Abstract
Each heartbeat that pumps blood throughout the body is initiated by an electrical impulse generated in the sinoatrial node (SAN). However, a number of disease conditions can hamper the ability of the SAN’s pacemaker cells to generate consistent action potentials and/or maintain an orderly conduction path, leading to arrhythmias. For symptomatic patients, current treatments rely on implantation of an electronic pacing device. However, complications inherent to the indwelling hardware give pause to categorical use of device therapy for a subset of populations, including pediatric patients or those with temporary pacing needs. Cellular-based biological pacemakers, derived in vitro or in situ, could function as a therapeutic alternative to current electronic pacemakers. Understanding how biological pacemakers measure up to the SAN would facilitate defining and demonstrating its advantages over current treatments. In this review, we discuss recent approaches to creating biological pacemakers and delineate design criteria to guide future progress based on insights from basic biology of the SAN. We emphasize the need for long-term efficacy in vivo via maintenance of relevant proteins, source-sink balance, a niche reflective of the native SAN microenvironment, and chronotropic competence. With a focus on such criteria, combined with delivery methods tailored for disease indications, clinical implementation will be attainable.
Subject terms: Biological pacemaker, sinoatrial node, stem cell, cardiac pacemaker cell extracellular matrix, action potential generation, source-sink balance
Keywords: biological pacemaker, stem cells, sinoatrial node
The sinoatrial node (SAN) is a small, heterogeneous structure with the enormous task of initiating the electromechanical activity of the entire heart. However, a number of disease conditions can prevent the SAN from working properly. As a means to better understand how such conditions lead to SAN failure, and as a potential treatment option itself, “biological pacemakers” have been proposed. Such pacemakers are created in vitro through stem cell differentiation or through direct reprogramming of various cardiac cell types to pacemaker-like cells that are capable of spontaneously pacing cardiac tissue. Numerous approaches for generating biological pacemakers have been published over the past two decades, however, clinical implementation has yet to be realized. Here we consider biological pacemakers in the context of notable design criteria and discuss how each criterion has been met or requires further discovery or technological innovation to achieve.
The Need for Biological Pacemakers
Every year, more than 350,000 pacemaker procedures are performed in the U.S. alone,1 with sinus node dysfunction accounting for over half of implantations.2 In general, pacemakers are effective and reliable and continue to receive iterative improvements in hardware and software. However, they carry several limitations. A critical issue is the potential for device failure, possibly due to loose or faulty leads, battery depletion, or electromagnetic interference. Incidence rates of post-implantation complications including those related to the lead, infection, and thoracic trauma are 9% within the first month and 6% in the following three years.3 While leadless pacemakers obviate problems related to pacing leads,4,5 encapsulation within heart tissue has been seen within 1–2 years of implantation, rendering device extraction unfeasible.6,7 Further, cardiac pacing is particularly risky in newborns and infants since their great vessels are still too narrow or not accessible due to congenital malformations.
The limitations of electronic pacemakers have thus spurred the development of biological pacemakers. A biological pacemaker is a group of cells, created through reprogramming of somatic cells or differentiation of human pluripotent stem cells (hPSCs), with the ability to generate spontaneous action potentials in order to pace cardiac muscle. These derived pacemaker cells offer the potential to avoid issues such as recurring procedures to address device repositioning, replacement, or failure. Additionally, these cells would be powered by the patients’ own metabolism, negating the need for a generator, which is especially beneficial for pediatric patients. Beyond a potential treatment strategy, biological pacemakers can also function as in vitro models in order to increase our understanding of the sinoatrial node, such as mechanisms of development, causes of malfunction, the effect of various diseases on pacing, and the effect of therapeutics on heart rhythm. For instance, derived pacemaker cells were used to gain insight of developmental processes, identifying a novel regulatory element for the SAN transcription factor Tbx3.8 Moreover, while many features of the SAN are conserved among vertebrates,9 a reliable human model of the cardiac pacemaker would be of great value. Biological pacemakers would allow for the study of potential treatments for SAN dysfunction that have only been previously studied in animals, such as inhibition of the muscarinic-gated potassium channel, found to be effective in treating tachy-brady syndrome in mice.10 Testing such genetic or pharmacological treatments in vitro on human cells would allow for a safer transition to clinical trials in humans.
Substantial progress has been made to generate biological pacemakers, which thus far may offer the ability to pace temporarily, yet challenges remain that limit clinical application for long-term pacing. As a means of evaluating the current state of the field and the most promising avenues forward, we define a successful biological pacemaker in terms of a series of design criteria important for reliable pacing (Figure 1).
Figure 1. Sinoatrial Node Features to be Replicated in Biological Pacemakers.
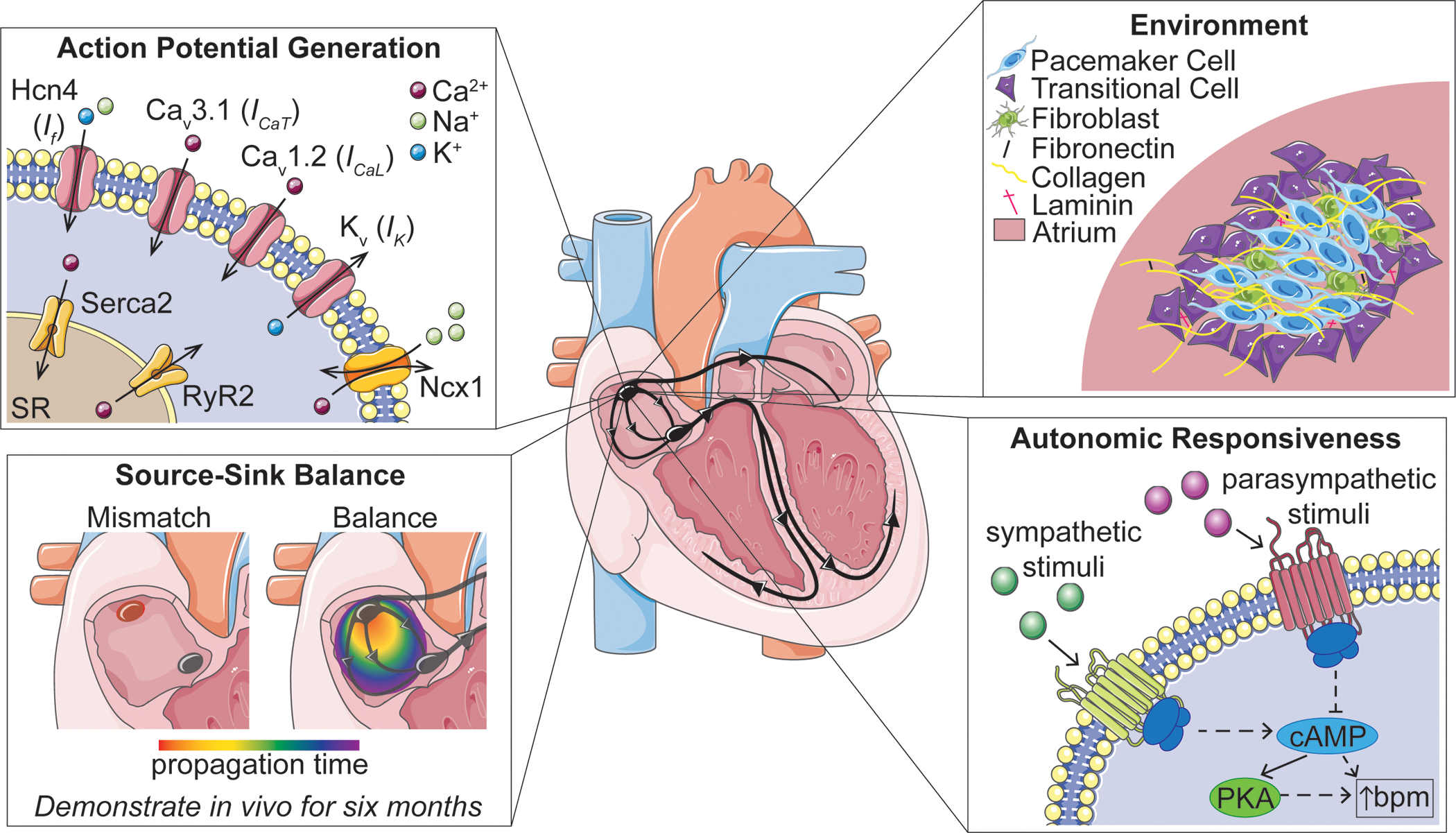
Membrane clock and calcium clock channel proteins will be expressed in biological pacemakers in order to generate action potentials consistently and reliably. Pacemaker cells will ideally be embedded in relevant extracellular matrix proteins and surrounded by transitional cells and other key cell types. Source-sink mismatch will be overcome, and action potentials will be propagated to surrounding myocardial tissue; cells will be able to pace for extended periods of time. Lastly, biological pacemakers should respond to autonomic input to ensure reliable pacing for patients. (Hcn4: hyperpolarization-activated cyclic nucleotide-gated channel 4, Ncx1: sodium-calcium exchanger 1, RyR2: ryanodine receptor 2, SR: sarcoplasmic reticulum, cAMP: cyclic adenosine monophosphate, PKA: protein kinase A, bpm: beats per minute)
The ideal biological pacemaker would:
Generate spontaneous action potentials originating from a coupled membrane and calcium clock system, with clock protein expression maintained at levels comparable to that of the native pacemaker.
Overcome source-sink mismatch to pace and drive large regions of tissue.
Provide reliable pacing over long periods of time.
Contain a relatively similar level of heterogeneous nodal cell types, as seen in the SAN.
Replicate nodal tissue architecture, with consideration for pacemaker and transitional cell patterning, extracellular matrix (ECM) composition, and culture substrates.
Demonstrate autonomic responsiveness, adjusting the rate of pacing based on the physiological needs of the body.
Fulfilling these criteria will build the foundation for clinical translation. Application of current knowledge of SAN structure, embryonic development of the SAN, and limitations of previous reports of biological pacemaker development, as discussed below, will be key for success.
Cardiac Pacemaker Cells and the Sinoatrial Node: Structure-Function Relationships
The SAN is distinguishable from the rest of the heart by its cell types, mechanical structure, and electrical activity. Pacing is performed by a relatively small number of nodal cardiomyocytes, generating rhythmic electrical impulses that propagate through the cardiac conduction system and activate the working myocardium. In addition to the nodal cardiomyocytes, transitional cells have been found between the nodal cardiomyocytes and the right atrium that share characteristics of both, aiding in electrical propagation from the SAN to the atrium. For instance, these transitional cells express Nkx2.5,11,12 a transcription factor found in the chamber cardiomyocytes, but not in pacemaker cells. It was shown that upon localized inactivation of Nkx2.5 in the SAN, the atrium failed to receive electrical signals from the fully-functional pacemaker cells, emphasizing the importance of the transitional cells.12 Surrounding the cells of the SAN is a dense, collagen-rich connective tissue that provides mechanical and electrical insulation, presumably protecting the SAN from the hyperpolarizing electrical load of the surrounding atrium.13 Further, the ECM has been found to be highly elastic, providing further mechanical insulation from the beating heart, with an abundance of basement membrane proteins, collagen VI, and fibronectin detected in the mouse SAN14 and elastin surrounding clusters of pacemaker cells in the pig SAN.15
The cardiomyocytes of the SAN are distinguishable from chamber cardiomyocytes in several ways. First, as they are not required to generate as much force as atrial or ventricular cardiomyocytes, pacemaker cells have less-organized contractile units and fewer mitochondria.16 Thus, they are typically described as spindle- or spider-shaped, with their cytoplasm significantly smaller than that of chamber cardiomyocytes. Pacemaker cells also harbor a unique set of proteins that varies from atrial and ventricular cardiomyocytes. For instance, gap junction proteins differ, with human pacemaker cells expressing the low-conductance connexin (Cx) 45 rather than the higher-conductance Cx43 or Cx40. Transcription factors, such as Shox2, Isl1, Tbx3, and Tbx18, and a notable lack of Nkx2.5, are important for SAN development and function. Expression of specific ion channels form two mechanisms suggested to create such automaticity, the calcium clock and the membrane clock, and many studies support an interplay between these two clocks.17–19
The membrane clock is composed of a series of cell-surface ion channels including hyperpolarization-activated cyclic nucleotide-gated (Hcn) channels, L- and T-type voltage-gated calcium channels, and delayed rectifier potassium channels. This clock runs primarily based on the unique biophysical properties of the Hcn channels – with Hcn4 being the dominant family member found on the sarcolemma of pacemaker cells.20,21 Hcn channels uniquely increase their probability of opening as cells polarize which results in a slow inward current during the resting (diastolic) phase of the cardiac cycle.22,23 The funny current (If), which moves through the Hcn channel, depolarizes the membrane potential toward the activation range of voltage-gated calcium channels, resulting in an action potential. Delayed rectifier potassium channels then initiate an outward current hyperpolarizing membrane potential back into the range at which Hcn channels reopen, restarting the cycle.24
The calcium clock, on the other hand, is driven by independent currents that interact and entrain with membrane current activation. Herein, sarcoplasmic reticulum release of calcium into the cytoplasm (via the cardiac ryanodine receptor, RyR2) induces a transient increase in intracellular calcium concentration.25–27 This activates the sodium-calcium exchanger (Ncx1) which expels one calcium ion for three sodium ions, resulting in a net depolarizing current.28 As with the function of Hcn, Ncx1 activity brings membrane potential up to the activation threshold for voltage gated calcium channels, which open and depolarize the cell.18,19,25–27,29 Cytoplasmic calcium is then reloaded into the sarcoplasmic reticulum via the sarco/endoplasmic reticulum Ca2+-ATPase 2 pump (Serca2),30 starting the process again.
The rate of action potential generation is highly regulated by the autonomic nervous system, through various mechanisms recently reviewed.31 Briefly, released neurotransmitters bind to G-protein coupled receptors (e.g. β-adrenergic receptors in the sympathetic system or muscarinic receptors in the parasympathetic system), initiating or inhibiting signaling pathways such as those involving cyclic adenosine monophosphate (cAMP) and protein kinase A (PKA). Both cAMP and PKA contribute to numerous intracellular responses involved in both clock mechanisms, ultimately leading to an increase in heart rate.
Together, these structural and functional properties of the sinoatrial node may provide key insights for biological pacemaker development. For instance, more elastic substrates may be ideal for pacemaker culture, and there may be a need to incorporate electrical and mechanical insulation for clinical translation. It is also critical that engineered pacemaker cells mimic the SAN’s unique expression pattern of ion channels and transcription factors, as described above, to function properly. To fully recapitulate these complex pacemaker cell properties, it is beneficial to understand their origination during embryonic development of the sinoatrial node.
SAN Development
Cardiac pacemaker cells rapidly acquire most of their defining genetic, functional, and structural characteristics during early embryonic development. This is in contrast to the majority of the working myocardium which continues to remodel and mature well into post-natal life. While many questions remain unanswered, particularly related to how pacemaker cells functionally integrate into the embryonic heart, the basic features of pacemaker cell development have already begun to provide foundational principles from which strategies to generate pacemaker-like cells have emerged.
Early SAN Formation
As with all cardiomyocytes, pacemaker cells are derived from the lateral plate mesoderm. Fate mapping studies conducted in the chick32 and mouse33,34 have demonstrated that following gastrulation, pacemaker progenitor cells occupy a discreet region of the mesoderm adjacent to progenitors of atrial and atrioventricular myocardium. Of interest, explant studies revealed that these progenitors appear to be largely specified to the pacemaker cell lineage at early post-gastrulation stages, well before the onset of cardiac morphogenesis, and prior to this region of the embryo expressing classical molecular markers of cardiac fate.32 These data suggest that the pacemaker cell fate is segregated from the working myocardium very early during development and that the initial cues that dictate lineage specification may be related to spatially restricted signaling events that occur during early mesodermal patterning. In support of this, several factors involved in the anterior-posterior patterning of the mesoderm, including retinoic acid (RA), bone morphogenic proteins (BMPs), and Wnt signaling, have been shown to promote pacemaker cell fate in the embryo.32,35–37
During the period following their specification but preceding functional differentiation, pacemaker cells activate a transcriptional program that differs significantly from the rest of the heart. While the upstream events that control this program remain to be determined, it is clear that from very early stages, pacemaker cells follow a unique developmental trajectory. For instance, as pacemaker cells are being specified in the early lateral plate mesoderm, they do not express classic markers of early heart development, such as Nkx2.5 and Isl1.32 Further, during heart tube stages, pacemaker cells begin expressing the second heart field marker Isl1, as well as the T-box transcription factor Tbx18.33,38–42 As morphogenesis proceeds, pacemaker cells become incorporated into the sinus venosus, which sits in a posterior position relative to the forming right atria. Concomitant with this, pacemaker cells begin expressing the transcription factors Tbx3 and Shox2.38,40,43–46 Of note, the tail region of the forming SAN can display hybrid transcription factor profiles (Nkx2.5+/Tbx18+ and/or Nkx2.5+/Shox2+),12,40 but current expression profiling and next generation sequencing analysis highlight that the developing head region of the SAN is predominantly occupied by a pacemaker cell population that expresses Tbx18, Isl1, Tbx3, and Shox2.11,42,47–49
Among the most intriguing aspects of pacemaker cell development is the immediacy with which these cells transition from being electrically inert to autonomously initiating high-rate, rhythmic depolarizations. This occurs within a matter of hours at the end of cardiac looping and corresponds with the expression of a series of ion handling proteins. These include genes required for the membrane and calcium clock systems that drive cardiac pacemaker cell automaticity, as described above. Of note, the membrane clock components Hcn4, Cav3.1, and Cav1.2, as well as the calcium clock components Ncx1, RyR2, and Serca2, are all turned on at the transcript level right as pacemaker cells become electrically active,32,50 and these factors remain associated with the pacemaker cell lineage through life.14 In many ways, the goals of creating biological pacemakers are to replicate the expression and maintenance of these factors in order to generate large numbers of cells capable of stable electrical oscillation.
Developmental Events that Influence Function
Beyond the ability of individual cells to upregulate the channels required for autonomous depolarization, there are several other important electrophysiological limits the embryonic heart must overcome in order to build a functional SAN. These limits mainly pertain to the ability of pacemaker cells to sustain their excitability and generate impulses while in electrical communication with large numbers of chamber cardiomyocytes. For instance, mature pacemaker cells only reach a maximal diastolic polarization around −55 to −65mV compared to the adjacent atrial myocardium which actively maintains resting membrane potentials of greater than −80mV.17,51–53 In theory, coupling these cells would elicit current flows toward the atrial cell that would delay or even suppress the ability of pacemaker cells to build up enough charge to fire an action potential. Furthermore, pacemaker cells represent only a small fraction of the total cellular volume of the heart, and the size discrepancy between the SAN and atrial myocardium represents a significant barrier to electrical propagation. Herein, two interdependent but related concepts are relevant, safety factor of propagation and source-sink interactions.53–56 In order for an electrical impulse to propagate, current flow into a polarized cell must exceed a threshold to activate enough voltage-gated ion channels and induce depolarization. Safety factor is a quantitative metric to assess whether the minimum requirements for action potential activation can be met.57 Safety factor is low when a small “source” of charge is connected to a large population of heavily coupled cells.53 Essentially, current from the source is not sufficient to activate local depolarization before it dissipates across the downstream field of cells, which act as an electrical “sink.” Such an imbalance between the size of upstream and downstream populations is referred to as a source-sink mismatch and has long been recognized as a feature of cardiac electrophysiology that pacemaker cells must overcome.56,58,59
These unfavorable electrochemical conditions with which the SAN is presented introduce the question of how pacemaker cells are able to reliably function. While perhaps not immediately intuitive, the probability of successful action potential generation and propagation is actually increased by creating barriers to conduction, which appears to be the strategy employed in the SAN. By lowering intercellular conductance and/or creating regions through which currents cannot pass, depolarizing charge is retained by pacemaker cells during the diastolic phase of the cardiac cycle. The SAN accomplishes this by physically isolating pacemaker cells via embedding them in an ECM-rich microenvironment and by suppressing the gap junctional linkages that are used for electrical communication to the remainder of the heart.59–63 This allows incoming charge to maximally influence the local membrane potential before it can be lost to downstream cells. Indeed, computational modelling studies have repeatedly demonstrated that lowering electrical communication between pacemaker cells and atrial cardiomyocytes lessens source-sink burdens.58,62,64
It appears that the means of protecting pacemaker cells from unfavorable source-sink conditions are established early during embryogenesis. As pacemaker cells differentiate relatively late during cardiac development, they must integrate into the preexisting cellular circuitry of the embryonic heart, which entails rapidly building the insulatory systems needed to sustain their function. From the earliest stages of pacemaker cell differentiation, these cells express diminished levels of Cx43 and Cx40 compared to the working myocardium. Indeed, the nodal-enriched transcription factors Tbx3 and Shox2 appear to suppress transcription of these high-conductance gap junctions,43,65 and Shox2 may also promote the expression of the low-conductance gap junction Cx45.66 By diminishing the absolute level of high-conductance gap junctions, pacemaker cells are thought to electrically insulate themselves. Therefore, the pacemaker cell genetic program is specialized from very early stages to optimize cell-cell interactions to protect against electrogenic suppression.
Consistent with reducing gap junctional coupling, the developing sinoatrial node also acquires structural features that promote electrical insulation. This can be seen in a dramatic remodeling of the pacemaker cell microenvironment that occurs shortly after these cells differentiate. During looping stages, the right inflow myocardium of the heart (in which pacemaker cells reside) closely resembles the morphology of the adjacent working myocardium. Cardiomyocytes are closely arranged, and little interstitial ECM is present. However, over the completion of cardiac looping and into early cardiac septation stages, a population of non-muscle mesenchymal cells invades the forming SAN.50 This results in formation of a collagen-rich ECM which physically remodels the SAN into clusters of loosely connected pacemaker cells. Cell tracing studies in both the chick and mouse embryo have demonstrated that at least one major source of this invading mesenchymal cell population is the proepicardium, which gives rise to the epicardium as well as a large population of cardiac fibroblasts, smooth muscle cells, and endothelium.50 Importantly, microsurgical removal of the proepicardium prior to non-muscle cell incorporation into the SAN results in failed ECM deposition in the forming SAN. Consequently, pacemaker cells acquire a cellular architecture that greatly resembles the chamber myocardium and displays molecular changes including the upregulation of Cx40.50 Functionally, disruption of SAN remodeling results in decreased rhythmicity in pacemaker cells and conduction block at the SAN-atrium junction.50 Indeed, the electrophysiological defects that emerge mimic computational modeling of cardiac source-sink imbalance.67 Collectively, these data highlight the significance that proper microenvironmental patterning plays in pacemaker cell function and indicate that protecting pacemaker cells from the working myocardium will be an important consideration for building biological pacemakers.
Moving forward, the design and implementation of strategies for creating cellular-based biological pacemakers for therapeutic purposes will most likely continue to borrow heavily from instructions provided by the embryo. As the genetic, cellular, and environmental conditions that build the native SAN are uncovered, they can serve as a powerful template for novel bioengineering approaches.
Approaches to Biological Pacing
Over the past two decades, the creation of pacemaker cells has been approached through various methods, broadly divided into two major categories: 1) functional and genetic reprogramming of chamber cardiomyocytes and non-myocytes from the heart and 2) directed differentiation of pacemaker cells from pluripotent stem cells (Figure 2). Both approaches have increasingly relied on learned developmental cues for pacemaker differentiation, and both have shown promise in vivo, as highlighted below, demonstrating potential for future translational success.
Figure 2. Timeline of Biological Pacemaker Creation and in vivo Testing.
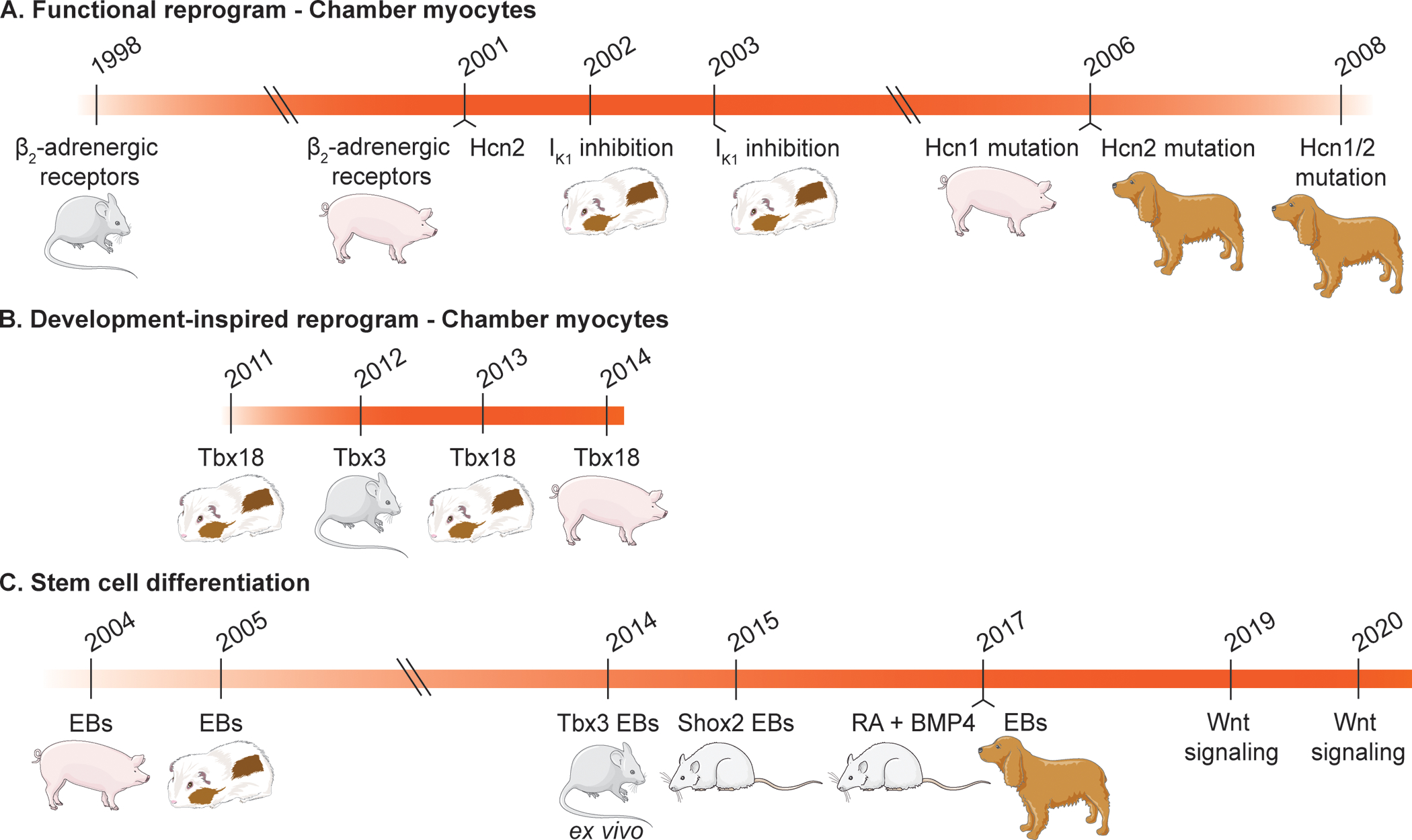
Methods have transitioned over time, beginning with atrial and ventricular reprogramming to induce pacemaker-like functions (A),68–75 transitioning to reprogramming with a developmental approach (B),76–80 and most recently, to stem cell differentiation via application of developmental cues (C).35–37,66,81–84 More recent studies lack evaluation of pacing efficacy in large animal models. (Hcn: hyperpolarization-activated cyclic nucleotide-gated channel, EBs: embryoid bodies, RA: retinoic acid, BMP4: bone morphogenic protein 4)
Reprogramming to pacemaker cells
Functional reprogramming
Early genetic therapies for biological pacemakers were largely focused on the functional reprogramming of chamber cardiomyocytes to obtain cells that behave like pacemaker cells, specifically with increased automaticity and autonomic responsiveness. This work included strategies such as upregulation of the β2-adrenergic receptors involved in heart rate regulation,68,69 dominant-negative suppression of inward rectifying current IK1 in ventricular cardiomyocytes to initiate spontaneous depolarization,70 and overexpression or functional mutations of Hcn channels.72–75
Although each of these strategies were successful in increasing automaticity, they were hampered by the artificial nature of their pacemaker phenotype. While upregulation of β2-adrenergic receptors led to an increased heart rate in vivo, the approach did not create de novo automaticity and concerns remained about the potential for arrhythmias due to the non-specific effects of this receptor and its downstream pathways. Further, long-QT phenotypes were seen in guinea pig left ventricles with suppression of IK1.71 Ectopic expression of Hcn mutations in canine failed to work as well, with one mutation only providing pacing for 2.5–3 weeks,73 and another causing tachycardia (>200 beats per minute).74 A separate induced Hcn1 mutation was able to generate a slow atrial rhythm in a porcine model of sick sinus syndrome; however use of Hcn1 is not ideal due to its low sensitivity to cAMP.75 These studies and other analogous approaches85,86 offered the first evidence for electrophysiological conversion of quiescent myocardium to spontaneous activity. However, the concept requires permanent genomic insertion of a transgene, a prerequisite burdened with inherent concerns of unintended genome modifications. The field has also begun to appreciate the complexity of the SAN and that the creation of functional pacemaker cells requires more than addition or subtraction of an ionic current. Thus, studies have turned to SAN development for inspiration.
Development-inspired reprogramming
Developmental studies have shown the critical role that transcription factors, such as Tbx3 and Tbx18, play in the formation of the cardiac conduction system. Tbx3 is required for proper development of the cardiac conduction system, impacting pre- and postnatal survival upon reduction in Tbx3 in a dosage-dependent manner.80 When Tbx3 is expressed ectopically in the developing myocardium, myocardial genes such as atrial natriuretic peptide and Cx40 could not be detected, but Hcn4 was not activated, indicating Tbx3 does not directly regulate Hcn4.40 When Tbx3 was expressed ectopically in the developing atria, spontaneous electrical activity was observed in both the right and left atria.43 It is important to note these studies were performed during embryonic development where ectopic expression of Tbx3 began at E9.540 and E10.5.43 Attempts to reprogram adult ventricular myocardium with inducible expression of Tbx3 failed to induce automaticity.79 Hcn4 was downregulated upon ectopic expression of Tbx3 in the ventricular myocardium, and no spontaneously oscillating action potentials were observed in the atrial or ventricular cardiomyocytes. Only when Tbx3 was expressed in the embryonic context, spontaneously active atrial cardiomyocytes could be identified.79 Hence, ectopic expression of Tbx3 does not induce pacemaker function in adult chamber cardiomyocytes, but provides potential when expressed from embryonic stages onwards in atrial cardiomyocytes.
Emerging before Tbx3 during development, Tbx18 is necessary for the development of the leading pacemaker region of the SAN.38 Lineage tracing studies have confirmed that Tbx18-expressing mesenchymal progenitor cells found in the inflow tract of the developing heart differentiate into the pacemaker lineage.38 Given this importance, Tbx18 was overexpressed in neonatal rat cardiomyocytes in vitro and adult guinea pig ventricles in vivo via adenovirus (AdvTbx18) transduction. In contrast to Tbx3, singular expression of Tbx18 was sufficient to reprogram ventricular cardiomyocytes to induced pacemaker cells in vitro and in vivo, as demonstrated by an increase in Hcn4 expression and If, repression of chamber myocardial ion channels and gap junctions, and oscillatory local Ca2+ release events.76,77 Further, ectopic pacemaker activity was induced in vivo.77 In a clinically relevant large animal model of complete heart block, AdvTbx18 was delivered to the AV junctional region to achieve septal pacing and ventricular synchrony. This resulted in significantly reduced dependence on backup electronic pacemakers and heart rate adaptation that correlated with the animals’ activity, without increase in arrhythmogenecity.78 However, biological pacing appeared to decline one to two weeks after gene transfer, possibly due to degradation of the adenoviral transgene or incomplete reprogramming of the host cells.
Stem cell differentiation to pacemaker cells
Human pluripotent stem cells have proved valuable to the field of regenerative medicine, providing an indefinite source of de novo cardiomyocytes and potentially unlimited tissue for human transplantation. While many studies have focused on increasing cardiomyocyte yield and/or maturation, relatively few studies have provided approaches for direct differentiation to nodal cardiomyocytes. In this section, we introduce several stem cell-based systems for the derivation of pacemaker-like cells and corresponding demonstrations of function in vivo.
Cardiac embryoid bodies
The first in vivo demonstration of an hPSC-derived biological pacemaker used spontaneously beating embryoid bodies (EBs) derived from human embryonic stem cells (ESCs) in the ventricles of pigs with complete heart block.81 Three-dimensional electrophysiological mapping showed that the engrafted human EBs successfully paced the hearts, providing proof-of-concept evidence that the injected cells can survive, integrate with host myocardium, and provide pacing. Further studies confirmed the ability of human ESC-derived EBs to integrate with and pace guinea pig hearts in vivo.82 The transplanted EBs were also found to be responsive to β-adrenergic stimulation and pharmacological agents, evidence of autonomic responsiveness. As human ESCs give rise to ethical concerns and the need for immunosuppression, Chauveau, et al., used EBs derived from human induced pluripotent stem cells (iPSCs). These EBs demonstrated biological pacemaker function in a canine model of complete heart block for up to 13 weeks.83
Though the cardiac EB approach shows evidence of pacing function in several large-animal models, the rate and robustness of pacing have yet to be optimized. Further, as EBs are formed simply by 3D culture with no biological cues for directed differentiation, there are multiple cell types present, likely in inconsistent proportions among differentiations. To date it remains unclear how the observed biological pacing of cardiac EB transplantation may be affected by differences in the derived cardiac subtype makeup present in the EB, but perhaps this assortment of subtypes could actually be beneficial given the heterogeneity of the SAN. Thus, this phenomenon begs the question of what cell subtype proportions will be optimal for robust and less arrhythmogenic stem cell-derived biological pacemakers. Nevertheless, it remains clear that pacemaker-specific differentiation protocols are needed for control over cellular composition.
Genetic programming for EB culture
In order to obtain a pacemaker cell-enriched population in cardiac EBs, Jung, et al., transfected mouse ESCs with Tbx3, generating EBs with 38.5% pacemaker-like cells and 38.8% “intermediate” cells.84 While this was an improvement compared to control EBs with 20% pacemaker cells, the authors introduced a Myh6-Neomycin cassette for antibiotic selection of cells expressing myosin heavy chain 6. Over 82% of these cells produced action potentials reflecting that of mature pacemaker cells and achieved beat rates comparable to that of the mouse SAN.
Ionta, et al., on the other hand, used Shox2 to improve EB differentiation to pacemaker cells.66 The Shox2 transcription factor is expressed in the sinus venosus region during development, and Shox2 null mutations in mouse models revealed bradycardia and significantly underdeveloped SAN tissue.44 The important role of Shox2 in SAN development and Nkx2.5 repression46 make it a suitable gene to target for direct differentiation. Exogenous overexpression of Shox2 during early stages of mouse ESC EB differentiation showed enhanced pacemaker differentiation.66 This exogenous overexpression activated endogenous expression of Shox2 post-differentiation, which, along with increased expression of proteins related to the pacemaker’s membrane (Hcn4) and calcium clocks (Ncx1), infer a strong pacemaker phenotype. Shox2-EBs displayed higher rates of automaticity in vitro and paced the rat heart following transplantation in vivo. Overall, while these two studies by Jung and Ionta are promising, they have yet to be translated to human cells and larger, more clinically relevant animal models.
Retinoic acid, BMP, and Wnt signaling
While inducing expression of transcription factors is encouraging, another approach to creating stem cell-derived pacemaker cells relies on the use of biological molecules important in pacemaker development. As discussed above, RA and BMPs play a key role in mesoderm patterning and the induction of pacemaker cell gene expression. Thus, hPSCs were treated with varying concentrations of RA and BMP4 at various time points to enrich for Nkx2.5-negative myocytes. An Nkx2.5 fluorescent reporter cell line allowed cell sorting to obtain a pure population of SAN-like pacemaker cells (SANLPCs).35 SANLPCs were found to contain several molecular and electrophysiological features of the native SAN, such as increased Shox2 expression, characteristic action potentials, and increased density of If, and demonstrated biological pacemaker function in a rat model of complete heart block. To circumvent the need for a transgenic line, which would limit clinical translation, further optimization was done to increase SANLPC yield via inhibition of fibroblast growth factor signaling. Future work in a large animal model will be needed to evaluate the robustness of biological pacing with these cells.
Wnt/β-catenin signaling has been shown to be important for the maintenance and proliferation of Tbx18+ and Nkx2.5- mesenchymal progenitors in sinus horn formation.87 Cardiac differentiation protocols begin with activation of Wnt signaling via inhibition of glycogen synthase kinase 3β (GSK3β) to induce expression of mesendodermal markers. Upon formation of the mesoderm, Wnt inhibitors are added in order to drive cardiac lineage specification.88 By adding Wnt activator CHIR99021 to hPSCs for two days following this inhibition, Ren, et al., were able to obtain a high proportion of cells expressing pacemaker genes that were able to pace 3D-bioprinted human ESC-derived cardiomyocytes.36 Alternatively, by omitting exogenous Wnt inhibitors altogether, Liang, et al., found differentiated mouse and human PSCs to have an increased yield of spontaneously beating cardiomyocytes with action potentials similar to native pacemaker cells.37 Further activation of Wnt/β-catenin signaling by exogenous addition of Wnt3a ligand increased the yield of pacemaker-like cells at the expense of total cardiomyocyte yield. Careful fine-tuning and manipulation of canonical Wnt signaling therefore appear to be critical for specific differentiation of pacemaker cells.
Structural considerations for overcoming source-sink mismatch
The unique microenvironment and heterogeneous cell mixture of the sinoatrial node may offer critical design considerations for successful biological pacing. One of the first experiments to shed light on microenvironment considerations for biological pacemakers involved enzymatically-isolated canine sinoatrial cells delivered to the right ventricle of dogs with complete heart block.89 Despite these sinoatrial cells representing a potentially ideal source for biological pacing, only about half of all ventricular contractions originated from the injection site. This lack of function emphasized the importance of recapitulating SAN architecture to overcome source-sink mismatch.
As the first of its kind, Grijalva, et al., created in vitro models of source-sink mismatch with Tbx18-reprogrammed pacemaker cells cultured in defined structures in order to test design features necessary for overcoming mismatch.90 Tbx18-induced pacemaker cells were aggregated to form three-dimensional spheres (sphTbx18), co-cultured with a monolayer of ventricular myocytes, and loaded with the calcium indicator Rhod-2 AM to determine the origin of electrical activation. Pace-and-drive was most successful when contact was limited to a single side of the ventricular monolayer, whereas when the sphTbx18 pacing source was surrounded on all sides by the ventricular syncytium, pacing was significantly limited. Interestingly, both pharmacological inhibition of electrical coupling between source and sink tissues and β-adrenergic stimulation significantly increased the pace-and-drive ability of sphTbx18 sources.90
Lastly, as excitability of minuscule cardiac tissues such as the SAN would easily be dominated by the hyperpolarizing myocardium, Sayegh, et al., studied parameters that would electrically shield a bioengineered pacemaker tissue from the surrounding myocardium. This study found that smaller tissue dimensions, perpendicular cell alignment relative to the electrical field, and increased fibroblast content independently raised the electrical capture threshold of the small cardiac tissue.91 Based on these findings, the ideal biological pacemaker would be small, exhibit isotropic cell alignment, and consist of a large non-myocyte population. Discrete exit pathways for electrical propagation92 should be tested as well.
To design such architecture and complexity, 3D bioprinting is a promising approach, capable of spatially organizing several cell types and relevant ECM components. Currently, printing multiple ink types together in a single print requires extrusion-based printing with multiple nozzles; however, light-based methods (e.g., stereolithography, digital light processing) offer higher resolution, which will be needed to recreate the micrometer-scale features of the SAN. Recent research using light-based printing has demonstrated simplified multi-material printing93 and submicron resolution.94 As the bioprinting field continues to rapidly expand, along with the field of proteomics for identifying components of the SAN, the structural complexity of the SAN may be recapitulated with high fidelity.
Delivery Strategies for Clinical Translation
Development of a safe and effective delivery strategy will be instrumental to the clinical translation of biological pacemakers. The method of delivery in the clinical setting will depend on the final design of the biological pacemaker and disease indications. Most proof-of-concept studies discussed here delivered the adenovirus, plasmid, or cells via injection into the myocardial wall following a thoracotomy.35,66,68,71,75–77,81–83 In attempt to be less invasive, others have used intracardiac catheters via subcutaneous venous access in large animal models.69,73,74,78 While injection, either by thoracotomy or via catheter, provides some degree of gene/cell retention, this method may not be compatible with biological pacemakers containing structurally organized tissues with multiple cell types and specific protein compositions. Thus, a “patch” option, as introduced with biologics for treating damaged myocardium, may be a more suitable approach..95,96 These patches have typically been delivered via open-chest surgery,94,97 but less-invasive methods are being developed,98,99 which will be similarly useful for the delivery of biological pacemakers. Emphasis on material selection98 and location of injection99 have circumvented the need for thoracotomies, while also demonstrating vascularization and imposed function on the heart. This work shows promise for long-term use and retention. In the end, the delivery method cannot be of high complexity relative to electronic pacemaker placement.
Key Opportunities
Through review of SAN structure, function, and development, as well as attempts to recapitulate these features in a dish, the need for further studies is clear. Revisiting our proposed design criteria, we emphasize several items for consideration in future studies.
Biological pacemakers must generate spontaneous action potentials originating from a coupled membrane and calcium clock system, with clock protein expression maintained at levels comparable to that of the native pacemaker.
As the generation of rhythmic action potentials is the basis of SAN pacing, this feature is crucial to have in derived pacing cells. Most of the studies detailed above were able to successfully produce spontaneous action potentials that reflect those of the SAN. What remains to be examined further, though, is the regularity of spontaneous activity and whether it can be maintained with long-term cultures. A lack of stable oscillations is a concern in generating pacemaker cells, which would be unacceptable for translation to the clinic. Further, the expression of membrane and calcium clock proteins in these cells is rarely measured, despite their importance in creating such oscillations. To have stable functionality, derived pacemaker cells must express relevant proteins (e.g. Hcn4 and Ncx1) and maintain this expression at levels comparable to that of native cells. An increased understanding of early pacemaker specification, particularly gene regulation, would be ideal for improving the derivation of pacemaker cells with relevant protein expression.
Biological pacemakers must overcome source-sink mismatch to pace and drive large regions of tissue.
While in vivo studies suggest a possible source-sink balance between the biological pacemaker and the myocardium, this phenomenon is modestly characterized. Electrical integration of biological pacemakers into the heart will need to be better understood prior to clinical implementation. For example, the formation of gap junction proteins between native and implanted tissues should be studied moving forward.
Biological pacemakers must provide reliable pacing over long periods of time.
Pacing the myocardium in vivo demonstrates the ability of the biological pacemaker to generate and propagate electrical activity. However, the longevity of these pacemakers has yet to be determined. Most studies described here only study in vivo functionality for days or a couple weeks, which are insufficient durations to validate these cells. Studies must continue for at least six months to ensure reliable long-term pacing. The success of this criterion will depend heavily on overcoming source-sink mismatch and maintaining pacemaker phenotype long-term.
Biological pacemakers would ideally contain a relatively similar level of heterogeneous nodal cell types, as seen in the SAN.
Many studies of the SAN have demonstrated the importance of its transitional cells; however, previous attempts to form biological pacemakers have neglected this important feature of the SAN. Thus, in addition to being able to form a population of pacing cells, it is likely there will also be a need to form transitional cells as well. With a concrete protocol for generating pacemaker cells will likely come an ability to generate these intermediate cells. Co-culture with other relevant cell types found in the SAN, including nerve, smooth muscle, macrophages, or fibroblasts, may also provide improvements to biological pacing.
Biological pacemakers would ideally replicate nodal tissue architecture, with consideration for pacemaker and transitional cell patterning, extracellular matrix composition, and culture substrates.
Once transitional cells can be derived, and the role of other cell types are delineated, studies will need to focus on the patterning of such cells. The cells will need to be positioned to maximize pacemaker function. Further, the important role of ECM proteins in electrical and mechanical insulation has been demonstrated, yet previous studies have not incorporated this concept in the creation of biological pacemakers. Recapitulating the role of the extracellular environment will be important, particularly inclusion of the collagen and other elastic proteins to help sustain the biological pacemaker long term. There are limited studies regarding the extracellular matrix of the SAN and the effect of age, disease, and development on this environment, presenting a large opportunity moving forward.
Biological pacemakers would ideally demonstrate autonomic responsiveness, adjusting the rate of pacing based on the physiological needs of the body.
Many studies have examined the effect of exogenous neurotransmitters, such as isoproterenol and acetylcholine, on derived pacemaker cells. However, such effects should also be examined in vivo. Specifically, studies should examine the impact of various conditions, particularly stress, on pacing the entire heart in order to demonstrate consistent chronotropic competence. Being able to withstand common modifiers of heart rate, such as environmental stressors, exercise, and adrenaline, is ideal for a successful biological pacemaker.
Given the current state of preclinical research and progress made in the field of biological pacing, clinical long-term replacement of electronic pacemakers is not yet achievable. Table 1 summarizes the status of the field and what needs to be accomplished for success in the clinic. Overall, many biological pacemaker studies have focused heavily on action potential generation, but the complexity of the SAN indicates much more is necessary. Further work may still be needed to unlock potential benefits of incorporating SAN-like structural features in cellular makeup and ECM. Despite these shortfalls, current biological pacemaker approaches show exceptional promise in their ability to pace in vivo and may provide a device-free alternative for temporary and short-term pacing in certain high-risk patient populations, such as patients with corrective surgery complications or device-related infections. With focus on the proposed design criteria in future work, we can move closer to implementing biological pacemakers as a long-term treatment.
Table 1.
Biological Pacemaker Design Criteria, Current and Future Work
| Design Criteria | Current Progress | Future Work |
|---|---|---|
| Generate spontaneous action potentials with a coupled clock | Most studies verify spontaneous generation of pacemaker-like action potentials. | Measure expression and maintenance of calcium and membrane clock proteins. |
| Overcome source-sink mismatch | In vivo studies aim to prove pacing is possible, and various mapping techniques have shown electrical propagation. | Electromechanical integration should be demonstrated. Incorporate design features such as electrical insulation and exit pathways. |
| Provide reliable pacing over long periods of time | Most in vivo studies are days to two weeks long. The longest study was 13 weeks.83 | Monitor for at least 6 months for potential failure or arrhythmias. |
| Contain heterogeneous nodal cell types | Studies using embryoid bodies contain multiple cell types, but this concept is not otherwise addressed. | A reliable method for generating nodal cells, as well as transitional cells, will need to be developed. Co-culture with multiple cell types may yield new insights. |
| Replicate the nodal tissue architecture | No design studies address the role of ECM,* cell environment, or culture substrates. | Determine the effect of cell patterning and incorporate ECM to tissue-engineered scaffolds. Further characterization of SAN† ECM is necessary. |
| Demonstrate autonomic responsiveness | Response to isoproterenol is frequently shown in vitro, and two in vivo studies monitor heart rate during daily activity.78,83 | Chronotropic competence must be studied thoroughly, particularly under high stress, such as exercise stress testing. |
extracellular matrix
sinoatrial node
Sources of Funding
This work was supported by the NIH T32GM008347 (E.R.K), R01HL137204 (E.R.K. and B.M.O.), F31HL149272-01A1 (D.W.W.), R01HL143065 (H.C.C.), R01HL111646 (H.C.C.), R01HL146626 (M.B.), R01HL157363 (H.C.C.), and the American Heart Association CDA34760248 (M.B.) and 20TPA35260085 (H.C.C.).
Nonstandard Abbreviations and Acronyms
- BMP
bone morphogenic protein
- cAMP
cyclic adenosine monophosphate
- Cx
connexin
- EBs
embryoid bodies
- ECM
extracellular matrix
- ESCs
embryonic stem cells
- Hcn
hyperpolarization-activated cyclic nucleotide-gated channel
- hPSCs
human pluripotent stem cells
- iPSCs
induced pluripotent stem cells
- Ncx1
sodium-calcium exchanger 1
- PKA
protein kinase A
- RA
retinoic acid
- RyR2
ryanodine receptor 2
- SAN
sinoatrial node
- SANLPCs
sinoatrial node-like pacemaker cells
- Serca2
sarco/endoplasmic reticulum Ca2+-ATPase 2
Footnotes
Disclosures
None.
References
- 1.Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, et al. Heart disease and stroke statistics-2021 update: A report from the American Heart Association. Circulation. 2021;143:e254–e743. [DOI] [PubMed] [Google Scholar]
- 2.Birnie D, Williams K, Guo A, Mielniczuk L, Davis D, Lemery R, Green M, Gollob M, Tang A. Reasons for escalating pacemaker implants. Am J Cardiol. 2006;98:93–97. [DOI] [PubMed] [Google Scholar]
- 3.Cantillon DJ, Exner D V, Badie N, Davis K, Gu NY, Nabutovsky Y, Doshi R. Complications and health care costs associated with transvenous cardiac pacemakers in a nationwide assessment. JACC Clin Electrophysiol. 2017;3:1296–1305. [DOI] [PubMed] [Google Scholar]
- 4.Reddy VY, Exner D V, Cantillon DJ, Doshi R, Bunch TJ, Tomassoni GF, Friedman PA, Estes NAM, Ip J, Niazi I, et al. Percutaneous implantation of an entirely intracardiac leadless pacemaker. N Engl J Med. 2015;373:1125–1135. [DOI] [PubMed] [Google Scholar]
- 5.Knops RE, Tjong FVY, Neuzil P, Sperzel J, Miller MA, Petru J, Simon J, Sediva L, De Groot JR, Dukkipati SR, et al. Chronic performance of a leadless cardiac pacemaker: 1-year follow-up of the LEADLESS trial. J Am Coll Cardiol. 2015;65:1497–1504. [DOI] [PubMed] [Google Scholar]
- 6.Tjong FVY, Stam OCG, van der Wal AC, Beenen LF, Bouma BJ, de Groot JR, Wilde AAM, Knops RE. Postmortem histopathological examination of a leadless pacemaker shows partial encapsulation after 19 months. Circ Arrhythmia Electrophysiol. 2015;8:1293–1295. [DOI] [PubMed] [Google Scholar]
- 7.Kypta A, Blessberger H, Kammler J, Lichtenauer M, Lambert T, Silye R, Steinwender C. First autopsy description of changes 1 year after implantation of a leadless cardiac pacemaker: Unexpected ingrowth and severe chronic inflammation. Can J Cardiol. 2016;32:1578.e1–1578.e2. [DOI] [PubMed] [Google Scholar]
- 8.van Eif VWW, Protze SI, Bosada FM, Yuan X, Sinha T, van Duijvenboden K, Ernault AC, Mohan RA, Wakker V, de Gier-de Vries C, et al. Genome-wide analysis identifies an essential human TBX3 pacemaker enhancer. Circ Res. 2020;127:1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhard S, van Eif V, Garric L, Christoffels VM, Bakkers J, Poelmann RE, Jongbloed MRM. On the evolution of the cardiac pacemaker. J Cardiovasc Dev Dis. 2017;4:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesirca P, Bidaud I, Briec F, Evain S, Torrente AG, Le Quang K, Leoni AL, Baudot M, Marger L, Chong ACY, et al. G protein-gated IKACh channels as therapeutic targets for treatment of sick sinus syndrome and heart block. Proc Natl Acad Sci U S A. 2016;113:E932–E941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodyer WR, Beyersdorf BM, Paik DT, Tian L, Li G, Buikema JW, Chirikian O, Choi S, Venkatraman S, Adams EL, et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ Res. 2019;125:379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Li D, Wang Y, Huang Z, Xu J, Yang T, Wang L, Tang Q, Cai C- L, Huang H, et al. Nkx2–5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development. 2019;146:dev178145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csepe TA, Kalyanasundaram A, Hansen BJ, Zhao J, Fedorov V V. Fibrosis: A structural modulator of sinoatrial node physiology and dysfunction. Front Physiol. 2015;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linscheid N, Logantha SJRJ, Poulsen PC, Zhang S, Schrölkamp M, Egerod KL, Thompson JJ, Kitmitto A, Galli G, Humphries MJ, et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat Commun. 2019;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluck JM, Herren AW, Yechikov S, Kao HKJ, Khan A, Phinney BS, Chiamvimonvat N, Chan JW, Lieu DK. Biochemical and biomechanical properties of the pacemaking sinoatrial node extracellular matrix are distinct from contractile left ventricular matrix. PLoS One. 2017;12:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James TN, Sherf L, Fine G, Morales AR. Comparative ultrastructure of the sinus node in man and dog. Circulation. 1966;34:139–163. [DOI] [PubMed] [Google Scholar]
- 17.Tsutsui K, Monfredi OJ, Sirenko-Tagirova SG, Maltseva LA, Bychkov R, Kim MS, Ziman BD, Tarasov K V., Tarasova YS, Zhang J, et al. A coupled-clock system drives the automaticity of human sinoatrial nodal pacemaker cells. Sci Signal. 2018;11:7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res. 2010;106:659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maltsev VA, Lakatta EG. Synergism of coupled subsarcolemmal Ca2+ clocks and sarcolemmal voltage clocks confers robust and flexible pacemaker function in a novel pacemaker cell model. Am J Physiol - Hear Circ Physiol. 2009;296:594–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brioschi C, Micheloni S, Tellez JO, Pisoni G, Longhi R, Moroni P, Billeter R, Barbuti A, Dobrzynski H, Boyett MR, et al. Distribution of the pacemaker HCN4 channel mRNA and protein in the rabbit sinoatrial node. J Mol Cell Cardiol. 2009;47:221–227. [DOI] [PubMed] [Google Scholar]
- 21.Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiFrancesco D, Ferroni A, Mazzanti M, Tromba C. Properties of the hyperpolarizing‐activated current (if) in cells isolated from the rabbit sino‐atrial node. J Physiol. 1986;377:61–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiFrancesco D The cardiac hyperpolarizing-activated current, if. Origins and developments. Prog Biophys Mol Biol. 1985;46:163–183. [DOI] [PubMed] [Google Scholar]
- 24.DiFrancesco D Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. [DOI] [PubMed] [Google Scholar]
- 25.Vinogradova TM, Zhou YY, Maltsev V, Lyashkov A, Stern M, Lakatta EG. Rhythmic ryanodine receptor Ca2+ releases during diastolic depolarization of sinoatrial pacemaker cells do not require membrane depolarization. Circ Res. 2004;94:802–809. [DOI] [PubMed] [Google Scholar]
- 26.Vinogradova T, Tagirova (Sirenko) S, Lakatta EG. Unique Ca2+-cycling protein abundance and regulation sustains local Ca2+ releases and spontaneous firing of rabbit sinoatrial node cells. Int J Mol Sci. 2018;19:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogdanov KY, Vinogradova TM, Lakatta EG. Sinoatrial nodal cell ryanodine receptor and Na+-Ca2+ exchanger: Molecular partners in pacemaker regulation. Circ Res. 2001;88:1254–1258. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann S, Lipp P, Wiesen K, Stieber J, Nguyen H, Kaiser E, Ludwig A. The cardiac sodium-calcium exchanger NCX1 is a key player in the initiation and maintenance of a stable heart rhythm. Cardiovasc Res. 2013;99:780–788. [DOI] [PubMed] [Google Scholar]
- 29.Bychkov R, Juhaszova M, Tsutsui K, Coletta C, Stern MD, Maltsev VA, Lakatta EG. Synchronized cardiac impulses emerge from heterogeneous local calcium signals within and among cells of pacemaker tissue. JACC Clin Electrophysiol. 2020;6:907–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders L, Rakovic S, Lowe M, Mattick PAD, Terrar DA. Fundamental importance of Na+-Ca2+ exchange for the pacemaking mechanism in guinea-pig sino-atrial node. J Physiol. 2006;571:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald EA, Rose RA, Quinn TA. Neurohumoral control of sinoatrial node activity and heart rate: Insight from experimental models and findings from humans. Front Physiol. 2020;11:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mommersteeg MTM, Domínguez JN, Wiese C, Norden J, De Gier-De Vries C, Burch JBE, Kispert A, Brown NA, Moorman AFM, Christoffels VM. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010;87:92–101. [DOI] [PubMed] [Google Scholar]
- 34.Domínguez JN, Meilhac SM, Bland YS, Buckingham ME, Brown NA. Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ Res. 2012;111:1323–1335. [DOI] [PubMed] [Google Scholar]
- 35.Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat Biotechnol. 2017;35:56–68. [DOI] [PubMed] [Google Scholar]
- 36.Ren J, Han P, Ma X, Farah EN, Bloomekatz J, Zeng XXI, Zhang R, Swim MM, Witty AD, Knight HG, et al. Canonical Wnt5b signaling directs outlying Nkx2.5+ mesoderm into pacemaker cardiomyocytes. Dev Cell. 2019;50:729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang W, Han P, Kim EH, Mak J, Zhang R, Torrente AG, Goldhaber JI, Marbán E, Cho HC. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem Cells. 2020;38:352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiese C, Grieskamp T, Airik R, Mommersteeg MTM, Gardiwal A, De Gier-De Vries C, Schuster-Gossler K, Moorman AFM, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. [DOI] [PubMed] [Google Scholar]
- 39.Christoffels VM, Mommersteeg MTM, Trowe M- O, Prall OWJ, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AFM, et al. Formation of the venous pole of the heart from an Nkx2–5–negative precursor population requires Tbx18. Circ Res. 2006;98:1555–1563. [DOI] [PubMed] [Google Scholar]
- 40.Mommersteeg MTM, Hoogaars WMH, Prall OWJ, de Gier-de Vries C, Wiese C, Clout DEW, Papaioannou VE, Brown NA, Harvey RP, Moorman AFM, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai C, Chen J, Evans S. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang X, Zhang Q, Cattaneo P, Zhuang S, Gong X, Spann NJ, Jiang C, Cao X, Zhao X, Zhang X, et al. Transcription factor ISL1 is essential for pacemaker development and function. J Clin Invest. 2015;125:3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoogaars WMH, Engel A, Brons JF, Verkerk AO, De Lange FJ, Wong LYE, Bakker ML, Clout DE, Wakker V, Barnett P, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. [DOI] [PubMed] [Google Scholar]
- 45.Ye W, Wang J, Song Y, Yu D, Sun C, Liu C, Chen F, Zhang Y, Wang F, Harvey RP, et al. A common Shox2–Nkx2–5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Dev. 2015;142:2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev Biol. 2009;327:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vedantham V New approaches to biological pacemakers: Links to sinoatrial node development. Trends Mol Med. 2015;21:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Eif VWW, Stefanovic S, van Duijvenboden K, Bakker M, Wakker V, de Gier-de Vries C, Zaffran S, Verkerk AO, Boukens BJ, Christoffels VM. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Dev. 2019;146:1–15. [DOI] [PubMed] [Google Scholar]
- 49.Galang G, Mandla R, Ruan H, Jung C, Sinha T, Stone NR, Wu RS, Mannion BJ, Allu PK, Chang K, et al. ATAC-seq reveals an Isl1 enhancer that regulates sinoatrial node development and function. Circ Res. 2020;127:1502–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bressan M, Henley T, Louie JD, Liu G, Christodoulou D, Bai X, Taylor J, Seidman CE, Seidman JG, Mikawa T. Dynamic cellular integration drives functional assembly of the heart’s pacemaker complex. Cell Rep. 2018;23:2283–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verkerk AO, Wilders R, Van Borren MMGJ, Peters RJG, Broekhuis E, Lam K, Coronel R, De Bakker JMT, Tan HL. Pacemaker current (If) in the human sinoatrial node. Eur Heart J. 2007;28:2472–2478. [DOI] [PubMed] [Google Scholar]
- 52.Monfredi O, Tsutsui K, Ziman B, Stern MD, Lakatta EG, Maltsev VA. Electrophysiological heterogeneity of pacemaker cells in the rabbit intercaval region, including the SA node: insights from recording multiple ion currents in each cell. Am J Physiol Circ Physiol. 2018;314:H403–H414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kléber AG, Rudy Y. Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiol Rev. 2004;84:431–488. [DOI] [PubMed] [Google Scholar]
- 54.Rohr S, Kucera JP, Fast VG, Kléber AG. Paradoxical improvement of impulse conduction in cardiac tissue by partial cellular uncoupling. Science. 1997;275:841–844. [DOI] [PubMed] [Google Scholar]
- 55.Rohr S, Kucera JP, Kléber AG. Slow conduction in cardiac tissue, I: Effects of a reduction in excitability versus a reduction of electrical coupling on microconduction. Circ Res. 1998;83:781–794. [DOI] [PubMed] [Google Scholar]
- 56.Unudurthi SD, Wolf RM, Hund TJ. Role of sinoatrial node architecture in maintaining a balanced source-sink relationship and synchronous cardiac pacemaking. Front Physiol. 2014;5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Circ Res. 1997;81:727–741. [DOI] [PubMed] [Google Scholar]
- 58.Joyner RW, van Capelle FJ. Propagation through electrically coupled cells: How a small SA node drives a large atrium. Biophys J. 1986;50:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouman LN, Jongsma HJ. Structure and function of the sino-atrial node: A review. Eur Heart J. 1986;7:94–104. [DOI] [PubMed] [Google Scholar]
- 60.Masson-Pevet MA, Bleeker WK, Besselsen E, Treytel BW, Jongsma HJ, Bouman LN. Pacemaker cell types in the rabbit sinus node: A correlative ultrastructural and electrophysiological study. J Mol Cell Cardiol. 1984;16:53–63. [DOI] [PubMed] [Google Scholar]
- 61.Honjo H, Boyett MR, Coppen SR, Takagishi Y, Opthof T, Severs NJ, Kodama I. Heterogeneous expression of connexins in rabbit sinoatrial node cells: Correlation between connexin isotype and cell size. Cardiovasc Res. 2002;53:89–96. [DOI] [PubMed] [Google Scholar]
- 62.Inada S, Zhang H, Tellez JO, Shibata N, Nakazawa K, Kamiya K, Kodama I, Mitsui K, Dobrzynski H, Boyett MR, et al. Importance of gradients in membrane properties and electrical coupling in sinoatrial node pacing. PLoS One. 2014;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47:658–687. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, Holden AV., Kodama I, Honjo H, Lei M, Varghese T, Boyett MR. Mathematical models of action potentials in the periphery and center of the rabbit sinoatrial node. Am J Physiol - Hear Circ Physiol. 2000;279:H397–H421. [DOI] [PubMed] [Google Scholar]
- 65.Hoogaars WMH, Tessari A, Moorman AFM, De Boer PAJ, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. [DOI] [PubMed] [Google Scholar]
- 66.Ionta V, Liang W, Kim EH, Rafie R, Giacomello A, Marbán E, Cho HC. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Reports. 2015;4:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fast VG, Kléber AG. Block of impulse propagation at an abrupt tissue expansion: evaluation of the critical strand diameter in 2- and 3-dimensional computer models. Cardiovasc Res. 1995;30:449–459. [PubMed] [Google Scholar]
- 68.Edelberg JM, Aird WC, Rosenberg RD. Enhancement of murine cardiac chronotropy by the molecular transfer of the human β2 adrenergic receptor cDNA. J Clin Invest. 1998;101:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edelberg JM, Huang DT, Josephson ME, Rosenberg RD. Molecular enhancement of porcine cardiac chronotropy. Heart. 2001;86:559–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miake J, Marbán E, Nuss HB. Biological pacemaker created by gene transfer. Nature. 2002;419:132–133. [DOI] [PubMed] [Google Scholar]
- 71.Miake J, Marbán E, Nuss HB. Functional role of inward rectifier current in heart probed by Kir2.1 overexpression and dominant-negative suppression. J Clin Invest. 2003;111:1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qu J, Barbuti A, Protas L, Santoro B, Cohen IS, Robinson RB. HCN2 overexpression in newborn and adult ventricular myocytes: distinct effects on gating and excitability. Circ Res. 2001;89:e8–e14. [DOI] [PubMed] [Google Scholar]
- 73.Bucchi A, Plotnikov AN, Shlapakova I, Danilo P, Kryukova Y, Qu J, Lu Z, Liu H, Pan Z, Potapova I, et al. Wild-type and mutant HCN channels in a tandem biological-electronic cardiac pacemaker. Circulation. 2006;114:992–999. [DOI] [PubMed] [Google Scholar]
- 74.Plotnikov AN, Bucchi A, Shlapakova I, Danilo P, Brink PR, Robinson RB, Cohen IS, Rosen MR. HCN212-channel biological pacemakers manifesting ventricular tachyarrhythmias are responsive to treatment with If blockade. Hear Rhythm. 2008;5:282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tse HF, Xue T, Lau CP, Siu CW, Wang K, Zhang QY, Tomaselli GF, Akar FG, Li RA. Bioartificial sinus node constructed via in vivo gene transfer of an engineered pacemaker HCN channel reduces the dependence on electronic pacemaker in a sick-sinus syndrome model. Circulation. 2006;114:1000–1011. [DOI] [PubMed] [Google Scholar]
- 76.Kapoor N, Galang G, Marbán E, Cho HC. Transcriptional suppression of connexin43 by Tbx18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J Biol Chem. 2011;286:14073–14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kapoor N, Liang W, Marbán E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu YF, Dawkins JF, Cho HC, Marbán E, Cingolani E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci Transl Med. 2014;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bakker ML, Boink GJJ, Boukens BJ, Verkerk AO, Van Den Boogaard M, Den Haan AD, Hoogaars WMH, Buermans HP, De Bakker JMT, Seppen J, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94:439–449. [DOI] [PubMed] [Google Scholar]
- 80.Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. PNAS. 2012;109:E154–E163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kehat I, Khimovich L, Caspi O, Gepstein A, Shofti R, Arbel G, Huber I, Satin J, Itskovitz-Eldor J, Gepstein L. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22:1282–1289. [DOI] [PubMed] [Google Scholar]
- 82.Xue T, Cho HC, Akar FG, Tsang SY, Jones SP, Marbán E, Tomaselli GF, Li RA. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes. Circulation. 2005;111:11–20. [DOI] [PubMed] [Google Scholar]
- 83.Chauveau S, Anyukhovsky EP, Ben-Ari M, Naor S, Jiang YP, Danilo P, Rahim T, Burke S, Qiu X, Potapova IA, et al. Induced pluripotent stem cell-derived cardiomyocytes provide in vivo biological pacemaker function. Circ Arrhythmia Electrophysiol. 2017;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jung JJ, Husse B, Rimmbach C, Krebs S, Stieber J, Steinhoff G, Dendorfer A, Franz WM, David R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Reports. 2014;2:592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kashiwakura Y, Cho HC, Barth AS, Azene E, Marbán E. Gene transfer of a synthetic pacemaker channel into the heart: A novel strategy for biological pacing. Circulation. 2006;114:1682–1686. [DOI] [PubMed] [Google Scholar]
- 86.Cho HC, Kashiwakura Y, Marbán E. Creation of a biological pacemaker by cell fusion. Circ Res. 2007;100:1112–1115. [DOI] [PubMed] [Google Scholar]
- 87.Norden J, Greulich F, Rudat C, Taketo MM, Kispert A. Wnt/β-catenin signaling maintains the mesenchymal precursor pool for murine sinus horn formation. Circ Res. 2011;109:e42–e50. [DOI] [PubMed] [Google Scholar]
- 88.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang H, Lau DH, Shlapakova IN, Zhao X, Danilo P, Robinson RB, Cohen IS, Qu D, Xu Z, Rosen MR. Implantation of sinoatrial node cells into canine right ventricle: Biological pacing appears limited by the substrate. Cell Transplant. 2011;20:1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grijalva SI, Gu J, Li J, Fernandez N, Fan J, Sung JH, Lee SY, Herndon C, Buckley EM, Park SJ, et al. Engineered cardiac pacemaker nodes created by TBX18 gene transfer overcome source–sink mismatch. Adv Sci. 2019;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sayegh MN, Fernandez N, Cho HC. Strength-duration relationship as a tool to prioritize cardiac tissue properties that govern electrical excitability. Am J Physiol - Hear Circ Physiol. 2019;317:H13–H25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li N, Hansen BJ, Csepe TA, Zhao J, Ignozzi AJ, Sul L V., Zakharkin SO, Kalyanasundaram A, Davis JP, Biesiadecki BJ, et al. Redundant and diverse intranodal pacemakers and conduction pathways protect the human sinoatrial node from failure. Sci Transl Med. 2017;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grigoryan B, Sazer DW, Avila A, Albritton JL, Padhye A, Ta AH, Greenfield PT, Gibbons DL, Miller JS. Development, characterization, and applications of multi-material stereolithography bioprinting. Sci Rep. 2021;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao L, Kupfer ME, Jung JP, Yang L, Zhang P, Da Sie Y, Tran Q, Ajeti V, Freeman BT, Fast VG, et al. Myocardial tissue engineering with cells derived from human-induced pluripotent stem cells and a native-like, high-resolution, 3-dimensionally printed scaffold. Circ Res. 2017;120:1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Zhu W, Radisic M, Vunjak-Novakovic G. Can we engineer a human cardiac patch for therapy? Circ Res. 2018;123:244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mei X, Cheng K. Recent development in therapeutic cardiac patches. Front Cardiovasc Med. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui H, Liu C, Esworthy T, Huang Y, Yu Z, Zhou X, San H, Lee S, Hann SY, Boehm M, et al. 4D physiologically adaptable cardiac patch: A 4-month in vivo study for the treatment of myocardial infarction. Sci Adv. 2020;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Montgomery M, Ahadian S, Davenport Huyer L, Lo Rito M, Civitarese RA, Vanderlaan RD, Wu J, Reis LA, Momen A, Akbari S, et al. Flexible shape-memory scaffold for minimally invasive delivery of functional tissues. Nat Mater. 2017;16:1038–1046. [DOI] [PubMed] [Google Scholar]
- 99.Zhu D, Li Z, Huang K, Caranasos TG, Rossi JS, Cheng K. Minimally invasive delivery of therapeutic agents by hydrogel injection into the pericardial cavity for cardiac repair. Nat Commun. 2021;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]


