Abstract
Most genome-wide association studies for obesity and body mass index (BMI) have so far assumed an additive mode of inheritance in their analysis, although association testing supports a recessive effect for some of the established loci, for example, rs1421085 in FTO. In two whole-genome sequencing (WGS) studies of asthmatic children and their parents (892 Costa-Rican trios and 286 North American trios), we discovered an association between a locus (rs9292139) in LOC102724122 and BMI that reaches genome-wide significance under a recessive model in the combined analysis. As the association does not achieve significance under an additive model, our finding illustrates the benefits of the recessive model in WGS analyses.
BACKGROUND
Obesity and asthma are two of the most important pediatric health problems and epidemiological studies suggest that both phenotypes are linked.1 Obese children with asthma are more likely to have a decreased response to inhaled steroids and therefore lower quality of life.1 Obesity has been hypothesized as a contributing factor for asthma risk, but the “obesity-asthma” link could also be attributable to non-genetic factors such as lifestyle and diet, or explained by reverse causality.
Although the first genome-wide association studies (GWASs) for body mass index (BMI) was conducted under a recessive model,2 most GWASs of obesity and asthma have only considered an additive genetic model in their analysis. However, this assumption can reduce the statistical power, as an additive association test performs poorly if the recessive mode of inheritance is indeed true. For example, the well-known loci for BMI such as FTO and INSIG2 were reported to have a recessive effect, but have less significant association test results under an additive model.2,3
Motivated by these observations and based on two family-based whole-genome sequencing (WGS) studies for childhood asthma, we tested for association between genetic loci and BMI under the recessive mode of inheritance, and for comparisons, under the additive mode.
METHODS
For the first part of the analysis, we utilized asthmatic trios from the Genetic Epidemiology of Asthma in Costa Rica Study (GACRS).4 To replicate findings from this analysis, we utilized asthmatic trios from the Childhood Asthma Management Program (CAMP).5 Both studies, GACRS and CAMP, used similar protocols for collection of phenotypic data, but GACRS is a cross-sectional and CAMP is a longitudinal study. Subject recruitment and study procedures for GACRS and CAMP have been previously described.4,5 WGS data for GACRS and CAMP were generated as part of the National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) WGS Program.6 Details on DNA sample handling, quality control, library construction, clustering and sequencing, read processing, and sequence data quality control are described on the TOPMed website (https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-methods-freeze-8). Variant calls were obtained from TOPMed data freeze 8 variant call format files aligned to the GRCh38 genome reference. In our analyses, we included only bi-allelic SNPs with a minimal depth of coverage of 10 reads that were marked as PASS in the VCF FILTER column. After sample quality control including removal of duplicates and pedigree mismatches, WGS and BMI data were available for 892 asthmatic GACRS trios and 286 asthmatic CAMP trios (European American: African American: Latinx: others = 216: 28: 19: 23). For additional variant quality control (QC), SNPs with > 3 Mendelian errors, ≥ 2% genotyping missing rate, minor allele frequency < 1%, or deviations from Hardy-Weinberg proportions (p < 10−8) were removed. After QC, 8,689,950 variants remained in the analysis. BMI adjusted for age, sex, and usage on both inhaled or systemic corticosteroids was analyzed under (1) a recessive and (2) an additive genetic model. Association tests were evaluated using the family-based association test (FBAT) software (version 2.04). We explored the cis-expression quantitative trait loci (eQTL) for the top variant in the GTEx RNA-seq data V8 (https://www.gtexportal.org/home) and the expression of potential gene based on the “HPA RNA-seq normal tissues” project, i.e., RNA-seq performed on 27 different tissue samples from 95 human individuals in NCBI data (https://www.ncbi.nlm.nih.gov/gene). A comparison of the main characteristics in children who were and were not overweight or obese was conducted using t-tests or Chi-square tests, as appropriate. Overweight or obesity were defined as an age- and sex-adjusted BMI Z score between the 85th and 95th percentiles or greater than the 95th percentile, respectively.
RESULTS
The main characteristics of the asthmatic children in our GACRS and CAMP analysis are shown in Table 1. Participants in GACRS and CAMP differed with respect to ethnicity but were otherwise similar regarding age, age of asthma onset, and BMI. In GACRS, overweight or obese subjects had higher age and height than non-overweight subjects. Previous usage on inhaled corticosteroids was suggestively associated with BMI (p = 0.098).
Table 1.
Comparisons of demographic characteristics and clinical features between analysis groups (mean ± SD or N (%))
| GACRS subjects |
Is overweight/obese*? | p value† | CAMP subjects |
||
|---|---|---|---|---|---|
| No | Yes | ||||
| N | 892 | 758 (84.98) | 134 (15.02) | 286 | |
| Age | 9.28 ± 1.86 | 9.19 ± 1.85 | 9.78 ± 1.84 | p<0.001 | 8.90 ± 2.16 |
| Onset age | 2.25 ± 2.20 | 2.27 ± 2.21 | 2.12 ± 2.16 | 0.4739 | 2.92 ± 2.38 |
| Male sex | 526 (58.97) | 448 (59.10) | 78 (58.21) | 0.9214 | 193 (67.48) |
| Height, m | 1.33 ± 0.12 | 1.32 ± 0.12 | 1.40 ± 0.11 | p<0.001 | 1.34 ± 0.14 |
| Body mass index, kg/m2 | 18.45 ± 3.92 | 17.23 ± 2.46 | 25.39 ± 3.36 | p<0.001 | 18.24 ± 3.28 |
| Spirometric measure | |||||
| FEV1 (% of predicted) | 98.87 ± 16.84 | 98.84 ± 16.76 | 99.00 ± 17.36 | 0.9224 | 92.82 ± 13.40 |
| FEV1/FVC, % | 84.36 ± 7.79 | 84.53 ± 7.73 | 83.37 ± 8.11 | 0.1257 | 79.69 ± 8.07 |
| Bronchodilator Response as % of baseline FEV1 | 5.58 ± 10.20 | 5.52 ± 10.10 | 5.97 ± 10.75 | 0.6564 | 10.86 ± 9.49 |
| Allergy | |||||
| Doctor-diagnosed eczema | 37 (4.15) | 34 (4.49) | 3 (2.24) | 0.3271 | 69 (24.13) |
| Doctor-diagnosed hay fever | 286 (32.06) | 243 (32.06) | 43 (32.09) | 1 | 109 (38.11) |
| Total serum IgE, IU/mL | 726.47 ± 899.57 | 738.79 ± 930.67 | 657.05 ± 697.76 | 0.2384 | 444.56 ± 797.47 |
| Eosinophil, count/mm2 | 554.56 ± 404.05 | 559.93 ± 411.04 | 523.72 ± 361.20 | 0.3050 | 511.96 ± 425.43 |
| Number of positive skin tests to allergens | 3.09 ± 1.82 | 3.09 ± 1.80 | 3.04 ± 1.92 | 0.7574 | 5.56 ± 4.20 |
| Severity | |||||
| Inhaled corticosteroid | 467 (52.35) | 393 (51.85) | 74 (55.22) | 0.5302 | 111 (38.81) |
| Oral corticosteroid | 700 (78.48) | 590 (77.84) | 110 (82.09) | 0.3220 | 102 (35.66)‡ |
| Shortness of breath from wheezing | 784 (87.89) | 664 (87.60) | 120 (89.55) | 0.6462 | 251 (87.76) |
| Chest tightness | 647 (72.53) | 543 (71.64) | 104 (77.61) | 0.2161 | NA |
| ER visit for asthma in last year | 742 (83.18) | 631 (83.25) | 111 (82.84) | 1 | 78 (27.27) |
Overweight was defined as an age- and sex-adjusted BMI between the 85th and 95th percentiles and obesity as an age- and sex-adjusted BMI equal to or above the 95th percentile.
The p value is for the comparison of overweight/obese versus non-overweight subjects in the GACRS.
Injected corticosteroid was also included.
BMI, body mass index; CAMP, Childhood Asthma Management Program; ER, emergency room; FEV1, forced expiratory volume in one second; FVC, forced vital capacity; GACRS, Genetic Epidemiology of Asthma in Costa Rica Study; IgE, Immunoglobulin E; NA, not available.
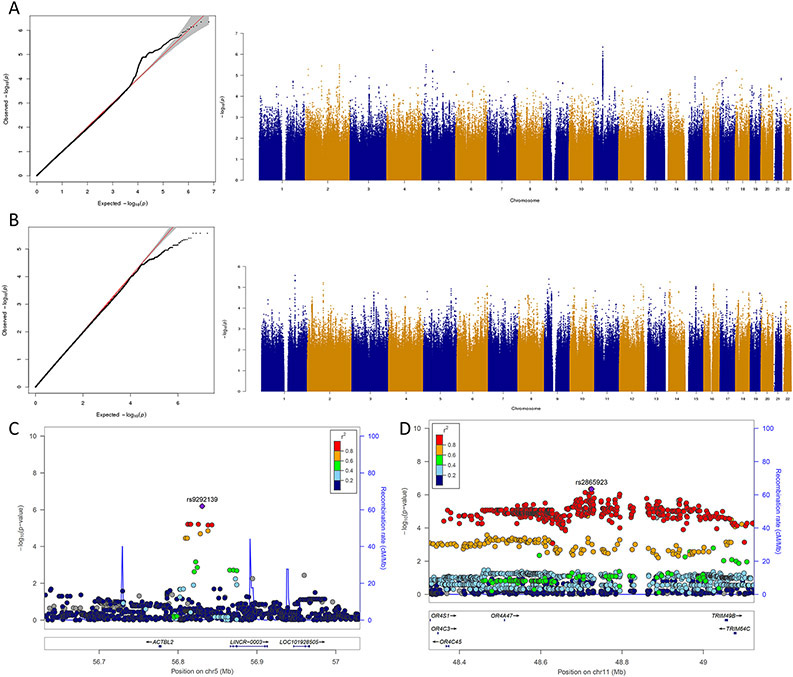
In our discovery GACRS analysis, SNP rs9292139 (located on LOC102724122 on chromosome 5q11.2 with a minor allele (G) frequency of 25.0%) and rs2865923 (located on chromosome 11p11.2 with a minor allele (G) frequency of 36.5%) were the most significant SNPs under a recessive model (p = 6.40 × 10−7 and 4.52 × 10−7, respectively, Figure 1A). Under an additive model, both SNPs were less significant (p = 1.29 × 10−3 and 0.209, respectively), and no other SNPs reached a p value below 10−6 (Figure 1B). In the ethnically diverse CAMP trios, SNP rs2865923 was not replicated (p = 0.381), but rs9292139 was associated with BMI at the baseline visit, assuming a recessive genetic model and adjusting for the same covariates as in GACRS analysis (p= 0.018). A combined FBAT analysis (GACRS and CAMP) between SNP rs9292139 and adjusted BMI reached genome-wide significance (p = 5.01 × 10−8). Moreover, a 4-year follow-up analysis of BMI in CAMP also demonstrated a consistent recessive effect of rs9292139 (Figure 2A). On the other hand, SNP rs9292139 was not significantly associated with asthma affection status, total IgE, and eosinophil count (padditive model = 0.723, 0.946, and 0.226, respectively; precessive model = 0.411, 0.170, and 0.854, respectively). Also, there was no significant interaction between rs9292139 and corticosteroid usage (inhaled or systemic) for adjusted BMI (p = 0.137). No cis-eQTL association for SNP rs9292139 was reported in the GTEx data, but a quantitative RNA expression analysis based on deep sequencing of LOC102724122 showed that this gene was highly expressed in adipose tissue (Figure 2B).7
Figure 1.
Quantile-quantile and Manhattan plots for the GWAS of adjusted BMI based on 892 asthmatic GACRS trios under a recessive (A) or an additive (B) genetic model. For the recessive model, p values for the minor allele are displayed. Regional plots for adjusted BMI-associated loci where purple diamond represents the top-ranked SNPs, rs9292139 (C) or rs2865923 (D), and other SNPs are colored according to their r2 value in relation to each SNP. Linkage disequilibrium information based on the 1000 genomes project EUR dataset.10 BMI, body mass index; GACRS, Genetic Epidemiology of Asthma in Costa Rica Study; GWAS, genome-wide association study; SNPs, single nucleotide polymorphisms.
Figure 2.
Longitudinal change of BMI from screening and randomisation to visit after 48 months in CAMP cohort, demonstrating the constant recessive effect of SNP rs9292139 (A). Quantitative transcriptomics analysis (RNA-seq) of LOC102724122 in tissue samples from 95 human individuals representing 27 different tissues to show fat tissue-specificity of LOC102724122 (B).7 BMI, body mass index; CAMP, Childhood Asthma Management Program; F/U, follow-up; SNP, single nucleotide polymorphism.
DISCUSSION & CONCLUSION
Our recessive family-based association analysis of two WGS studies for childhood asthma identified rs9292139 as a new genetic risk locus for BMI in asthmatic children. As the association between rs9292139 and BMI did not reach genome-wide significance under an additive model, our finding illustrates the benefit of including a secondary recessive analysis into the standard analysis protocols of WGS studies. SNP rs9292139 is located on LOC102724122, an uncharacterized gene encoding a long non-coding RNA gene (lncRNA, a class of non-protein coding RNAs of >200 nucleotides in length). Although little is known about lncRNAs, increasing evidence suggests that lncRNAs may be key regulators in obesity-related biological processes through diverse mechanisms.8 We did not observe a significant association between rs9292139 and asthma affection status, or other asthma-related phenotypes. Further analyses are needed to investigate this association’s dependency on the ascertainment condition of childhood asthma. Finally, we note that the PRKCA gene (SNP rs2244497), previously reported in the context of genetic associations with BMI in the GACRS cohort using a recessive model,9 also obtained a nominal significant p value in our combined GACRS and CAMP analysis (precessive model = 0.011 vs. padditive model = 0.281).
ACKNOWLEDGEMENTS
Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Genome Sequencing for "NHLBI TOPMed: Genetic Epidemiology of Asthma in Costa Rica" (phs000988.v4.p1) was performed at the Northwest Genomics Center (HHSN268201600032I, 3R37HL066289-13S1). Genome Sequencing for "NHLBI TOPMed: Childhood Asthma Management Program (CAMP)" (phs001726.v1.p1) was performed at the Northwest Genomics Center (HHSN268201600032I). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The TOPMed Banner Authorship list can be found online (https://www.nhlbiwgs.org/topmedbanner-authorship).
Funding:
This work was supported by the Industrial Core Technology Development Program (20000134) by the Ministry of Trade, Industry and Energy (MOTIE, Korea); the Bio-Synergy Research Project (NRF-2017M3A9C4065964) of the Ministry of Science, ICT and Future Planning through the National Research Foundation; Cure Alzheimer's Fund; the National Human Genome Research Institute (R01HG008976, 2U01HG008685); and the National Heart, Lung, and Blood Institute (P01HL132825, U01HL089856, U01HL089897, P01HL120839).
Footnotes
Competing interests
JCC has received research materials from Merck and GSK (inhaled steroids) and Pharmavite (vitamin D and placebo capsules), in order to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. Patient consent for publication Not required. Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Ahmadizar F, Vijverberg SJ, Arets HG, et al. Childhood obesity in relation to poor asthma control and exacerbation: a meta-analysis. Eur Respir J 2016;48(4):1063–73. doi: 10.1183/13993003.00766-2016 [published Online First: 2016/09/03] [DOI] [PubMed] [Google Scholar]
- 2.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science 2006;312(5771):279–83. doi: 10.1126/science.1124779 [published Online First: 2006/04/15] [DOI] [PubMed] [Google Scholar]
- 3.Namjou B, Keddache M, Marsolo K, et al. EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front Genet 2013;4:268. doi: 10.3389/fgene.2013.00268 [published Online First: 2013/12/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunninghake GM, Soto-Quiros ME, Avila L, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007;119(3):654–61. doi: 10.1016/j.jaci.2006.12.609 [published Online First: 2007/03/06] [DOI] [PubMed] [Google Scholar]
- 5.Covar RA, Fuhlbrigge AL, Williams P, et al. The Childhood Asthma Management Program (CAMP): Contributions to the Understanding of Therapy and the Natural History of Childhood Asthma. Curr Respir Care Rep 2012;1(4):243–50. doi: 10.1007/s13665-012-0026-9 [published Online First: 2013/01/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taliun D, Harris DN, Kessler MD, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. 2019:563866. doi: 10.1101/563866/JbioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600 [published Online First: 2013/12/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Cui Q, Zhang X, et al. Long non-coding RNAs regulation in adipogenesis and lipid metabolism: Emerging insights in obesity. Cell Signal 2018;51:47–58. doi: 10.1016/j.cellsig.2018.07.012 [published Online First: 2018/08/03] [DOI] [PubMed] [Google Scholar]
- 9.Murphy A, Tantisira KG, Soto-Quiros ME, et al. PRKCA: a positional candidate gene for body mass index and asthma. Am J Hum Genet 2009;85(1):87–96. doi: 10.1016/j.ajhg.2009.06.011 [published Online First: 2009/07/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]