Abstract
Almost 80% of people confronting COVID-19 recover from COVID-19 disease without any particular treatments. They experience heterogeneous symptoms; a wide range of respiratory symptoms, cough, dyspnea, fever, and viral pneumonia. However, some others need urgent intervention and special treatment to get rid of this widespread disease. So far, there isn't any unique drug for the potential treatment of COVID 19. However, some available therapeutic drugs used for other diseases seem beneficial for the COVID-19 treatment. On the other hand, there is a robust global concern for developing an efficient COVID-19 vaccine to control the COVID-19 pandemic sustainably. According to the WHO report, since 8 October 2021, 320 vaccines have been in progress. 194 vaccines are in the pre-clinical development stage that 126 of them are in clinical progression. Here, in this paper, we have comprehensively reviewed the most recent and updated information about coronavirus and its mutations, all the potential therapeutic approaches for treating COVID-19, developed diagnostic systems for COVID- 19 and the available COVID-19 vaccines and their mechanism of action.
Keywords: COVID-19, Coronavirus, Diagnosis, Treatment, Vaccine
Graphical Abstract
1. Introduction
A novel coronavirus called SARS‐CoV‐2 is the reason for today's pandemic of COVID-19 pneumonia. Coronaviruses are classified into four genera (α, β, γ, and δ), and they can be diagnosed in an extensive range of animal species, such as humans [1], [2]. Coronaviruses are small single-strand RNA viruses encapsulated with spike-like projections on their surface. Their outer excrescence resembles a crown when seen under the electron microscope [3]. Human coronaviruses HKU1, 229E, NL63, and OC43 are four different coronaviruses that cause moderate respiratory illness [4] SARS-CoV, MERS-CoV, and SARS-CoV-2 can give rise to severe disease [5]. The intense scourge of coronavirus-caused respiratory disorder began in December 2019 in Wuhan [6], [7]. 11 January 2020 was when the first deadly case was reported [4]. At 4 October 2021 (12:14 GMT), the worldwide spread accounted for 235,810,070 confirmed coronavirus cases, 4,817,796 deaths (2.21%), 212,693,277 recovered cases (97.79%) and 18,298,997 active cases in the world [7], [8], [9]. This disease is now affecting all countries around the world [10].
In the beginning, the Coronavirus appeared to be mainly a respiratory disease. Patients infected with Coronavirus manifested various symptoms like chills, fever, fatigue, weakness, coughing, etc. Surprisingly, some patients were asymptomatic. The severe cases displayed hypoxia with ARDS(acute respiratory distress syndrome) or pneumonia and required mechanical ventilation and intensive care. Since April, doctors have diagnosed and added many other symptoms like fever, sore throat, muscle aches and chills, and gastrointestinal problems, like nausea and diarrhea, to the CDC's primary symptoms of COVID-19. Moreover, one of the significant symptoms of infection may be a sudden, intense decrease in one's sense of smell and taste. Young adults and teenagers, in some cases, have shown painful purple and red lesions on their fingers and toes — named "COVID toe" — in addition to few other critical symptoms [11].
WHO declared that most patients with developing symptoms(about 80%) recover from the infection and do not need hospital therapy, 15% get very ill, requiring oxygen, and 5% become seriously sick, necessitating intensive care. Symptoms like Acute respiratory distress syndrome (ARDS), thromboembolism, and multi-organ failure, including injury to the liver, heart, or kidneys, are usually displayed in severe patients and can result in death. Children can develop severe inflammatory syndrome a few weeks following infection in some uncommon circumstances [12]. Age above three years old, and underlying diseases, like hypertension, diabetes mellitus, respiratory and cardiovascular disease, are positively related to a severe illness or death from COVID-19 [10]. China's National Health Commission (NHC) primarily estimates that patients over 60 with underlying medical conditions and health issues like cardiovascular disease and diabetes make up almost 80% of death cases by COVID-19 [13]. Because the endemic coronaviruses (eCoVs) and SARS-CoV-2 have an analogy in their sequence, they have similar antigens. They may reply to the same immune response, but that doesn't confirm the idea of immunologic memory made by the former infection with eCoV, for SARS-CoV-2 [14]. There have been many studies on the clinical features, pathology, virology, and radiology of COVID-19, but there have been few comprehensive reviews. The purpose of this review is to focus on the clinical features, pathogenesis, diagnosis, and treatment of COVID19, as well as various aspects of currently available vaccines.
1.1. Coronavirus variants and mutations
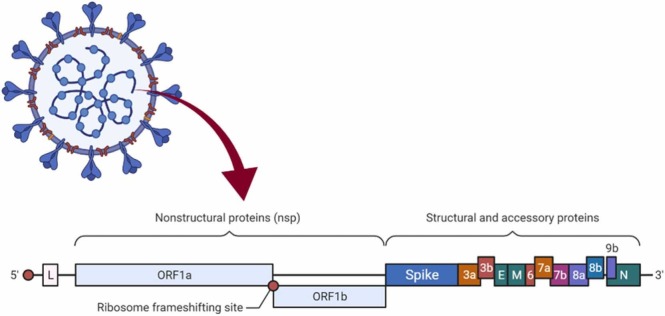
Each Coronavirus has almost 30,000 RNA letters. The virus uses this genetic information to infect cells and constrain them to make new viruses ( Fig. 1).
Fig. 1.
Diagram of the Coronavirus genome.
Mutations occur when the Coronavirus amplifies in the host cell. If the arisen mutation doesn't cause excessive differences, lineages are formed. A new strain is constructed if diverse mutations occur in a lineage and materially change the viral epidemiology. COVID-19 is caused by the new Coronavirus, which belongs to the SARS-CoV-2 strain. Scientists are concerned about various SARS-CoV-2 variants produced throughout the outbreak because they can prolong the pandemic or reduce the vaccine's effectiveness [1]. In Table 1 , the SARS-CoV-2 different variants and mutations are summarized.
Table 1.
SARS-CoV-2 variants and mutations [1].
| Date | Variants |
|---|---|
| April 20 | As a variant of interest, B.1.617, a "double mutant" now common in India, was added. |
| 5 March | In a sample from Portland, Oregon, scientists discovered the E484K mutation. |
| 23 February | The B.1.526 variant, which is spreading in New York City, has been added. |
| 23 February | According to studies, the California-discovered variation is more contagious. |
| 17 February | The P.1 variety has been confirmed in Maryland for the first time. |
| 16 February | The B.1.351 variation has been confirmed in Massachusetts for the first time. |
| 15 February | The Q677 spike mutation, discovered in multiple lineages in the United States, has been added. |
| 15 February | In a Connecticut resident hospitalized in New York City, B.1.351 variant has been confirmed. |
| 13 February | According to studies, B.1.1.7 is more dangerous than other circulating forms. |
| 11 February | The B.1.351 variation has been confirmed in Illinois and North Carolina for the first time. |
| 7 February | South Africa has abandoned the use of AstraZeneca's B.1.351 vaccine. |
| 7 February | In the United States, the B.1.1.7 variant doubles every ten days. |
Many different variants of this novel virus have occurred, but scientists are more concerned about four more dangerous and five designated variants of interest. Table 2 presents these four variants, and Table 3 shows four currently designated concerns of interest. In the following section, variants that will introduce in Table 2 and Table 3 will be reviewed.
Table 2.
Variants of concern.
| Variant name | Lineage | Status |
|---|---|---|
| Alpha | B.1.1.7 | It first appeared in the United Kingdom in December, and it is estimated to be 50% more contagious. Now it is the dominant variant in the US. |
| Beta | B.1.351 | In December, it appeared in South Africa.Some vaccines' effectiveness is reduced. |
| Gamma | P.1 | In late 2020, it appeared in Brazil.These variants’ mutations are comparable to those found in B.1.351. |
| Delta | B.1.617.2 | It is very prevalent in India,Among all other variants, only this variant carries the L452R spike mutation. |
Table 3.
Variants of interest [15].
| Variant name | Lineage | Status |
|---|---|---|
| Eta | B.1.525 | In December 2020, it appeared in multiple countries. |
| Iota | B.1.526 | It appeared in November 2020, and it covers two notable mutations (E484K, S477N). |
| Lambda | C.37 | It appeared in August 2020. Alias of B.1.1.1.37, lineage in Peru, Chile, USA, and Germany |
| Mu | B.1.621 | It appeared in January 2021. Lineage predominantly in Colombia with several spike mutations |
1.1.1. A deadlier form of COVID: UK
1.1.1.1. Name: 20I/501Y.V1, VOC 202012/01, B.1.1.7
1.1.1.1.1. Notable mutation: N501Y
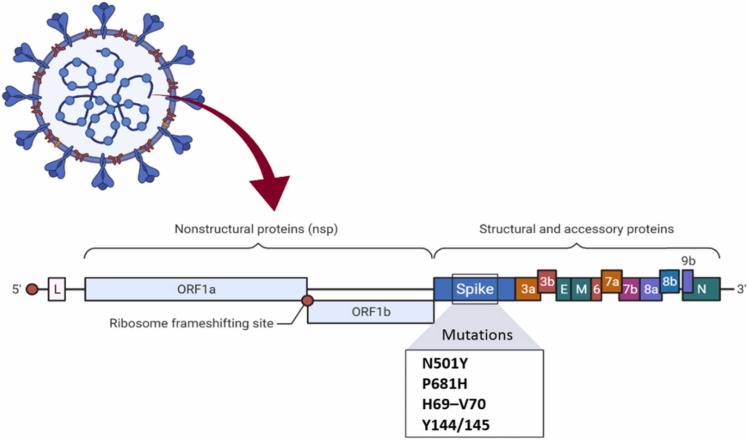
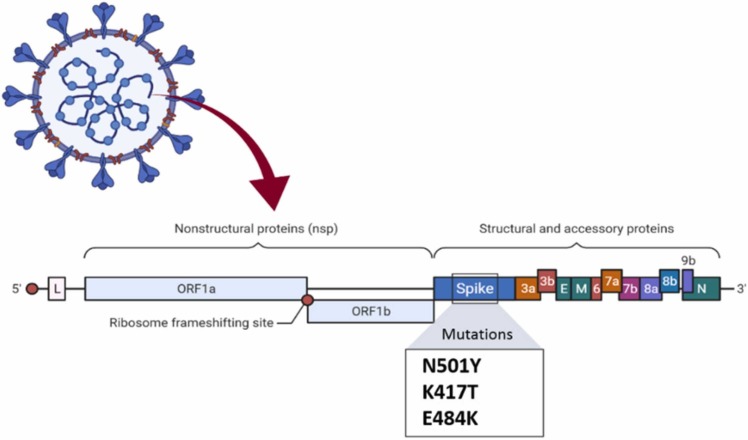
This variant was first discovered in the United Kingdom, and it was named Variant of Concern 202012/01. 20I/501Y.V1, or simply B.1.1.7. according to the new name system based on greek letters established by the World Health Organization (WHO) on 1 June 2021, it was named B.1.1.7 Alpha [16]. People infected with B.1.1.7 are at more risk of death than other circulating variants, regardless of their sex, age, and pre-existing health problems. That's why this particular variant is significant [17]. British prime minister Boris Johnson announced that the B.1.1.7 variant is 30% more fatal than the covid19 original virus [18]. The predominant coronavirus variant in the United Kingdom is B.1.1.7, and it's rapidly expanding throughout Europe. According to the experts, this virus variation might cause a more lethal pandemic than earlier strains without vaccines and other control measures [17]. The virus is spreading rapidly across the country. The epidemiological data indicate 50% more transmission than the initial form of the virus. The United Kingdom's worldwide travel prohibitions and lockdown procedures have been strengthened. The B.1.1.7 variant has 17 mutations; many have occurred in the spike protein and made the virus more infectious [19]. It should be noticed that B.1.1.7 has been diffused in over 90 countries now [16]. N501Y, P681H, H69–V70, and Y144/145 deletions are the four mutations of the spike protein shown in Fig. 2 .
Fig. 2.
Mutations in the B.1.1.7 lineage.
1.1.2. South Africa
1.1.2.1. Names: 20H/501Y.V2, B.1.351
1.1.2.1.1. Notable mutations: E484K, N501Y, K417N
In December, a variant of the Coronavirus B.1.351 lineage known as 20H/501Y.V2 was discovered in South Africa. It was named Beta by the World Health Organization. Scientists are more worried about another mutation named E484K that emerged in the South Africa version. The genetic change may help the virus evade the vaccines and the immune system [19].
This mutation has troubled scientists because People who have survived other variations cannot fight B.1.351. In other words, their antibodies aren't strong enough to capture the viruses [16].
Researches indicate that the Oxford-AstraZeneca COVID-19 vaccine provides "minimal" protection for this variant. After receiving the Oxford-AstraZeneca vaccine, patients infected with the B.1.351 coronavirus experienced mild or moderate illness. Though it is not proven that the B.1.351 variant can cause more intense sickness, it might cause people who survived the original variant of Coronavirus one more period of moderate or mild COVID-19 [20].
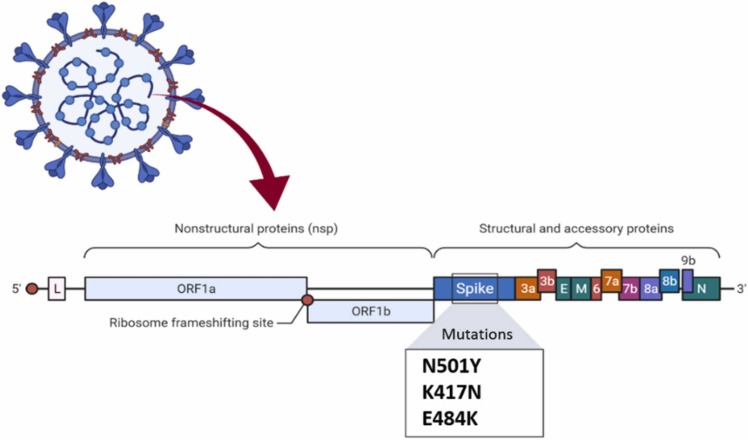
There are three mutations in the spike protein located at the top point, including N501Y, K417N, and E484K, shown in Fig. 3.
Fig. 3.
Mutations in the B.1.351 lineage.
1.1.3. Brazil
1.1.3.1. Name: B.1.1.28, VOC202101/02, 20J/501Y.V3, P.1
1.1.3.1.1. Notable mutation: E484K, K417N/T, N501Y
The P.1 lineage, an offshoot of the broader B.1.1.28 lineage, has a variety known as 20J/501Y.V3. 20J/501Y.V3 variant was primarily reported in 4 cases in japan. They had confronted p.1 when visiting Brazil. The Gamma lineage first arose in Manaus, Brazil. It soon became the most common variety in that city and other South American cities [16].
This variant has three alterations in the gene coding spike protein, including N501Y, K417T, and E484K, shown in Fig. 4 .
Fig. 4.
Mutations in the P.1 lineage.
1.1.4. India
1.1.4.1. Name: B.1.617, double mutation
1.1.4.1.1. Notable mutation: E484Q, L452R
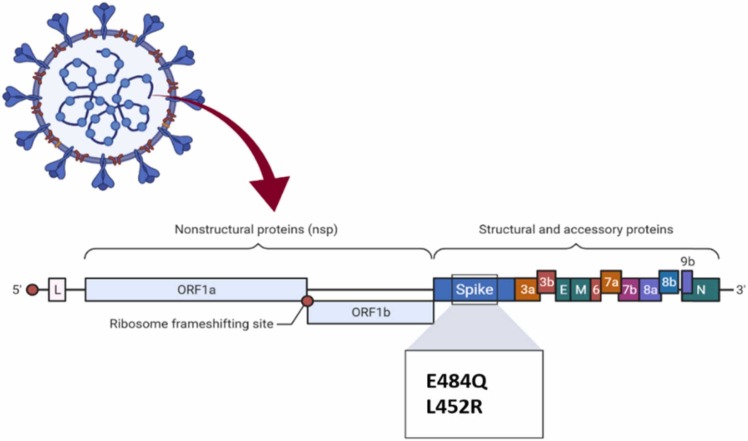
B.1.617 is a new variant with two eminent mutations (named "double mutations”): E484Q and L452R, which is displayed in Fig. 5., that is responsible for a mortal wave of Coronavirus in India. India is the second country in the world with the newest/death cases. The B.1.617 lineage has been divided into two separated lineages: B.1.617.1 or Kappa, and B.1.617.2 or Delta. It has emerged as a virus that is rapidly spreading, exceeding other strains of concern.
Fig. 5.
Mutations in the B.1.617 lineage.
The B.1.617 variety primarily emerged in October in Maharashtra/India, and now it's the most frequent variant there. This variant was also detected in the United Kingdom, the United States, and most recently in Israel [21].
1.1.5. Eta variant, which founded in multiple countries
1.1.5.1. Name: B.1.525, Eta
1.1.5.1.1. Notable mutation: E484K, Q677H
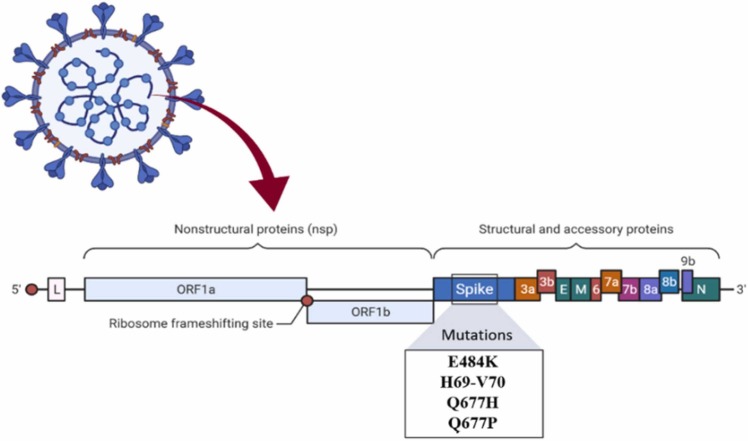
In New York, the B.1.525 lineage, also known as Eta, was separated. This variant has the same previous mutations as the B.1.1.7 lineage or Alpha, such as E484K and the H69-V70 deletion. It is also affected by the Q677H mutation ( Fig. 6). This variant was discovered for the first time in December 2020 [22].
Fig. 6.
Mutations in the B.1.525 lineage.
1.1.6. United States of America
1.1.6.1. Name: B.1.526 or Iota
1.1.6.1.1. Notable mutation: E484K, S477N
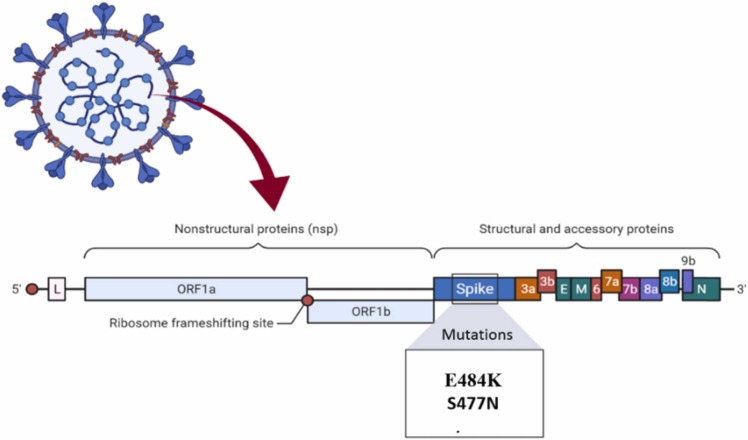
The Iota, or B.1.526, has been rapidly spreading in New York City. This type comes in two varieties. The first is the E484K mutation, which can assist the virus in evading antibodies. The other is the S477N mutation ( Fig. 7), allowing this variant to bind very tightly to human cells. This variant first appeared in November 2020, and by mid-February 2021, it accounted for approximately 27% of New York City [22].
Fig. 7.
Mutations in the B.1.526 lineage.
1.1.7. Peru
1.1.7.1. Name: C.37, Lambda
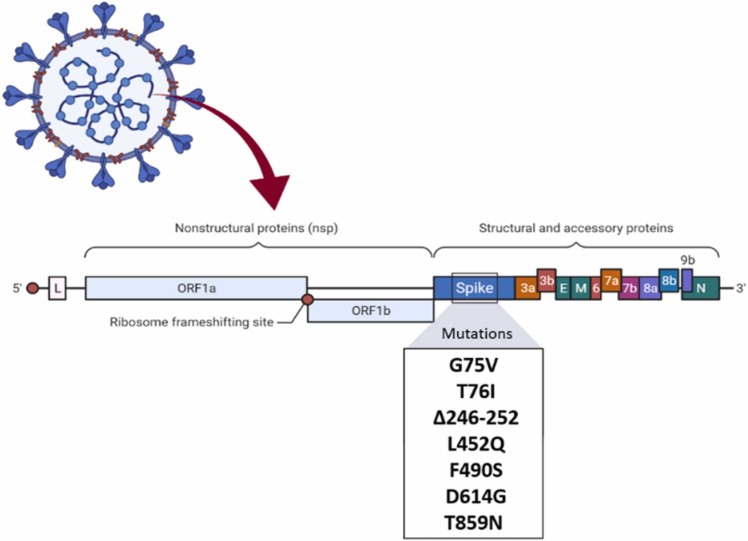
1.1.7.1.1. Notable mutation: G75V, T76I, Δ246-252, L452Q, F490S, D614G, T859N
The Lambda variant of SARS-CoV-2, known as C.37, was first detected in Peru in August 2020. This variant has spread to at least 30 countries worldwide, and it is well-known to be more resistant to neutralizing antibodies than other variants. Also, the Lambda variant could be more infectious to vaccines than the Alpha and Gamma variants. The Lambda spike protein code includes G75V, T76I, Δ246–252, L452Q, F490S, D614G, and T859N [23] ( Fig. 8).
Fig. 8.
Mutations in the C.37 lineage.
1.1.8. Colombia
1.1.8.1. Name: B.1.621, VUI-21JUL-1, Mu
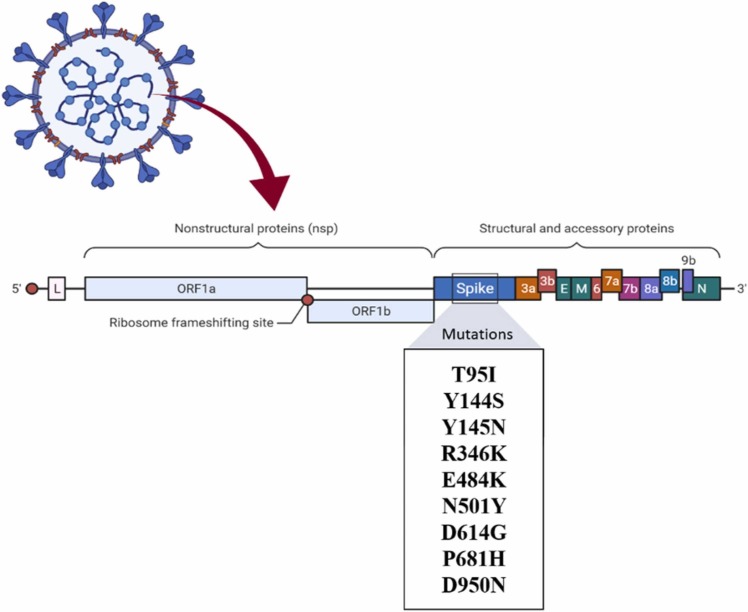
1.1.8.1.1. Notable mutation: T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N
B.1.621, also known as VUI-21JUL-1 or Mu, is one of the most dangerous variants introduced to date. This variant was discovered for the first time in Colombia in January 2021.
WHO identified Mu as one of the variants of interest. This variant contains a total of 21 mutations, nine of which are amino. T95I, Y144S, Y145N, R346K, E484K or the escape mutation, N501Y, D614G, P681H, and D950N the nine amino acid mutations that code ( Fig. 9). Many resources stated that the Mu variant has mutations that indicate a risk of resistance to current vaccines and that more research was needed to understand it better. Mu was regarded as a variant of interest but not yet a variant of concern [23].
Fig. 9.
Mutations in the B.1.621 lineage.
1.2. Established protocols to control COVID-19
The main objectives for all nations are to restrain and manage COVID-19 by diminishing the infection s contagion rate and foreseeing the associated ailments and mortality. The response to covid19 should differ based on the local environment and the spreading of the disease. Above all, national protocols for fighting against covid and breaking the person-to-person transmission chain should be done by providing clinical care for all patients, monitoring, isolating, and segregating new cases. Social methodologies like school systems, governments, and communities can help achieve these goals [30], [31]. Widespread misinformation and misconceptions concerning COVID-19 prevalence have created difficulties in communicating open wellbeing proposals [32]. The virus is transmitted between people close to each other through nose and mouth droplets and aerosols. The transmission of infection particles is particularly risky under a roof with inadequate ventilation [33]. Since the infection spreads through respiratory beads? And close contact thus, it is essential to avoid doing some activities. Although the virus can survive briefly on a few surfaces, it is impossible to spread through residential or universal mailing items or packages. This might come to mind that you can be infected with covid19 by touching your mouth or nose after contacting contaminated objects or surfaces, but this is not the major way for the virus to spread [34].
Evidence indicates that staying at home is as effective as social distancing and using a face mask to decrease infection transmissions. For masks to be effective, they must be used consistently and correctly [35]. According to the WHO statistics, nearly 80% of COVID-19 cases shall care for themselves at home [36]. Thus, a few recommendations for therapy and avoidance of SARS-Cov-2 are given in this part, such as stress reduction and self-care [1]. Two-year-old and older children should wear a mask. It is essential to be noticed that other preventive measures such as washing your hands habitually and remaining at least 6 feet away from others must be remembered [34]. Masks, as a fraction of the comprehensive methodology of measures, are necessary to block out covid19 transmission. Using a mask alone isn't enough to ensure your safety against covid19. If you are in an area that COVID-19 cases are increased, take care of your health by paying attention to these points: social distancing, wearing a mask, providing good ventilation in buildings, avoiding crowds, washing your hands repeatedly, and covering your mouth when coughing or sneezing [37].
There is a considerable argument among professionals about the best choice between masks, for example, the KN95, N95, surgical masks, and cloth masks. Surgical masks provide the wearer with only a little protection from breathing in airborne particles, and the viruses might leak out from the edge of the mask when the sick user breathes out. N95 masks have minor leakage and filter up to 95% of the air particles when inhaling, but the ones with exhalation valves allow respiratory droplets containing viruses to escape. The last option would be cloth masks. Cloth masks have never been tested or regulated, and each type of cloth has a different level of filtration, but they are acceptable for general public use. However, even if KN95s do not satisfy US productivity standards, they may still provide better protection against COVID-19 than a surgical or cotton mask. However, it is fundamentally vital to note that none of the masks are effective unless worn appropriately [38]. Another undeniable matter is how children should use a mask. Healthy children can wear a non-medical or cloth mask to control the spreading of the virus if they are infected and protect themselves. The cloth mask should be appropriate and cover the nose to the chin of the child. Both children and adults with underlying health problems such as immunodeficiency, cystic fibrosis, or cancer are proposed to wear a medical mask to simultaneously control the spreading of the infection and take care of their health [37]. In Table 4 , a brief comprehension of how to choose the best mask based on people's needs is illustrated [39].
Table 4.
How to choose the best mask [24].
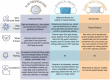 |
Drawing attention to the importance of wearing masks, here we have summarized the essentials of wearing a mask:
-
1.
wash your hands repeatedly, especially whenever you touch your mask, before and after you wear it, and when you want to take it off.
-
2.
check out if the mask is covering your face from nose to chin thoroughly.
-
3.
if you are using a medical mask, dispose of it in a trash bin. But if you use a fabric mask, put it in a clean plastic bag to keep it clean whenever taking it off. You can even wash it every day.
-
4.
pay attention that n95 masks with valves are dangerous for other people because the viruses can easily escape. So please don't use them [37].
Using disposable gloves is recommended to prevent infection dispersion but not obligated because just washing your hands is enough [40]. COVID-19 is a respiratory illness, and it spreads mainly between people nearby each other through the respiratory particles or droplets containing the virus that are inhaled or come into direct contact with the mouth, nose, or eyes. In addition, Coronaviruses cannot increase in food– they require a living creature for multiplying. The best sanitizer recommended for infectivity of encapsulated viruses like COVID-19 on surfaces or hands is alcohol-based disinfectants. Ethanol or propanol can be used in concentrations of 70–80% and a minimum of 1-minute presence for best decontamination. Another chemical for surface disinfection is sodium hypochlorite [41]. Several individual and social responsibilities are needed to be done to overcome this global crisis. Some of them are listed below In Table 5.
Table 5.
The most remarkable responsibilities and tasks.
| Individual duties | Social duties | Food safety |
|---|---|---|
|
|
|
2. Methods for virus detection
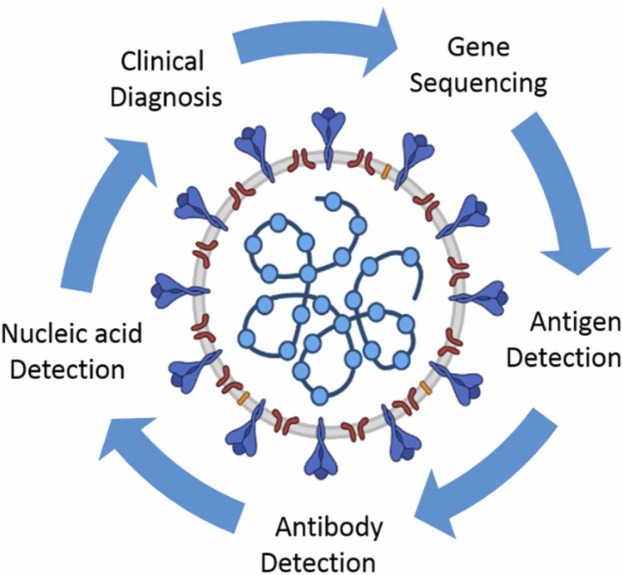
Methods applied for detecting the identified deadly human coronaviruses (MERS-CoV, SARS-CoV-1, and SARS-CoV-2) are summarized in Fig. 10.
Fig. 10.
Different methods for coronavirus diagnosis.
Many techniques were developed for virus detection that varies according to the virus type and its individual particle's properties. In our previous studies, we have discussed diverse methods of detecting COVID-19, summarized in Table 6.
Table 6.
Summery of COVID-19 detection methods.
| Corona virus detection methods: |
| Immunofluorescence Methods [1] |
| Nucleic Acid Amplification Tests (NAATs) [29] |
| Isothermal Amplification [30], [31], [32] |
| Next-Generation Sequencing (NGS) [33] |
| Electron Microscopy (E.M.) [34] |
| Cell Culture [35] |
| Nanoelectromechanical Devices [36] |
| Nanotechnology-based methods |
| Artificial intelligence |
| CRISPR/Cas based methods |
| a real-time reverse-transcription polymerase chain reaction (RT-PCR) assay |
| thin-slice chest CT [2] |
In recent years, health centers and clinics have used reaction (RT-PCR) assay for COVID-19 detection. Though RT-PCR is the Gold standard for covid19 detection [37], the false-negative rate [38] and its unavailability in the initial stages of the covid19 disease have prevented fast recognition of infected patients. it is good to note that thin-slice chest CT is an essential method in detecting the covid19 virus [39].
Lately, gene sequencing has been used to detect Coronavirus separated from COVID-19 cases in different countries. HTGS (High-throughput gene sequencing) is an excellent sequencing technology that provides the critical elements on complete pathogenic sequences. HTGS has aided virus discovery by making it faster and applicable for wide-ranging detection. Besides that, by connecting detection methods with sequence analysis, HTGS offers opportunities to upgrade public health disease supports and monitoring diseases [40].
Furthermore, the Loop-mediated isothermal amplification (LAMP) approach that we previously discussed [2] works on the Watson-Crick base pairing origin of nucleic acid amplification. The primers were used to mark the amplified RNA and DNA strands, then examined by gel electrophoresis. The LAMP assay was used to detect SARS by choosing the ORF1b region of SARS-CoV. The ORF1b area was then amplified. Afterward, it was labeled with primer and finally identified by gel electrophoresis [41]. In a study by Mohammed et al., the leucocrystal violet dye was used to detect the COVID-19 viral sequence, Claustral X, amplified with LAMP primers [40].
Another option is Rapid tests. Rapid antigen tests or RDT (rapid diagnostic test) identify viral antigens in throat or nose samples obtained with a swab. But it has its pros and cons: they are not as definite as PCR tests, but they are cheap and provide fast results [42].
2.1. Diagnosis of coronavirus infections using nanotechnology
COVID-19 infections can be reduced in their spread and morbidity by using a covert and precise detection technique. As previously stated, there are numerous techniques available, but most of them have limitations such as low sensitivity, high costs, time-consuming procedures, and late detection [43]. The recent standard technique for COVID-19 detection, RT-PCR, is a time-consuming procedure requiring expert users and complex equipment. Novel detection assays based on nanotechnology are getting popular. Because of the ease of functionalization, high surface area, and special optical properties of NPs, they have found use as sensitive, rapid, and cost-effective diagnostic systems [44], [45]. NPs have been used extensively in the design of various nanobiosensors to detect infectious diseases over the last few decades [45].
2.1.1. COVID-19 detection biosensors based on nanotechnology
A biosensor is a device used to detect analytes in liquids, body fluids, and solutions when combined with a biorecognition element and a physical transducer. When an analyte interacts with a biological element, a signal is produced that the physical transducer converts into a measurable and quantifiable entity. The biorecognition element includes nucleic acids such as DNA or RNA, proteins such as antibodies or enzymes, biological and organic receptors, tissues, and whole cells [46], [47].
2.1.1.1. Nucleic acid-based biosensors
A plasmonic biosensor for the sensitive detection of SARS-CoV-2 nucleic acid has been developed. The device consists of a two-dimensional gold nano-island integrated chip with a cDNA receptor for RdRp, ORF1ab, or the E gene. This biosensor's performance in clinical samples has yet to be determined [48].
Another assay combined LAMP amplification, multiplex analysis, and reverse transcription with a nanoparticle-based lateral flow immunoassay biosensor in an integrated approach (COVID-19 RT-LAMP-LFB). This platform's LOD was found to be 12 copies per reaction, with no cross-reactivity. Two LAMP primer sets, an ORF1 antibody, and the SARS-CoV-2N gene were amplified simultaneously and detected with streptavidin-coated polymer nanoparticles [49].
2.1.1.2. Aptamer-based biosensors
Aptamers have recently gained popularity in virus detection. These are made up of ssRNA or ssDNA that binds conformationally to multiple targets and detects them with high sensitivity and specificity. Aptamer-based sensors can distinguish between infected and uninfected host cells by modifying the designed assays. Aptamer-based detection has several advantages over antibodies, including ease of synthesis via a systematic evolution of ligands by exponential enrichment (SELEX) method, high stability at a wide range of temperatures and situations, and easy modification to meet the needs of the assay [50].
An aptamer-based approach was used in one study to detect SARS-CoV N-protein in blood serum. The SELEX method was used to screen RNA aptamers 1 and 2, which bind specifically to the C-terminal region of the SARS-CoV N-protein. Aptamer 1 has a higher binding affinity than aptamer 2 (K D 1.65 nM for aptamer 1 vs. K D 2.1 μM for aptamer 2), and it also binds more strongly than an antibody. An ELISA was designed to detect N-protein using a streptavidin-coated plate for aptamer immobilization. This method's LOD is comparable to that of ELISA, which uses both polyclonal and monoclonal antibodies. Furthermore, an antibody was used to design a nanoarray chip, and the fluorescently labeled secondary antibody was shown to be 10 times more sensitive than the chemiluminescence assay [51], [52].
A novel RBD aptamer (anti-RBD-SARS-CoV-2) was recently screened using a SELEX and ACE-2 competition-based selection strategy. A machine learning screening algorithm was also used in this development. Flow cytometry was used to assess the enrichment of interacted aptamers with SARS-CoV-2-RBD using histidine-tagged RBD-modified nickel beads. Finally, two aptamers with a high affinity for binding were chosen, with KD values of 19.9 nM and 5.8 nM, respectively [53].
2.1.1.3. Antibody-based biosensors
A novel antibody-based biosensor was reportedly used to detect COVID-19 spike protein. COVID-19 antibody was coated on graphene sheets of a field-effect transistor (FET) using clinical patients' nasopharyngeal swab samples as antigens. The LOD of the developed biosensor was determined to be 100 and 1 fg/mL in a universal transport medium and phosphate-buffered saline, respectively. This sensor detected the virus in the culture medium as well. With a detection limit of 2.42 102 copies/mL, the COVID-19 FET sensor can distinguish between infected and healthy individuals [54].
IgG and IgM antibodies were observed utilizing an ELISA kit developed and tested on patients based on recombinant N- and S-protein. Among 214 patients, N-protein-based IgM and IgG ELISAs were used to detect 146 (68.2%) and 150 (70.1%), respectively, while spike protein-based ELISAs were used to identify 165 (77.1%) and 159 (74.3%). The positive rate for both kits was 82.2% for S-protein and 80.4% for N-protein, respectively [55].
2.1.2. Diagnosis of coronavirus infections using microfluidic devices
Microfluidic devices are another approach for use as a proof-of-concept test. These are palm-sized chips with micrometer-sized channels and reaction chambers imprinted on them. They have advantages such as miniaturized size, a short detection time, and a small sample volume requirement. The basic idea behind these microfluidic chips is that they use capillary action and electrokinetic properties to mix and separate liquid samples [56].
When tested on 96 patients in Rwanda, the platform demonstrated specificity and sensitivity of 87% and 100%, respectively. Different antibodies against three sexually transmitted infections can be detected using microfluidic devices and a smartphone application attachment. These devices can be modified to detect SARS-CoV-2 RNA or proteins due to their simplicity and dependability [57].
2.1.3. Diagnosis of coronavirus infections using colorimetric assay
Colorimetric analytical systems using silver nanoparticles as colorimetric substrates have been developed to detect MERS-CoV nucleic acids. Similarly, gold nanoparticles and quantum dot-based immunosensors have been designed to detect Avian coronavirus infection. These immunosensor-based methods outperform ELISA in terms of accuracy, sensitivity, and turnaround time [58].
AuNPs have gained attention in the development of biosensors because of their fascinating optical properties related to surface plasmon resonance (SPR) absorption. The prevalence rate of light on a metal surface causes collective coherent oscillation of conduction electrons. A UV-Vis spectrometer can be used to measure an AuNp's color or LSPR band [59], [60].
The anatomy of SARS-CoV-2 was studied using a novel technique that takes advantage of the optical properties of AuNPs. Thiol-modified antisense oligonucleotides (ASOs) for the N phosphoprotein were used for conjugation. When ASOs were added to an AuNP (ASO) aggregate, the surface plasmon resonance changed. When RNaseH is added, the RNA strand is cleaved from the RNA–DNA hybrid, resulting in a visually detectable precipitate facilitated by AuNP agglomeration. In the presence of MERS-CoV viral RNA, the LOD was 0.18 ng/L of RNA with a SARS-CoV-2 viral load [61].
2.2. Diagnosis of coronavirus infections using artificial intelligence
The rapid rise in COVID-19 cases is putting a strain on healthcare services around the world. Precise diagnosis of COVID19-infected patients is critical to providing appropriate treatment and avoiding overburdening the healthcare system due to high costs and a scarcity of resources [62]. Within certain constraints, chest CT-based diagnosis is the quickest way to diagnose COVID-19 in suspected patients clinically. A recent study used a deep learning-based model for COVID-19 to distinguish between community-acquired pneumonia and COVID-19 on its own. With a scarcity of point-of-care diagnostics, AI-driven tools could supplement the transmission dynamics of SARS-CoV-2 among different population groups, and the risk of disease spread [63], [64]. At the moment, the active learning process in AI devices is said to boost confidence during decision-making processes. As a result, protocols must be standardized to develop AI-based devices that can be used in the event of these disasters. Deep learning methods based on chest CT and X-ray images are helpful in diagnosing COVID-19 positive cases [65], [66] Recent reports state that chest CTs of COVID-19 patients demonstrated usual radiographic aspects like ground-glass opacities, multifocal patchy consolidation, and interstitial changes with the peripheral distribution. With an overall accuracy of 86.7%, a CNN-based approach distinguished between COVID-19 and influenza A infected pneumonic lungs [67], [68]. A transfer learning process of designing a CNN-based approach to generate disease diagnosis from patients' chest anterior-posterior radiographs could predict with 96.3% accuracy and high sensitivity and specificity. A meta-analysis of published research with independent datasets on CT scans and other imaging techniques for COVID-19 diagnosis revealed an excellent accuracy rate [64], [67].
Aside from symptoms, the current COVID-19 diagnostic approach, which employs real-time reverse-transcriptase polymerase chain reaction (rRT-PCR), is more trustable. Even so, while this testing method is analytically sensitive enough to detect viral nucleic acid in samples, sample collection, sample quality, and experimental errors may have a significant impact on the rRT-PCR test accuracy. In rRT-PCR, a false negative rate of up to 20% has been reported [63], [69].
It was also discovered that when complemented by an AI-based deep learning computer-aided diagnostic methods, some initially false-negative rRT-PCR results were later determined positive. Thus, complementary combined AI-based approaches, such as CT-derived deep learning methods in conjunction with rRT-PCR, can provide a more reliable diagnosis of COVID-19. As the interpretation of the lung CT assisted by the deep learning-based method via a computer-aided diagnostic system, which has been pre-trained with a large number of CT scans of COVID-19 patients, could improve diagnosis accuracy and cover events missed by the use of rRT-PCR alone [64], [70], [71]. AI-based algorithms that combine chest CT findings with clinical symptoms, laboratory testing, and exposure history are as sensitive as radiological disease diagnosis. Many studies have discovered AI-enhanced radiological image findings to differentiate between infected and non-infected people. Another study used robust deep learning models in 2D and 3D for patient monitoring and automated detection. With 98.2% sensitivity and 92.2% specificity, the deep learning CT image analysis of 157 international patients demonstrated accurate disease progression measurement. It was also shown that when combined with AI-based image analysis, high accuracy in COVID-19 diagnosis could be achieved in a timely manner [70].
A complete and accurate machine learning approach was used to create a CRISPR-based nucleic acid detection system for SARS-CoV-2 and symptomatically related pathogens that could be used for diagnostic and surveillance purposes. The CRISPR-Cas13 detection system was put through its paces using a lateral-flow assay to demonstrate its speed and sensitivity with synthetic targets [72]. As a result, AI approaches that work in tandem with existing molecular diagnostic procedures could aid in disease diagnosis and early control. Through AI/mL algorithms that have been well-tested and verified in several disease outbreaks, AI-based tools can provide speed in the healthcare setup amid the COVID-19 pandemic crisis [73].
2.3. Point-of-Care diagnosis
In comparison to RT-PCR, point-of-care testing is a cutting-edge diagnostic process that requires less time. It aids medical personnel and physicians develop appropriate quarantine methods for positive patients, allowing them to receive immediate medical attention and preventing the disease from spreading further. Point-of-Care techniques can be categorized as serological antigen or molecular diagnostic methods. The former detects COVID-19 in blood by lateral flow immunoassay and colloidal gold immunochromatography, and the latter detecting COVID-19 in the nasopharyngeal swab, saliva, throat swab, and nasal swab via Pcr assay.
Based on current evidence, the WHO (World Health Organization) now recommends using these new point-of-care immunodiagnostic tests only in research settings. They should not be used in other situations, such as clinical decision-making, unless adequate evidence substantiates their use [74], [75].
2.4. Diagnosis of coronavirus infections using CRISPR/Cas system
With the expansion of the COVID-19 virus worldwide, every scientist with diverse specialties tried to find a way to diagnose this disease in its early phase and cure its early phase and cure it. Different diagnostic methods are currently used; one of them is RT-PCR. At present, RT-PCR is the most popular molecular test used for COVID-19 diagnosis, but it has its weak points. The amount of the COVID-19 virus varies in different phases of the illness, so RT-PCR may not be able to discover the low loads of the virus [76].
Another recently proposed COVID-19 diagnostic test is the Viral RNA test based on CRISPR (clustered regularly interspaced short palindromic repeats)/Cas method. CRISPR is also called “molecular scissors.” CRISPR/Cas system is a prokaryotic (archaea and bacteria) immune system against the virus (bacteriophages) invasions. With the help of the CRISPR/Cas system, the bacteria identifies the invading virus's nucleic acid sequence (DNA/RNA) and breaks it [77].
Scientists working on CRISPR/Cas system found out that CRISPR can also be used for COVID-19 diagnosis. In diagnostic tests based on CRISPR, nucleic acids are recognized as biomarkers. CRISPR/Cas-based diagnostic tests are applicable by routine lab reagents that greatly reduce the cost for patients and labs. Unlike qRT-PCR, the CRISPR method can target a singular and particular target sequence in less than an hour with no need for complicated laboratory equipment or setups [76].
After sample collection (mostly nasopharyngeal swabs), COVID-19 RNA is extracted by special virus RNA extraction kits. Afterward, RNA is amplified through loop-mediated isothermal amplification (LAMP) or recombinase polymerase amplification (RPA) methods to increase the limit of detection(LOD). LAMP and RPA are two superior replacements for PCR in the RNA amplification process because they are cheaper and faster. Unlike qRT-PCR, a single temperature is required for virus RNA amplification, so there is no need for special equipment that provides thermal cycling. Following that, the unique nucleotide sequence matching the guide RNA will be detected by the CRISPR/Cas system, and the Cas protein will do the cleavage [78].
Another fact about using the CRISPR/Cas method for COVID-19 diagnosis is that they are applicable in any place because they can be done using simple devices such as paper-based LFA (lateral flow assay) strips. This is good news for cities with poor economic conditions with no facilitated labs. CRISPR/Cas is also more specific and sensitive (100%) in comparison to qRT-PCR (90% specificity), which has made it much more valuable and attractive as a diagnostic approach [76].
2.4.1. CRISPR/Cas system classes
CRISPR method is classified into two main groups (class1 and class 2) and six types summarized in Table 7 [76].
Table 7.
Different classes of CRISPR/Cas system.
| Class 1 | Application | Class 2 | Application |
|---|---|---|---|
| Type 1 – Cas3 | Cutting DNA (nuclease) | Type 2-Cas9 | Cutting DNA (Endonuclease) |
| Type 3 - Cas10 | Cutting RNA (nuclease) | Type 5-Cas12 | Cutting DNA |
| Type 4 | Type 6-Cas13 | Cutting RNA |
Up to now, CRISPR/Cas has been popularly used for both sickness detection and cure through genetic alteration. Specifically, CRISPR/Cas3, CRISPR/Cas12a, CRISPR/Cas13a, CRISPR/Cas12b, based systems have been used for COVID-19 detection since its start. CRISPR/Cas-based systems used for COVID-19 detection are summarized in Table 8.
Table 8.
CRISPR/Cas based systems used for COVID-19 detection.
| System | Cas type | Ref. |
|---|---|---|
| SENA | 12a | [79] |
| CRISPR-Cas12 based | 12a | [80] |
| CRISPR-Cas12a-NER | 12a | [81] |
| CRISPR-FDS | 12a | [82] |
| ITP-CRISPR | 12a | [83] |
| AIOD-CRISPR | 12a | [84] |
| DETECTR | 12a | [85,86] |
| iSCAN | 12a | [87] |
| VaNGuard | 12a | [88] |
| STOP-COVID | 12b | [89] |
| CASdetec | 12b | [90] |
| CRISPR-COVID | 13a | [91] |
| SHERLOCK | 13a | [92] |
| CREST | 13a | [93] |
| SHINE | 13a | [94] |
| CONAN | 3 | [95] |
Cas12 (DNA nuclease) and Cas13 (RNA nuclease) both recognize and link to their target nucleic sequence through their guide RNA (gRNA) and become activated. It is good to note that when Cas12 and Cas13 are activated, they start to cut every DNA/RNA near them randomly. Cas13 and Cas12 can be modified and engineered for targeting the COVID-19 genome [77].
2.4.2. CRISPR based methods
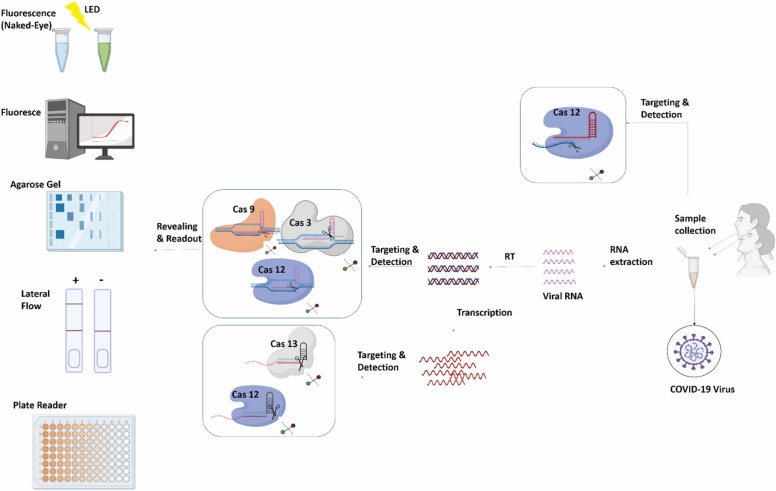
One of the most challenging problems in COVID-19 diagnosis is its high mutation rate. With the help of the CRISPR/Cas system and selecting specific SgRNA, distinguishing between various strains of COVID-19 has been made possible [78]. DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) and SHERLOCK (sensitivity enzymatic reporter unlocking) are two CRISPR/Cas mediated methods authorized and commercially available for COVID-19 diagnosis [76]. General CRISPR/Cas based methods for COVID-19 detection are reviewed in Fig. 11. SHERLOCK and CRISPR/Cas13 were used for targetting COVID-19, S, and ORF1ab genes in a study by Zhang et al. In this procedure, Cas13 recognizes and fixes to S and ORF1ab RNA sequences, and afterward, the arbitrary fractionation of surrounding ssRNA molecules is initiated. Alongside, a ssRNA with blocked fluorescent is utilized as a reporter (Biotin-RNA-FITC). When the CRISPR system recognizes the virus RNA, Cas is activated, and the reporter RNAs are spliced. Consequently, fluorescent signals are detected. This method can detect the COVID-19 genome at attomolar quantity with 93% sensitivity [76].
Fig. 11.
Illustrates the most common CRISPR/Cas based methods for COVID-19 diagnosis (sample to report).
The other method, DETECTR, is used to identify the presence of N and E genes in COVID-19 through CRISPR/Cas12 system [77]. When Cas12a is activated, alike Cas13 demonstrates nuclease activity. However,cas12 cuts DNA. DETECTRs exactness is comparable to qRT-PCR. Like qRT-PCR, DETECTR has standard protocols and commonly accessible analytes. Still, unlike QRT-PCR, there is no need for a thermo-cycler because it has high incorporation ability with other reporting methods, for instance, Lateral flow strips. Other advantages of DETECTR over qRT-PCR are faster process completion and high target specificity (even single nucleotides matter) [76].
Another more complicated method is SHERLOCK v.2. SHERLOCK. In this method, Csm6 ribonuclease and Cas13 are utilized together to find various matching sequences accurately. As a result, any infection with COVID-19 can be diagnosed. Further, any DNA/RNA sequence can be recognized using this method [96].
Recently, another Cas9 family protein named FnCas9 was found in Francisella novicida. It was used in a new detection method FELUDA (FnCas9 editor linked uniform detection assay) for COVID-19 detection. FELUDA was found a fast, cheap nucleic acid detection method with high specificity and sensitivity, even in distinguishing between two strains of COVID-19 virus with one nucleotide point mutation. This method doesn’t need special lab equipment, and like other CRISPR/Cas-based methods mentioned above, even a very small amount of virus loads can be detected through lateral flow kits [97].
Still, one of the main reasons for the complexity of CRISPR/Cas-based detection methods is its two-step operation system. First, DNA/RNA should be amplified, and then CRISPR/Cas system is added for diagnosis. To reduce the complexity, most recently, Jun et al. designed a new diagnostic one-step COVID-19 diagnosis method named CRISPR-top (CRISPR mediated testing in one-pot). In this method, nucleic acid amplification and CRISPR/Cas detection steps are done in one reaction tube. CRISPR-top advantages over other processes are its simplicity, remarkable specificity (100%), and the high ability to detect COVID-19 virus in low loads (10copies). This method's sensitivity in COVID-19 virus detection was similar to qRT-PCR (65%). Results can be understood by lateral flow assays or visual fluorescence [98].
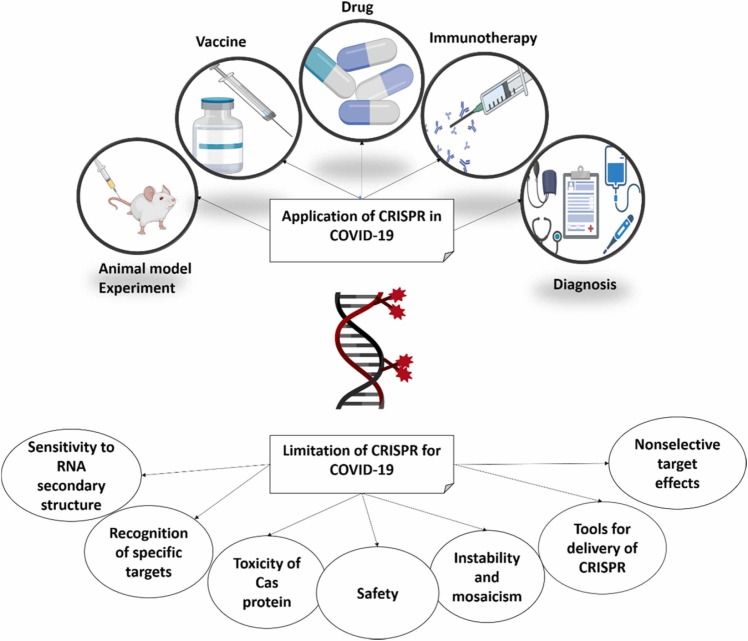
2.4.3. CRISPR challenges
Like any other newly designed method, CRISPR has its challenges in disease diagnostics, specially COVID-19. For instance, Each CRISPR/Cas system has a unique gRNA that can bind only to its target sequence. So, each system can detect one target gene at a time (Rahimi). Gootenburg and coworkers designed the first multiplex sensing system by using different classes of Cas effectors for detecting four different sequences in one reaction. They used different Cas proteins (PsmCas13b, LwaCas13a, CcaCas13b, and AsCas12a). scale-up is impossible because of the limit in Cas systems. The main limit in this procedure was the intervention between the verifying agents and the probability of irritating reaction [99].
Another point is that CRISPR-based detection methods are qualitative. In other words, they can maintain whether the target genome is present or not; no information about its quantity is given. Cas protein is the fundamental part of this system that ascertains the properties and application of each CRISPR/Cas system. For example, Some CRISPR systems acquire PAM sequences for the correct binding of gRNA and the target sequence. Besides, PAM sequences maintain mismatch tolerance. And the number of mismatches acceptable entirely depends on the Cas protein class [77]. A summary of CRISPR challenges for COVID-19 detection is shown in Fig. 12.
Fig. 12.
CRISPR challenges for COVID-19 detection.
3. Treatment of a patient with COVID-19
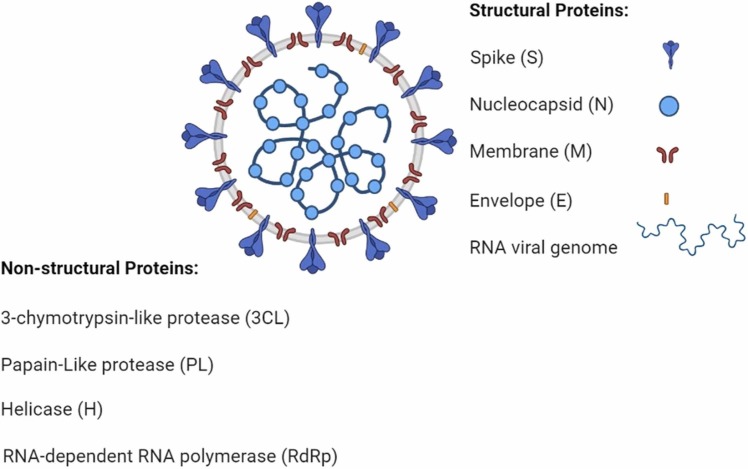
New Coronavirus (SARS COVID-19) has killed more than a million people and affects thousands daily. Thus it is an important target for the rapid development of drugs [100]. We have thoroughly investigated the ordinary medicines like home remedies, homegrown pharmaceuticals for COVID-19 alongside chemical drugs, plasma therapy/medication, and vaccines, discussed before [1]. Clinical trials are testing different drugs to find a potential therapeutic cure for covid19. Different anti-viral treatment targets are Clinical trials performed. Clinical trials are being carried out in which potential anti-viral treatment targets are tried, such as restraining viral proteins identified for genome replication or blocking viral sections into human cells [101]. There are numerous pharmacological strategies to fight against COVID19:Small-molecule drugs, immunizations, Plant-based Molecules, Nasal drugs, Stem cell therapies, peptides, monoclonal antibodies, and nanotechnology-based drugs [102]. Effective solutions against Coronavirus can be categorized based on their active component [103]: Those that act on viral proteins and proteins in this way anticipating RNA replication and union Those that act on the viral auxiliary proteins, restraining self-assembly or blocking the infection. The S protein could be a primary target for vaccine improvement [104]. Different particles of the virus, like the envelope antigens, nucleocapsid, or even the membrane, can be targeted by the synthesized drugs. A schematic illustration of the SARS-CoV-2 anatomy and some of its structural proteins is presented in Fig. 13.
Fig. 13.
SARS-CoV-2 and a few of its molecular protein targets.
3.1. Drugs
Vaccines are protective, but they are not entirely effective in preventing serious illness or the spread of the virus, especially in newer variants. Some of the drugs that are being applied as investigational medicines against COVID-19 are repurposed medications designed to cure other viral diseases. Successful pharmacotherapeutic approaches against SARS-CoV-2 can either be tied down to using particular treatments to block viral connection like Peptide fusion inhibitors or utilize the wide-ranging antiviral drugs like protease inhibitors that break the bonds between long strings of protein sub-units and are used by the coronavirus to make the proteins it needs to replicate within human cells. Without them, the virus cannot multiply [105]. anti-SARS-CoV-2 neutralizing monoclonal antibodies and anti-ACE2 monoclonal antibodies are conceivable pharmacotherapeutic alternatives [105]. Clinical trial reports indicate that a few antiviral drugs, such as Favipiravir, Remdesivir, and Molnupiravir, are helpful in some cases. Molnupiravir is a new antiviral with illustrated activity against SARS-CoV-2. It has been proven that the drug is safe and has antiviral activity in humans, it should be noted that the phase 3 trial has been initiated in non-hospitalized patients at risk of severe disease progression. Results will not be known before the end of the year 2021 [106]. Favipiravir is a promising drug for treating COVID-19 that may reduce hospitalization and their need for mechanical ventilation. However, it should be noted that only patients with mild to moderate COVID-19 had a better prognosis than patients with severe disease [1], [2], [30], [31]. Favipiravir probably has no beneficial effect on mortality in the general group of patients with mild to moderate COVID-19. We need to keep in mind that it is too late to use antiviral drugs when they show symptoms, which may explain their low efficacy in clinical settings [1]. In October 2020, the FDA approved the antiviral drug RamedSivir for the treatment of COVID-19. It may be used to treat adults and children 12 years of age and older who weigh at least 88 pounds and have been hospitalized for COVID-19. Clinical trials show that in these patients, RamedSivier can moderately accelerate recovery time. Also, Ribavirin is highly available and low cost; thus, it supports its potential for positive impact in the treatment of COVID-19 infections. Nitazoxanide may work by inhibiting both cell entry and viral particle assembly and seemed to improve viral clearance in patients with symptomatic COVID-19 [107]. Readers are encouraged to follow our previous review paper around this subject for more information about valuable drugs and medications against COVID-19infection [1].
3.2. Other potential drugs
Dexamethasone is a glucocorticoid drug used to treat rheumatic problems, some skin conditions, severe allergies, asthma, chronic obstructive pulmonary disease, brain swelling, eye pain after eye surgery, and antibiotics for tuberculosis. Remdesivir and Dexamethasone are said to be two therapies that are effective against Sars-Cov-2-related diseases [106]. Dexamethasone reduced the incidence of death in ventilated patients [108]. Recombinant ACE-2 is helpful because for entering the cells, the corona virus must first unlock them - this is done by attaching to a human protein called ACE-2. Scientists have developed a synthetic ACE-2 protein that can prey and repel coronavirus from vulnerable cells. Recombinant ACE-2 proteins have shown positive results in experiments on cells and animals but not yet in humans. PF-07321332 is a completely different drug that prevents the virus from multiplying inside cells. Pfizer developed a drug in the early 2000s as a potential treatment for SARS caused by the SARS-CoV corona virus. At the beginning of the COVID-19 epidemic, they retrained it against SARS-CoV-2, which has similar biology. In addition, they changed the drug to pills. When the drug was given orally to mice, it was sufficient to block the coronavirus. PF-07321332, as the drug is now known, conducted clinical trials in March 2021, followed by the more significant Phase 3 trial in July. The results of this study are expected by October.
3.3. Plasma
Existing documents show that hyper-immune (or convalescent) plasma from patients who have got back on their feet from different viral infections can be valuable for the sickness treatment without special warnings. Hyper immune (or convalescent) plasma has already been utilized as a last shot in patients with primary SARS-CoV pneumonia not answering back to maximal treatment [109]. Diverse studies on SARS-CoV pneumonia showed that hyper immune plasma was successful in decreasing hospitalization and fatality. A meta-analysis of 32 studies on SARS-CoV illness and severe/acute influenza found a statistically significant reduction in the pooled odds of mortality following healing plasma treatment compared to placebo or no therapy. A protocol to determine the application of hyper immune plasma in curing patients influenced by MERS-CoV pneumonia was built up in 2015. Thus far, a few case reports of COVID- 19 patients treated with hyper immune plasma were released that displayed profitable/favorable outcomes, most probably due to its antiviral action. These case reports suggest that COVID-19 patients treated with convalescent plasma showed less viral loads in serum, and most showed no infection signs 3 days after treatment [110]. A Chinese randomized clinical trial was just published. In this study, a populace of 103 severely ill subjects with life-threatening COVID-19 infection underwent treatment with covalent plasma. There was no actual superiority in treated patients' conditions in terms of clinical improvement, mortality rate, and time to discharge, compared to standard treatment in 28days. In another study, a subgroup of seriously sick but not basic patients treated with convalescent plasma displayed faster clinical change (p = 0.03). It is important to note that the study was underpowered because the patients’ registration was ended early due to contamination in China. It did not reach the target of 200 patients. It is important to mention that, predictably, convalescent plasma was very satisfactory in the early viremia stages of the disease. In this case, it was dealt with highly late after the onset of negative effects. (middle of 30 days). For these reasons, it would be beneficial to assess hyper immune plasma adequacy in the early stages of COVID-19 infection to see if earlier administration is associated with better outcomes [111].
3.4. Home remedy
As mentioned many times before, almost 80% of COVID-19 patients have to care for themselves at home [1]. Our previous study thoroughly discussed self-care, reducing stress, and home treatments for sore throats and sinus blockage [1]. It is recommended to drink lots of fluids and stay hydrated. Also, it is important to rest and avoid physical activities; gargle salt water can be helpful. Although there isn’t any scientifically proven about it, many people believe that salt water helps their sore throat [100]. Generally, what is genuinely vital here, is to boost the immune system and prevent infection.
3.4.1. Herbal medicine
Plants have been extensively studied since ancient times, and many important chemical compounds with tremendous therapeutic potential have been identified. Attacks by microorganisms, including viruses and bacteria, can be controlled by an efficient immune system, and therefore, stimulating the body's defense mechanism against infections is a helpful method. Plant products and their biologically active molecules modulate immune responses by stimulating and modifying lymphocytes, macrophages, and cytokine production. Many plant extracts contain a number of active principles, including polysaccharides, terpenoids, alkaloids, flavonoids, glycosides, and essential oils, which have the extraordinary ability to maintain or stimulate the immune system primarily by modulating nonspecific immune responses. The use of herbal medicines has increased due to their therapeutic value compared to allopathic medicines since these biological compounds show fewer side effects [112]. Plant molecules are less expensive to maintain and are prone to large-scale production [113], [114]. In addition, although some synthetic compounds and monoclonal antibodies are available as immune regulators, their use has been associated with some limitations, including side effects. Immunomodulators of different plants, their extracts, plant active ingredients, and plant products have been studied [115]. Amparo et al. reported that potential anti-SARS-CoV-2 compounds are mainly phenolic compounds [116]. Herbal plants include Artemisia kermanensis, Eucalyptus caesia, Mentha spp., Rosmarinus officinalis, Satureja hortensis, Thymus spp., and Zataria multiflora are typical examples of rich sources of phenol [117]. Bioflavonoids from Torreya nucifera prevent the replication of SARS-CoV 3CLpro [118]. Another group of secondary metabolites which are highly antiviral and are beneficial against coronaviruses is the alkaloids. The bisbenzylisoquinoline alkaloids, tetrandrine, fangchinoline, and cepharanthine from roots of Stephania tetrandra inhibit expression of the human coronavirus, HCoV-OC43, spike, and nucleocapsid proteins; thus, they can provide sufficient immunomodulation [119], [120]. lycorine from G. glabra and Lycoris radiate, respectively, displayed positive activity against SARS-CoV [121]. Glycyrrhizin has also shown good potential against the COVID-19 virus SARS-CoV-2 [121]. It may emerge as an alternative drug for its treatment [122], [123]. Based on these findings, a novel mixture of vitamin C, curcumin, and glycyrrhizic acid (VCG plus) has been proposed to treat coronavirus infections [124]. Here are some other plants that may have antiviral effects due to their chemical compounds. Curcumin, which has polyphenolic compounds, showed activity against various important human pathogens, including influenza virus, HCV, HIV, and SARS-CoV-2 [125]. Qing Fei Pai Du Tang and or Ma Xing Shi Gan Tang (MXSGT) are two popular Chinese medications that have been used during the corona virus outbreak in China. Qing Fei Pai Du Tang has been actionable for healing sick people with any clinical side effects of severe to a mild range of COVID-19 [126]. Ma Xing Shi Gan Tang (MXSGT) is famous for its anti-fever effects and is commonly useful for treating respiratory diseases such as pneumonia and influenza [127]. Lianhua Qingwen Capsule (LHQW) is a plant-based drug; it has been clinically proven to treating COVID-19 disease. Several exposed LHQW components in humans play an important role in combating SARS-CoV-2 and play a significant role in binding ACE2 and S proteins, which is an important way to inhibit virus infection. Some studies provided direct chemical and biochemical evidence related to the molecular mechanisms of clinical use of LHQW for the prevention and treatment of COVID-19. In addition, human exposure-based trials have been performed to identify active medicinal components in an herbal drug with beneficial therapeutic effects [128]. Cryptolepis Sanguinolenta is a Ghanaian herbal medicine locally known as Nibima used for clinical trials against COVID-19. Alkaloids from Cryptolepis sanguinolenta could serve as lead compounds as cure for the corona virus disease [129] . In addition, researchers from Hue University have recently investigated the essential oil from Garlic (Allium sativum L.) against COVID-19 in molecular docking studies [130], [131]. The docking results illustrate that the organosulfur compounds (e.g., allyl disulfide and allyl trisulfide) effectively prevent the host's angiotensin-converting enzyme 2 (ACE2; membrane glycoprotein) and targeting the PDB6LU7 protein, the main protease of SARS-CoV-2. This prevention regulates the PDB6LU7 protein maturation of the COVID-19 and the further spread of infection [130], [131]. Besides, some Indian spices are suggested to be helpful. In India, such as Cinnamomum cassia (cinnamon), Piper nigrum (dark pepper), Syzygium aromaticum (clove), Ocimum basilicum L. (basil), Allium sativum L. (garlic), Tinospora Cordifolia (giloy), Azadirachta indica (neem), turmeric, and Zingiber officinale (ginger). However, the inordinate and overmuch consumption of spices and herbs may cause diverse secondary effects, to be specific, acid reflux, heartburn, constipation, diarrhea ulcers within the mouth, blood pressure fluctuation, and so on [132]. It should be noted that lots of these medications are valuable for preventing the disease. In a few Asian nations it is common to do traditional Chinese medicine (TCM) food treatment, TCM tea and foot bath, these are also considered as the non-medicine treatments advised for a diverse population to clean up and restore lungs, give strength to the spleen, and stomach, helping to resist and heal disease. These are some of the traditional drugs that are still in use in China and Southeast Asia, but their efficacy needs to be investigated further. In our previous study, we have entirely reviewed Chinese herbal medicine [1]. Nano-sized herbal drugs have been developed as nanophytomedicines based on their unique nature. Different nanotechnology-based systems such as polymeric NPs, solid lipid NPs, magnetic NPs, metal and inorganic NPs, nanospheres, nanocapsules, quantum dots, nanoemulsions, polymeric micelles, liposomes, and dendrimers have been used for the successful delivery of natural products from traditional drugs. This brings potential plant-based pharmaceutical carriers as an alternative and supplementary medicine to the modern system, pushing the fight against many chronic and pandemic global issues like COVID-19 one step forward [133]. The combination between traditional medicine and nanomedicine will be novel, safe, and effective. Also it is very supportive for a pandemic crisis like COVID-19 [134]. Spherical NPs can deliver plant metabolites and body parts of microorganisms as a potential strategy for antiviral therapies [135]. Glycyrrhizic acid, a substance in the Chinese herb licorice, has a known anti-SARS-CoV effect. Still, its application is limited due to cytotoxicity, poor water and bio-fluid solubility, and low bioavailability. Synthesizing highly biocompatible glycyrrhizic acid NPs significantly improved antiviral and anti-inflammatory effects in vitro and in vivo [136]. A typical Indonesian natural product administration culture, called jamu, is commonly practiced to relieve pain and inflammation from acute and chronic disorders. The efficacy and the value of jamu have been improved using various nanotechnology approaches such as nanosuspension, nanoemulsion, nanoencapsulation, and nanofiber fabrication [137]. At Alfaisal University researchers mixed AgNPs with a black tea extract (theaflavin) and attained a potent viral replication inhibition effect that can assist in the combat against COVID-19 by slowing the viral reproduction rate in a host also it is useful to decrease the severity of infections symptoms [137].
3.5. Novel and helpful treatments
The body's normal reaction to viral disease is to produce a wide range of antibodies that can directly intrude with the virus's capability/potentiality for replication. Analysts recognized the most attainable and resistant antibodies against the Coronavirus and reproduced them in great quantity [138]. The subsequent 'monoclonal antibodies have been used in a different state of affairs as medications for COVID-19. Treatment with monoclonal antibodies should be started as soon as possible after the patient receives a positive result on a SARS-CoV-2 antigen or nucleic acid amplification test within ten days of symptom onset. The use of anti-SARS-CoV-2 monoclonal antibodies should be considered for patients with mild to moderate COVID-19 or those who are hospitalized for a reason other than COVID-19. Anti-SARS-CoV-2 monoclonal antibodies are not currently authorized for use in patients hospitalized with severe COVID-19; However, they may be available through widespread access programs for patients who have not developed an antibody response or are not expected to develop an effective immune response to SARS-CoV-2 infection. Three anti-SARS-CoV-2 monoclonal antibody products currently have Emergency Use Authorizations (EUAs) from the Food and Drug Administration (FDA) to treat mild to moderate COVID-19 in nonhospitalized patients with laboratory-confirmed SARS-CoV-2 infection who are at high risk for progressing to severe disease and hospitalization. It should be mentioned that the issuance of a EUA does not constitute FDA approval. These products are: Bamlanivimab plus etesevimab are neutralizing monoclonal antibodies that bind to different but overlapping epitopes in the spike protein of SARS-CoV-2 [139]. The distribution of bamlanivimab plus etesevimab was stopped on June 25, 2021, because both the Gamma (P.1) and Beta (B.1.351) variants of concern are currently circulating in the United States have reduced susceptibility to bamlanivimab and etesevimab [140]. Casirivimab plus imdevimab are recombinant human monoclonal antibodies that bind to nonoverlapping epitopes of the spike protein RBD [141]. Sotrovimab monoclonal antibody was originally identified in 2003 from a SARS-CoV survivor. It targets an epitope in the RBD of the spike protein that is mutual between SARS-CoV and SARS-CoV-2 [142]. The U.S. Food and Drug Administration issued an emergency use authorization (EUA) for casirivimab and imdevimab to be administered together to treat mild to moderate COVID-19 in adults and pediatric patients with positive results of COVID-19 viral test and those who are at high risk for progressing to severe COVID-19. This includes those who are 65 years of age or older or who have certain chronic medical conditions. In a clinical trial of patients with COVID-19, casirivimab and imdevimab, utilized together, to reduce COVID-19-related hospitalization or emergency room visits in patients at high risk for disease progression within 28 days after treatment when compared to placebo [141]. The safety and efficiency of this therapies for COVID-19 infection continues to be evaluated. Casirivimab and imdevimab are not authorized for patients who are hospitalized due to COVID-19 or require oxygen therapy due to COVID-19. Merit of casirivimab and imdevimab treatment has not been illustrated in hospitalized patent due to COVID-19 [141]. Nasal sprays are used to deliver drugs locally in the nasal cavities. They are used for conditions such as nasal congestion and allergic rhinitis. Nasal drugs have shown to be effective in decreasing COVID-19 transmission [143]. Iota-Carrageenan is a sulfated polymer from red algae. It happens to be a broadly active anti-viral compound. It is effective and safe for the prevention and treatment of respiratory infections [130], [144]. a nasal spray of Iota-Carrageenan showed significant efficacy in preventing SARS-Cov-2 infection in hospital personnel who work beside patients with COVID-19 disease [145]. According to research by an Argentinian group, an Iota-Carrageenan-based nasal spray showed significant efficacy in preventing COVID-19 among hospital staff who were in touch with COVID-19 infected people [130]. In addition, nitric oxide (NO) is a free radical gas molecule involved in innate immunity, and it is also beneficial in terms of wound healing, vasodilation, neurotransmission, and angiogenesis [146], [147]. Treatment with NONS was effective in reducing the viral load in patients with mild, symptomatic SARS-CoV-2 infection. NONS may reduce symptom, duration, infectivity period, hospital admissions of COVID-19 infected patients. They also have experience lower disease severity [147]. The World Health Organization acknowledged that clinical trials are going to evaluate nasal spray vaccines for COVID-19. The vaccines are currently available and offer strong protection against severe forms of COVID-19 but are less reliable at preventing the spread of the virus. Generally, they are intranasal administration of an engineered IgM, which can improve efficacy and reduce resistance also simplify the treatment of COVID-19 [148]. The vaccinated animals displayed less amount of the virus, so they are not contagious anymore, and that's one of the advantages of nasal spray. this nasal spray could serve as a booster shot to fight against SARS-CoV-2. Another novel Therapeutic approach is a Stem cell. Mesenchymal stem cells(MSCs) have received specific attention since they have the ability to help the immune system and prevent inflammation caused by cytokine storms caused by SARS-CoV-2 infection. These new therapies may reduce mortality rates in patients with COVID19 [149]. Studies have shown that mesenchymal stem cells improve lymphocyte populations, especially through dendritic cells and shifting immune system cells [149], [150]. research showed that the injection of MSCs decreased inflammation in patients with severe COVID-19. Also it Improves lung damage [149], [151]. It should be mentioned here that the timing of stem cell injections in case of severe COVID19 is critical. Patients with this situation in the inflammatory phase respond better to cell therapy [149]. findings acknowledged that MSC treatment not only significantly repair lung damage, but also improves patient recovery. Proposed mechanisms for MSC action in patients with severe COVID-19 has some steps. At first, SARS-CoV-2 primarily occupies the respiratory tract including the lung; the infiltration of immune cells (neutrophils, monocytes/macrophages, NK, CD4+ T, CD8+ T, Th17, and B cells) increases; then cytokine storms (including IFN-α, IL-1, IL-6 and TNF-α) occur. Hyaline membrane formation, the release of cellular fibromyxoid exudates, and pneumocyte desquamation are also observed. After stem-cell infusion, the number of infiltrated immune cells decreases significantly, and the damaged lung tissue is getting improved. MSCs play an important role in regeneration and immune regulation [151]. Currently, several clinical trials are undergoing for cell-based therapeutic approaches to prevent or treat COVID-19 patients. Recently, MSCs have become noticeable for clinical trials according to their immunomodulatory and regenerative aspects. They could preserve alveolar epithelial cells, repair the pulmonary microenvironment, inhibit pulmonary fibrosis, and help cure damaged tissues of the lungs [152]. Despite many completing and ongoing clinical trials, there is no clear conclusion regarding some issues related to MSCs limitations, including their safety, tumorigenicity, profibrogenicity, and heterogeneity [153]. The potential risks of mesenchymal stem cells are multiplying or changing into inappropriate cell types, product contamination, growth of tumors, infections, thrombus formation, and administration site reactions. Furthermore, it was reported that several stem cell-based clinical trials received approval for treating COVID-19 patients with acute respiratory disease. Thus., stem cells can be played a key role in the control of this viral infection. Next-generation cell-based therapies can be utilized for each person to treat their specific diseases [154]. Nanotechnology is the design and application of materials that are < 100 nm [155]. Nowadays, nanotechnology is playing a key role in antiviral therapy for COVID-19 [156]. Nanoparticles have been developed specifically for the delivery of biotherapy drugs across physiological barriers [58]. It is also utilizing in the development of air purifiers to inhibit transmission of the SARS-CoV-2 virus [157]. A wide range of nanodevices, such as nanosensors, nano-based vaccines, and smart nanomedicines, offers significant overview for fighting against different mutated strains of coronaviruses. Nanomaterials, such as silver colloid, titanium dioxide, and diphyllin nanoparticles, are antiviral agents and drug-delivery platforms for the effective control of coronavirus infection [58]. An appropriately designed nanoparticle-antiviral drug combination can be expected to enhance the effect of the compounds in several ways (e.g. facilitate interaction with the viral particles, disrupts their entry into cells, increases bioavailability and formula stability, and releases antiviral agents in a controlled manner [158], [159], [160]). Biocompatible nanoparticles may show great antiviral activity [161]. nanomaterials can attach to viral particles and inhibit their interaction with the host cell. For instance, carbon quantum particles prevent the entry of another human coronavirus (HCoV-229E strain) into the host cells by interacting with the S protein of the virus and do not allow the virus to bind with the host cells. These nanomaterials may also useful in case of SARS-CoV-2 [162] Bioavanta LLC/Bosti Trading Ltd has promoted Novochizol, a chitosan-based aerosol nanoparticle that is formulated for stick to lung epithelial tissues and ensures sustained release without systemic distribution, so it is an ideal intrapulmonary delivery system [157]. SARS-CoV-2 and nanomaterials have same size, they make direct contact for combat SARS-CoV-2; additionally, nanomaterials are able to reduce the side effects of antiviral drugs they also can co-deliver multiple drugs and they enhance the stability of mRNA vaccines and nanotechnology facilitates the drug release [163]. SiRNAs are highly efficient in reducing the replication of RNA viruses, such as coronaviruses. In Nano-based gene therapy Nano carriers can improve siRNA stability by preventing enzymatic degradation. Polymer / lipid Nano carriers have shown promising results for loading inhalable antiviral siRNA and delivering aerosol-based antiviral siRNA to the lungs. In nano-based immunotherapy, nanoparticles, such as dendrimers, liposomes, carbon nanotubes, polymer-based materials, and mineral nanoparticles, can be combined with several antigens to activate the immune system more strongly. Nanoparticles have shown promising results in modulating the performance of safety components and reducing immunomodulation-related toxicity [58].
3.6. Antibiotics
Antibiotics are recommended for people who are assumed or confirmed to have COVID-19 infection. Antimicrobial therapy should be evaluated daily for stepping/scaling down [61]. The course of experimental antibiotic treatment should be as short as possible. Generally, 5–7 days. Increased antibiotics consumption during an epidemic may cause terrible reactions, such as Clostridioids difficile infections, with clinical infections ranging from diarrhea and fever to colitis [89]. Antibiotic care programs should be implemented or continued among COVID-19 patients [90]. In summary, the decision to use antibiotics relied more on laboratory markers of inflammation than clinical markers of COVID-19.
4. Vaccines
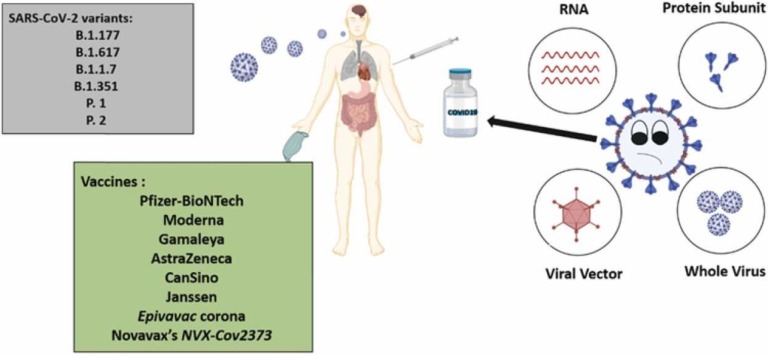
On 11 January 2020, the genetic sequence of the Coronavirus that causes COVID-19 (SARS-Cov-2) was discovered. This discovery motivated a global race initiation for making an efficient vaccine for this disease. The COVID-19 pandemic's impact on humanity and the economy drive evaluation of next-generation vaccine technology to speed up advancement. The first COVID-19 vaccine entered human clinical analysis on 16 March 2020 [164]. According to the WHO report, since 8 October 2021, 320 vaccines have been in progress. 194 vaccines are in the pre-clinical development stage that 126 of them are in clinical progression [165].
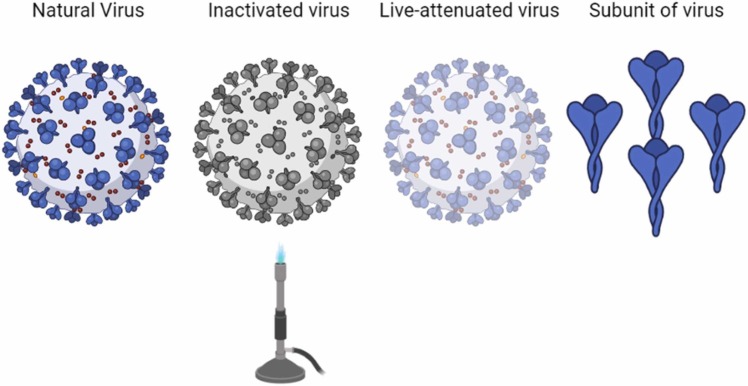
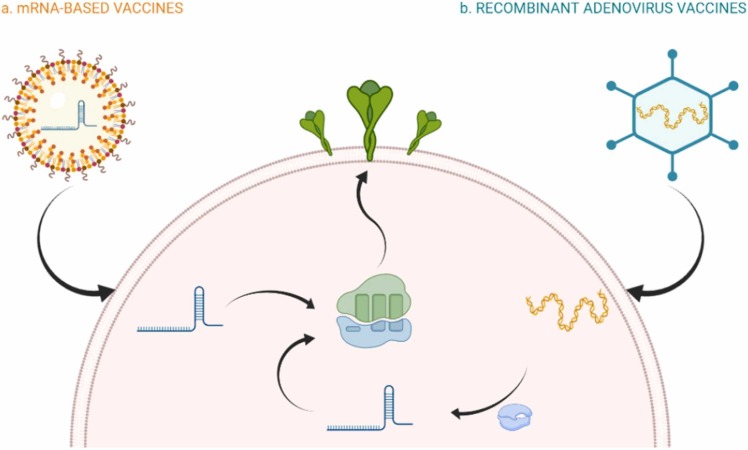
Vaccines that have recently acquired/attained clinical approval by national and international authorities vary by their design: deactivated vaccine, live-attenuated vaccine, subunit vaccine, nucleic acid vaccine, viral vector vaccine [166]. Inactivated or killed virus (KV) vaccines refer to a type of vaccine that chemicals have inactivated the native pathogen (virus) (e.g., Formaldehyde, Glutaraldehyde), PH, heat treatment, and radiation (e.g., gamma irradiation). They can replicate in human or animal bodies [167]. The live-attenuated vaccines consist of the weakened forms of the virus that can poorly replicate in the cell but can't cause any severe disease. These vaccines have been used for billions of people and usually provide immunity for decades [168]. Another type of vaccine is subunit vaccines which are based on virus particles. These vaccines typically contain adjuvants, molecules that increase the magnitude and moderate the immune response quality. About the conjugated vaccines, polysaccharide-based antigens are carried by protein carriers, so this type of vaccine can be classified as a subclass of subunit vaccines [169]. Vaccines based on nucleic acid are being developed to combine live-attenuated and subunit vaccines to use their respective advantages. Viral vectors (e.g., adenoviruses, poxviruses), recombinant bacteria, plasmid DNA and RNA, are different kinds of these vaccines [170]. RNA or DNA-based vaccines are expressed in the host cell instead of direct injection of the antigen or complete virus particles [171].
Vaccines are potentially designed to make numerous diverse antibodies to identify especial parts of the virus. So if one particle of the virus mutates, the antibodies could figure out another part of the virus. Understandably, there will be a variety of antibodies that may decrease vaccine viability, so the vaccine companies are making a new class of vaccines that ought to work against the latest strains of SARS-CoV-2 [172].
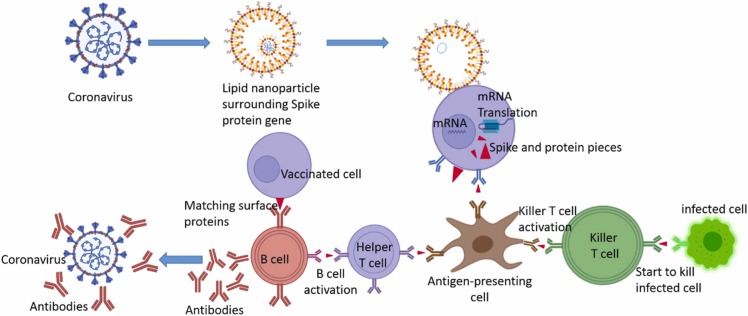
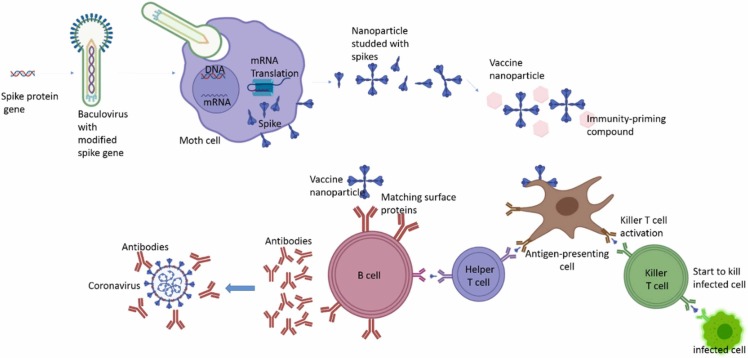
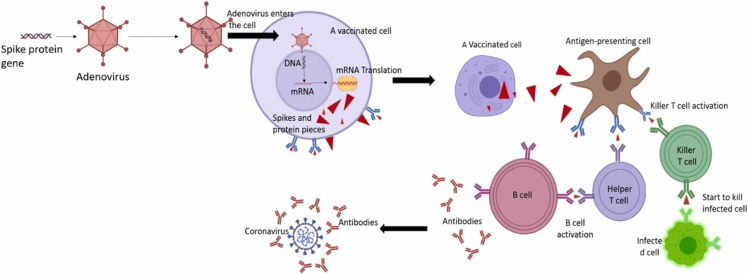
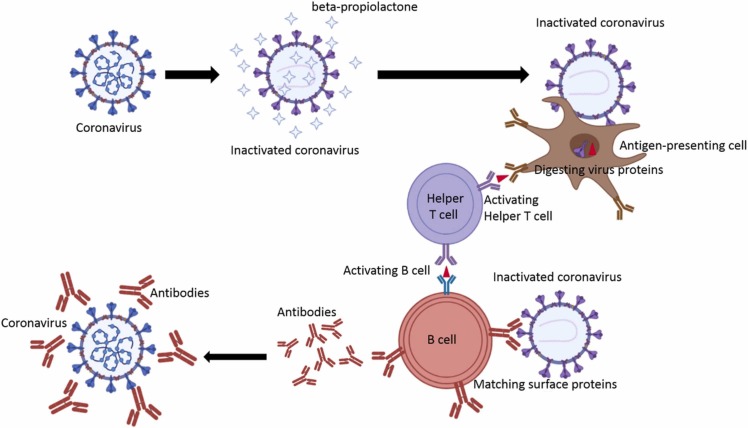
Table 8 the comparison between vaccines available. These types of vaccines consist of lipid nanoparticles shown in Fig. 14 and Fig. 15 .
Fig. 14.
The differences between the vaccines are based on the inactivated virus, live-attenuated virus, and subunit of a virus with the natural virus.
Fig. 15.
(a) These types of vaccines consist of a lipid nanoparticle carrying mRNA molecule. The ribosome would translate this mRNA to make S proteins of SARS-CoV-2, which cause an immune response in the human body. (e.g., Pfizer-BioNTech, Moderna) (b). Recombinant adenovirus vaccines are adenovirus vectors that carry a DNA molecule (the S protein gene) to the host cell. The RNA polymerase would transcribe the DNA in these viral vectors to generate an mRNA. Ribosomes translate this mRNA molecule to produce S proteins of SARS-CoV-2 (e.g., Gamaleya, AstraZeneca-Oxford, Cancino, Janssen). Graphics are created with Biorender.com.
4.1. Pfizer-BioNTech vaccine
The vaccine developed by Pfizer and BioNTech is a nucleic acid vaccine that comprises a nucleoside-modified mRNA encapsulated in lipid nanoparticles. This modified mRNA encodes immunogens taken from the spike glycoprotein (S) of SARS-CoV-2 [173]. These developers are testing four vaccine candidates simultaneously [174]. BNT162b1 encodes a dissolvable, discharged trimerized receptor-binding domain (known as the RBD–folded). BNT162b2 encodes the full-length transmembrane S glycoprotein, coated in its prefusion configuration by the transposition of two residues with proline (S(K986P/V987P); from now on, S(P2) (known as P2 S as well)) [173]. According to FDA’s fact sheet about Pfizer-BioNTech vaccine, this vaccine contains the following ingredients: mRNA, lipids ((4-hydroxybutyl) azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate), 2 [(polyethylene glycol)−2000]-N,N-di-tetradecyl acetamide, 1,2-Distearoyl-sn-glycerol-3- phosphocholine, and cholesterol, potassium chloride, monobasic potassium phosphate, sodium chloride, dibasic sodium phosphate dihydrate, and sucrose [175]. Efficacy analysis of the Pfizer-BioNTech vaccine demonstrates that starting 28 days after the first dose, BNT162b2 is 95% successful in resistance to the SARS-CoV-2 virus. This efficacy was 94% in people above 65 years old. These data consist of 43,000 participants of various ages, gender, race, and ethnicity [175]. this vaccine should be stored in an ultra-cold freezer between − 80 °C and − 6 °C for long term use. But for short term use that should not exceed 2 weeks, it can also be stored in the freezer between − 25 °C and − 15 °C [175].
It is important to mention that no deaths were reported because of the vaccination; only mild to moderate resultants were observed. Generally, this variation had a proficiency of up to 95% [176]. The new variants of the virus provide the elevated capability to spread expeditiously, but no change in the severity of the disease was reported. The current vaccines can recognize and neutralize some of the new variations of the virus [172]. Commonly detailed side effects in non-anaphylaxis allergic responses included pruritus, rash, itchy and scratchy sensations within the throat, and gentle respiratory symptoms [177]. Furthermore, a few Antagonistic Events Pain such as swelling, redness, fever, weakness, headache, chills, vomiting, diarrhea, muscle pain, joint pain, lymphadenopathy, shoulder injury, right axillary lymphadenopathy, paroxysmal ventricular arrhythmia, syncope, and right leg paresthesia has been reported [178], [179]. In general, the side effects associated with the Pfizer-BioNTech COVID-19 vaccine are similar to those associated with most previous vaccinations, and most of them are tolerable [180], [181]. Moreover, the manufacturer and the FDA Facts Sheet [182]. Anyway, it is required for some individuals to be monitored for a brief time instantly after getting their vaccine dosages [183].
4.2. Moderna vaccine
Moderna was one of the first vaccine developers who launched the clinical trials of vaccines against SARS-CoV-2. Like the Pfizer-BioNTech vaccine, Moderna's vaccine is also based on mRNA technology [184]. This vaccine consists of an mRNA molecule that is encapsulated by lipid nanoparticles. This (mRNA-1273) generates a full-length spike protein of SARS-CoV-2 with stabilized permeation/infiltration and induces an immunological response in human cells. 30,420 volunteers took part in this clinical trial. 50% of them received two injections (each included 100 µg of mRNA) 28 days apart. According to the results, the vaccine's efficacy is 94.1%, and no severe adverse events were reported [185]. The best long-term storage temperature for the vaccine is between − 25 °C and − 15 °C in the freezer. And for short-term storage(less than 30 days), between 2 °C−8 °C in the refrigerator [174].
In vaccination trials, moderate side effects such as low-grade fever, chills, and a general feeling of malaise were seen [186]. The antibody reactions to the mRNA-1273 vaccination were related to the vaccine dosage utilized. mRNA-1273's most frequent side effects are weakness, chills, headache, myalgia, and injection site pain. The overall effectiveness of this vaccine was 94.1% in escaping COVID-19 sickness [176]. This vaccine immunogenicity continued no less than 3 months. The second vaccine is provided 90 days after the previous vaccination [101]. There are a few negative occurrences. Swelling, redness at the vaccine site, and other areas of pain caused by the Moderna vaccine [178], [186] weakness, headache, chills, vomiting, arthralgia, myalgia, urticarial. These clinical symptoms were mild and tolerable after the primary dosage of the vaccine. However, after the second dose of the vaccine, the undesirable effects were moderate to deadly [102], [178]. Besides, facial swelling and Bell's paralysis have also been reported [178], [187]. The Moderna vaccine is much easily transported compared to the Pfizer vaccine because it is easily transported at − 4 Fahrenheit [178].
Pfizer-BioNTech and Moderna have shown the most effective leading antibodies using nanotechnology with the mRNA stage [103]. Two dosages of the Pfizer-BioNTech and Moderna COVID-19 vaccines are essential for good resistance [172]. It should be highlighted that the hazard of neurological complications or any other opposing impact related to COVID-19 immunization is low at both the individual and social levels, and the advantages of vaccination overcome any potential risks or side effects [104].
4.3. Gamaleya vaccine
Russian vaccine, which is based upon two adenovirus vectors, was industrialized by the Gamaleya National Center of Epidemiology and Microbiology (Moscow, Russia). Russa was the premier country in the world that authorized a COVID-19 vaccine by approving the Gamaleya vaccine (referred to as Sputnik V/Gam-COVID-Vac) [105]. This endorsement was given after the phase I trial (38 people) without any published results report [188]. The efficacy of Sputnik V is 91.6% based on 19,866 volunteers who received two separate doses (with a 21 days interval) [189]. The first dose consists of recombinant adenovirus 26 (rAd26), while the second one is rAd5. Both recombinant adenovirus vectors contain the gene for the full-length glycoprotein S of SARS-CoV-2 [190]. Developers expect to have more immune responses by using two different vectors. This vaccine can be kept in 2–8 °C in the refrigerator, which makes this vaccine easy to transport and distribute [191].
Studies show that Sputnik V has around 92% efficiency in the phase 3 trial without significant side effects. Side effects of this vaccine, even minor complications, are registered in less than 0.1% of cases that take Sputnik V, counting even slight body temperature reaction, slight pain around the injection site, weakness, and low energy. In the trial phase of this vaccine, 4 deaths were reported. Evidence shows that none of them was related to the Sputnik V vaccine [192], [193].
4.4. AstraZeneca-Oxford vaccine
The British-Swedish Company AstraZanca and Oxford University developed a vaccine based on a recombinant chimpanzee adenovirus vector (ChAdOx1 nCoV-19 (AZD1222)). Studies reported that the efficacy of this vaccine (Vaxzevria) is 82.4% after receiving the second dose with 21 days' interval. In contrast, the effectiveness is 76% after 90 days, while there is no second injection. It is not clear how long this first dose can protect the body against SARS-CoV-2 (24). The AstraZeneca-Oxford vaccine is cheap in comparison to Pfizer-BioNTech and Moderna mRNA vaccines. This vaccine can be stored in a refrigerator for transportation and distribution [191]. In adults of all ages, the advantages of Vaxzevria outweigh the dangers; nonetheless, infrequent occurrences of blood clots and low blood platelets have been reported following vaccination.
The most prevalent side effects include mild-to-moderate symptoms like headache, fatigue, muscle or joint pain, fever, chills, and nausea. In addition, some vaccination components may cause an allergic response. The symptoms may contain hives, swelling, a rash, and respiratory symptoms. Anaphylaxis, a severe and potentially life-threatening allergic reaction, has also been reported in a few cases. Anaphylaxis, on the other hand, is a highly unusual occurrence [21].
4.5. CanSino’s Ad5-nCoV vaccine
Chinese one-shot vaccine is based on recombinant adenoviruses, like Gamaleya and AstraZeneca-Oxford vaccines. The Ad5-nCoV vaccine from CanSino uses adenovirus to deliver the SARS-CoV-2 genome through the cells [194]. The efficacy of this vaccine is 65.7% only by receiving one dose to prevent symptomatic cases. This vaccine was 90.98% successful in severe disease prevention. According to clinical trials, this is being applied in the following countries: Pakistan, Mexico, Russia, Argentina, and Chile [175].
This vaccine is effective with a single dose and can achieve dual protection, humoral and cellular immunity simultaneously. This vaccine is impressive for older adults. Because no serious side effects were monitored based on phases two and three of the trials that involved elderly participants. CanSino’s Ad5-nCoV vaccine has 65.7% efficacy against symptomatic COVID-19 cases and 90.98% against severe diseases. Rare side effects are reported for this vaccine, for example, fever, headaches, and pain in the injection site [21], [195].
4.6. Janssen vaccine
Janssen (the pharmaceutical wing of Johnson & Johnson) developed a vaccine based on recombinant human adenovirus (Ad26. COV2). Janssen launched two separate phases III, one of them with two doses and only one shot, to evaluate the effectiveness of separate injections [196]. COVID-19 vaccine of Johnson & Johnson is a single dose vaccine and should be stored at 2–8 °C in a dark place [175]. FDA and EUA have approved Janssen for individuals aged 18 years and older [197].
At first, the Johnson & Johnson vaccine was specified to generate and release antibodies against SARS-CoV-2 in 90% of people by receiving the first dose of the vaccine. The antibodies quantity was higher for those who obtained two doses of the vaccine. Results reported by Johnson & Johnson show that the first dose of the Johnson & Johnson vaccine was 66% effective in prohibiting moderate to severe COVID-19% and 100% effective in defending against COVID-19–related hospitalization and death [110].
Primary evidence indicates that the asymptomatic infection, in which a person is infected with the COVID-19 virus but does not become sick, may be prohibited by the Janssen vaccine. However, this vaccine has some secondary effects too. The most common side effects include mild-to-moderate symptoms: pain at the injection site, headache, fatigue, muscle aches, and nausea. The majority of these adverse effects occurred within 1–2 days after vaccination and remained for 1–2 days [198].
4.7. EpiVacCorona
The second leading Russian vaccine has developed by the vector institute in Novosibirsk named EpiVacCorona. EpiVacCorona, instead of using complete viruses, relies on synthetic peptide antigens subset of SARS-CoV-2 antigens. This process, including recreating the spike protein, uses virus fragments to elicit an immune response [111]. The vaccine (EpiVacCorona) is an intramuscular suspension containing chemically synthesized immunogen peptides of the S antigen of the SARS-CoV-2 Coronavirus joined to a carrier protein adsorbed on aluminum hydroxide. This vaccine should be used in two separate injections with 21–28 days intervals [199]. According to the phase, I and II of clinical trials, local vaccine administration responses were moderate, except for a brief discomfort at the injection site. There was no proof of local or systemic developing side effects. In 100% of the volunteers, the two-dose vaccination system triggered the development of unique antibodies for the antigens. Seroconversion with a neutralizing antibody titer of less than 1:20 was observed in 100% of the volunteers 21 days after the injection [200]. The results for phase III of the clinical trial have not been published yet. This study contains 2250 volunteers who will be vaccinated twice by a dose of 0.5 mL and 750 volunteers who will receive the placebo twice at a dose of 0.5 mL [198].
For this vaccine, minimum side effects have been reported. Volunteers in Phase I and Phase II of the trials have shown minor soreness around the injection area, which passed rapidly. No other adverse symptoms were reported [198].
4.8. Novavax's NVX-Cov2373 vaccine
Novavax's NVX-Cov2373 vaccine induces an immune response by using a stabilized version of the coronavirus spike protein in 2 doses [201]. Novavax used recombinant nanoparticle technology to generate antigens acquired from the coronavirus spike protein. Additionally, this company utilized its Matrix-M™ as an adjuvant. Pre-clinical studies have shown that this adjuvant helps the vaccine bind more efficiently to human receptors targeted by SARS-CoV-2 [193]. Matrix M1 is a saponin-based adjuvant. Saponins are derived from the Quillaja saponaria Molina tree bark and are formulated separately with phospholipids and cholesterol to form matrix fragments (40 nm cage-like structures) [202]. The adjuvant (Matrix M1) is dose-sparing since the lower 5 µg dosage of N.V.X. CoV2373 performs similarly to the higher 25 µg dose. NVX-Cov2373 should be saved at 2–8 °C that facilitates its transportation [198]. Novavax SARS-CoV-2 vaccine is 95.6% effective against the original variant of SARS-CoV-2. This vaccine is also 85.6% and 60% effective against B.1.1.7 and B.1.351 variants separately [9] ( Table 9).
Table 9.
The comparison of 8 leading vaccines.
| Vaccine | Technology | Efficacy | Dosing | Route of Administration | Storage condition | Status |
|---|---|---|---|---|---|---|
 |
mRNA | 95% | 2 Doses (28 days interval) | Intramuscular | -80 °C to − 60 °C Or -25 °C to − 15 °C (2 weeks) | Approved in U.S., other countries. Emergency use in E.U., other countries. |
 |
mRNA | 94.10% | 2 Doses (28 days interval) | Intramuscular | -25 °C to − 15 °C Or 2–8 °C (30 Days) | Approved in Switzerland. Emergency use in U.S., E.U., other countries. |
 |
Recombinant Adenovirus | 91.60% | 2 Doses (21 days interval) | Intramuscular | 2–8 °C | Emergency use in Russia, other countries. |
 |
Recombinant Adenovirus | 82.40% | 2 Doses (21 days interval) | Intramuscular | 2–8 °C | Approved in Brazil. Emergency use in U.K., E.U., other countries. |
 |
Recombinant Adenovirus | 65.70% | 1 Dose | Intramuscular | -25 °C to − 15 °C | Approved in China. Emergency use in other countries. |
 |
Recombinant Adenovirus | 77% | 1 Dose | Intramuscular | 2–8 °C | Emergency use in U.S., E.U., other countries. |
 |
Protein | – | 2 Doses (21 days interval) | Intramuscular | 2–8 °C | Approved in Turkmenistan. Early use in Russia. |
| Protein | 95.60% | 2 Doses(21–28 days interval) | Intramuscular | 2–8 °C | – |
5. Mechanisms of vaccines
FDA has approved different types of vaccines that are being used in various countries. Each vaccine's functional mechanism in the body is unique. In this section, we briefly introduce these mechanisms.
5.1. mRNA vaccine
The dynamic element that encodes the viral spike glycoprotein (S) of SARS-CoV-2 is the nucleoside modified messenger RNA (modRNA). This mRNA acts as a template for producing the specific protein that initiates the host’s immune response against the virus. The mRNA in mRNA vaccines is wrapped in LNPs, which aids in the RNA transport and protects it from degradation with salts that function as a buffer. Another consideration is sucrose, which serves as a cryoprotectant [103]. The LNP-mRNA cargos enter muscle cells via endocytosis shortly after infusion, and then the mRNA is transcribed. Subsequently, invading antigen-presenting cells(APCs) can be recruited by a network of blood arteries next to the muscles [203]. The vaccine molecules then collide with the membrane and penetrate the cells. Afterward, the mRNA is released and then translates inside the host to produce the SARS-CoV-2 S protein [204], [205]. spike proteins are then assembled to form spikes that travel to the cell’s surface and stick out their tips from the membrane. Besides, the vaccinated cells start fragmentation of a few spike proteins so that the immune system can recognize them. When an immunized cell dies, it leaves a lot of spike proteins and protein fragments behind, which can be picked up by special immune cells called antigen-presenting cells. The antigen-presenting cells have to exhibit the pieces of the uptaken spike protein on its surface. After that, helper T cells, another type of Immune cells, detect these pieces and blow the whistle for the B cells and offer assistance to them for fighting the infection. When B cells contact coronavirus spikes on the surface of vaccinated cells or free-floating spike protein, they attach to it, draw it inside and show spike protein fragments on its surface. If these B cells are activated by helper T cells at that point, they will multiply and produce antibodies against the spike protein. These antibodies target coronavirus spikes, label the virus for destruction and prevents sickness by preventing the spikes from joining alternative cells. Antigen-presenting cells can also activate a type of immune cell known as a killer T cell, which will hunt out and destroy any coronavirus-infected cells with spike protein fragments on their surfaces [206]. The mechanism is shown in Fig. 16 .
Fig. 16.
Mechanism of mRNA-based vaccines.
5.2. Protein-based vaccines
To produce the vaccine, researchers used a modified spike gene. Researchers inserted the gene into another virus, a baculovirus, which can infect moth cells at the first step. This method of growing and harvesting viral proteins is already used to make Influenza and HPV vaccines. Then, the spike proteins are collected and linked to nanoparticles. Afterward, the nanoparticles imitate the molecular structure of a Coronavirus that cannot replicate or cause disease. Then the vaccine is ready for injection. Each injection into the arm muscles includes many spike nanoparticles coupled with a special compound drawn out from the soapbark tree, which is very practical in the food and drug industry. This compound absorbs immune cells to the muscles of the arm and makes them respond highly to the vaccine. Antigen-presenting cells, another immune cell, identifies the nanoparticles for the virus and take them up. An antigen-presenting cell chops the spike proteins and exhibits some of the fragments on its surface. The helper T cell has some surface proteins that can identify the spike fragments, and if a particle fits into the surface proteins, the helper T cell becomes active. Now the helper T cell can warn other immune cells to feedback to the nanoparticles. Then for making antibodies, B cells may also identify the vaccine nanoparticles. B cells have various surface proteins as well, and some of them have the right shape to attach to the spike protein so it can swallow the particle inside and unfold the spike protein fragments on its surface. If an activated helper T cell is attached to one of the fragments, it activates the B cell. Then the B cells increase in number and produce large amounts of antibodies into the environment. Antibodies are similar to surface proteins. As a result, if vaccinated people are later exposed to the Coronavirus in any way, their antibodies can attach to the spike proteins. Thus the Coronavirus cannot enter cells, and the infection is blocked. This vaccine can also stimulate another kind of protection by killing infected cells. When coronavirus infects a cell, they unfold fragments of the spike protein on their surface. Also, Antigen-presenting cells can activate killer T cells, which can identify covid19-infected cells and kill them as soon as possible before they become able to make new viruses [207] ( Fig. 17).
Fig. 17.
Mechanism of protein-based vaccines.
5.3. Adenovirus-based vaccines
This vaccine has been produced by adding the coronavirus spike protein gene into different adenoviruses, such as Ad26 and Ad5. These adenoviruses can invade cells but can’t replicate. In the Gamaleya vaccine, both of these adenoviruses have been used. In the CanSino vaccine, the coronavirus spike protein gene was converted to Ad5, and in Janssen, Ad26 was utilized. However, the Oxford-AstraZeneca team utilized ChAdOx1, a modified variant of a chimpanzee adenovirus.
After vaccination, these adenoviruses strike towards cells and plug into their surface proteins. Then the cell swallows the virus. Afterward, the virus migrates to the nucleus, and the cell starts producing spike proteins. Some spike proteins get fragmented and go to the cells' surface and stick out their tips.
As said above, the antigens are recognized by special immune cells (antigen-presenting cells, T helper cells, and B cells), and antibodies are produced. If the virus invasion happens again, antibodies can lock onto coronavirus spikes, label the virus for destruction and prevent infection [199].
The mechanism of this vaccine is shown in Fig. 18 .
Fig. 18.
Mechanism of Adenovirus-based vaccines.
The critical issue concerned researchers is that immune systems could reply to an adenovirus vaccine by producing antibodies, making the second dose ineffective. The Russian vaccine has solved this problem by utilizing two adenoviruses, Ad26 for the first injection and Ad5 for the second dose.
COVID-19 Adenovirus-based vaccinations are more durable than Pfizer and Moderna’s mRNA vaccines. Because, one, The adenovirus’s strong protein shield helps safeguard the genetic information (DNA) within and two, RNA is much more sensitive and delicate than DNA. As a result, adenovirus-based vaccines can be stored in the refrigerator and do not require minimum storage temperatures (−80c) [198].
5.4. Inactivated virus vaccine
Viruses can be inactivated and though non-infectious through various chemical or physical methods. Defective viruses contain different viral proteins for the immune system to recognize and make antibodies against them. So, effective vaccines against viral diseases like influenza can be developed by using defective viruses [208]. Many companies like Sinopharm, Sinovac, Sinopharm-Wuhan, and Bharat Biotech use inactivated Methode in SARS-CoV-2 vaccine production.
For inactivating the virus, chemicals, heat, or radiation are used to destroy the genetic material of the virus, which will stop its replication. For example, the BBIBP-CorV vaccine that is constructed by Sinopharm company is developed using the inactivated COVID-19. Scientists choose 1 out of 3 covid samples from three individuals for the basis of the vaccine. Beta-propiolactone is the chemical used for the coronavirus's inactivation. It remodels the virus’s genetic substance not to replicate, but the virus's proteins, including spike, remained intact. Other companies have also utilized very similar methods in producing COVID-19 inactivated vaccines. These companies produced vaccines containing an adjuvant called aluminum hydroxide to improve the vaccine's effectiveness [193].
As previously stated, the humoral immune system is activated following the injection. Antigen-presenting cells capture inactivated viruses that have entered the body and display fragments of the viruses' spike proteins on their surface. Following that, the T helper cells activate the B cells, resulting in the production of antibodies. The mechanism has indicated in Fig. 19 .
Fig. 19.
Mechanism of inactivated virus vaccine.
After the vaccination is completed, with the help of the antibodies that cover the invaders’ antigens, the immune system can respond to active coronavirus infection. Antibodies that bind to the spike protein can stop the virus from infecting cells and stop the disease [198].
5.5. Other types of COVID-19 vaccines
Tobacco plants were initially used to generate antibodies to create vaccinations. The United States Department of Agriculture (USDA) has authorized the world's first plant-based vaccination for the Newcastle disease virus (NDV) in poultry. COVID-19 plant-based vaccines can be made by producing the antigenic component of SARS-CoV-2 to induce active immunity or by expressing the virus's antibodies to offer passive protection [209]. Several proof-of-concept studies have looked at the possibilities of using plant expression systems to develop vaccines for respiratory diseases such as SARS, influenza, tuberculosis, and anthrax. The sustained S protein (S1) production in tomato and low-nicotine tobacco plants was used in a plant-based vaccination against SARS. Preclinical investigations on mice revealed that the plant-derived vaccination elicited an antibody response. Recombinant SARS-CoV N protein generated transiently in N. benthamiana was found to be immunogenic in another research. After the third parental injection, the tobacco-produced recombinant N protein substantially stimulated the humoral immune response. Previous research can aid in the design and development of an efficient plant-based vaccine against SARS-CoV-2. Since the virus sequence was made, there are 6 plant-based vaccines in pre-clinical studies and 2 in clinical studies (As of 13 August 2021) [210].
Intranasal vaccination is one of several ways of improving immunity in mucosal organs such as the oral, pulmonary, conjunctiva, rectal, and vaginal mucosa. Intranasal vaccinations are used to stimulate the respiratory tract's mucosal immune system in principle [211]. SARS-CoV-2 is a mucosal virus that infects people through the nose and mouth in most cases. Its most efficient transmission mode is by breathing droplets, which are subsequently passed on to other people. Compared to systemic, IgG-only humoral and T-cell responses, a vaccination that also induces protective mucosal responses mediated by IgA is more likely to decrease transmission [212]. Mucosal lymphoid tissues (MALT) of the nasopharynx (also known as nasopharynx-associated lymphoid tissue—NALT) and bronchus-associated lymphoid tissue (BALT) of the lungs are examples. Intranasal vaccination of mice with a recombinant adenovirus-based vaccine expressing Middle East respiratory syndrome coronavirus (MERS-CoV) spike proteins resulted in T lymphocytes in the respiratory airway and lungs [211].
5.6. Application of nanotechnology in COVID-19 vaccine development
An efficient packing mechanism is essential to avoid RNA degradation before delivery to the host translational machinery. Cationic lipid nanoparticles, initially designed for nucleic acid delivery in cancer immunotherapy and other vaccine applications, can help meet this need. Using cationic lipids, mRNA is efficiently condensed into solid lipid nanoparticles that host APCs may take up through endocytic or phagosomal routes. On the other hand, full-length spike protein or viral components can be encapsulated or self-assembled into nanoparticles as an option to using nanoparticles for CoV-2 vaccine development [213]. Nanoparticles not only protect the antigen's native structure but also enhance antigen delivery and presentation to antigen-presenting cells (APCs). Many biological systems, including viruses (particularly SARS-CoV-2) and proteins, are nanosized. Therefore vaccination nanocarriers provide some advantages [214]. Nanoparticles can be delivered by oral and intranasal routes and subcutaneous and intramuscular injections, overcoming tissue barriers and targeting critical sites such as lymph nodes, mucosal, and epithelial barriers (airway, nasal, gastrointestinal, etc.) [215], [216]. Another strategy of applying nanotechnology in vaccine development is VANs (vaccine adjuvant nanoparticles), which are considered to increase the overall effectiveness and safety of the immune response induced. Vaccine adjuvants are essential, especially in the case of the COVID-19 pandemic, for decreasing the needed antigen dosage (dose-sparing), allowing for the manufacture of more units, and making it available to a broader population [213].
6. COVID-19 global vaccination
The current coronavirus disease (COVID-19) vaccination program seeks to attain worldwide vaccination coverage, which will aid in pandemic control. As a result, individuals who refuse to be vaccinated or forego COVID-19 vaccination may slow overall vaccination coverage, resulting in lower vaccination rates, and may obstruct global efforts to control the spread of SARS-CoV-2 unvaccinated individuals can act as SARS-CoV-2 reservoirs, causing more outbreaks [217]. According to the WHO, vaccine hesitancy is one of the most serious threats to global health [218]. The importance of social attitudes in developing herd immunity cannot be overstated. It's also worth noting that, to ensure a successful and effective vaccination program, governments and social media should motivate individuals to collaborate to address the public health problem while also giving information to alleviate their fears. Survey research in many countries have revealed varied vaccine acceptance percentages, indicating that vaccine hesitancy must be addressed from the start of immunization programs. Concerns about the distribution of vaccinations and how they are delivered are also significant concerns for COVID-19 immunization. People are sorted into groups based on their vaccination priority. First, this is critical for ensuring a fair and equitable distribution of vaccinations worldwide to respond effectively to the SARS-CoV-2 pandemic [219]. A proper vaccination distribution, according to the Centers for Disease Control and Prevention (CDC), will contain the following: Phase 1a involves vaccination of healthcare personnel and long-term care residents; Phase 1b involves essential frontline employees and persons over the age of 75, and Phase 1c affects people aged 65–74 or 16–64 with underlying medical problems and other essential workers. Following that, when vaccination doses become available, more categories will be included.
Nonetheless, scientists estimate that distributing COVID-19 vaccinations to the general public would take 6–12 months, involving various challenging conditions to manage [220]. The affluent nations have already secured 60% of the entire coronavirus disease (COVID-19) vaccine supply for their populations. Vaccine dosages adequate to vaccinate their people several times have been preordered in some of these nations. Only by guaranteeing equitable access to COVID-19 vaccines will global vaccination coverage be attained. COVAX is a worldwide movement organized by WHO, the Coalition for Epidemic Preparedness Innovations, and Gavi, the Vaccine Alliance, that guarantees equitable access to COVID-19 vaccines [221].
7. Conclusion
COVID-19 is a lethal pandemic disease that needs urgent global attention to prevent more disasters, including significant social, psychological, and economic dilemmas. Coronavirus can cause severe permanent respiratory syndromes and other organs malefunctionality and in some cases death. Thus, it is vital to develop fast, cheap, reliable, and accurate diagnostic systems and efficient therapeutic approaches for treating COVID-19. There are no specific antiviral drugs available at the moment for COVID-19, but many clinical trials are going on, and lots of potential herbal/chemical treatments are being used. Here, in this comprehensive review, we have tried to gather the most recent and updated information about different coronavirus mutations, approaches for the detection of coronavirus, approved chemical and herbal drugs for COVID-19 treatment and developed vaccines and their mechanism. Although, among all mentioned therapies, vaccination is the most promising approach to reduce mortality and permanent side effects of this disease. But, some people refuse to be vaccinated cause slowing overall vaccination coverage, resulting in lower vaccination rates, and may obstruct global efforts to control the spread of SARS-CoV-2. Therefore, governments and social media should motivate individuals to collaborate to address the public health problem meanwhile giving information to alleviate their fears. Hope this paper could pave the way of researchers to find more efficient solutions to control this pandemy.
Ethical approval and consent to participate
Not applicable for this study.
Funding
To perform this study, Matin Mahmoudifard received grant support from the National Institute of Genetic Engineering and Biotechnology (NIGEB) of The Islamic Republic of Iran.
Consent for publication
All authors have read and approved the final version of the manuscript.
CRediT authorship contribution statement
Matin Mahmoudifard has a role in the design and collection of data. All authors contributed equally in drafting the first version of the manuscript. All authors participated in writing modified versions and read and approved the final manuscript.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was made possible by a grant from the National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran.
Availability of data and materials
Not applicable for this study.
References
- 1.Barati F., Pouresmaieli M., Ekrami E., Asghari S., Ziarani F.R., Mamoudifard M. Potential drugs and remedies for the treatment of COVID-19: a critical review. Biol. Proced. Online. 2020;22(1):1–17. doi: 10.1186/s12575-020-00129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ekrami E., Pouresmaieli M., Barati F., Asghari S., Ziarani F.R., Shariati P., Mamoudifard M. Potential diagnostic systems for coronavirus detection: a critical review. Biol. Proced. Online. 2020;22(1):1–18. doi: 10.1186/s12575-020-00134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richman D.D., Whitley R.J., Hayden F.G. John Wiley & Sons; 2020. Clinical Virology. [Google Scholar]
- 4.Chawla D., Chirla D., Dalwai S., Deorari A.K., Ganatra A., Gandhi A., Kabra N.S., Kumar P., Mittal P., Parekh B.J., Sankar M.J., Singhal T., Sivanandan S., Tank P. Perinatal-neonatal management of COVID-19 infection—guidelines of the Federation of Obstetric and Gynaecological Societies of India (FOGSI), National Neonatology Forum of India (NNF), and Indian Academy of Pediatrics (IAP) Indian Pediatr. 2020;57(6):536–548. doi: 10.1007/s13312-020-1852-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F., Scarlata S., Agrò F.E. COVID‐19 diagnosis and management: a comprehensive review. J. Intern. Med. 2020;288(2):192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piombino P., Committeri U., Norino G., Vaira L.A., Troise S., Maglitto F., Mariniello D., De Riu G., Califano L. Facing COVID-19 pandemic: development of custom-made face mask with rapid prototyping system. J. Infect. Dev. Ctries. 2021;15(01):51–57. doi: 10.3855/jidc.13384. [DOI] [PubMed] [Google Scholar]
- 8.John Hopkins University and Medicine – Coronavirus Resource Center, 2020 COVID-19 case tracker. [cited 2020 10th September]. 〈https://coronavirus.jhu.edu/map.html〉.
- 9.Worldometers, 2021 [cited 2021 9th March]. 〈https://www.worldometers.info/coronavirus/〉.
- 10.He J., Guo Y., Mao R., Zhang J. Proportion of asymptomatic coronavirus disease 2019: a systematic review and meta‐analysis. J. Med. Virol. 2021;93(2):820–830. doi: 10.1002/jmv.26326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.nytimes news. [cited 2021 9th March]. 〈https://www.nytimes.com/news-event/coronavirus〉.
- 12.WHO. [cited 2021 9th March]. 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/coronavirus-disease-COVID-19#:~:text=symptoms〉.
- 13.worldometers. [cited 2021 13th March]. 〈https://www.worldometers.info/coronavirus/〉.
- 14.Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Investig. 2021;131(1) doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. 2021. 〈www.who.int/en/activities/tracking-SARS-CoV-2-variants〉.
- 16.nytimes news. 2021 [cited 2020 10th March]. 〈https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html〉.
- 17.Nature. 2021 [cited 2021 10th March]. 〈https://www.nature.com/articles/d41586–020-00502-w〉.
- 18.fortune.com. 2021 [cited 2022 10th March]. 〈https://fortune.com/2021/01/22/uk-virus-strain-more-lethal/〉.
- 19.scientificamerican.com. 2021 [cited 2021 10th March]. 〈https://www.scientificamerican.com/article/the-most-worrying-mutations-in-five-emerging-coronavirus-variants/〉.
- 20.hopkinsmedicine. 2021 [cited 2021 10th March]. 〈https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/a-new-strain-of-coronavirus-what-you-should-know〉.
- 21.2021 [cited 2021 30th April]. 〈https://www.ema.europa.eu/en/news/astrazenecas-COVID-19-vaccine-benefits-risks-context〉.
- 22.nytimes. 2021. 〈https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html#Q677〉.
- 23.WHO. 2021 [cited 2021 3rd October]. 〈www.who.int〉.
- 24.Masks-FaceCoverings. 2021 [cited 2021 20th March]. 〈https://easa.com/Portals/0/Images/COVID19/Masks-FaceCoverings_Know-the-Difference_USArmy〉.
- 25.World Health Organization, Coronavirus disease (COVID-19) advice for the public 2022, 2021 [cited 2021 20th March]. 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public〉.
- 26.WHO. 2021 [cited 2021 14th March]. 〈https://www.who.int/docs/default-source/coronaviruse/who_risk-management_visiting-care-facility.pdf?sfvrsn=1dffe1e6_7〉.
- 27.who.int 2021 [cited 2021 20th March]. 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/coronavirus-disease-COVID-19-food-safety-for-consumers%20march%208th%20202〉.
- 28.Finegan O., Abboud D., Fonseca S., Malgrati I., Morcillo Mendez M.D., Burri J.M., Guyomarc’h P. International Committee of the Red Cross (ICRC): cemetery planning, preparation and management during COVID-19: a quick guide to proper documentation and disposition of the dead. Forensic Sci. Int. 2020;316 doi: 10.1016/j.forsciint.2020.110436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007;20(1):49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15(2):62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7) doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent M., Xu Y., Kong H. Helicase‐dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al Rwahnih M., Daubert S., Urbez-Torres J.R., Cordero F., Rowhani A. Deep sequencing evidence from single grapevine plants reveals a virome dominated by mycoviruses. Arch. Virol. 2011;156(3):397–403. doi: 10.1007/s00705-010-0869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richert-Pöggeler K.R., Franzke K., Hipp K., Kleespies R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019;9:3255. doi: 10.3389/fmicb.2018.03255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt N. Diagnostic Procedures for Viral and Rickettsial Infections. fourth ed. American Public Health Association, Inc.; New York, NY: 1969. Tissue culture technics for diagnostic virology; pp. 81–178. [Google Scholar]
- 36.Bisht R., Mandal A., Mitra A.K. Emerging Nanotechnologies for Diagnostics, Drug Delivery and Medical Devices. Elsevier; 2017. Micro-and nanotechnology-based implantable devices and bionics; pp. 249–290. [Google Scholar]
- 37.Wong H.Y.F., Lam H., Fong A.H., Leung S.T., Chin T.W., Lo C., Lui M.M., Lee J., Chiu K.W., Chung T.W., Lee E., Wan E., Hung I., Lam T., Kuo M.D., Ng M.Y. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. 2020;296(2):E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Committee, G.O.o.N.H., Office of state administration of traditional Chinese medicine. Notice on the issuance of a program for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (trial version 6) [text in Chinese], 2020.
- 40.Benzigar M.R., et al. Current methods for diagnosis of human coronaviruses: pros and cons. Anal. Bioanal. Chem. 2020:1–20. doi: 10.1007/s00216-020-03046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyrc K., Milewska A., Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J. Virol. Methods. 2011;175(1):133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WHO. 2021 [cited 2021 25th March]. 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/coronavirus-disease-COVID-19〉.
- 43.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186(4):1–10. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abedi M., Bathaie S., Mousavi M. Interaction between DNA and some salicylic acid derivatives and characterization of their DNA targets. Electroanalysis. 2013;25(11):2547–2556. [Google Scholar]
- 45.Arca-Lafuente S., Martínez-Román P., Mate-Cano I., Madrid R., Briz V. Nanotechnology: a reality for diagnosis of HCV infectious disease. J. Infect. 2020;80(1):8–15. doi: 10.1016/j.jinf.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Gupta R., Sagar P., Priyadarshi N., Kaul S., Sandhir R., Rishi V., Singhal N.K. Nanotechnology-based approaches for the detection of SARS-CoV-2. Front. Nanotechnol. 2020;2:6. [Google Scholar]
- 47.Zou X., Wu J., Gu J., Shen L., Mao L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wandtke T., Woźniak J., Kopiński P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7(2):751–780. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn D.-G., Jeon I.J., Kim J.D., Song M.S., Han S.R., Lee S.W., Jung H., Oh J.W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst. 2009;134(9):1896–1901. doi: 10.1039/b906788d. [DOI] [PubMed] [Google Scholar]
- 52.He Q., Du Q., Lau S., Manopo I., Lu L., Chan S.W., Fenner B.J., Kwang J. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods. 2005;127(1):46–53. doi: 10.1016/j.jviromet.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92(14):9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 54.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 55.Hendriksen R.S., Le Hello S., Bortolaia V., Pulsrikarn C., Nielsen E.M., Pornruangmong S., Chaichana P., Svendsen C.A., Weill F.X., Aarestrup F.M. Characterization of isolates of Salmonella enterica serovar Stanley, a serovar endemic to Asia and associated with travel. J. Clin. Microbiol. 2012;50(3):709–720. doi: 10.1128/JCM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foudeh A.M., Fatanat Didar T., Veres T., Tabrizian M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip. 2012;12(18):3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- 57.Laksanasopin T., Guo T.W., Nayak S., Sridhara A.A., Xie S., Olowookere O.O., Cadinu P., Meng F., Chee N.H., Kim J., Chin C.D., Munyazesa E., Mugwaneza P., Rai A.J., Mugisha V., Castro A.R., Steinmiller D., Linder V., Justman J.E., Nsanzimana S., Sia S.K. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015;7(273):273. doi: 10.1126/scitranslmed.aaa0056. p. 273re1-273re1. [DOI] [PubMed] [Google Scholar]
- 58.Bidram E., Esmaeili Y., Amini A., Sartorius R., Tay F.R., Shariati L., Makvandi P. Nanobased platforms for diagnosis and treatment of COVID-19: from benchtop to bedside. ACS Biomater. Sci. Eng. 2021;7:2150–2176. doi: 10.1021/acsbiomaterials.1c00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priyadarshini E., Pradhan N. Gold nanoparticles as efficient sensors in colorimetric detection of toxic metal ions: a review. Sens. Actuators B Chem. 2017;238:888–902. [Google Scholar]
- 60.Sun J., Lu Y., He L., Pang J., Yang F., Liu Y. Colorimetric sensor array based on gold nanoparticles: design principles and recent advances. TrAC Trends Anal. Chem. 2020;122 [Google Scholar]
- 61.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arora G., Joshi J., Mandal R.S., Shrivastava N., Virmani R., Sethi T. Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19. Pathogens. 2021;10(8):1048. doi: 10.3390/pathogens10081048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mei X., Lee H.C., Diao K.Y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rakita A., Nikolić N., Mildner M., Matiasek J., Elbe-Bürger A. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., Lang G., Li Y., Zhao H., Liu J., Xu K., Ruan L., Sheng J., Qiu Y., Wu W., Liang T., Li L. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito R., Iwano S., Naganawa S. A review on the use of artificial intelligence for medical imaging of the lungs of patients with coronavirus disease 2019. Diagn. Interv. Radiol. 2020;26(5):443–448. doi: 10.5152/dir.2019.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaid S., Kalantar R., Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int. Orthop. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gozes O., et al. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv Prepr. arXiv:2003. 05037. 2020 [Google Scholar]
- 71.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Metsky H.C., et al. CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. BioRxiv. 2020 [Google Scholar]
- 73.Malik Y.S., et al. How artificial intelligence may help the Covid‐19 pandemic: pitfalls and lessons for the future. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Asdaq S.M.B., Ikbal A.M.A., Sahu R.K., Bhattacharjee B., Paul T., Deka B., Fattepur S., Widyowati R., Vijaya J., Al mohaini M., Alsalman A.J., Imran M., Nagaraja S., Nair A.B., Attimarad M., Venugopala K.N. Nanotechnology integration for SARS-CoV-2 diagnosis and treatment: an approach to preventing pandemic. Nanomaterials. 2021;11(7):1841. doi: 10.3390/nano11071841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gadwal A., Roy D., Khokhar M., Modi A., Sharma P., Purohit P. CRISPR/Cas-new molecular scissors in diagnostics and therapeutics of COVID-19. Indian J. Clin. Biochem. 2021:1–9. doi: 10.1007/s12291-021-00977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rahimi H., Salehiabar M., Barsbay M., Ghaffarlou M., Kavetskyy T., Sharafi A., Davaran S., Chauhan S.C., Danafar H., Kaboli S., Nosrati H., Yallapu M.M., Conde J. CRISPR systems for COVID-19 diagnosis. ACS Sens. 2021;6(4):1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 78.Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang W., Yu L., Wen D., Wei D., Sun Y., Zhao H., Ye Y., Chen W., Zhu Y., Wang L., Wang L., Wu W., Zhao Q., Xu Y., Gu D., Nie G., Zhu D., Guo Z., Ma X., Niu L., Huang Y., Liu Y., Peng B., Zhang R., Zhang X., Li D., Liu Y., Yang G., Liu L., Zhou Y., Wang Y., Hou T., Gao Q., Li W., Chen S., Hu X., Han M., Zheng H., Weng J., Cai Z., Zhang X., Song F., Zhao G., Wang J. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lucia C., Federico P.-B., Alejandra G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. BioRxiv. 2020 [Google Scholar]
- 81.Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G., Yang Z., Huang X., Liu M. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020;65(17):1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(47):29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding X., Yin K., Li Z., Liu C. All-in-One dual CRISPR-cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. BioRxiv. 2020 doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Metsky H.C., Freije C.A., Kosoko-Thoroddsen T.S.F., Sabeti P.C., Myhrvold C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. BioRxiv. 2020 [Google Scholar]
- 87.Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., Pain A., Alofi F.S., Alsomali A., Hashem A.M., Khogeer A., Almontashiri N.A.M., Abedalthagafi M., Hassan N., Mahfouz M.M. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ooi K.H., et al. A CRISPR-based SARS-CoV-2 diagnostic assay that is robust against viral evolution and RNA editing. bioRxiv. 2020 [Google Scholar]
- 89.Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.W., Kim N.G., Yu X., Li J., Walker B.D., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. MedRxiv. 2020 [Google Scholar]
- 90.Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6(1):1–4. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16(8) doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang F., Abudayyeh O.O., Gootenberg J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. 2020;8 [Google Scholar]
- 93.Rauch J.N., Valois E., Solley S.C., Braig F., Lach R.S., Audouard M., Ponce-Rojas J.C., Costello M.S., Baxter N.J., Kosik K.S., Arias C., Acosta-Alvear D., Wilson M.Z. A scalable, easy-to-deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. J. Clin. Microbiol. 2021;59(4):e02402–e02420. doi: 10.1128/JCM.02402-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arizti-Sanz J., Freije C.A., Stanton A.C., Boehm C.K., Petros B.A., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.S.F., Kemball M.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Lemieux J.E., Sabeti P.C., Myhrvold C. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv. 2020 doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.K. Yoshimi et al., Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3, 2020.
- 96.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Azhar M., Phutela R., Ansari A.H., Sinha D., Sharma N., Kumar M., Aich M., Sharma S., Rauthan R., Singhal K., Lad H., Patra P.K., Makharia G., Chandak G.R., Chakraborty D., Maiti S. Rapid, field-deployable nucleobase detection and identification using FnCas9. BioRxiv. 2020 [Google Scholar]
- 98.Li S., Huang J., Ren L., Jiang W., Wang M., Zhuang L., Zheng Q., Yang R., Zeng Y., Luu L.D.W., Wang Y., Tai J. A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): application for the diagnosis of COVID-19. Talanta. 2021;233 doi: 10.1016/j.talanta.2021.122591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chakravarti R., Singh R., Ghosh A., Dey D., Sharma P., Velayutham R., Roy S., Ghosh D. A review on potential of natural products in the management of COVID-19. RSC Adv. 2021;11(27):16711–16735. doi: 10.1039/d1ra00644d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Buchanan W., Luke C.J., Ledgerwood J.E., Mascola J.R., Graham B.S., Beigel J.H., mRNA-Study G. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2021;384(1):80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Noor R. Developmental status of the potential vaccines for the mitigation of the COVID-19 pandemic and a focus on the effectiveness of the Pfizer-BioNTech and moderna mRNA vaccines. Curr. Clin. Microbiol. Rep. 2021:1–8. doi: 10.1007/s40588-021-00162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waheed S., Bayas A., Hindi F., Rizvi Z., Espinosa P.S. Neurological complications of COVID-19: Guillain-Barre syndrome following Pfizer COVID-19 vaccine. Cureus. 2021;13(2):13426. doi: 10.7759/cureus.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burki T.K. The Russian vaccine for COVID-19. Lancet Respir. Med. 2020;8(11):e85–e86. doi: 10.1016/S2213-2600(20)30402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oksuz E., et al. Cost-effectiveness analysis of remdesivir treatment in COVID-19 patients requiring low-flow oxygen therapy: payer perspective in turkey. Adv. Ther. 2021:1–14. doi: 10.1007/s12325-021-01874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.tracker, J.H.U.a.M.C.R.C.C.-c. 2020 [cited 2020 10th March]. 〈http://coronavirus.jhu.edu/map.html〉.
- 108.COVID-19. 2021 [cited 2021 3rd October]. 〈https://COVID-19.sciensano.be/sites/default/files/Covid19/COVID-19_InterimGuidelines_Treatment_ENG.pdf〉.
- 109.Soiza R.L., Scicluna C., Thomson E.C. Efficacy and safety of COVID-19 vaccines in older people. Age Ageing. 2021;50(2):279–283. doi: 10.1093/ageing/afaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Livingston E.H., Malani P.N., Creech C.B. The Johnson & Johnson vaccine for COVID-19. Jama. 2021;325(15):1575. doi: 10.1001/jama.2021.2927. p. 1575-1575. [DOI] [PubMed] [Google Scholar]
- 111.Baraniuk C. COVID-19: what do we know about Sputnik V and other Russian vaccines? bmj. 2021;372:743. doi: 10.1136/bmj.n743. [DOI] [PubMed] [Google Scholar]
- 112.Soleimani V., Delghandi P.S., Moallem S.A., Karimi G. Safety and toxicity of silymarin, the major constituent of milk thistle extract: an updated review. Phytother. Res. 2019;33(6):1627–1638. doi: 10.1002/ptr.6361. [DOI] [PubMed] [Google Scholar]
- 113.N. Mahmood, S. Nasir, K. Hefferon, Plant-Based Drugs and Vaccines for COVID-19. Vaccines, 2021; 9: 15. 2020, s Note: MDPI stays neu-tral with regard to jurisdictional clai-ms in …. [DOI] [PMC free article] [PubMed]
- 114.Mohammadinejad R., Shavandi A., Raie D.S., Sangeetha J., Soleimani M., Shokrian Hajibehzad S., Thangadurai D., Hospet R., Popoola J.O., Arzani A., Gómez-Lim M.A., Iravani S., Varma R.S. Plant molecular farming: production of metallic nanoparticles and therapeutic proteins using green factories. Green Chem. 2019;21(8):1845–1865. [Google Scholar]
- 115.Divya M., Vijayakumar S., Chen J., Vaseeharan B., Durán-Lara E.F. A review of South Indian medicinal plant has the ability to combat against deadly viruses along with COVID-19? Microb. Pathog. 2020;148 doi: 10.1016/j.micpath.2020.104277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Amparo T.R., Seibert J.B., Almeida T.C., Costa F., Silveira B.M., da Silva G.N., Dos Santos O., de Souza G. In silico approach of secondary metabolites from Brazilian herbal medicines to search for potential drugs against SARS‐CoV‐2. Phytother. Res. 2021;35:4297–4308. doi: 10.1002/ptr.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma L., Yao L. Antiviral effects of plant-derived essential oils and their components: an updated review. Molecules. 2020;25(11):2627. doi: 10.3390/molecules25112627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tohidi B., Rahimmalek M., Arzani A. Essential oil composition, total phenolic, flavonoid contents, and antioxidant activity of Thymus species collected from different regions of Iran. Food Chem. 2017;220:153–161. doi: 10.1016/j.foodchem.2016.09.203. [DOI] [PubMed] [Google Scholar]
- 119.Kim D., Min J., Jang M., Lee J., Shin Y., Park C., Song J., Kim H., Kim S., Jin Y.H., Kwon S. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules. 2019;9(11):696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu T., Liu X., Li W. Tetrandrine, a Chinese plant-derived alkaloid, is a potential candidate for cancer chemotherapy. Oncotarget. 2016;7(26):40800–40815. doi: 10.18632/oncotarget.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.P.K. Mukherjee, Antiviral evaluation of herbal drugs, in: Quality Control and Evaluation of Herbal Drugs, 2019, pp. 599–628.
- 122.Bailly C., Vergoten G. Glycyrrhizin: an alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol. Ther. 2020;214 doi: 10.1016/j.pharmthera.2020.107618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van de Sand L., Bormann M., Alt M., Schipper L., Heilingloh C.S., Steinmann E., Todt D., Dittmer U., Elsner C., Witzke O., Krawczyk A. Glycyrrhizin effectively inhibits SARS-CoV-2 replication by inhibiting the viral main protease. Viruses. 2021;13(4):609. doi: 10.3390/v13040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen L., Hu C., Hood M., Zhang X., Zhang L., Kan J., Du J. A novel combination of vitamin C, curcumin and glycyrrhizic acid potentially regulates immune and inflammatory response associated with coronavirus infections: a perspective from system biology analysis. Nutrients. 2020;12(4):1193. doi: 10.3390/nu12041193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gupta H., Gupta M., Bhargava S. Potential use of turmeric in COVID‐19. Clin. Exp. Dermatol. 2020;45:902–903. doi: 10.1111/ced.14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao J. Investigating mechanism of Qing-Fei-Pai-Du-Tang for treatment of COVID-19 by network pharmacology. Chin. Tradit. Herb. Drugs. 2020:829–835. [Google Scholar]
- 127.Li Q., Bai C., Yang R., Xing W., Pang X., Wu S., Liu S., Chen J., Liu T., Gu X. Deciphering the pharmacological mechanisms of Ma Xing Shi Gan Decoction against COVID-19 through integrating Network pharmacology and experimental exploration. Front. Pharmacol. 2020;11:1761. doi: 10.3389/fphar.2020.581691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., Chen J., Zhang L., Lv L., Zhang G., Yuan Y., Chai Y., Zhu M., Wu C. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm. Sin. B. 2021;11(1):222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Borquaye L.S., Gasu E.N., Ampomah G.B., Kyei L.K., Amarh M.A., Mensah C.N., Nartey D., Commodore M., Adomako A.K., Acheampong P., Mensah J.O., Mormor D.B., Aboagye C.I. Alkaloids from Cryptolepis sanguinolenta as potential inhibitors of SARS-CoV-2 viral proteins: an in silico study. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/5324560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mehta P.P., Dhapte-Pawar V.S. Novel and evolving therapies for COVID-19 related pulmonary complications. Am. J. Med. Sci. 2021;361:557–566. doi: 10.1016/j.amjms.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thuy B.T.P., My T., Hai N., Hieu L.T., Hoa T.T., Thi Phuong Loan H., Triet N.T., Anh T., Quy P.T., Tat P.V., Hue N.V., Quang D.T., Trung N.T., Tung V.T., Huynh L.K., Nhung N. Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega. 2020;5(14):8312–8320. doi: 10.1021/acsomega.0c00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu X., Cheng Z., Fan H., Ai S., Han R. Electrochemical detection of avian influenza virus H5N1 gene sequence using a DNA aptamer immobilized onto a hybrid nanomaterial-modified electrode. Electrochim. Acta. 2011;56(18):6266–6270. [Google Scholar]
- 133.Ansari S., Islam F., Sameem M. Influence of nanotechnology on herbal drugs: a review. J. Adv. Pharm. Technol. Res. 2012;3(3):142–146. doi: 10.4103/2231-4040.101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karuppath S., Pillai P., Nair S.V., Lakshmanan V.K. Comparison and existence of nanotechnology in traditional alternative medicine: an onset to future medicine. Nanosci. Nanotechnol. 2018;8(1):13–25. [Google Scholar]
- 135.Carvalho A.P.A., Conte‐Junior C.A. Recent advances on nanomaterials to COVID‐19 management: a systematic review on antiviral/virucidal agents and mechanisms of SARS‐CoV‐2 inhibition/inactivation. Glob. Chall. 2021;5(5) doi: 10.1002/gch2.202000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao Z., Xiao Y., Xu L., Liu Y., Jiang G., Wang W., Li B., Zhu T., Tan Q., Tang L., Zhou H., Huang X., Shan H. Glycyrrhizic acid nanoparticles as antiviral and anti-inflammatory agents for COVID-19 treatment. ACS Appl. Mater. Interfaces. 2021;13(18):20995–21006. doi: 10.1021/acsami.1c02755. [DOI] [PubMed] [Google Scholar]
- 137.Kurniawan D.W., Ikhsanudin A. Potential of Jamu in nanotechnology perspective as an alternative treatment for COVID-19. Pharm. Sci. Res. 2020;7(3):1. [Google Scholar]
- 138.Yaqinuddin A., Shafqat A., Kashir J., Alkattan K. Effect of SARS-CoV-2 mutations on the efficacy of antibody therapy and response to vaccines. Vaccines. 2021;9(8):914. doi: 10.3390/vaccines9080914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.D.-G. Ahn et al., Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19), 2020. [DOI] [PMC free article] [PubMed]
- 140.Public Health Emergency, Pause in the distribution of bamlanivimab, 2021. 〈https://www.phe.gov/emergency/events/COVID19/investigation-MCM/Bamlanivimab-etesevimab/Pages/bamlanivimab-etesevimab-distribution-pause.aspx〉.
- 141.FDA. 2021. 〈https://www.fda.gov/news-events/press-announcements/coronavirus-COVID-19-update-fda-authorizes-monoclonal-antibodies-treatment-COVID-19〉.
- 142.Food and Drug Administration, Fact sheet for healthcare providers: emergency use authorization (EUA) of sotrovimab, 2021. Available from: 〈https://www.fda.gov/media/149534/download〉.
- 143.Sivaraman K., et al. Can povidone Iodine gargle/mouthrinse inactivate SARS-CoV-2 and decrease the risk of nosocomial and community transmission during the COVID-19 pandemic? An evidence-based update. Jpn. Dent. Sci. Rev. 2021 doi: 10.1016/j.jdsr.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Morokutti-Kurz M., Unger-Manhart N., Graf P., Rauch P., Kodnar J., Große M., Setz C., Savli M., Ehrenreich F., Grassauer A., Prieschl-Grassauer E., Schubert U. The saliva of probands sucking an Iota-Carrageenan containing Lozenge inhibits viral binding and replication of the most predominant common cold viruses and SARS-CoV-2. Int. J. Gen. Med. 2021;14:5241–5249. doi: 10.2147/IJGM.S325861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Figueroa J.M., et al. Efficacy of a nasal spray containing Iota-Carrageenan in the prophylaxis of COVID-19 in hospital personnel dedicated to patients care with COVID-19 disease. A pragmatic multicenter, randomized, double-blind, placebo-controlled trial (CARR-COV-02) medRxiv. 2021 doi: 10.2147/IJGM.S328486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moncada S., Higgs E. Endogenous nitric oxide: physiology, pathology and clinical relevance. Eur. J. Clin. Investig. 1991;21(4):361–374. doi: 10.1111/j.1365-2362.1991.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 147.Winchester S., John S., Jabbar K., John I. Clinical efficacy of nitric oxide nasal spray (NONS) for the treatment of mild COVID-19 infection. J. Infect. 2021;83:237–279. doi: 10.1016/j.jinf.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ku Z., Xie X., Hinton P.R., Liu X., Ye X., Muruato A.E., Ng D.C., Biswas S., Zou J., Liu Y., Pandya D., Menachery V.D., Rahman S., Cao Y.A., Deng H., Xiong W., Carlin K.B., Liu J., Su H., Haanes E.J., Keyt B.A., Zhang N., Carroll S.F., Shi P.Y., An Z. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 2021;595(7869):718–723. doi: 10.1038/s41586-021-03673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saleh M., Vaezi A.A., Aliannejad R., Sohrabpour A.A., Kiaei S., Shadnoush M., Siavashi V., Aghaghazvini L., Khoundabi B., Abdoli S., Chahardouli B., Seyhoun I., Alijani N., Verdi J. Cell therapy in patients with COVID-19 using Wharton’s jelly mesenchymal stem cells: a phase 1 clinical trial. Stem Cell Res. Ther. 2021;12(1):1–13. doi: 10.1186/s13287-021-02483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., Shan G., Meng F., Du D., Wang S., Fan J., Wang W., Deng L., Shi H., Li H., Hu Z., Zhang F., Gao J., Liu H., Li X., Zhao Y., Yin K., He X., Gao Z., Wang Y., Yang B., Jin R., Stambler I., Lim L.W., Su H., Moskalev A., Cano A., Chakrabarti S., Min K.J., Ellison-Hughes G., Caruso C., Jin K., Zhao R.C. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11(2):216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., Hu X., Chen J., Liu B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Golchin A., Seyedjafari E., Ardeshirylajimi A. Mesenchymal stem cell therapy for COVID-19: present or future. Stem Cell Rev. Rep. 2020;16(3):427–433. doi: 10.1007/s12015-020-09973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Golchin A., et al. Cell Biology and Translational Medicine, Volume 4. Springer; 2018. Promotion of cell-based therapy: special focus on the cooperation of mesenchymal stem cell therapy and gene therapy for clinical trial studies; pp. 103–118. [DOI] [PubMed] [Google Scholar]
- 154.Basiri A., et al. Stem cell therapy potency in personalizing severe COVID-19 treatment. Stem Cell Rev. Rep. 2021:1–21. doi: 10.1007/s12015-020-10110-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choi Y.H., Han H.-K. Nanomedicines: current status and future perspectives in aspect of drug delivery and pharmacokinetics. J. Pharm. Investig. 2018;48(1):43–60. doi: 10.1007/s40005-017-0370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tavakol S., Zahmatkeshan M., Mohammadinejad R., Mehrzadi S., Joghataei M.T., Alavijeh M.S., Seifalian A. The role of Nanotechnology in current COVID-19 outbreak. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh P., Singh D., Sa P., Mohapatra P., Khuntia A., K Sahoo S. Insights from nanotechnology in COVID-19: prevention, detection, therapy and immunomodulation. Nanomedicine. 2021;16(14):1219–1235. doi: 10.2217/nnm-2021-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Abo-Zeid Y., Urbanowicz R.A., Thomson B.J., Irving W.L., Tarr A.W., Garnett M.C. Enhanced nanoparticle uptake into virus infected cells: could nanoparticles be useful in antiviral therapy? Int. J. Pharm. 2018;547(1–2):572–581. doi: 10.1016/j.ijpharm.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 159.Hu C.J., Chen Y.T., Fang Z.S., Chang W.S., Chen H.W. Antiviral efficacy of nanoparticulate vacuolar ATPase inhibitors against influenza virus infection. Int. J. Nanomed. 2018;13:8579–8593. doi: 10.2147/IJN.S185806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Łoczechin A., Séron K., Barras A., Giovanelli E., Belouzard S., Chen Y.T., Metzler-Nolte N., Boukherroub R., Dubuisson J., Szunerits S. Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl. Mater. Interfaces. 2019;11(46):42964–42974. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., Endy D., La Russa M.F., Lewis D.B., Qi L.S. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876. e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jindal S., Gopinath P. Nanotechnology based approaches for combatting COVID-19 viral infection. Nano Express. 2020;1 [Google Scholar]
- 163.Tang Z., Zhang X., Shu Y., Guo M., Zhang H., Tao W. Insights from nanotechnology in COVID-19 treatment. Nano Today. 2021;36 doi: 10.1016/j.nantod.2020.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 165.WHO. 2021. 〈www.who.int/publications/m/item/draft-landscape-of-COVID-19〉.
- 166.Chung Y.H., Beiss V., Fiering S.N., Steinmetz N.F. COVID-19 vaccine frontrunners and their nanotechnology design. ACS nano. 2020;14(10):12522–12537. doi: 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- 167.Delrue I., Verzele D., Madder A., Nauwynck H.J. Inactivated virus vaccines from chemistry to prophylaxis: merits, risks and challenges. Expert Rev. Vaccines. 2012;11(6):695–719. doi: 10.1586/erv.12.38. [DOI] [PubMed] [Google Scholar]
- 168.Pulendran B., Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12(6):509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vartak A., Sucheck S.J. Recent advances in subunit vaccine carriers. Vaccines. 2016;4(2):12. doi: 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11(6):885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 171.Schleef M., Blaesen M., Schmeer M., Baier R., Marie C., Dickson G., Scherman D. Production of non viral DNA vectors. Curr. Gene Ther. 2010;10(6):487–507. doi: 10.2174/156652310793797711. [DOI] [PubMed] [Google Scholar]
- 172.Livingston E.H. Necessity of 2 doses of the Pfizer and Moderna COVID-19 vaccines. JAMA. 2021;325(9):898. doi: 10.1001/jama.2021.1375. p. 898-898. [DOI] [PubMed] [Google Scholar]
- 173.Vogel A.B., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021:1–7. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 174.2021. 〈https://www.cdc.gov/vaccines/COVID-19/info-by-product/moderna/index.html〉.
- 175.2021 [cited 2021 3th April]. 〈https://www.cdc.gov/vaccines/COVID-19/info-by-product/pfizer/downloads/storage-summary.pdf〉.
- 176.Doroftei B., Ciobica A., Ilie O.D., Maftei R., Ilea C. Mini-Review Discussing The Reliability And Efficiency of COVID-19 vaccines. Diagnostics. 2021;11(4):579. doi: 10.3390/diagnostics11040579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Calendar, L.C., Communicating About COVID-19 Vaccination.
- 178.Meo S.A., Bukhari I.A., Akram J., Meo A.S., Klonoff D.C. COVID-19 vaccines: comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna Vaccines. Eur. Rev. Med. Pharmacol. Sci. 2021;25(3):1663–1669. doi: 10.26355/eurrev_202102_24877. [DOI] [PubMed] [Google Scholar]
- 179.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck RW Jr, Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C., Clinical Trial G. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.van Balveren-Slingerland L., Rümke H.C., Kant A.C. Reported adverse events following influenza vaccination. Ned. Tijdschr. Geneeskd. 2014;158:6841. p. A6841-A6841. [PubMed] [Google Scholar]
- 181.W. Zhou, S.S. Ellenberg, Surveillance for Safety After Immunization: Vaccine Adverse Event Reporting System (VAERS)—. Morbidity and Mortality Weekly Report: MMWR. Surveillance Summaries. Surveillance summaries, 2003. [PubMed]
- 182.Pfizer COVID-19 vaccine EUA fact sheet for healthcare providers administering vaccine 2021 [cited 2021 31th March]. 〈https://www.fda.gov/media/144413〉.
- 183.El-Shitany N.A., Harakeh S., Badr-Eldin S.M., Bagher A.M., Eid B., Almukadi H., Alghamdi B.S., Alahmadi A.A., Hassan N.A., Sindi N., Alghamdi S.A., Almohaimeed H.M., Mohammedsaleh Z.M., Al-Shaikh T.M., Almuhayawi M.S., Ali S.S., El-Hamamsy M. Minor to moderate side effects of Pfizer-BioNTech COVID-19 vaccine among Saudi residents: a retrospective cross-sectional study. Int. J. Gen. Med. 2021;14:1389–1401. doi: 10.2147/IJGM.S310497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cohen J. Vaccine designers take first shots at COVID-19. Am. Assoc. Adv. Sci. 2020 doi: 10.1126/science.368.6486.14. [DOI] [PubMed] [Google Scholar]
- 185.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study G. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Malayala S.V., Mohan G., Vasireddy D., Atluri P. Purpuric rash and Thrombocytopenia after the mRNA-1273 (Moderna) COVID-19 vaccine. Cureus. 2021;13:3. doi: 10.7759/cureus.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Moderna COVID-19 Vaccine. Facts Sheets. 2021 [cited 2021 30th April]. 〈https://www.idsociety.org/〉 COVID-19-real-time-learning-network/vaccines/moderna-COVID-19-vaccine/.
- 188.Mahase E. COVID-19: Russia approves vaccine without large scale testing or published results. BMJ Br. Med. J. 2020;370:m3205. doi: 10.1136/bmj.m3205. [DOI] [PubMed] [Google Scholar]
- 189.Mahase E. COVID-19: Russian vaccine efficacy is 91.6%, show phase III trial results. Br. Med. J. Publ. Group. 2021 doi: 10.1136/bmj.n309. [DOI] [PubMed] [Google Scholar]
- 190.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Egorova D.A., Shmarov M.M., Nikitenko N.A., Gushchin V.A., Smolyarchuk E.A., Zyryanov S.K., Borisevich S.V., Naroditsky B.S., Gintsburg A.L., Gam-COVID-Vac Vaccine Trial G. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Uddin K.N. Corona vaccine. BIRDEM Med. J. 2021;11(1):1–5. [Google Scholar]
- 192.2021 [cited 2021 29th April]. 〈https://www.news18.com/news/india/russias-sputnik-v-vaccine-approved-for-emergency-use-in-india-all-you-need-to-know-3632546.html〉.
- 193.2021 [cited 2021 21th April]. 〈https://tass.com/society/1274565〉.
- 194.Sharma O., Sultan A.A., Ding H., Triggle C.R. A review of the progress and challenges of developing a vaccine for COVID-19. Front. Immunol. 2020;11:2413. doi: 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.2021 [cited 2021 30th April]. 〈https://www.globaltimes.cn/page/202102/1215499.shtml〉.
- 196.Soiza R.L., Scicluna C., Thomson E.C. Efficacy and safety of COVID-19 vaccines in older people. Age Ageing. 2020;49:934–935. doi: 10.1093/ageing/afaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.2021 [cited 2021 22th April]. 〈https://www.medicalnewstoday.com/articles/johnson-johnson-COVID-19-vaccine-what-are-the-side-effects#How-does-the-vaccine-work〉.
- 198.2021 [cited 2021 20th April]. 〈https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-COVID-19/janssen-COVID-19-vaccine〉.
- 199.[cited 2021 21th April]. 〈https://www.precisionvaccinations.com/vaccines/epivaccorona-vaccine〉.
- 200.Ryzhikov A., et al. A single blind, placebo-controlled randomized study of the safety, reactogenicity and immunogenicity of the “EpiVacCorona” Vaccine for the prevention of COVID-19, in volunteers aged 18–60 years (phase I–II) Russ. J. Infect. Immun. 2021;11(2):283–296. [Google Scholar]
- 201.A. Fernandes et al., COVID-19 vaccine, Endocr. Pract. (2021). [DOI] [PMC free article] [PubMed]
- 202.Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C., Smith G.E., Frieman M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32(26):3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Park J.W., Lagniton P., Liu Y., Xu R.H. mRNA vaccines for COVID-19: what, why and how. Int. J. Biol. Sci. 2021;17(6):1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M., Dold C., Provine N.M., Aboagye J., Fowler J., Silk S.E., Alderson J., Aley P.K., Angus B., Berrie E., Bibi S., Cicconi P., Clutterbuck E.A., Chelysheva I., Folegatti P.M., Fuskova M., Green C.M., Jenkin D., Kerridge S., Lawrie A., Minassian A.M., Moore M., Mujadidi Y., Plested E., Poulton I., Ramasamy M.N., Robinson H., Song R., Snape M.D., Tarrant R., Voysey M., Watson M., Douglas A.D., Hill A., Gilbert S.C., Pollard A.J., Lambe T., Oxford COVID Vaccine Trial G. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27(2):270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 205.Vaccines and Related Biological Products Advisory Committee Meeting. [cited 2021, 30th April]. 〈https://www.fda.gov/media/〉.
- 206.2021 [cited 2021 26th April]. 〈http://www.nytimes.com/interactive/2020/health/moderna-COVID-19-vaccine.html〉.
- 207.Jonathan Corum, C.Z. 2021 [cited 2021 26th April]. 〈https://www.nytimes.com/interactive/2020/health/novavax-COVID-19-vaccine.html〉.
- 208.Speiser D.E., Bachmann M.F. COVID-19: mechanisms of vaccination and immunity. Vaccines. 2020;8(3):404. doi: 10.3390/vaccines8030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dhama K., Natesan S., Iqbal Yatoo M., Patel S.K., Tiwari R., Saxena S.K., Harapan H. Plant-based vaccines and antibodies to combat COVID-19: current status and prospects. Hum. Vaccines Immunother. 2020;16(12):2913–2920. doi: 10.1080/21645515.2020.1842034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shanmugaraj B., Siriwattananon K., Malla A., Phoolcharoen W. Potential for developing plant-derived candidate vaccines and biologics against emerging coronavirus infections. Pathogens. 2021;10(8):1051. doi: 10.3390/pathogens10081051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Annas S., Zamri-Saad M. Intranasal vaccination strategy to control the COVID-19 pandemic from a veterinary medicine perspective. Animals. 2021;11(7):1876. doi: 10.3390/ani11071876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rice A., Verma M., Shin A., Zakin L., Sieling P., Tanaka S., Balint J., Dinkins K., Adisetiyo H., Morimoto B., Higashide W., Anders Olson C., Mody S., Spilman P., Gabitzsch E., Safrit J.T., Rabizadeh S., Niazi K., Soon-Shiong P. Intranasal plus subcutaneous prime vaccination with a dual antigen COVID-19 vaccine elicits T-cell and antibody responses in mice. Sci. Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-94364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nel A.E., Miller J.F. Nano-enabled COVID-19 vaccines: meeting the challenges of durable antibody plus cellular immunity and immune escape. ACS nano. 2021;15(4):5793–5818. doi: 10.1021/acsnano.1c01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS nano. 2020;14(7):7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- 215.Ballester M., Nembrini C., Dhar N., de Titta A., de Piano C., Pasquier M., Simeoni E., van der Vlies A.J., McKinney J.D., Hubbell J.A., Swartz M.A. Nanoparticle conjugation and pulmonary delivery enhance the protective efficacy of Ag85B and CpG against tuberculosis. Vaccine. 2011;29(40):6959–6966. doi: 10.1016/j.vaccine.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 216.Slütter B., Bal S., Keijzer C., Mallants R., Hagenaars N., Que I., Kaijzel E., van Eden W., Augustijns P., Löwik C., Bouwstra J., Broere F., Jiskoot W. Nasal vaccination with N-trimethyl chitosan and PLGA based nanoparticles: nanoparticle characteristics determine quality and strength of the antibody response in mice against the encapsulated antigen. Vaccine. 2010;28(38):6282–6291. doi: 10.1016/j.vaccine.2010.06.121. [DOI] [PubMed] [Google Scholar]
- 217.Dhama K., et al. COVID-19 vaccine hesitancy–reasons and solutions to achieve a successful global vaccination campaign to tackle the ongoing pandemic. Hum. Vaccines Immunother. 2021:1–5. doi: 10.1080/21645515.2021.1926183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Griffith J., Marani H., Monkman H. COVID-19 vaccine hesitancy in Canada: content analysis of tweets using the theoretical domains framework. J. Med. Internet Res. 2021;23(4) doi: 10.2196/26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.García-Montero C., Fraile-Martínez O., Bravo C., Torres-Carranza D., Sanchez-Trujillo L., Gómez-Lahoz A.M., Guijarro L.G., García-Honduvilla N., Asúnsolo A., Bujan J., Monserrat J., Serrano E., Álvarez-Mon M., De León-Luis J.A., Álvarez-Mon M.A., Ortega M.A. An updated review of SARS-CoV-2 vaccines and the importance of effective vaccination programs in pandemic times. Vaccines. 2021;9(5):433. doi: 10.3390/vaccines9050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mahase E. COVID-19: vaccine roll out could take a year and will require difficult prioritisation decisions. Br. Med. J. Publ. Group. 2020:m3846. doi: 10.1136/bmj.m3846. [DOI] [PubMed] [Google Scholar]
- 221.K. Sharun, K. Dhama, COVID-19 vaccine diplomacy and equitable access to vaccines amid ongoing pandemic, Arch. Med. Res. (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable for this study.