome using STAR (17). R (version 4.0.2) and DESeq2 package (version 1.28.1) were utilized for differential gene expression analysis (18).
Gene expression levels were quantified and estimated using RSEM (19). Gene set enrichment analysis (GSEA; version 4.1.0) and Hallmarker gene set were used for the GSEA based on transcripts per million (TPM). Pathways with P < 0.05 and false discovery rate (FDR) < 0.25 were considered significantly enriched.
Dot plots of enriched pathways, heat maps of genes, and volcano plots were generated using the Pheatmap, ggplot2, and EnhancedVolcano in R (version 4.0.2).
In vitro cell line flow cytometry
Cells were plated on tissue culture dishes. Indicated concentrations of compound were added into medium the next day. After 6 hours of treatment, cells were collected as cell suspension and filtered through 70-μm cell strainer (Thermo Fisher Scientific). Then, cells were stained with a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Molecular Probes) and resuspended in PBS supplemented with 2% FBS. Anti-human HER2 antibody (clone 24D2, BioLegend) was used to estimate the HER2 level on the cell surface. Cells were imaged on a BD Bioscience LSRFortessa and analyzed with FlowJo software.
Tumor-infiltrating immune cell isolation and flow cytometry analysis
Mice were euthanized, and lungs were perfused with sterile PBS through heart perfusion from the left ventricle after collection of bronchoalveolar lavage (BAL) fluid. Whole lung was minced and digested in collagenase D (11088866001, Roche) and DNase I (10104159001, Roche) in Hank's Balanced Salt Solution at 37°C for 30 minutes. After incubation, digested tissue was filtered through a 70-μm cell strainer (Thermo Fisher Scientific) to obtain single-cell suspensions. Separated cells were treated with 1 × RBC lysis buffer (BioLegend) to lyse red blood cells. Live cells were determined with a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Molecular Probes). Cell pellets were resuspended in PBS with 2% FBS for FACS analysis. Cells were stained with the indicated cell-surface markers and fixed/permeabilized using a Fixation/Permeabilization Kit (eBioscience). Cells were imaged on a BD Biosciences LSRFortessa and analyzed with FlowJo software. The gating strategy was described previously (20).
Flow antibodies
Isolated lung infiltrating immune cells were stained with fluorescently labeled anti-mouse antibody CD45 (clone 30-F11, BioLegend), CD3 (clone 17A2, BioLegend), CD49b (clone DX5, BioLegend), CD19 (clone 6D5, BioLegend), CTLA-4 (clone UC10-4B9, Invitrogen), CD69 (clone H1.2F3, BioLegend), TIM3 (clone RMT3-23, Invitrogen), FOXP3 (clone FJK-16s, eBioscience), PD-1 (clone 29F.1A12, BioLegend), KI-67 (clone 16A8, BioLegend), CD4 (clone GK1.5, BioLegend), CD8 (clone 53-6.7, BioLegend), H-2 (clone M1/42, BioLegend), Ly6G (clone 1A8, BioLegend), CD103 (clone 2E7, BioLegend), F4/80 (clone BM8, BioLegend), CD86 (clone GL-1, BioLegend), PD-L1 (clone 10F.9G2, BioLegend), Ly6C (clone HK1.4, BioLegend), CD206 (clone C068C2, BioLegend), CD11B (clone M1/70, BioLegend), CD11C (clone N418, BioLegend), CD4 (clone RM4-4, BioLegend), CD8 (clone 53-6.7, BioLegend), GZMB (clone GB11, BD Horizon), CD62 L (clone MEL-14, BioLegend), and CD44 (clone IM7, BioLegend).
Statistical analysis
Student t test (two-tailed) was utilized for statistical analysis in GraphPad Prism (Version 8), and Kaplan–Meier method was used for PFS under log-rank (Mantel–Cox) test. A P value of less than 0.05 was set as statistically significant, and standard error of the mean was used for the error bars.
Results
HER2 exon 20 insertion mutations are sensitive to mobocertinib
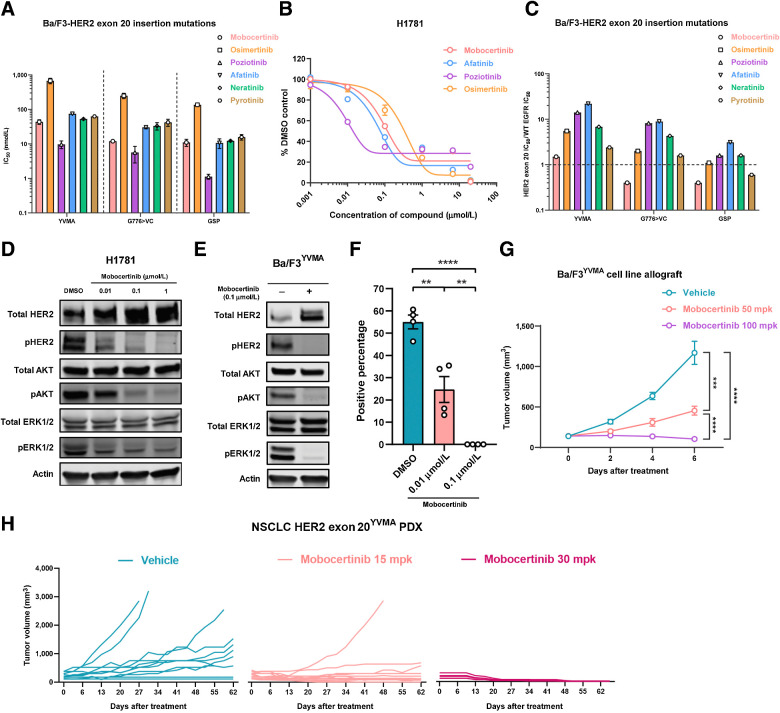
We first tested the ability of mobocertinib to inhibit HER2 exon 20 insertions in vitro. Cell lines harboring insertion mutations of HER2 exon 20, which are located in the tyrosine kinase domain were utilized for in vitro testing, including HER2 A775_G776insYVMA (YVMA), G776>VC, and P780_Y781insGSP (GSP). The Ba/F3 cell line is a commonly utilized system for investigating kinases signaling and targeted inhibitors in EGFR and HER2-mutant lung cancer (21, 22). Mobocertinib showed a higher half-maximal inhibitory concentration (IC50) than poziotinib, comparable with or slightly lower than afatinib, neratinib, or pyrotinib, and much lower than osimertinib on Ba/F3 HER2 exon 20 insertion–mutant cell lines (Fig. 1A). Of note, we observed that different types of HER2 exon 20 insertion mutations responded differently to mobocertinib. In our case, HER2 exon 20YVMA–mutant cell lines have higher IC50 than HER2 exon 20G776>VC and HER2 exon 20GSP cell lines (Fig. 1A). This indicated that HER2 exon 20 insertion mutations are sensitive and may respond distinctively to mobocertinib. In addition, we performed an IC50 experiment on the H1781 cell line, a human lung adenocarcinoma cell line with HER2G776>VC deletion insertion. Consistent with in Ba/F3 cell lines, mobocertinib has a moderate IC50 (0.12 μmol/L), which is less potent than poziotinib, similar to afatinib, and more effective than osimertinib in H1781 (Fig. 1B).
Figure 1.
HER2 exon 20 insertion mutations are sensitive to mobocertinib. A, IC50 of mobocertinib, osimertinib, neratinib, poziotinib, afatinib, and pyrotinib on Ba/F3-HER2 exon 20 insertion–mutant cell lines. B, IC50 of mobocertinib, poziotinib, osimertinib, and afatinib on H1781 (HER2G776>VC) cell line. C, The ratio of Ba/F3-HER2 exon 20 insertion–mutant IC50 to EGFR WT IC50. D, Western blot of HER2 signaling (total HER2, pHER2, total AKT, pAKT, total ERK, pERK, and actin) of H1781 (HER2 Exon 20G776>VC) cell line treated with 0.01 μmol/L, 0.1 μmol/L, and 1 μmol/L mobocertinib for 6 hours. E, Western blot of HER2 signaling (total HER2, pHER2, total AKT, pAKT, total ERK, pERK, and actin) of Ba/F3-HER2 exon 20YVMA cell line treated with 0.1 μmol/L of mobocertinib for 6 hours. F, Quantification of colony formation assay on H1781 (HER2 exon 20G776>VC) cell line treated with DMSO control, 0.01 μmol/L, and 0.1 μmol/L of mobocertinib. G, Tumor volumes of Ba/F3-HER2 exon 20YVMA allograft treated with vehicle, mobocertinib 50 and 100 mpk. H, Tumor volume change of NSCLC HER2 exon 20YVMA PDX treated with vehicle, 15 and 30 mpk mobocertinib. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we explored the effect of mobocertinib and other TKIs on WT EGFR, as its inhibition is the main contributor of clinical toxicity with skin rash and diarrhea observed. The WT EGFR-dependent A431 was used as the study model to investigate the WT EGFR inhibition. Compared with other TKIs in the class, mobocertinib has a moderate IC50 against WT EGFR and poziotinib has an extremely low IC50 against WT EGFR, which is almost 50 times lower than that of mobocertinib (Supplementary Fig. S1A). We used the ratio of the HER2 insertion mutation IC50 to WT EGFR IC50, which is able to demonstrate the extent of selectivity on mutant HER2 rather than WT EGFR. We found that mobocertinib had the lowest ratio of all three HER2 exon 20 insertion mutations compared with osimertinib, poziotinib, afatinib, neratinib, and pyrotinib, showing selective sensitivity on HER2 exon 20 insertion mutations (Fig. 1C). In contrast, poziotinib had relatively high ratios on all three HER2 exon 20 insertion–mutant cell lines, consistent with the reported toxicity in the clinics (23). Although mobocertinib presented a manageable safety profile, it was observed a higher incidence of gut toxicity rather than traditional skin rash reported with other TKIs.
HER2 signaling in H1781 and Ba/F3-HER2 exon 20YVMA-mutant cell lines was inhibited by mobocertinib at 0.1 μmol/L with significantly decreased phosphorylations of HER2, AKT, and ERK1/2 in a dose-dependent fashion (Fig. 1D and E). We performed a colony formation assay using the H1781 cell line, and found that mobocertinib significantly inhibited the growth of H1781 cells at 0.01 μmol/L (Fig. 1F; Supplementary Fig. S1B). To understand the inhibitory profile of mobocertinib, Ba/F3-HER2 exon 20YVMA cell lines were implanted subcutaneously in mice, and dosed with vehicle, 50 milligrams per kilogram (mpk), or 100 mpk of mobocertinib for 7 days. Mobocertinib (50 mpk) was able to inhibit the tumor, and a higher dosage (100 mpk), induced tumor regression (Fig. 1G). Although we observed body weight loss (Supplementary Fig. S1C) in the 100 mpk treated mice, this was manageable (less than 20%) and had no effect on their general health in the long-term monitoring.
Furthermore, we used the PDXs ST31 harboring HER2 exon 20YVMA. Mobocertinib showed robust antitumor efficacy (Fig. 1H) and was well tolerated. These data indicate that HER2 exon 20 insertion–mutant cell lines and subcutaneous models (exon 20YVMA) including PDX can be inhibited by mobocertinib.
HER2 exon 20G776>VC genetically engineered mouse model showed sustained response to mobocertinib
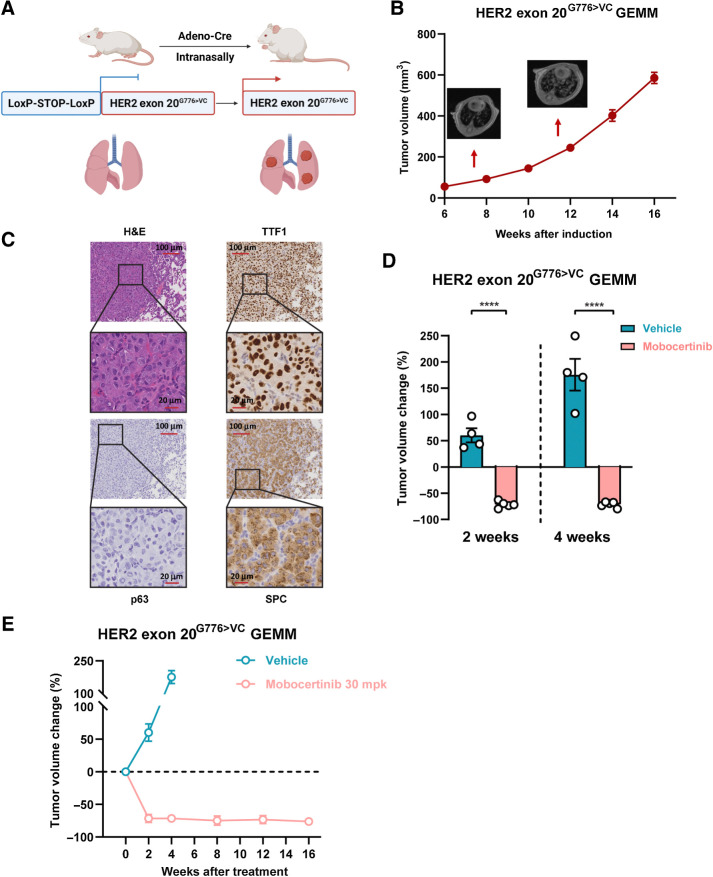
We next tested mobocertinib in orthotopic lung cancer genetically engineered mouse models (GEMM) to better evaluate cognate human malignancies with tissue-specific immunity. GEMM harboring human HER2 exon 20G776>VC mutation was generated using the LoxP-STOP-LoxP system for the first time. We intranasally delivered adeno-Cre virus into adult HER2 Exon 20G776>VC mice to initiate HER2 exon 20G776>VC expression specifically in murine lung epithelial cells (Fig. 2A). MRI showed that lung tumors appeared 8 weeks after viral delivery (Fig. 2B). According to IHC, HER2 exon 20G776>VC GEMM tumors were confirmed as adenocarcinoma histology with a strong positive staining of adenocarcinoma markers TTF1 and SPC, along with a negative squamous cell carcinoma marker p63 (Fig. 2C).
Figure 2.
HER2 exon 20G776>VC GEMM showed sustained response to mobocertinib. A, Working model of genetically engineered mice with HER2 exon 20G776>VC insertion mutation using the LoxP-STOP-LoxP system. B, Different time points of tumor volume quantification on MRI of HER2 exon 20G776>VC–mutant mice after induction. C, Representative image of hematoxylin and eosin (H&E) staining and immunochemistry of TTF1, p63, and SPC on HER2 exon 20G776>VC–mutant tumors. D, Dot plot of tumor volume changes on MRI from HER2 exon 20G776>VC insertion–mutant GEMMs treated with mobocertinib for 2 and 4 weeks. E, Tumor volume change for up to 16 weeks on MRI from HER2 exon 20G776>VC insertion–mutant GEMMs treated with mobocertinib. ****, P < 0.0001.
In the HER2 exon 20G776>VC–mutant GEMM model, mobocertinib dosed at 30 mpk demonstrated a rapid tumor regression after just 2 weeks of treatment with close to complete response after 4 weeks of treatment (Fig. 2D; Supplementary Fig. S2A). Sustained response was observed through up to 16 weeks of treatment monitoring (Fig. 2E). In addition, 3 days of daily mobocertinib treatment was sufficient to completely abolish HER2 downstream signaling as monitored by phosphorylated-ERK1/2 staining of HER2 exon 20G776>VC tumors (Supplementary Fig. S2B).
HER2 exon 20YVMA GEMM initially responded to mobocertinib but became acquired resistant upon continuous treatment
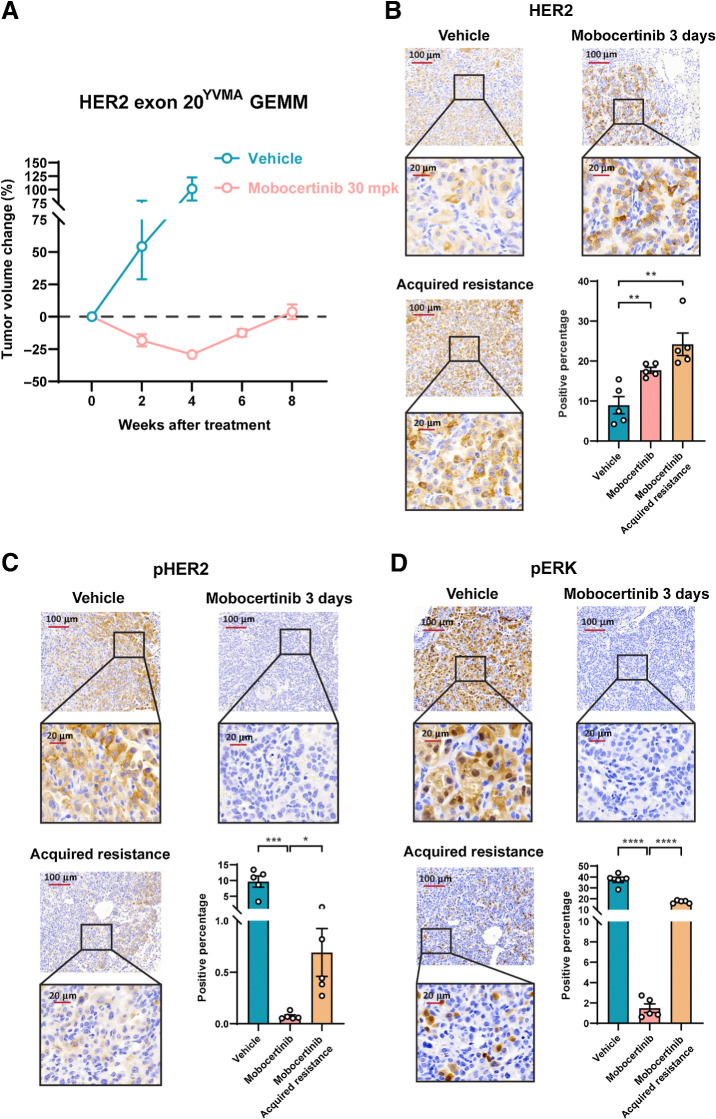
In addition to the HER2 exon 20G776>VC–mutant GEMM model, we used another mouse model with a HER2 exon 20YVMA mutation that was previously generated (24). We investigated the effects of mobocertinib on HER2 exon 20YVMA–insertion tumors. Partial response of about 30% tumor shrinkage was seen after 4 weeks of mobocertinib treatment (Fig. 3A; Supplementary Fig. S2C). IHC results confirmed that a 3-day treatment of mobocertinib effectively abolished p-HER2 and inhibited its major downstream signaling target pERK, whereas a significant increase of total HER2 was also seen (Fig. 3B–D), supporting the on-target efficacy of mobocertinib against HER2 exon 20YVMAin vivo. When we continued the treatment for 6 to 8 weeks, tumors then relapsed. We stained IHC on the relapsed tumors for total HER2, p-HER2, and pERK1/2. This showed significantly higher levels of pERK1/2 and p-HER2, along with an increase in total HER2 expression compared with treatment at 3 days (Fig. 3B–D).
Figure 3.
HER2 exon 20YVMA GEMM initially responded to mobocertinib but became acquired resistant upon continuous treatment. A, Tumor volume change on MRI from HER2 exon 20YVMA insertion–mutant GEMMs treated with mobocertinib. B, Representative immunochemistry image and quantification of HER2 protein on HER2 exon 20YVMA insertion–mutant GEMMs treated with vehicle or mobocertinib together with the acquired resistant tumors. C, Representative immunochemistry image and quantification of pHER2 protein on HER2 exon 20YVMA insertion–mutant GEMMs treated with vehicle or mobocertinib together with the acquired resistant tumors. D, Representative immunochemistry image and quantification of pERK protein on HER2 exon 20YVMA insertion–mutant GEMMs treated with vehicle or mobocertinib together with the acquired resistant tumors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
These data suggest that mobocertinib showed potent on-target efficacy in both HER2 exon 20G776>VC and HER2 exon 20YVMA GEMMs, but had differential outcomes after long-term treatment.
Pathways enriched in HER2 exon 20YVMA–acquired resistant tumors
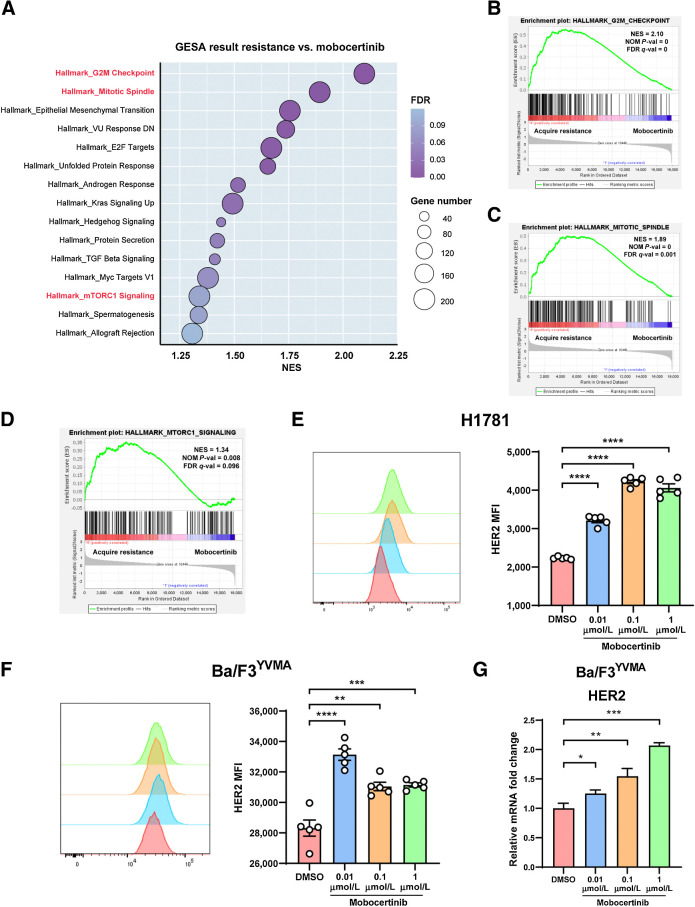
To compare mobocertinib-acquired resistance (continuous treatment and confirmed relapse by MRI) and early response (3 days of treatment) on HER2 exon 20YVMA tumors (three mice each), and probe pathways leading to acquired resistance, we performed bulk RNA-seq on tumor nodules from HER2 exon 20YVMA tumor–bearing mice. Using GSEA, we found that 15 pathways of the Hallmark gene set were significantly enriched in acquired resistant tumors as determined by cutoff value (P < 0.05 and FDR <0.25; Fig. 4A; Supplementary Table S1). Three of them enlightened us to the candidates for combinational therapies, which are “G2M Checkpoint” (NES = 2.10; NOM P = 0; FDR q-value = 0), “Mitotic Spindle” (NES = 1.89; NOM P = 0; FDR q-value = 0.001) and “mTORC1 Signaling” (NES = 1.34; NOM P = 0.008; FDR q-value = 0.096; Fig. 4B–D). Also, significantly differentially expressed (P < 0.05) genes involved in these three pathways were listed in Supplementary Fig. S3A–S3C. To note, no second mutations and/or amplification on HER2 site were detected based on the Sanger sequencing and quantitative PCR of HER2 in genomic DNA extracted from treatment-naïve and acquired resistant tumors (Supplementary Fig. S3D and S3E).
Figure 4.
Pathways enriched in HER2 exon 20YVMA acquired resistant tumors and mobocertinib increased HER2 expression at the cell membrane. A, Dot plot of enriched pathways (NOM P < 0.05 and FDR q < 0.25) in GSEA result from mobocertinib acquired resistant tumors versus response (treated for 3 days) tumor nodules. B, Enrichment plot of “G2M Checkpoint” gene set. C, Enrichment plot of “Mitotic Spindle” gene set. D, Enrichment plot of “mTOR1 Signaling” gene set. E, Mean fluorescence intensity of surface HER2 receptor on H1781 (HER2G776>VC) cell line treated with 0.01, 0.1, and 1 μmol/L mobocertinib using flow cytometry. F, Mean fluorescence intensity of surface HER2 receptor on the Ba/F3 HER2 exon 20YVMA cell line treated with 0.01, 0.1, and 1 μmol/L mobocertinib using flow cytometry. G, Relative mRNA change of HER2 on Ba/F3-HER2 exon 20YVMA cell line treated with 0.01, 0.1, and 1 μmol/L mobocertinib using real-time PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Mobocertinib increases HER2 expression at the cell membrane
It is worth noting that total HER2 protein level increased when treated with mobocertinib both on H1781 and Ba/F3-HER2 exon 20YVMA cell lines (Fig. 1D and E) and on HER2 exon 20YVMA tumors (Fig. 3B). To further delineate if HER2 expression increased on cell surface, flow cytometry was performed on H1781 and Ba/F3-HER2 exon 20YVMA cell lines. When treated with various concentrations of mobocertinib for 6 hours, both cell lines showed significantly higher level of HER2 mean fluorescent intensity compared with control (Fig. 4E and F). Using reverse transcription polymerase chain reaction (RT-PCR), we showed that HER2 mRNA levels increased with treatment after 6 hours (Fig. 4G). In addition, from RNA-seq result, mobocertinib-treated nodules showed elevated HER2 TPM (Supplementary Fig. S3F).
Mobocertinib and T-DM1 is the most effective among all three combinations
Based on the pathway analysis described above and the increasing level of HER2 on cell surface, 3 candidate drugs were further assessed for potential synergy with mobocertinib: alisertib (aurora kinase A inhibitor), sapanisertib (mTOR inhibitor), and T-DM1 (HER2 ADC). Aurora kinase A is involved in the G2–M process and mitotic spindle assembly and in regulating resistance to EGFR TKI (25). Sapanisertib acts through inhibition of the mTOR pathway (26). T-DM1 acts as a cytotoxic HER2 therapy as well as an inhibitor of mitotic spindle signaling (27).
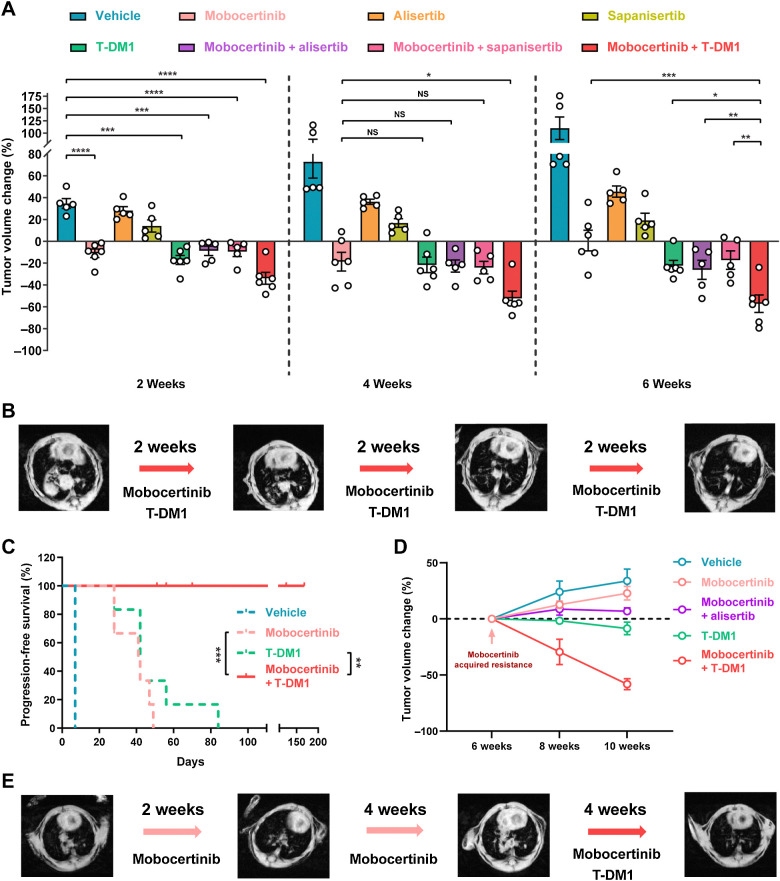
We first tested these 3 candidate drugs with HER2 exon 20YVMA mice in a first-line setting with 8 cohorts. After 2 weeks of treatment, mice in the mobocertinib and T–DM1 monotherapy groups had significant reduction in tumor volume compared with the vehicle group. That was not seen in the alisertib and sapanisertib monotherapy groups (Fig. 5A). All 3 combination therapies significantly decreased the tumor volume compared with vehicle (Fig. 5A).
Figure 5.
Mobocertinib and T-DM1 combination is the most effective among all three combinations. A, Dot plot of tumor volume change on MRI at the time point of weeks 2, 4, and 6 from HER2 exon 20YVMA insertion–mutant GEMMs treated with monotherapy (mobocertinib, T-DM1, alisertib, and sapanisertib) and combinations (mobocertinib combined with T-DM1, alisertib, and sapanisertib). B, Representative MRI image of HER2 exon 20YVMA insertion–mutant GEMMs treated with mobocertinib and T-DM1 combination. C, Survival curve of HER2 exon 20YVMA insertion–mutant GEMMs treated with vehicle, mobocertinib, T-DM1, and combination. D, Tumor volume change of acquired resistant HER2 exon 20YVMA insertion–mutant tumors treated with monotherapies (mobocertinib and T-DM1) and combinations (mobocertinib plus alisertib or T-DM1). E, Representative MRI image of acquired resistant HER2 exon 20YVMA insertion–mutant tumors treated with mobocertinib and T-DM1 combination therapy. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, nonsignificant.
After 6 weeks of treatment, mobocertinib and T-DM1 combination had the highest efficacy on HER2 exon 20YVMA compared with mobocertinib or T-DM1 monotherapy and other combinational therapies with mobocertinib (Fig. 5A and B). Importantly, mobocertinib and T-DM1 combinational treatment was able to prolong the PFS of HER2 exon 20YVMA mice (Fig. 5C). Of note, mice treated with mobocertinib and T-DM1 combination showed sustained response for up to 24 weeks.
Next, in a mobocertinib acquired resistance setting, we investigated if monotherapy-resistant tumors showed similar responses to combinational therapies. Mice were treated with single-agent mobocertinib for 6–8 weeks until acquired resistance developed. Alisertib and T-DM1 were then added to mobocertinib. Although combination with alisertib showed stable disease, treatment with mobocertinib and T-DM1 showed significant shrinkage (Fig. 5D and E). This effect was not seen in monotherapy with T-DM1–only sequential therapy (Fig. 5D).
Taken together, this demonstrates that mobocertinib and T-DM1 combinational therapy was not only able to show robust efficacy for untreated HER2 exon 20YVMA tumors and preventing acquired resistance, but could also provide promising efficacy in tumors that developed resistance.
M1 macrophage infiltration and CD4+ T-cell activation contributed to the synergic effect of mobocertinib and T-DM1 combination
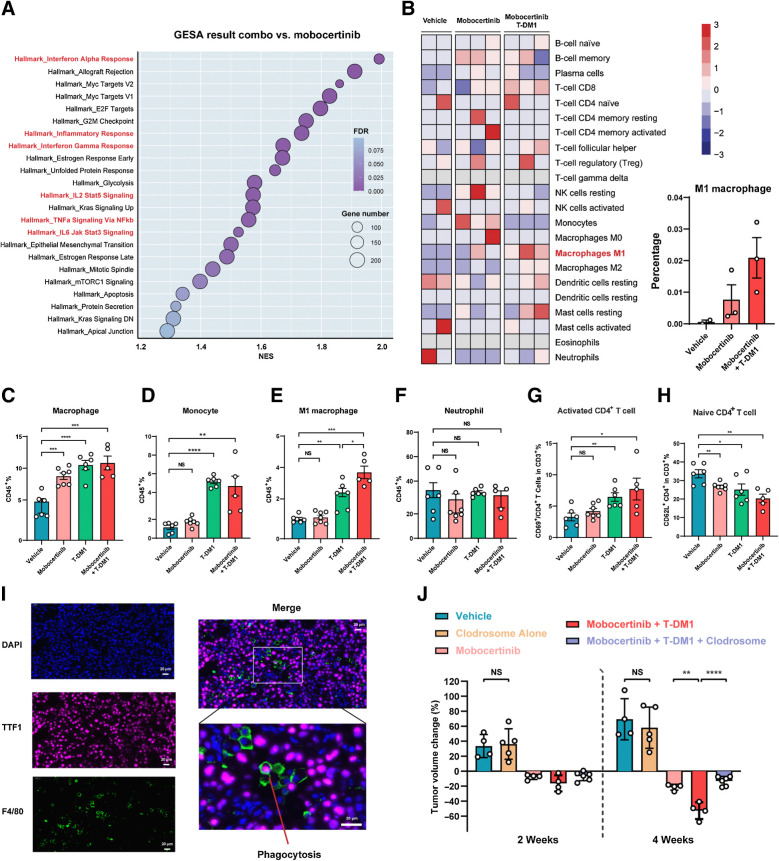
To elucidate the mechanisms involved in the enhanced efficacy of mobocertinib and T-DM1 combination, we performed bulk RNA-seq and GSEA pathway analysis on the tumor nodules treated with the mobocertinib and T-DM1 combination after 3 days. Intriguingly, 6 immune-related pathways were enriched in the tumors treated with combination therapies, including “Interferon Alpha Response,” “Inflammatory Response,” “Interferon Gamma Response,” “IL2 Stat5 Signaling,” “TNFα Signaling Via NFκb,” and “IL-6 Jak Stat3 Signaling” (Fig. 6A; Supplementary Table S2). Enrichment plots of immune-related enriched pathways are shown in Supplementary Fig. S4A.
Figure 6.
M1 macrophage infiltration and CD4+ T-cell activation contributed to the synergic effect of mobocertinib and T-DM1 combination. A, Dot plot of enriched pathways (NOM P < 0.05 and FDR q < 0.25) in GSEA resulting from mobocertinib and T-DM1–treated tumors versus response (treated for 3 days) tumor nodules. B, Heat map of 22 different immune cell populations from RNA-seq data of tumor nodules treated with vehicle, mobocertinib, and mobocertinib plus T-DM1 combination using CIBERSORT analysis. C–H, Comprehensive immuno-profiling of HER2 exon 20YVMA insertion-mutant GEMMs treated with vehicle, mobocertinib, T-DM1, and combination therapy using multiparameter flow cytometry. I, Multi-immunofluorescence of DAPI, TTF1, and F4/80 on mobocertinib and T-DM1–treated tumor. J, Dot plot of tumor volume change on MRI from HER2 exon 20YVMA insertion–mutant GEMMs treated with vehicle, Clodrosome, mobocertinib, mobocertinib plus T-DM1, and mobocertinib combined with T-DM1 plus Clodrosome. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, nonsignificant.
Using the CIBERSORT method (28), a well-established method for analyzing distinct immune cell populations from RNA-seq, tumor nodules from combination therapy with mobocertinib and T-DM1 showed a substantially stronger M1 macrophages transcriptional signature compared with that of mobocertinib monotherapy (Fig. 6B).
Considering the upregulation of immune-related pathways and the increasing trend of M1 macrophage in CIBERSORT analysis, we then used multiparameter flow cytometry to directly characterize changes in immune cell populations during treatment with mobocertinib and T-DM1. Analysis of myeloid lineage showed significant increase in macrophage and monocytes (Fig. 6C and D). In particular, M1 macrophages in the combination group were increased compared with monotherapy (Fig. 6E). In contrast, no changes were seen in neutrophil infiltration (Fig. 6F). Analysis of lymphocytes showed no difference in quantities of T-cell or B-cell populations (Supplementary Fig. S4B–S4E). However, combinational therapy increased activated CD4+ T cells and decreased in naïve CD4+ T cells (Fig. 6G and H).
Trastuzumab (antibody part of T-DM1) was proved to trigger antigen-dependent cellular phagocytosis (ADCP) by interacting with Fcγ receptors on macrophages (29). Then we performed multi-immunofluorescence staining using DAPI (nucleus), TTF1 (tumor cells), and F4/80 (macrophage) in mobocertinib and T-DM1–treated mouse tumors. There was abundant macrophage infiltration along with triple-positive cells, suggesting a role for macrophages in ADCP (Fig. 6I).
Based on these findings, we suggested that mobocertinib contributes to the accumulation of HER2 on the cancer cell surface, which binds to T-DM1, leading to both direct anticancer effect and infiltration of macrophages, which function as ADCP. Meanwhile, M1 macrophages led to increased CD4+ T-cell activation. Thus, the immune tumor microenvironment following this mobocertinib and T-DM1 combinational treatment exhibits a more favorable antitumor phenotype.
The immune-profiling data indicated that macrophages play an essential role in tumor nonautonomous mechanism of mobocertinib and T-DM1 combinational therapy. To further confirm this finding, we performed a macrophage-depletion treatment experiment using the agent Clodrosome (liposomal clodronate; Encapsula NanoSciences LLC), a liposome suspension encapsulated with clodronate, which selectively leads to macrophage apoptosis (30). Naïve tumors showed significantly more positive area of F4/80 using IHC staining than Clodrosome-treated ones, which indicated that Clodrosome successfully depleted macrophages (Supplementary Fig. S4F). Triple combination treatment with Clodrosome, mobocertinib, and T-DM1 in GEMM HER2 exon 20YVMA–mutant mice abrogated the efficacy of mobocertinib and T-DM1 combination (Fig. 6J).
In addition to mobocertinib and T-DM1, the therapeutic strategy to further enhance the function of macrophages as the third combination will be a future direction.
Discussion
No targeted therapies are currently approved for HER2 exon 20 insertion mutation–driven NSCLC. Mobocertinib is a novel small-molecule TKI designed to selectively target EGFR and HER2 exon 20 insertion mutations and has been granted Breakthrough Therapy Designation by the US Food and Drug Administration for patients with metastatic EGFR exon 20 insertion–mutant NSCLC based on a phase I/II clinical trial (43% confirmed objective response rate; ref. 14). Considering the outstanding efficacy on EGFR exon 20 insertion–mutant lung cancer, it is necessary to explore the role of mobocertinib in HER2 exon 20 insertion–mutant lung cancer in both the preclinical and clinical fields (31). Preclinical data reported here represent a robust evaluation of mobocertinib in HER2 exon 20 insertion–mutant lung adenocarcinoma. These data indicate that HER2 exon 20 insertion–mutant cell lines, PDX and GEMMs can be robustly inhibited by mobocertinib. A limitation of our study is that we utilized one HER2 exon 20–mutant cell line (only one available) and PDX. It will be extremely important to further investigate in human tumor settings.
Several other TKIs have demonstrated preclinical activity and are also under clinical investigation. Afatinib, a pan-HER TKI, does not show the expected potential for disease control in patients (32). Tarloxotinib, a hypoxia-activated prodrug of a pan-HER TKI, exhibits antitumor activity in HER2 activating-mutant NSCLC patient (33). DS-2087b, a selective inhibitor of EGFR/HER2 exon 20 insertions, demonstrated in vitro and in vivo inhibition in preclinical assays (34). Poziotinib demonstrated some efficacy in patients with HER2 mutations, but it was associated with high rates of WT EGFR-mediated toxicity (23, 35). In our study, mobocertinib demonstrates robust antitumor activity in HER2 exon 20 insertion–mutant preclinical models and manageable toxicity in patients with EGFR exon 20 insertion–mutant lung adenocarcinoma in clinical trial (14).
Intriguingly, HER2 exon 20 insertion mutations respond differently to mobocertinib. In this study, we generated a new GEMM harboring HER2 exon 20G776>VC mutation, which is a robust method to investigate the difference in response to mobocertinib among various types of insertions. Among the most frequent mutations, HER2 exon 20G776>VC tumors showed deeper and more sustained response than HER2 exon 20YVMA tumors. A recent published study suggested that YVMA insertion in the C-helix of the HER2 kinase domain generated a more rigid structure, resulting in less amenable inhibition by TKI compounds (36).
In addition, further studies are needed to analyze the effect of comutations (such as TP53) on response to mobocertinib in HER2 exon 20 insertion–mutant patients (37, 38). From a clinical perspective, the next-generation sequencing results of both HER2 subtypes and comutation status should be examined.
Notably, we discovered the striking synergic effect of mobocertinib and T-DM1 on HER2 exon 20YVMA lung cancer for both first-line and second–line settings when tumors developed acquired resistance. Even though the HER2 exon 20G776>VC tumors showed sustained complete responses to mobocertinib for more than 16 weeks, acquired resistance inevitably occurred. It will be also essential to elucidate if the mobocertinib and T-DM1 combo can convert this resistance in HER2 exon 20G776>VC tumors.
Previous studies indicate that lapatinib and poziotinib are capable of increasing HER2 expression on cancer cell surfaces through decreasing ubiquitination and then synergizing with the trastuzumab-conjugated cytotoxic drug to provide extra benefits (35, 39). In our setting, HER2 mRNA was elevated after mobocertinib treatment, which suggested that it is likely to be through a transcriptional mechanism. However, whether posttranscriptional mechanisms are involved is worthy of future investigation. Also, irreversible HER2 TKI neratinib was found to markedly increase T-DM1 internalization and enhance activity (40). Mobocertinib is an irreversible HER2 TKI, and we found that mobocertinib is able to increase total HER2 expression. The combination with T–DM1 has synergic benefits on HER2 exon 20YVMA tumors, which accounts for the majority of HER2 exon 20 insertion–mutant lung adenocarcinoma patients.
As little is known about how the immune microenvironment reacts to a small molecular TKI plus ADC, our study indicated that the infiltration of M1 macrophages and CD4+ T-cell activation are the major drivers in mobocertinib and T-DM1 combinational therapy. This phenotype is consistent with previous findings that M1 macrophages were proved able to activate T cells (41–43). In our case, M1 macrophage is a critical component in the immune microenvironment upon mobocertinib and T-DM1 combinational treatment. The additional treatment to enhance macrophage's function on top of mobocertinib and T-DM1 combination is a therapeutic direction to explore.
In conclusion, HER2 exon 20G776>VC tumors responded to mobocertinib, whereas HER2 exon 20YVMA tumors showed initial partial response but developed acquired resistance after continuous treatment. Using pathway analysis, we demonstrated that mobocertinib combined with T-DM1 showed potent efficacy in HER2 exon 20YVMA–untreated and –resistant tumors. Furthermore, we investigated the mechanism involved in the synergic effect of mobocertinib and T-DM1 and elucidated that in addition to cancer cells' autonomous effect, M1 macrophage infiltration and T-cell activation may play vital parts. Above all, this paper provides a meaningful preclinical rationale for the development of mobocertinib clinical trials for patients with HER2 exon 20 insertion–mutant NSCLC.
Finally, given the prevalence of HER2 mutations across multiple cancer types, it will be vital to see if the combination of mobocertinib and HER2 antibody–drug conjugate (e.g., T-DM1 and the new emerging drug DS-8201) will remain effective on other cancer models. If so, this will support the clinical development of mobocertinib and HER2 ADC combinational treatments in patients with other HER2 mutation-driven cancer types.
Authors' Disclosures
T.E. Baker reports personal fees from Takeda, personal fees from Amathus Therapeutics, and personal fees from MOMA Therapeutics outside the submitted work. V.M. Rivera reports other support from ARIAD Pharmaceuticals and other support from Theseus Pharmaceuticals outside the submitted work. S. Vincent reports other support from Takeda during the conduct of the study. K.K. Wong reports grants from Takeda during the conduct of the study and grants from Bristol Myers Squibb, grants from Mirati, grants from Merus, grants from Alkermes, grants from Ansun, grants from Tvardi, grants from Delfi, grants from Dracen, other support from Recursion Pharmaceuticals, other support from Prelude, grants and other support from Rentals, and grants and other support from Janssen outside the submitted work. K.K. Wong is a founder and equity holder of G1 Therapeutics. No disclosures were reported by the other authors.
Supplementary Material
GSEA analysis of mobocertinib acquired resistant tumors versus response tumors Genes involved in "G2M CHECKPOINT", "MITOTIC SPINDLE" and "MTORC1 SIGNALING" pathways of HALLMARK gene sets.
GSEA analysis of mobocertinib and T-DM1 treated tumors versus mobocertinib-only treated tumors Genes involved in "INTERFERON ALPHA RESPONSE", "INFLAMMATORY RESPONSE", "INTERFERON GAMMA RESPONSE", "IL2 STAT5 SIGNALING", "TNFA SIGNALING VIA NFKB" and "IL6 JAK STAT3 SIGNALING" pathways of HALLMARK gene sets.
Figure S1
Figure S2
Figure S3
Figure S4
Acknowledgments
This study is supported by NIH grants CA154303 and CA098101 as well as by Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited.
The authors thank Teodor G. Paunescu for the great help with writing and coordination. The authors thank Minal Metha, Eric N. Churchill, Ashish Gulati, and Michael J. Humphries for reviewing the manuscript. The authors thank Yaoyu Ning for contributions to this study. Editorial support was provided by Peloton Advantage, LLC, an OPEN Health company, Parsippany, NJ, and funded by Millennium Pharmaceuticals, Inc., Cambridge, MA, a wholly owned subsidiary of Takeda Pharmaceutical Company Limited.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
H. Han: Conceptualization, data curation, software, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. S. Li: Conceptualization, data curation, writing–review and editing. T. Chen: Conceptualization, resources, writing–review and editing. M. Fitzgerald: Conceptualization, writing–review and editing. S. Liu: Resources, writing–review and editing. C. Peng: Writing–review and editing. K.H. Tang: Conceptualization, visualization, writing–review and editing. S. Cao: Writing–review and editing. J. Chouitar: Conceptualization and data curation. J. Wu: Writing–review and editing. D. Peng: Methodology, writing–review and editing. J. Deng: Writing–review and editing. Z. Gao: Methodology, writing–review and editing. T.E. Baker: Conceptualization and data curation. F. Li: Methodology, writing–review and editing. H. Zhang: Conceptualization, writing–review and editing. Y. Pan: Methodology, writing–review and editing. H. Ding: Writing–review and editing. H. Hu: Methodology, writing–review and editing. V. Pyon: Data curation. C. Thakurdin: Data curation. E. Papadopoulos: Data curation. S. Tang: Data curation. F. Gonzalvez: Conceptualization, data curation, writing–review and editing. H. Chen: Supervision, writing–review and editing. V.M. Rivera: Supervision, writing–review and editing. R. Brake: Conceptualization and data curation. S. Vincent: Conceptualization, resources, data curation, supervision, project administration, writing–review and editing. K.-K. Wong: Conceptualization, resources, supervision, methodology, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Recondo G, Facchinetti F, Olaussen KA, Besse B, Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694–708. [DOI] [PubMed] [Google Scholar]
- 4. Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tomizawa K, Suda K, Onozato R, Kosaka T, Endoh H, Sekido Y, et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer 2011;74:139–44. [DOI] [PubMed] [Google Scholar]
- 6. Mazieres J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997–2003. [DOI] [PubMed] [Google Scholar]
- 7. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642–6. [DOI] [PubMed] [Google Scholar]
- 9. Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018;554:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mazieres J, Barlesi F, Filleron T, Besse B, Monnet I, Beau-Faller M, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol 2016;27:281–6. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y, Jiang T, Qin Z, Jiang J, Wang Q, Yang S, et al. HER2 exon 20 insertions in non-small-cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann Oncol 2019;30:447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prelaj A, Bottiglieri A, Proto C, Lo Russo G, Signorelli D, Ferrara R, et al. Poziotinib for EGFR and HER2 exon 20 insertion mutation in advanced NSCLC: results from the expanded access program. Eur J Cancer 2021;149:235–48. [DOI] [PubMed] [Google Scholar]
- 13. Neal J, Doebele R, Riely G, Spira A, Horn L, Piotrowska Z, et al. P1.13-44 safety, PK, and preliminary antitumor activity of the oral EGFR/HER2 exon 20 inhibitor TAK-788 in NSCLC. J Thorac Oncol 2018;13:S599. [Google Scholar]
- 14. Riely GJ, Neal JW, Camidge DR, Spira AI, Piotrowska Z, Costa DB, et al. Activity and safety of mobocertinib (TAK-788) in previously treated non-small cell lung cancer with EGFR exon 20 insertion mutations from a phase 1/2 trial. Cancer Discov 2021;11:1688–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akbay EA, Moslehi J, Christensen CL, Saha S, Tchaicha JH, Ramkissoon SH, et al. D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and neurodegeneration in mice. Genes Dev 2014;28:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012;483:613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 2013;49:503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A 2006;103:7817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimamura T, Ji H, Minami Y, Thomas RK, Lowell AM, Shah K, et al. Non-small-cell lung cancer and Ba/F3 transformed cells harboring the ERBB2 G776insV_G/C mutation are sensitive to the dual-specific epidermal growth factor receptor and ERBB2 inhibitor HKI-272. Cancer Res 2006;66:6487–91. [DOI] [PubMed] [Google Scholar]
- 23. Prelaj A, Bottiglieri A, Proto C, Lo Russo G, Signorelli D, Ferrara R, et al. Poziotinib in advanced NSCLC with EGFR or HER2 exon 20 insertion mutation: initial results from a single site expanded access program [abstract 1388P]. Ann Oncol 2020;31:S882. [Google Scholar]
- 24. Perera SA, Li D, Shimamura T, Raso MG, Ji H, Chen L, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A 2009;106:474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med 2019;25:111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012;485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res 2008;68:9280–90. [DOI] [PubMed] [Google Scholar]
- 28. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi Y, Fan X, Deng H, Brezski RJ, Rycyzyn M, Jordan RE, et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcgamma receptors on macrophages. J Immunol 2015;194:4379–86. [DOI] [PubMed] [Google Scholar]
- 30. van Rooijen N, Sanders A, van den Berg TK. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J Immunol Methods 1996;193:93–9. [DOI] [PubMed] [Google Scholar]
- 31. Gonzalvez F, Vincent S, Baker TE, Gould AE, Li S, Wardwell SD, et al. Mobocertinib (TAK-788): a targeted inhibitor of EGFR exon 20 insertion mutants in non-small cell lung cancer. Cancer Discov 2021;11:1672–87. [DOI] [PubMed] [Google Scholar]
- 32. Dziadziuszko R, Smit EF, Dafni U, Wolf J, Wasag B, Biernat W, et al. Afatinib in NSCLC with HER2 mutations: results of the prospective, open-label phase II NICHE trial of European Thoracic Oncology Platform (ETOP). J Thorac Oncol 2019;14:1086–94. [DOI] [PubMed] [Google Scholar]
- 33. Liu SV, Villaruz LC, Lee VHF, Zhu VW, Baik CS, Sacher A, et al. First analysis of RAIN-701: study of tarloxotinib in patients with non-small cell lung cancer (NSCLC) EGFR Exon 20 insertion, HER2-activating mutations & other solid tumours with NRG1/ERBB gene fusions [abstract LBA61]. Ann Oncol 2020;31:S1189. [Google Scholar]
- 34. Nagamoto Y, Miyamoto M, Togashi N, Taira T, Jimbo T, Isoyama T, et al. Preclinical evaluation of DS-2087b, a novel and selective inhibitor of EGFR/HER2 exon 20 insertions [abstract 11P]. Ann Oncol 2020;31:S248. [Google Scholar]
- 35. Robichaux JP, Elamin YY, Vijayan RSK, Nilsson MB, Hu L, He J, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell 2019;36:444–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao S, Fang W, Pan H, Yang Y, Liang Y, Yang L, et al. Conformational landscapes of HER2 exon 20 insertions explain their sensitivity to kinase inhibitors in lung adenocarcinoma. J Thorac Oncol 2020;15:962–72. [DOI] [PubMed] [Google Scholar]
- 37. Yuan B, Zhao J, Zhou C, Wang X, Zhu B, Zhuo M, et al. Co-Occurring alterations of ERBB2 exon 20 insertion in non-small cell lung cancer (NSCLC) and the potential indicator of response to afatinib. Front Oncol 2020;10:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang W, Zhao S, Liang Y, Yang Y, Yang L, Dong X, et al. Mutation variants and co-mutations as genomic modifiers of response to afatinib in HER2-mutant lung adenocarcinoma. Oncologist 2020;25:e545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene 2009;28:803–14. [DOI] [PubMed] [Google Scholar]
- 40. Li BT, Michelini F, Misale S, Cocco E, Baldino L, Cai Y, et al. HER2-mediated internalization of cytotoxic agents in ERBB2 amplified or mutant lung cancers. Cancer Discov 2020;10:674–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li F, Huang Q, Luster TA, Hu H, Zhang H, Ng WL, et al. In Vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in Kras-mutant lung adenocarcinoma. Cancer Discov 2020;10:270–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mills CD. Anatomy of a discovery: m1 and m2 macrophages. Front Immunol 2015;6:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W, Marinis JM, Beal AM, Savadkar S, Wu Y, Khan M, et al. RIP1 kinase drives macrophage-mediated adaptive immune tolerance in pancreatic cancer. Cancer Cell 2018;34:757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GSEA analysis of mobocertinib acquired resistant tumors versus response tumors Genes involved in "G2M CHECKPOINT", "MITOTIC SPINDLE" and "MTORC1 SIGNALING" pathways of HALLMARK gene sets.
GSEA analysis of mobocertinib and T-DM1 treated tumors versus mobocertinib-only treated tumors Genes involved in "INTERFERON ALPHA RESPONSE", "INFLAMMATORY RESPONSE", "INTERFERON GAMMA RESPONSE", "IL2 STAT5 SIGNALING", "TNFA SIGNALING VIA NFKB" and "IL6 JAK STAT3 SIGNALING" pathways of HALLMARK gene sets.
Figure S1
Figure S2
Figure S3
Figure S4