Abstract
Reprogramming non-cardiomyocytes (non-CMs) into cardiomyocyte (CM)-like cells is a promising strategy for cardiac regeneration in conditions such as ischemic heart disease. Here, we used a modified mRNA (modRNA) gene delivery platform to deliver a cocktail, termed 7G-modRNA, of four cardiac-reprogramming genes—Gata4 (G), Mef2c (M), Tbx5 (T), and Hand2 (H)—together with three reprogramming-helper genes—dominant-negative (DN)-TGFβ, DN-Wnt8a, and acid ceramidase (AC)—to induce CM-like cells. We showed that 7G-modRNA reprogrammed 57% of CM-like cells in vitro. Through a lineage-tracing model, we determined that delivering the 7G-modRNA cocktail at the time of myocardial infarction reprogrammed ∼25% of CM-like cells in the scar area and significantly improved cardiac function, scar size, long-term survival, and capillary density. Mechanistically, we determined that while 7G-modRNA cannot create de novo beating CMs in vitro or in vivo, it can significantly upregulate pro-angiogenic mesenchymal stromal cells markers and transcription factors. We also demonstrated that our 7G-modRNA cocktail leads to neovascularization in ischemic-limb injury, indicating CM-like cells importance in other organs besides the heart. modRNA is currently being used around the globe for vaccination against COVID-19, and this study proves this is a safe, highly efficient gene delivery approach with therapeutic potential to treat ischemic diseases.
Keywords: cardiac reprogramming, cardiovascular reprogramming, hindlimb ischemia, cardiac repair, modified mRNA, gene therapy
Graphical abstract
Using direct reprogramming with modified mRNA, we induce vascular regeneration in a mouse model of ischemic muscle injury.
Introduction
The adult mammalian heart has a very limited regeneration capacity; therefore, upon ischemic injury, large numbers of CMs die and are replaced by non-contracting, collagen-rich cardiac scar tissue that builds for several weeks after injury in a process called remodeling. Researchers are now evaluating whether directly reprogramming the scar cells (i.e., non-CMs) into functional CMs could overcome this lack of CMs and addition of non-CMs in the left ventricle (LV) after ischemic injury. In 2010, Ieda et al.1 identified 3 (Gata4, Mef2C, and Tbx5 [GMT]) out of 14 transcription factors that can reprogram cardiac or tail-tip fibroblasts into CM-like cells. Since then, numerous publications have established that GMT can induce cardiac reprogramming.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 Several research groups have shown that adding Myocardin (Myocd) to GMT is essential for successful human cardiac reprogramming.3,4,7, 8, 9,14,17 Moreover, other studies have demonstrated that including basic helix-loop-helix transcription factor Hand2 to GMT (GMTH) improves reprogramming efficiency.2,5,6,9,10,13,15,16
To date, there are two major obstacles in cardiac reprogramming: one is the poor efficiency of GMT and GMTH, and the other is the use of viral transfection (mostly retro- or lentiviruses) and small molecules, which can have detrimental side effects and regulatory safety concerns. The initial study of CM reprogramming with GMT indicated a 4.8% reprogramming efficiency (cTnT+ cells) in vitro,1 and a lineage-tracing mouse MI model showed 12% conversion into CM-like cells (α-myosin heavy chain [αMHC]+ cells) in vivo.11 More specifically, supplementing GMT with Hand2, Notch inhibitor, and AKT-kinase raised reprogramming efficiency to 70% in mouse embryonic fibroblasts (MEFs),2 while combining GMTH with several different microRNAs (miRs) and small molecules led to about 60% reprogramming efficiency.16 In the case of adult cardiac fibroblasts, however, the highest reprogramming efficiency was only ∼30%.7 In vivo reprogramming with GMT or GMTH converted 12% or 6.5%, respectively, of non-CMs to CM-like cells,11,13 though in both cases this moderate efficiency did improve MI outcome.11,13 These and other reprogramming studies have used lenti- or retroviral-delivery systems to deliver reprogramming genes to non-CMs.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Unfortunately, these methods cause long-term increased expression in cardiac development-related genes, which are transiently expressed under normal conditions, and thus may generate untoward effects over longer time periods. Gata4 overexpression can induce cardiac hypertrophy,18 and elevated Hand2 expression is associated with heart defects.19 Thus, a transient, non-immunogenic gene delivery reprogramming method that does not compromise genomic integrity would have advantages over current methodologies.
Modified mRNA (modRNA) is a transient, safe, non-immunogenic gene delivery method with no risk of genome integration. Our group and others have used modRNA to deliver genes into the heart after injury.20 The pharmacokinetics of modRNA in the heart allow gene expression to jumpstart promptly after delivery and persist for up to 10 days.20, 21, 22 modRNA has been used directly in vitro to reprogram human fibroblasts into hepatocytes and mesenchymal stem cells into neural-like cells;23 however, no groups have yet reported using modRNA for direct reprogramming in vivo.
In this study, we gauge the efficacy of a combinatory modRNA cocktail in directly reprogramming cardiac non-CMs during ischemic conditions. We evaluate the benefits of partial cardiac reprogramming in the heart and skeletal muscle in vivo. Our study demonstrates efficient, safe partial cardiac reprogramming that promotes vascular regeneration by inducing paracrine angiogenic factors.
Results
Seven-gene modRNA cocktail (7G) can induce high-efficiency cardiac reprogramming in mouse or human non-CMs in vitro
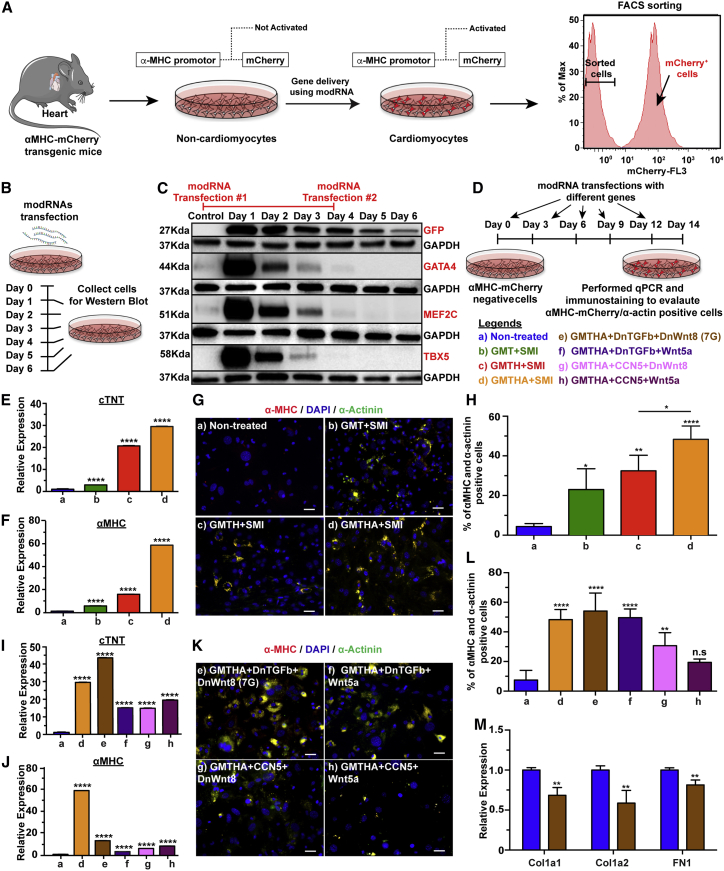
To show that modRNA technology can reprogram adult mouse non-CMs into CM-like cells, we employed a previously published in vitro lineage-tracing model1,11 based on αMHC-mCherry transgenic mice. Isolated mCherry-negative cells were plated and modRNA transfection began once the cells reached partial confluency. Successful cardiac reprogramming in vitro was confirmed by mCherry reporter gene expression in the non-CM subset that upregulates αMHC, a CM-specific cell marker (Figure 1A). As modRNA leads to transient gene expression, repeated administrations could replenish gene expression in the transfected cells. To optimize GMT transfection efficiency for reprogramming, we transfected non-CM cells with GFP or GMT modRNA (flag tagged) and measured their expression at different time points (day 1 through 6) using western blot (Figure 1B). We observed GMT expression until day 3, but not 4, after modRNA transfection (Figure 1C). Thus, we transduced non-CMs isolated from αMHC-mCherry mice with GMT modRNA every 3 days for 14 days before we evaluated cardiac reprogramming with qPCR and immunostaining (Figure 1D).
Figure 1.
Reprogramming strategy and screen for genes to induce CM-like cell reprogramming
(A) Schematic illustration of the approach used to test candidate cardiomyocyte-inducing factors. Cardiac cells isolated from transgenic mice were sorted for mCherry-negative cells to eliminate cardiomyocytes. (B) Experimental timeline for western blot (WB). (C) WB analysis for mCherry-negative cells transfected with GFP, Gata4, Mef2C, and Tbx5 modRNA to assess the transfection strategy for reprogramming non-cardiomyocytes into cardiomyocytes. (D) Schematic representation of timeline used to transfect mCherry-negative cells with different reprogramming genes. (E and F) Graphs showing expression of major cardiac genes (cTNT and α-MHC) determined by quantitative polymerase chain reaction (qPCR) of RNA extracted from non-CMs reprogrammed for 14 days with Gata4, Mef2c, and Tbx5 (GMT) or GMT plus Hand2 (GMTH) or GMTH plus AC (GMTHA) and small molecule inhibitors (SMI) (a, n = 10; b, n = 7; c, n = 5; and d, n = 6). (G) Representative immunostaining images of cells transfected with reprogramming genes plus SMI for cardiac markers α-MHC/α-actinin. (H) Percentage quantification of (G). (I and J) The mRNA expression of cTNT and α-MHC in mCherry-negative cells evaluated 14 days after GMTHA+SMI and GMTHA plus replacement gene transfections, determined by qPCR (a, n = 10; d, n = 6; e, n = 8; f, n = 4; g, n = 4; and h, n = 4). (K) Immunofluorescent staining for α-actinin in mCherry-negative cells 14 days after first transfection with GMTHA+SMI and GMTHA plus replacement genes. (L) Percentage quantification of (K). (M) Relative expression of fibroblast markers in non-CMs after 7G transfection for 14 days (Luc n = 4, 7G n = 4). One-way ANOVA, Tukey’s multiple comparison test for (E) and (F), (I) and (J), (H) and (L). Unpaired two-tailed t test for (M). ∗∗∗∗p < 0.0001; ∗∗p < 0.01; N.S., not significant. Scale bar = 20 μm.
GMT- or GMTH-driven cardiac reprogramming could be enhanced by inhibiting TGF-β and/or WNT pathways.2,6,7 In addition, as the reprogramming process and repeated use of transfection reagents can increase cell death rate, we designed a modRNA backbone that reduces apoptosis and stress. Strelow et al. have shown that acid ceramidase (AC) overexpression protects cells from elevated cell death and curtails cellular stress,24 and Hadas et al. have demonstrated that AC modRNA overexpression is sufficient and necessary to induce cardioprotection after MI.25 Therefore, as shown in Figure 1D, we compared untreated non-CMs to groups treated with either GMT only or GMT with Hand2 or AC (GMTH or GMTHA) modRNA cocktails, together with small molecules (SMI) that inhibit TGF-β and WNT pathways (SB431542 and XAV939, respectively), as well as to groups treated with GMTHA with gene modRNA that inhibits TGF-β (dominant-negative [DN] of TGFβ or CCN5) and WNT pathways (DN-Wnt8 or Wnt5a). Groups treated with either GMT, GMTH, or GMTHA modRNA cocktail together with SB431542 and XAV939 show cardiac reprogramming activity 14 days after first transfection, as demonstrated by upregulated troponin T (cTNT) and αMHC (Figures 1E and 1F). Notably, GMTHA produced the highest reprogramming rate: 48% of CM-like cells (αMHC+ and αActinin+ cells, Figures 1G and 1H).
We also demonstrated that the reprogramming helper small molecules SB431542 and XAV939 can both be replaced by reprogramming helper modRNA (DN-TGFβ or CCN5 and DN-Wnt8 or Wnt5a modRNA, respectively) without losing reprogramming efficiency (Figures 1I–1L). Our data suggest that 7-gene (7G) modRNA cocktail (GMTHA with DN-TGFβ and DN-Wnt8) significantly elevates cTNT and αMHC (Figures 1I and 1J), resulting in 57% CM-like cells 14 days after first transfections in vitro (Figures 1K and 1L). In addition, the collagen and fibronectin gene expression levels in reprogramed CM-like cells were significantly lower than in untreated non-CMs in vitro (Figure 1M). Importantly, repeated transfection for 21 or 28 days produced an insignificant number of contracting CM-like cells with mature sarcomere structures and complete cardiac reprogramming (Figure S1 and Movie S1).
We next assessed whether 7G modRNA induces cardiac reprogramming in normal human ventricle cardiac fibroblasts (NHCF-Vs). First, we conducted protein expression analysis to confirm that GMT modRNA kinetics in NHCF-Vs are similar to those in mouse non-CMs (Figures S2A and S2B). We also investigated different transfection reagents and, like in mouse non-CMs, the RNAiMAX transfection reagent gave the highest (∼80%) transfection efficiency, as compared to other commercially available transfection reagents (Figures S2C–S2E). Since previous reports indicate that SV40 pretreatment4,10,12,14 and additional Myocd4,10,14 improve cardiac reprogramming, we pretreated NHCF-Vs with SV40 modRNA 3 days before transfection with or without 6-gene modRNA (GMT+ Myocd + Hand2 + AC [GMTMHA]) along with reprogramming helper small molecules SB431542 and XAV939 or modRNA for different genes that can inhibit TGF-β and WNT pathways (Figure S2F). Our in vitro results show that GMTMHA + DN-TGFβ and DN-WNT8 or GMTMHA + DN-TGFβ and WNT5a modRNA cocktails significantly elevated cTNT and αMHC (Figures S2G and S2H), reduced collagen expression (Figure S2I), and resulted in 30%–40% CM-like cells 14 days after the first cardiac reprogramming transfection (Figures S2J and S2K).
Screening additional genes improves the reprogramming efficiency of 7G
Having established that 7G modRNA can lead to efficient cardiac reprogramming in vitro, we next evaluated whether adding one or several selected candidate genes can further enhance 7G cardiac reprogramming. We pre-selected genes that have either the potential to increase cardiac reprogramming (Zfpm2,4 Tbx6,26 DN-SNAI1,8 Mesp1,3 Ets1,27 Ets2,27 Esrrg,4 SMarcd3,3 or Srf3) or known functions in choreographing cardiac contractility (SUMO1), cell survival (Ad52E4), neonatal cardiac metabolism (Pkm221), or telomere size (hTERT). While all 7G modRNA cocktails with additional candidate modRNA genes led to significant cardiac reprogramming (Figures S3A–S3D), none of them produced similar or significantly higher cardiac reprogramming activity than 7G alone (Figures S3E and S3F).
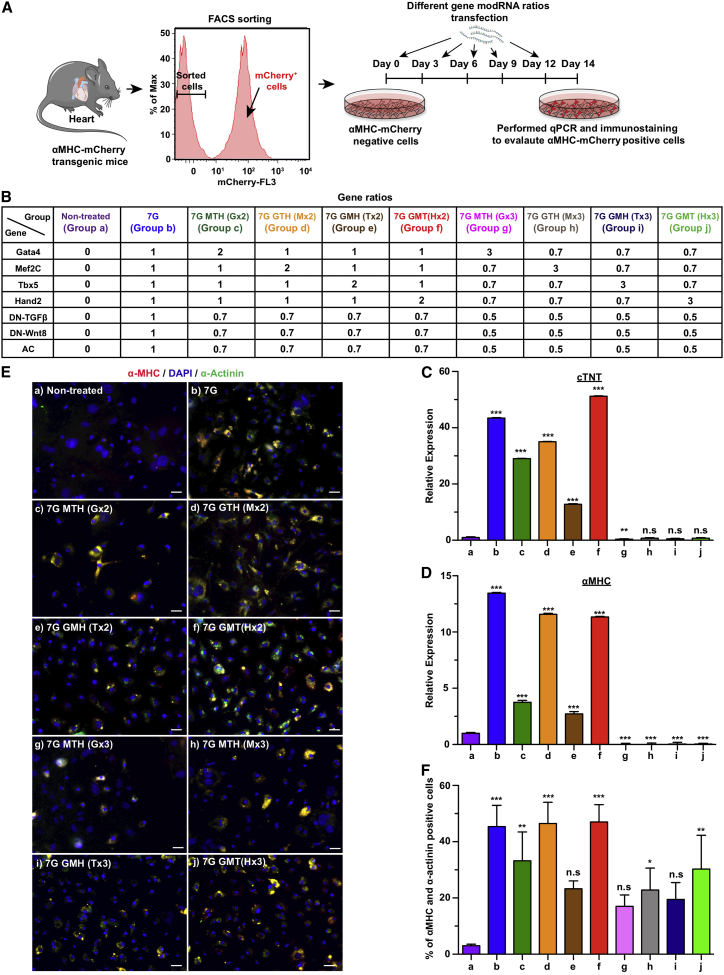
7G-modRNA cocktail with equal ratio or higher concentrations of either Hand2 or Mef2c yields high levels of cardiac reprogramming activity in mouse non-CM in vitro
One advantage modRNA gene delivery has over viral methods is the capability to control the amount of mRNA being delivered into cells. Therefore, we investigated how different ratios of reprogramming gene modRNA (GMTH) in the 7G modRNA cocktail would affect cardiac reprogramming (Figures 2A and 2B). As shown in Figure 2, our results indicated that double but not triple concentrations of Hand2 or Mef2C, compared to Tbx5 or Gata4, lead to similar cardiac reprogramming activity as 7G in vitro (Figures 2E and 2F). We thus concluded that the 7G-modRNA cocktail alone and its two enhanced versions, i.e., Hand2 7G-modRNA cocktail (7G GMT [HX2]) and Mef2c 7G-modRNA cocktail (7G G(MX2) TH), all have ∼50% cardiac reprogramming efficiency 14 days post transfection, in vitro.
Figure 2.
Varied stoichiometry among seven genes influences the efficiency of reprogramming cardiac fibroblasts to CM-like cells in vitro
(A) Schematic representation of isolating mCherry-negative cells from α-MHC transgenic mice, using FACS sorting, and transfecting the cells at 3-day intervals with different ratios of seven genes for cardiac reprogramming experiments. (B) Table showing transfection groups with different ratios of seven genes. (C and D) The mRNA relative expression of cTNT and αMHC, respectively, evaluated in mCherry-negative cells 14 days post transfection with different gene ratios, determined by qPCR (a, n = 10; b, n = 8; c, n = 2; d, n = 2; e, n = 2; f, n = 2; g, n = 2; h, n = 2; I, n = 2; and j, n = 4). (E) Immunostaining images of mCherry-negative cells that exhibited αMHC-mCherry and α-actinin after 2 weeks of transfection with reprogramming genes. (F) Percentage quantification of (E). One-way ANOVA, Tukey’s multiple comparison test for (C) and (D) and (F). ∗∗∗p < 0.001; ∗∗p < 0.01; N.S., not significant. Scale bar = 10 μm.
7G or 7G GMT (HX2) modRNA cocktails improve outcomes after MI
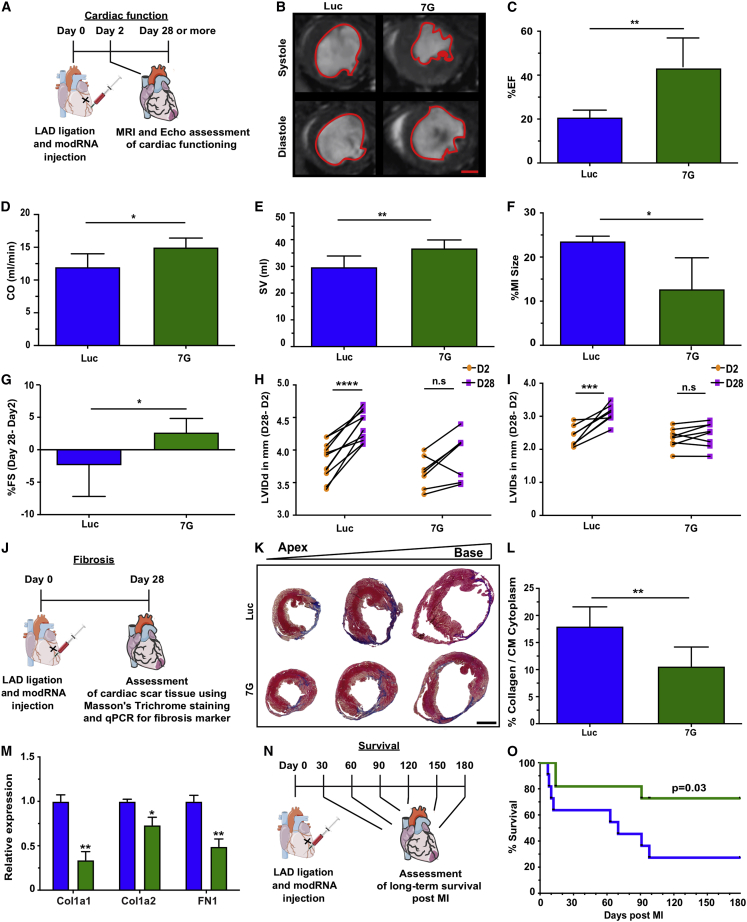
In addition to evaluating cardiac reprogramming in vitro, we assessed the 7G-modRNA cocktail’s ability to improve functional outcomes after MI. To do so, we used a mouse model of MI and measured cardiac function at days 2 and 28 post MI (Figure 3). MRI evaluation 28 days after MI with Luc or 7G modRNA delivery (Figures 3B–3F) showed significantly improved percentile ejection fraction (%EF, 42% for 7G versus 20% for control modRNA [luciferase (Luc)]), cardiac output (CO, 15% for 7G versus 12% for control), stroke volume (SV, 37% for 7G versus 29% for control), and MI size (12% for 7G versus 23% for control) (Movies S2 and S3). To confirm that these improvements are not due to MI induction differences, we measured the fractional shortening (%FS) of the left ventricular internal dimension (LVID) of both end-diastole (LVIDd) and end-systole (LVIDs) on days 2 and 28 post MI and treatment with various modRNA cocktails. The percent FS difference was significantly higher (2.5% for 7G in comparison to −2.4% for control) (Figure 3G). Moreover, while both LVIDd and LVIDs varied significantly between day 2 and day 28 in the control group (Figures 3H and 3I), these alterations were not significant when 7G-modRNA cocktails were delivered. We also evaluated cardiac scar formation and the expression of extracellular markers such as collagen and fibronectin 28 days after MI and different treatments (Figures 3J–3M). We showed that cardiac scars in mice treated with 7G modRNA cocktail were significantly smaller (10%) than control (17%) and had lower expression of both collagen and fibronectin (Figure 3M).
Figure 3.
7G modRNA improves cardiac function and outcome post MI
(A) Experimental timeline to evaluate cardiovascular function after delivery of Luc and 7G-modRNA in an acute MI mouse model. (B) Magnetic resonance imaging (MRI) assessments of left ventricular systolic and diastolic function, performed 28 days post MI. Representative images show the left ventricular chamber (outlined in red) during diastole and systole. (C–F) Percentage evaluation of ejection fraction (EF), cardiac output (CO,) and myocardial infarction (MI) size measured by MRI experiments in (B) (Luc, n = 5; 7G, n = 5). (G) Percentage of fractioning shortening differences, obtained by echocardiography, between day 2 (baseline) and day 28 post MI (Luc, n = 17; 7G, n = 8). (H and I) Measurement of differences in left ventricular internal diameter diastole (LVIDd) and systole (LVIDs), evaluated by echocardiography, between day 2 (baseline) and day 28 post-MI (Luc, n = 17; 7G, n = 8). (J) Experimental plan to isolate hearts for scar tissue assessment and qPCR analysis for fibrosis markers. (K) Representative histological sections with Masson’s trichrome staining to evaluate scar size 28 days post MI. (L) Percentage quantification of left ventricle area occupied by scar versus viable tissue 28 days post MI, based on (J) (Luc, n = 3; 7G, n = 3). (M) qPCR panel showing relative expression of fibrosis markers comparing Luc treatment with 7G-modRNA, 28 days post MI. (N and O) Long-term survival curve after myocardial infarction and 7G and 7G modRNA compared to Luc treatment (Luc, n = 12; 7G, n = 12). Unpaired two-tailed t test for (C)–(G) and (L) and (M). Two-way ANOVA, Bonferroni post hoc test for (H) and (I). Mantel-Cox log-rank test for (O). ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; N.S., not significant. Scale bar = 1 mm (B and K).
Importantly, 7G with higher Mef2c (7G G(Mx2) TH) concentration did not improve cardiac scarring or cardiac function, in comparison to control (Figure S4, Movie S4), while increased Hand2 (7G GMT (HX2) modRNA) showed a beneficial effect similar to that of 7G-modRNA alone. This finding indicates that, in vivo, higher concentration of Hand2, but not Mef2c, enhances outcomes after MI. Lastly, we show that 7G-modRNA cocktail significantly improved mouse survival 6 months after MI (Figures 3N and 3O).
7G-modRNA cocktails upregulate angiogenic paracrine factor secretion in the LV and promote cardiovascular regeneration post MI
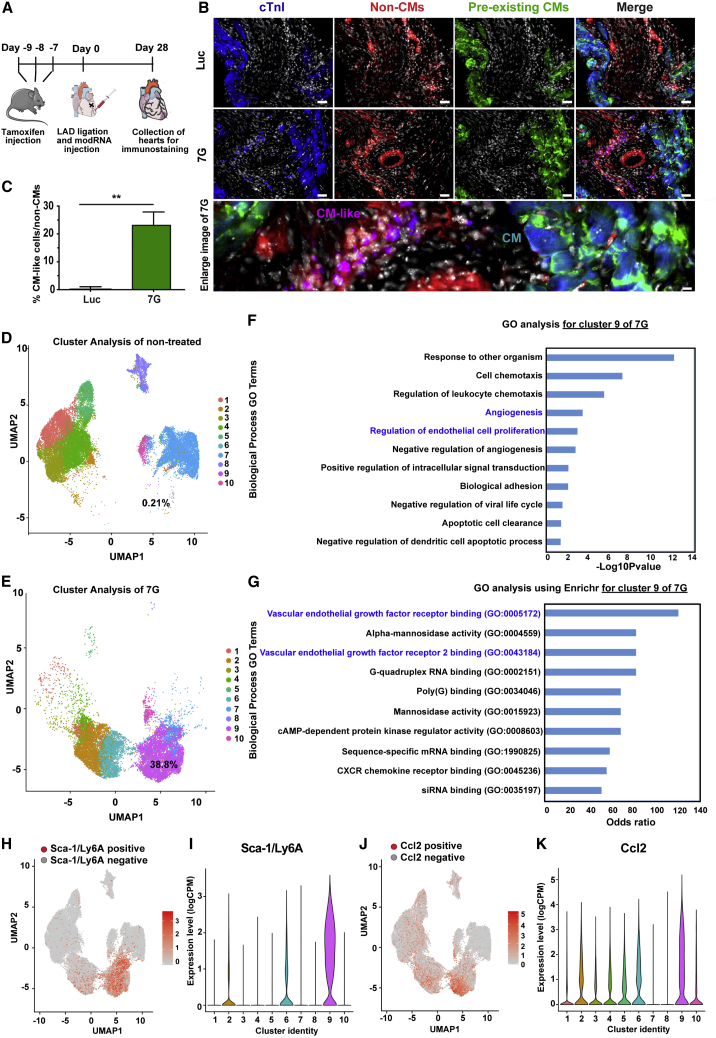
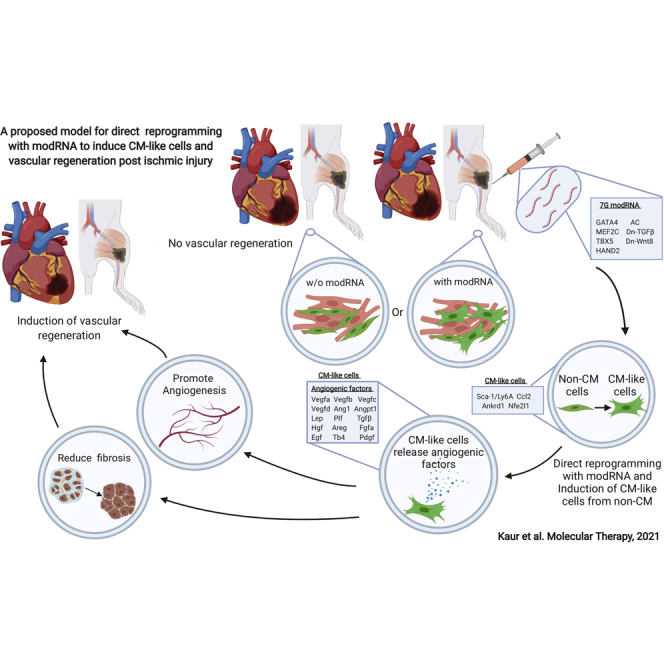
Because the 7G-modRNA cocktail promoted CM-like cell formation and improved post-MI outcomes in vitro, we evaluated its ability to induce CM-like cells in vivo. To this end, we used a lineage-tracing model (TnnTCre/mTmG mice, Figures S5A–S3C), which, unlike αMHC-mCherry mice that express mCherry under the cardiac-specific αMHC promoter (Figures S5D and S5E), pre-labels most (∼95%) preexisting CMs (via the cardiac-specific TnnT promoter) in the heart in green (GFP), while all non-CMs are labeled in red (tdTomato+). Our results showed that 4 weeks after MI and 7G modRNA cocktail delivery into the LV, 24% of total non-CMs (tdTomato+ cells) were CM-like cells (tdTomato+ and cTnI+ cells) (Figures 4A–4C). Notably, these CM-like cells lacked a mature sarcomere structure and were much smaller than preexisting mature CMs (Figure 4B). It was very unlikely that the immature, partially reprogrammed CM-like cells formed by the 7G-modRNA cocktail contributed to cardiac muscle regeneration. Therefore, we decided to assess the reprogrammed cells using unbiased single-cell RNA-seq to seek out beneficial mechanisms of action. We observed that 21 days of repeatedly (every 3 days) transfecting non-CMs with 7G-modRNA resulted in distinct cell clusters that separated 7G-treated cells from non-treated cells (Figure S6A). Batch effect evaluation showed similar clustering among different batches in proximity to each other and according to their treatment (n = 3, Figure S6B). As expected, treated cells had upregulated reprogramming genes (Figures S6C–S6H). Gene ontology (GO) enrichment analysis to define the elevated biological processes in the 7G-treated cells showed significantly enriched cardiac pathways (Figure S6I, in red), namely those involved in cardiac muscle proliferation, hypertrophy, looping, and cardiac cell apoptosis prevention. Importantly, one of the most upregulated biological processes was angiogenesis (Figure S6I, in blue). As we did not see any significant new muscle formation in vitro (Figure S1) or in vivo (Figure 4B), and because four of our reprogramming genes regulate cardiac muscle function (Gata4, Hand2, Mef2c, Tbx5), we decided to reanalyze our data without the 7G upregulated by our modRNA reprogramming genes. This new analysis (Figures S6J–S6L) indicated that treated and non-treated cells clustered separately and cell batches clustered in proximity to one another. Importantly, GO analysis excluding 7G indicates that angiogenesis, but not cardiac muscle pathways, were significantly upregulated (Figure S6L, in blue). Furthermore, cluster analysis of non-treated and treated cells (Figures 4D and 4E), without 7G, revealed enrichment (from 0.21% to 38.8%) of one cell cluster (cluster #9) in the treated cells (Figures 4D and 4E). GO analysis of cell cluster #9 showed elevated angiogenesis and regulation of endothelial cell proliferation processes (Figure 4F). Furthermore, GO analysis using Enrichr for cell cluster #9 uncovered increased VEGF receptor 1&2 binding (Figure 4G). A complete gene analysis showed that cell cluster #9 upregulated several known pro-angiogenic genes and transcription factors that are expressed in mesenchymal stromal cells (Sca-1/Ly6A, Ccl2, Ankrd1, Nfe2l1; Figures 4H–4K and S7).28, 29, 30 In addition, we saw elevated Fmr1, which regulates endothelial cell proliferation and angiogenesis,31 and higher Zc3 h15 expression that correlates with expression of the pro-angiogenic protein a-fetoprotein (AFP).32,33 Taken together, these results suggest the 7G-modRNA cocktail’s beneficial mechanism of action may occur via angiogenesis and paracrine effects to induce cardiovascular regeneration post MI.
Figure 4.
7G-modRNA delivery resulted in a subpopulation of CM-like cells expressing mesenchymal stromal cell markers and pro-angiogenic pathways
(A) Experimental plan for lineage tracing using tamoxifen injection, LAD ligation, and modRNA delivery followed by isolation of hearts from transgenic mice. (B) Representative immunostaining images of in vivo CM-like cells (in magenta) collected from Luc and 7G-modRNA-injected mice, showing positive staining for cardiomyocyte marker cTnI (blue) and Tdtomato-cre (red). (C) Quantification of CM-like cells depicted by immunostaining in (B) (Luc, n = 3; 7G, n = 3). (D and E) Single-cell RNA analysis to identify single cells without (D) or with (E) 21 days of 7G treatment (repeat transfection every 3 days), visualized in different colors using uniform manifold approximation and projection (UMAP) plots of 42,747 cells. (F and G) List of upregulated biological processes with GO and enriched GO functions (using Enricher) in cells treated with 7G for 21 days. (H–K) Top differentially expressed genes in cell cluster 9. Feature plots and violin plots show the relative gene expression strength of Sca-1/Ly6a (H and I) and Ccl2 (J and K), respectively. Unpaired two-tailed t test for (C). ∗∗p < 0.01. Scale bar = 50 μm and 5 μm (B).
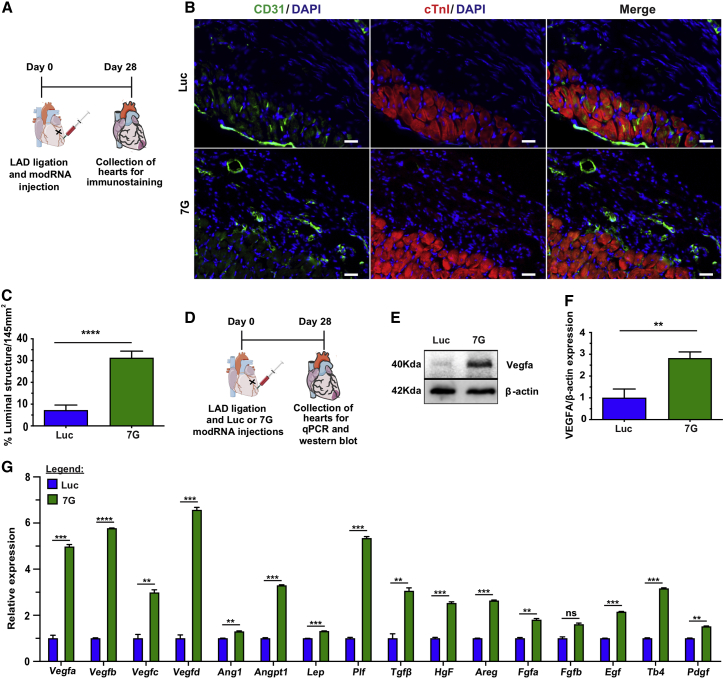
To test this hypothesis about 7G-modRNAs mechanism of action, we evaluated LV capillary density 28 days post MI and delivery of different modRNAs. We showed that, compared to control, both 7G and 7G GMT (Hx2) modRNA cocktail significantly promoted capillary density in the LV (Figures 5A–5C and S8A–S8F). Since Vegfa protein is a key regulator of cardiac vascularization22,34 and was activated in cell cluster #9 (Figure 4G), we used western blot analysis to further evaluate Vegfa levels 21 and 28 days after MI and different modRNA treatments. We found Vegfa levels to be significantly higher in the LV 28, but not 21, days following MI (Figures 5D–5F). Moreover, using qPCR, we investigated 16 pre-selected key angiogenic factors known to increase blood vessel formation and maturation. We assessed Vegfa,35 Vegfb,36 Vegfc,37 Vegfd,38 Ang1,39 Angpt1,40 Lep,41 Plf,42 Tgfβ,43 Hgf,44 Areg,45 Fgfa,46 Fgfb,46 Egf,47 Tb4,48 and Pdgf49 at 1, 2, 3, and 4 weeks post MI and delivery of 7G-modRNA cocktail, compared to Luc control modRNA (Figures 5G, S8G, S8H, and S9). Of the 16 angiogenic factors, 15 rose significantly between 1 and 4 weeks after MI; FGF-2 was the exception. Similar results were seen when 7G GMT (Hx2) modRNA cocktail was delivered post MI (Figure S9). In addition, qPCR evaluation of LV at different post-MI time points, with various treatments, indicated that 7G or 7G GMT (Hx2) modRNA cocktails reduced Col1a2 and fibronectin levels, especially in week 2 after MI (Figures S9E and S9F).
Figure 5.
7G improves capillary density and upregulates Vegfa and other pro-angiogenic factors
(A) Experimental model for analyzing capillary density. (B) Representative immunofluorescence showing luminal structures (green) in the scar area 28 days after Luc and 7G-modRNA delivery. (C) Percentage quantification of capillary density based on (E) (Luc, n = 5; 7G, n = 4). (D) Strategic plan for injecting modRNA and collecting hearts for mRNA and protein analyses post MI. (E) Exemplary western blot images showing Vegfa protein in cardiac tissue isolated 4 weeks post MI and subsequent Luc and 7G-modRNA injections. (F) Quantification of western blot (n = 2). (G) qPCR quantification of major angiogenic gene expression as determined from RNA extracted from cardiac tissue 28 days post modRNA treatment (n = 2). Unpaired two-tailed t test for (C), (F), and (G). ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; N.S., not significant. Scale bar = 25 μm.
As angiogenic factors upregulated following 7G treatment while fibroblastic markers downregulated, we explored whether the pro-angiogenic outcome of 7G-modRNAs effects on gene expression in cardiac fibroblasts also occurred other cell types (e.g., endothelial cells or CMs). To investigate this, we sorted cardiac cells into cardiac fibroblasts (by sorting for CD90+), endothelial cells (by sorting for CD31+), and CM (by using a neonatal CM isolation kit). We transfected the cells with Luc or 7G-modRNA on days 0 and 3 and collected the cells on day 7 (Figure S10A). Our analyses showed that the majority of cells involved in upregulating pro-angiogenic factors are cardiac fibroblasts and not endothelial cells or CMs (Figures S10B–S10D).
As highly unregulated Vegfa levels in the heart post MI can have deleterious side effects, such as edema and angioma, and may lead to leaky blood vessels,22,50 we investigated these factors in the heart, at different time points, after MI and different modRNA cocktail treatments. As shown in Figure S11, 7G and 7G GMT (Hx2) modRNA cocktails did not notably change heart weight to body weight ratio (Figure S11B) or heart size (Figure S11C) and did not induce angioma formation in the myocardium (Figure S11D) or leaky vessels (Figure S11E), as compared to Luc control modRNA 28 days post MI. In addition, in order to determine whether cardiac reprogramming of non-CMs, from not only mouse but also human hearts in vitro, could also induce pro-angiogenic pathways, we evaluated Vegfa expression 14 days following transfections with or without modRNA cocktails. Both mouse (Figures S12A and S12B) and human (Figures S12C and S12D) cardiac reprogramming led to significantly increased Vegfa expression, indicating that converting human non-CM to human CM-like cells may also turn on a pro-angiogenic program that facilitates this reparative process.
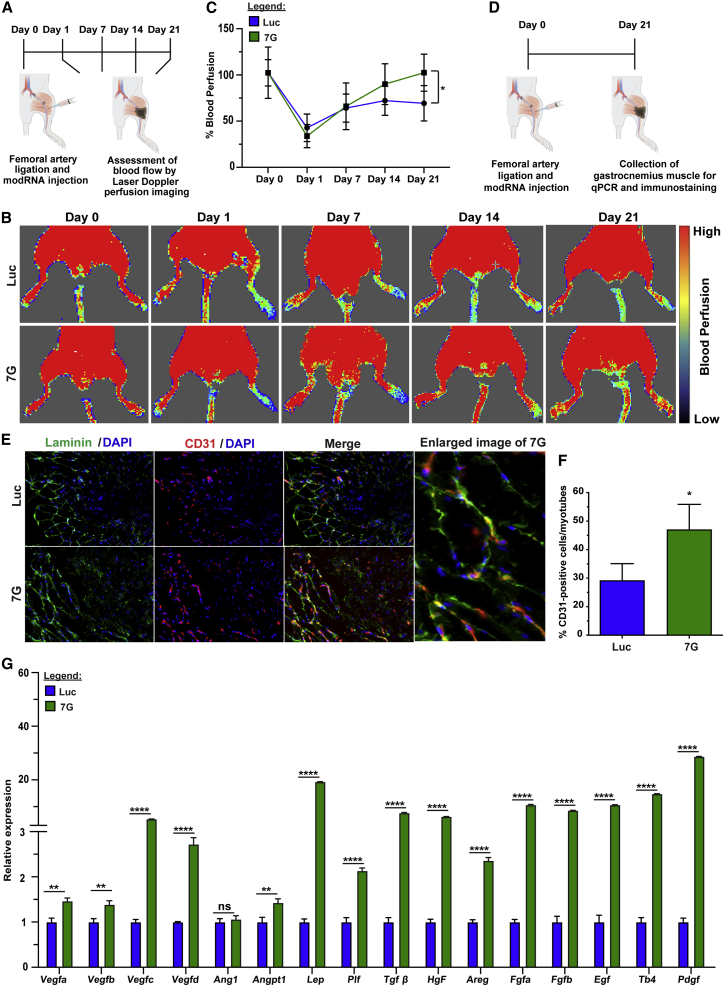
To further evaluate this pro-angiogenic program in non-CMs, we tested whether injecting 7G-modRNA into a non-cardiac (e.g., skeletal) muscle under ischemic conditions could enhance vascularization after injury. An ApoE−/− mouse hindlimb ischemia model has been established as a preferred approach for examining vascular regeneration in skeletal muscle.51 We injected 7G-modRNA cocktail or Luc modRNA immediately after femoral artery ligation and used laser Doppler perfusion imaging to evaluate blood perfusion in the foot region on day 0, 1, 7, 14, and 21 post injury (Figure 6A). Our data showed that 7G-modRNA cocktail improved blood perfusion significantly more than Luc modRNA (Figures 6B and 6C). These data were corroborated by more numerous CD31-positive cells in the ischemic area of mice that received 7G-modRNA treatment (Figures 6E and 6F). Also, key angiogenic factors expression was significantly unregulated, similar to what we observed in cardiac tissue, following muscular ischemic injury and 7G-modRNA cocktail delivery, as compared to Luc control modRNA (Figure 6G). In conclusion, our data show that the 7G-modRNA cocktail enriches a population of reprogramed cardiac fibroblasts (cell cluster #9) that exhibit pro-angiogenic mesenchymal stromal cell gene markers as well as transcription factors that support (cardiac and skeletal) muscular vascular systems. While we observed no contribution to newly formed CM and cardiac regeneration per se, we do observe a boost in angiogenic factors, which is not associated with detrimental angiogenic overexpression effects, such as edema or hemangioma (Figure S11).
Figure 6.
7G enhances blood perfusion and angiogenesis in ApoE−/− mouse hindlimb ischemia model
(A) Experimental plan for femoral artery ligation and modRNA delivery followed by blood perfusion analysis in ApoE−/− mice. (B) Representative laser Doppler perfusion imaging (LDPI) mapping dynamic blood perfusion changes in the foot region of a mouse which received Luc and 7G-modRNA after femoral artery ligation; LDPI shows days 0, 1, 7, 14, and 21. (C) Quantification of foot perfusion as measured by LDPI, based on (C), where the LDPI ratio is calculated by comparing blood perfusion between the ischemic limb to that in the contralateral hind limb (Luc, n = 5; 7G, n = 5). (D) Experimental timeline for performing qPCR and immunostaining on the mouse gastrocnemius muscle. (E) Representative immunofluorescence showing luminal structure (red) in the scar area 28 days after Luc and 7G-modRNA delivery in the gastrocnemius muscle. (F) Quantification based on (E). (G) qPCR panel showing upregulated angiogenic genes in ischemic muscle isolated from a mouse injected with Luc and 7G following critical limb ischemia. Two-way ANOVA, Bonferroni post hoc test for (C). Unpaired two-tailed t test for (E). ∗∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05; N.S., not significant.
Discussion
CMs occupy ∼70%–85% of the mammalian heart.52 Following cardiac ischemic injury, large numbers of CMs die and are replaced by non-CM cells. One approach to overcome this imbalanced CM ratio is to reprogram non-CMs to act like CMs and thereby generate de novo CMs. The main obstacles in optimizing such reprogramming for cardiac repair are the low efficacy of cardiac reprogramming and the possible detrimental side effects of viral and small-molecule delivery methods. Our work, as summarized in this manuscript, showed that delivering reprogramming genes and helper genes via modRNA technology eliminates the need for viral transfection or small molecules and leads to high numbers of CM-like cells in vitro and in vivo. The newly formed CM-like cells were not functional or beating CMs and were unable to replace the lost endogenous CMs. Nevertheless, our research sheds light on a different mechanism of action for direct cardiac reprograming. Specifically, our results revealed that CM-like cells upregulate pro-angiogenic mesenchymal stromal cell markers and transcription factors that lead to physiological secretion of numerous angiogenic factors. This mechanism resulted in better vascular regeneration post (cardiac and skeletal) muscle ischemic injury. To date, the highest reprogramming efficiency reported 14 days post transfection yielded ∼30% CM-like cells.7,9 Here, we demonstrated that using 7G or 7G GMT (Hx2) modRNA cocktails increased reprogramming activity and produced ∼50% CM-like cells in vitro (Figures 1 and 2). We also showed that using 7G + Myocd or GMTMHA + DN-TGFβ and WNT5a modRNA cocktails lead to 30%–40% reprogramming efficiency in human ventricle cardiac fibroblasts (Figure S2). These results are similar to previous reports of human cardiac reprogramming with GHTM, miR-1, and miR133a.10 Importantly, the findings that repeated transfections for 21 or 28 days did not produce significant numbers of contracting CMs in vitro (Figure S1 and Movie S1) and 7G delivery in vivo did not result in functional, matured CMs (Figures 4A–4C) lead us to conclude that the CM-like mechanism of action occurs not via reprogramming non-CMs to functional, beating CM-like cells but rather via secreting paracrine factors (Figure 5). We were unable to identify any additional gene or alternative reprogramming gene modRNA ratios that generated higher reprogramming levels (Figure 3). However, we did show that elevating any cardiac reprogramming gene 3-fold abolished the reprogramming process (Figure 2), indicating that each reprogramming gene plays an essential role and that lack or low levels of any single gene alone prevents reprogramming.
Using a lineage tracing in vivo model, we confirmed that modRNA successfully promoted the formation of ∼25% CM-like cells in the LV 28 days post MI and delivery of 7G modRNA cocktail (Figures 4A–4C). This is a higher cardiac in vivo reprogramming efficiency than the previously published 12%.53 Nonetheless, we were unable to detect any functional CMs with mature sarcomeres among the CM-like cell population. This could be because short-term (∼10 days) modRNA delivery was insufficient for effectively reprogramming non-CMs to mature contracting CMs, as has been shown with viral delivery systems that have long-term expression patterns.13 However, even when viral vectors were used to deliver cardiac reprogramming genes, the number of functional CMs observed in the heart was very low.13 This may indicate that the most dominant mechanism of action for direct cardiac reprogramming is not via newly formed functional CMs, but rather angiogenic paracrine secretion and vascular regeneration.
Even without forming de novo adult CMs, our protocol led to significantly improved cardiac function, scar size, and long-term survival 28 days post MI and delivery of 7G or 7G GMT (HX2) modRNA cocktails (Figures 3 and S4). Using single-cell RNA-seq (Figures 4D–4K, S6, and S7), we demonstrated that 7G-modRNA enriched for a cardiac fibroblast subpopulation (cell cluster #9), significantly upregulated angiogenic biological processes, and expressed pro-angiogenic mesenchymal stromal cells markers (Sca-1/Ly6A and Ccl2) and transcription factors (Ankrd1 and Nfe2l1).28, 29, 30 In addition, we showed that 7G-modRNA enhanced capillary density in the heart post MI and upregulated 15 key angiogenic factors without causing edema or angioma (Figures 5 and S8–S11). These results suggest that the elevation in these factors was in the physiological range, as excessive amount could detrimentally affect the heart. Lastly, we determined that 7G-modRNA can promote a pro-angiogenic program outside cardiac tissue (e.g., in skeletal muscle, see Figure 6), thereby indicating that four of the genes (Gata4, Mef2c, Tbx5, and Hand2), all of which are related to cardiac development, may also support angiogenesis in non-CMs outside the heart. Gata4 is a necessary regulator of cardiac gene expression, hypertrophy, stress compensation, and myocyte viability,54 while MEF2c is a major player in cardiac gene regulatory networks. Mef2c also regulates atrial and secondary heart fields, and its overexpression can lead to dilated cardiomyopathy.55 TBX5 is a member of the T-box transcription factor family and is primarily known for its role in cardiac and forelimb development.56 Hand2 is robustly expressed within the secondary heart field pharyngeal mesoderm that underlies and contributes to the growing heart tube.57 Additionally, Hand2 is a crucial downstream transcriptional effector of endocardial Notch signaling during both cardiogenesis and coronary vasculogenesis.58 This overwhelming evidence strongly signals that modRNA-directed cardiac reprogramming makes no contribution to mature CMs but rather promotes cardiac protection and vascular regeneration post ischemic injury via a paracrine effect. In this regard, our findings are similar to previous attempts to deliver angiogenic factor-secreting cells (mostly bone marrow-derived cells) to the LV in order to promote cardiovascular regeneration.59, 60, 61, 62, 63 As these earlier approaches produced only marginal benefits, our modRNA method might lay the foundation for improving upon earlier, ineffective cytokine delivery attempts to ignite cardiac repair. Although Vegfa modRNA has been shown to enhance blood vessel regeneration in ischemic cardiac or skeletal muscle mouse models before,22,64 here we present a 7G-cocktail that induces non-CMs to secrete, at physiologically significant levels, angiogenic factors long after modRNA translation ceases (∼day 10) and support vascular regeneration in vivo. For these reasons, our approach represents an advanced strategy for treating ischemic injuries.
Since 2002, bone-marrow-derived cells have been used in many clinical trials to promote vascular regeneration in patients with acute MI, chronic cardiac ischemia, or chronic critical limb ischemia disease.60,65 Years of clinical trials have shown this treatment is safe and leads to modest improvements in physiologic and anatomic parameters, above and beyond conventional therapies.60,65 However, this method has fundamental problems: injected cells have poor survival rates and the number of ex vivo-cultured cells, typically from the same donor, must increase over time.59, 60, 61, 62, 63 Our approach, by contrast, uses off-the-shelf modRNA cocktails that can convert non-CMs in the scar area to pro-angiogenic mesenchymal stromal cells (similar to bone marrow-derived cells) without engraftment issues or the need to culture cells ex vivo.
It is important to note that modRNA gene delivery maintains DNA integrity and does not endanger insertional mutagenesis. In addition, spike modRNA delivery has been used successfully by Moderna and Pfizer-BioNTech to vaccinate against COVID-19. We therefore expect that, going forward, clinical gene delivery will increasingly employ modRNA rather than lenti- or retroviral systems. Unlike modRNA, these viral gene transfer methods have serious disadvantageous side effects. modRNA can also eliminate the need for small molecules to inhibit TGF-β and WNT pathways during reprogramming. As we have shown, all modRNA cocktails can enhance cardiac reprogramming, expand capillary density, and bolster cardiovascular regeneration post ischemic injury.
Although we show that modRNA’s angiogenic effect does accrue in human cardiac reprogramming (Figure S11), there is a need to quantify modRNA cocktail’s ability to induce vascular regeneration in large animal models (e.g., swine) in order to progress this approach toward clinical translation. Taken together, the experiments in our study reveal the therapeutic angiogenic function of our 7G-modRNA cocktail following ischemic muscle injury, and our results may encourage others to create different, novel, modRNA cocktails for other types of multicomponent drug discovery.
Methods and materials
Mice
All animal procedures were performed under protocols approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (IACUC). For in vitro experiments, cardiac fibroblasts were isolated from P0-P4 α-myosin heavy chain-mCherry (α-MHC-mCherry) transgenic neonate hearts (mice purchased from Jackson Laboratories). For lineage-tracing studies, Tnnt2MerCreMer/+/R26mTmG/+ mice were generated by crossing TnnT-Cre mice (gifted by Dr. Chen-Leng Cai) and Rosa26mTmG (Jackson Laboratory) mice. When mice were 11 weeks old, they received injections of tamoxifen dissolved in sesame oil for 3 consecutive days at 0.1 mg tamoxifen per 1 g body weight. Myocardial infarction (MI) was induced by permanently ligating the left anterior descending (LAD) artery 1 week after tamoxifen injections. After MI induction, male and female mice were randomized into four different groups and treated with four different modRNA types (Luc, 7G, 7G GMT(Hx2), and 7G G(Mx2)TH), injected directly into the myocardium during open chest surgery. For Echo, MRI, and long-term survival analyses, 8- to 10-week-old CFW animals were allowed to recover for 6 months in the animal facility. Deaths were monitored and documented. Limb ischemic studies used 4-month-old ApoE−/− male and female mice. Unilateral hindlimb ischemia was induced by isolating the femoral artery from the femoral nerve and vein, ligating the left femoral artery and then cutting at the level of the internal iliac and popliteal arteries. Following artery ligation, randomized mice received Luc and 7G injections at three different sites in the gastrocnemius muscle.
modRNA synthesis
All modRNA was generated our laboratory by in vitro transcription of plasmid templates (GeneArt, Thermo Fisher Scientific). The full list of open reading frame sequences used to make modRNA for this study can be found in Table S1. The transcription step involved a customized ribonucleotide blend of anti-reverse cap analog; 30-O-Me-m7G(50) ppp(50)G (6 mM, TriLink Biotechnologies); guanosine triphosphate (1.5 mM, Life Technologies); adenosine triphosphate (7.5 mM, Life Technologies); cytidine triphosphate (7.5 mM, Life Technologies) and N1-methylpseudouridine-5-triphosphate (7.5 mM, TriLink Biotechnologies). Next, modRNA was purified with the Megaclear kit (Life Technologies) and treated with Antarctic Phosphatase (New England Biolabs). To eliminate any remaining impurities, modRNA was re-purified with the Megaclear kit and quantified using a Nanodrop spectrometer (Thermo Scientific). Lastly, modRNA was precipitated with ethanol and ammonium acetate and resuspended in 10 mM Tris-HCl and 1 mM EDTA.
modRNA transfection
For in vitro transfection, cells plated in 24-well and 6-well plates were transfected with 2.5 μg of mRNA and 10 μg of mRNA, respectively, encoding various genes using RNAiMAX transfection reagent (Life Technologies) in accordance with the manufacturers’ instructions. In vivo gene delivery was performed according to previously published methods.27 modRNA was delivered using sucrose citrate buffer containing 20 μL of sucrose in nuclease-free water (0.3 g/mL), with 20 μL of citrate (0.1 M [pH 7]; Sigma) mixed with 20 μL of different modRNA concentrations in saline to a total volume of 60 μl. The transfection mixture was directly injected into heart muscle surrounding the MI (two on either side of the ligation and one in the apex), with 20 μL at each site.
Cell culture
Mouse cardiac fibroblast culture
Hearts isolated from neonate mice were chopped into small pieces, approximately 1 mm3, and digested for 20 min on a rocker with collagenase type II in PBS and 0.25% (wt/vol) trypsin. After digestion, the clumpy heart tissue was centrifuged at 600 g for 2 min and plated in a 10cm dish (3-5 hearts per dish) in fibroblast explant media (Iscove’s modified Dulbecco medium with 20% FBS (IMDM)) at 37°C. After 30 min of incubation, the plate was washed with PBS and cells were quenched with fresh media. When confluent, attached cells were washed with PBS, collected with 5-min 0.05% trypsin treatment and quenched with fibroblast explant media. Cells were then filtered through a 70-μm filter and pellet was collected. Thereafter, the cells were sorted by fluorescence-activated cell sorting (FACS) for mCherry-negative cells, plated onto 6-well gelatin-coated plates at a concentration of 10000 cells/cm2 and used fresh for all reprogramming studies. After 6 h of modRNA transfection with RNAimax, media was replaced with cardiomyocyte induction medium (iCM) comprising DMEM:M199 (4:1), 10% FBS, 1× non-essential amino acids (NEAA), and 1× penicillin/streptomycin. Thereafter, SMI SB431542 (2.6 μM) and XAV939 (5 μM) were added 24 h and 48 h post infection, respectively.
Human cell line
Normal human cardiac fibroblasts-ventricular (NHCF-V) were purchased from Lonza (CC-2904) and grown in Cardiac Fibroblast Growth Medium (Lonza, CC-4526). Once semi-confluent, cells were transfected with SV40-Large T modRNA 3 days before transfection with reprogramming genes to obtain a stable immortalized cell line. Cells were transfected with different modRNA and SMI (wherever mentioned) for 6 consecutive days for WB and 14 consecutive days for cardiomyocyte reprogramming assay. For WB experiments, cells were collected every day during days 1–6, while reprogramming analyses (qPCR and ICC) were performed on day 14.
Immunofluorescence
28 days post MI, mice hearts were harvested, and excess blood was removed by injecting 1 mL PBS into the right ventricular chamber. Hearts were fixed via overnight incubation in 4% PFA, with subsequent PBS washings for at least an hour. Next, hearts were placed in 30% sucrose solution at 4°C overnight. The following day, hearts were fixed in OCT and frozen at −80°C. Transversal 10 μm-thick sections were made by cryostat and rehydrated in PBS for 5 min for immunostaining. All staining was performed on 3–8 hearts/group, with 2–3 sections/heart. To immunostain mCherry-negative neonatal mouse non-CMs following modRNA treatment, cells were fixed on coverslips with 4% PFA for 15 min at room temperature, then washed three times with PBST. Cells/heart and skeletal muscle tissues were permeabilized in PBS with 0.1% Triton X-100 (PBST) for 7 min followed by overnight staining with primary antibodies. We used the recommended concentrations of sacromeric α-actinin (Abcam, #9465), cardiac troponin I (Abcam, #7003), laminin (Abcam, #11575), and CD31 (R&D Systems, #3628) diluted in PBST, GFP (Abcam, #13970), and tdTomato (Origene, #8181-200). The next day, slides were washed with PBST (five times, 4 min each) followed by incubation with a secondary antibody (Invitrogen, 1:200) diluted in PBST for 2 h at room temperature. To remove the secondary antibody, the samples were further washed with PBST (three times, 5 min each) and stained with Hoechst 33342 (1 μg/mL) diluted in PBST for 7 min. After five 4-min washes with PBST, slides were mounted with mounting medium (Vectashield) for imaging. Stained slides were stored at 4°C. The fluorescent images were taken on Zeiss fluorescent microscopy at 10×, 20×, and 40× magnifications.
Masson’s trichome staining
Masson’s trichome staining was performed to evaluate scar size in the LV post MI and modRNA treatments. The OCT frozen transverse heart sections were air-dried for 30 min to 1 h at room temperature before proceeding to staining. Slides were pre-stained with Bouin’s solution for 45 min at 55°C. Next, slides were kept in Weigert’s iron hematoxylin, Biebrich scarlet-acid fucshin, phosphotungstic/phosphomolybdic acid solution, and aniline blue solution for the times suggested by manufacturers. Thereafter, tissue samples were differentiated with acetic acid for 2 min and dehydrated through 95% ethyl alcohol and absolute ethyl alcohol. After being cleared using xylene, slides were mounted with Permount mounting medium (Fisher Scientific). Images were collected using a bright field microscope and scar size analysis was conducted using ImageJ software.
H&E staining
H&E staining was performed according to standard protocol. The OCT frozen transverse heart sections were air-dried for 30 min to 1 h at room temperature, then hydrated in PBS for 10 min. The slides were kept in hematoxylin solution for 2 min and washed with tap water for 5 min. Thereafter, the sections were stained using eosin solution for 1 min and washed with tap water for 5 min. The slides were transferred to PBS for 5 min. Sections were then dehydrated in 100% ethanol and xylene for 1 min each. Finally, sections were mounted with Permount mounting medium (Fisher Scientific). The images were taken on a bright field microscope.
Western blotting
Total protein from respective cells or thawed tissues was isolated at given time points. 20 μg of protein from each sample was resolved using an SDS-PAGE electrophoresis system in 4%–15% Mini-PROTEAN TGX stain-free gels (Bio-Rad). The resulting bands were transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad). The membranes were blocked (5% BSA in Tris-buffered saline Tween 20 (TBS; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1%Tween 20) for 1 h at room temperature and then incubated with primary antibodies diluted in 5% BSA in TBST overnight at 4°C. We used anti-Flag (1:1,000, Sigma, #A8592), anti-Vegfa (1:1,000, Abcam, #51745), anti-GAPDH (horseradish peroxidase [HRP] conjugate 1:3,000, Cell Signaling, #8884), and mouse monoclonal anti-β-actin (horseradish peroxidase [HRP] conjugate 1:3,000, Cell Signaling, #12262) antibodies. Anti-rabbit and anti-mouse HRP-conjugated secondary antibodies were purchased from Cell Signaling. Antigen or antibody complexes were visualized with the ChemiDoc Touch imaging system (Bio-Rad).
RNA isolation and gene expression profiling using real-time PCR
Quick RNA kit (Zymo Research) was used to isolate total RNA from the cells and ischemic mouse tissue at the aforementioned time points. Isolated RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio Rad) according to the manufacturer’s instructions. Real-time qPCR analyses were performed on a Mastercycler Realplex 4 Sequence Detector (Eppendorf) using SYBR Green (PerfeCTa SYBR Green FastMix, QuantaBio). Data were normalized to GAPDH (in vitro experiments and in vivo limb tissue experiments), 18 s (in vivo experiments for cardiac tissue), and B2M (for human experiments). Fold changes in gene expression were determined by the ∂∂CT method and presented relative to an internal control. PCR primer sequences are shown in Table S2.
Echocardiography (Echo)
Transthoracic two-dimensional echocardiography was conducted 2 and 28 days after MI to assess LV dimensions and function using a GE Cares InSite (V7R5049) equipped with a 40 MHz mouse ultrasound probe. In a double-blind study (i.e., neither the surgeon nor the echography technician was aware of the treatment), Luc, 7G, or 7G GMT (Hx2) was injected into CFW mice (8–12 weeks old). Mice were anesthetized with 1%–2% isoflurane in air and imaging was performed. The ejection fraction and fractional shortening were calculated as percentages from the diastolic volume (EDV) and end-systolic volume (ESV) dimensions on an M-mode ultrasound scan. Percent of fractional shortening was calculated as (left ventricular internal dimension at end-diastole (LVIDd) – left ventricular internal dimension at end-systole (LVIDs))/ LVIDd∗100. Echocardiograms were performed on 3–10 hearts/treatment group.
Magnetic resonance imaging (MRI)
In a double-blind study, CFW mice (8 weeks old) treated with Luc, 7G, or 7G GMT (Hx2) modRNA underwent MRI assessment on day 28 post LAD ligation. We obtained delayed-enhancement CINE images on a 7-T Bruker Pharmascan with cardiac and respiratory gating (SA Instruments). For imaging, mice were anesthetized with 1%–2% isoflurane in air. To monitor optimal temperature during ECG, respiratory and temperature probes were placed on the mouse. Imaging was performed 10 to 20 min after IV injection of 0.3 mmol/kg gadolinium-diethylene triamine pentaacetic acid. A stack of 8 to 10 short-axis slices spanning from the hearts apex to its base were acquired with an ECG-triggered and respiratory-gated FLASH sequence with the following parameters: echo time (TE) 2.7 msec with 200 μm × 200 μm resolution; 1 mm slice thickness; 16 frames per R-R interval; 4 excitations with flip angle at 60°. After imaging, the obtained data were analyzed to calculate % ejection fraction, cardiac output, stroke volume, and % MI size.
Immunodetection methods
Leaky vessel detection was performed on heart tissues isolated from mice 28 days post MI and modRNA injection. Isolectin B4 (0.5 mg/mL, Vector Lab) was used to stain endothelial cells in cryosections to determine capillary density. To evaluate blood vessel leakiness, a mixture of 250 μl of isolectin B4 and 250 μl 70 kD FITC-dextran beads (50 mg/mL, Sigma) was injected into the mouse tail vein. Hearts were collected 30 min after injection and fixed overnight using 4% PFA. After four washes with PBS, hearts were placed in 30% sucrose overnight and frozen in OCT the following day. Sectioned heart tissue was evaluated for vessel leakiness under microscope.
Blood flow analysis in ischemic limb model
Using the laser Doppler perfusion imaging (LDPI) analyzer (Moor Instruments), blood flow measurement was performed in the ligated and control limbs at predetermined time points. Hind limb fur was removed and the animals were kept on a heating pad at 37°C to minimize temperature variation. Blood flow was recorded on days 0 (before ligation), 1, 7, 14, and 21 after ischemic induction and modRNA administration. % blood perfusion was calculated by comparing blood flow in the ischemic limbs to that in the control hind limbs.
Single-cell sequencing
cDNA library preparation and sequencing
Single-cell RNA-seq was performed using the Chromium platform (10x Genomics) with the 3′ gene expression (3′ GEX) V3 kit and an input of ∼10,000 cells from a debris-free suspension. Briefly, Gel-Bead in Emulsions (GEMs) were generated on the sample chip in the Chromium controller. Barcoded cDNA was extracted from the GEMs by Post-GEM RT-cleanup and amplified for 12 cycles followed by fragmentation of amplified cDNA, end-repair, poly A-tailing, adaptor ligation, and 10×-specific sample indexing following the manufacturer’s protocol. Libraries were quantified using QuBit (ThermoFisher) and Bioanalyzer (Agilent). Libraries were sequenced in paired end mode on a NextSeq550 instrument (Illumina) targeting a depth of 50,000–100,000 reads per cell.
Sequence alignment and sample aggregation
Raw sequences of 10× chromium single-cell 3′ v3 libraries were aligned to the custom references with 10× Genomics Cell Ranger 5.0.1. We further down-sampled and aggregated the matrices using 10× Genomics Cell Ranger 6.0.0.
Cell clustering and differential gene expression analysis
We used Seurat v3 toolkit66 for quality control and differential gene expression analysis. To remove poorly expressed genes, only genes expressed in at least five cells were kept. Cells with fewer than 750 genes were suspected to be debris and removed. To clean potential doublets, cells with more than 15,000 were also removed. Cells with more than 10% mitochondrial genes were suspected to be apoptotic and removed. The read counts were normalized using the SC transform procedure. Principal Component Analysis (PCA) was employed for dimensionality reduction, evaluating the highly variable genes and keeping the 50 most significant PCs. For sharing nearest neighbor (SNN) cluster analysis, the resolution was set to 1.2 and k parameter to 600, while mitochondrial genes were regressed out. Differentially expressed genes between clusters or treatments were identified using the Wilcox test. Significantly differentially expressed genes were defined as genes with adjusted p value of 0.05 or less and a log fold change of 0.25 or more. Enrichment for gene ontology terms were identified by g:Profiler67 and Enrichr68 using significantly upregulated genes in the relevant condition.
Data sharing statement
All modified mRNA (modRNA) vectors containing genes of interest noted in this paper will be made available to other investigators. My institution and I will adhere to the NIH Grants Policy on Sharing of Unique Research Resources including the “Sharing of Biomedical Research Resources: Principles and Guidelines for Recipients of NIH Grants and Contracts” issued in December 1999 (http://ott.od.nih.gov/NewPages/Rtguide_final.html). Specifically, material transfers will be made with no more restrictive terms than in the Simple Letter Agreement or the UBMTA and without reach-through requirements. Should any intellectual property arise which requires a patent, we would ensure that the technology remains widely available to the research community in accordance with the NIH Principles and Guidelines.
Statistical analysis
Statistical significance was determined by unpaired two-tailed t test, one-way ANOVA, Tukey’s multiple comparison test, one-way ANOVA, Bonferroni post hoc test, or log-rank (Mantel-Cox) test for survival curves, as detailed in the respective figure legends. p value < 0.05 was considered significant. All graphs represent average values, and values were reported as mean ± standard error of the mean. Unpaired two-tailed t test was based on assumed normal distributions. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; N.S, Not Significant.
Acknowledgments
The authors acknowledge Okino Tomotaka, Koji Shinozaki, Tomoyuki Bando, Tetsuya Sugiyama, and Matsushita Yuichiro for their help with this manuscript.
This work was mostly funded by a sponsor agreement given by Ono Pharmaceutical Co., Ltd. (Osaka, Japan) to the Zangi lab. It was also partially funded by a cardiology start-up grant awarded to the Zangi laboratory and by NIH grants R01 HL142768-01 and R01 HL149137-01. Y.H. and D.P. were supported by grants of the National Institute of Mental Health (NIMH) (R01-MH109715, D.P. and R21-MH105881, D.P).
Declaration of interests
L.Z. and K.K. are inventors of a Patent Cooperation Treaty application WO2021050877A1 (Compositions including molecules of modified mRNA and methods of using the same), which covers the results in this manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.07.014.
Supplemental information
References
- 1.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abad M., Hashimoto H., Zhou H., Morales M.G., Chen B., Bassel-Duby R., Olson E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Reports. 2017;8:548–560. doi: 10.1016/j.stemcr.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christoforou N., Chellappan M., Adler A.F., Kirkton R.D., Wu T., Addis R.C., Bursac N., Leong K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE. 2013;8:e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu J.D., Stone N.R., Liu L., Spencer C.I., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B.G., Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirai H., Katoku-Kikyo N., Keirstead S.A., Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc. Res. 2013;100:105–113. doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ifkovits J.L., Addis R.C., Epstein J.A., Gearhart J.D. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE. 2014;9:e89678. doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohamed T.M., Stone N.R., Berry E.C., Radzinsky E., Huang Y., Pratt K., Ang Y.S., Yu P., Wang H., Tang S. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M., Nakashima H., Akiyama M., Wada R., Inagawa K. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam Y.J., Lubczyk C., Bhakta M., Zang T., Fernandez-Perez A., McAnally J., Bassel-Duby R., Olson E.N., Munshi N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141:4267–4278. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J.A., DiMaio J.M., Baker L.A., Bassel-Duby R., Olson E.N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh V.P., Mathison M., Patel V., Sanagasetti D., Gibson B.W., Yang J., Rosengart T.K. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J. Am. Heart Assoc. 2016;5:5. doi: 10.1161/JAHA.116.003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Kaneda R., Suzuki T., Kamiya K. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamakawa H., Muraoka N., Miyamoto K., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Umei T., Akiyama M., Kuishi Y. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Reports. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y., Londono P., Cao Y., Sharpe E.J., Proenza C., O’Rourke R., Jones K.L., Jeong M.Y., Walker L.A., Buttrick P.M. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addis R.C., Ifkovits J.L., Pinto F., Kellam L.D., Esteso P., Rentschler S., Christoforou N., Epstein J.A., Gearhart J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Q., De Windt L.J., Witt S.A., Kimball T.R., Markham B.E., Molkentin J.D. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 19.Tamura M., Hosoya M., Fujita M., Iida T., Amano T., Maeno A., Kataoka T., Otsuka T., Tanaka S., Tomizawa S., Shiroishi T. Overdosage of Hand2 causes limb and heart defects in the human chromosomal disorder partial trisomy distal 4q. Hum. Mol. Genet. 2013;22:2471–2481. doi: 10.1093/hmg/ddt099. [DOI] [PubMed] [Google Scholar]
- 20.Sultana N., Magadum A., Hadas Y., Kondrat J., Singh N., Youssef E., Calderon D., Chepurko E., Dubois N., Hajjar R.J., Zangi L. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017;25:1306–1315. doi: 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magadum A., Singh N., Kurian A.A., Munir I., Mehmood T., Brown K., Sharkar M.T.K., Chepurko E., Sassi Y., Oh J.G. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation. 2020;141:1249–1265. doi: 10.1161/CIRCULATIONAHA.119.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simeonov K.P., Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS ONE. 2014;9:e100134. doi: 10.1371/journal.pone.0100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strelow A., Bernardo K., Adam-Klages S., Linke T., Sandhoff K., Krönke M., Adam D. Overexpression of acid ceramidase protects from tumor necrosis factor-induced cell death. J. Exp. Med. 2000;192:601–612. doi: 10.1084/jem.192.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadas Y., Vincek A.S., Youssef E., Żak M.M., Chepurko E., Sultana N., Sharkar M.T.K., Guo N., Komargodski R., Kurian A.A. Altering Sphingolipid Metabolism Attenuates Cell Death and Inflammatory Response After Myocardial Infarction. Circulation. 2020;141:916–930. doi: 10.1161/CIRCULATIONAHA.119.041882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadahiro T., Isomi M., Muraoka N., Kojima H., Haginiwa S., Kurotsu S., Tamura F., Tani H., Tohyama S., Fujita J. Tbx6 Induces Nascent Mesoderm from Pluripotent Stem Cells and Temporally Controls Cardiac versus Somite Lineage Diversification. Cell Stem Cell. 2018;23:382–395.e5. doi: 10.1016/j.stem.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islas J.F., Liu Y., Weng K.C., Robertson M.J., Zhang S., Prejusa A., Harger J., Tikhomirova D., Chopra M., Iyer D. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H.K., Kim H.S., Kim J.S., Kim Y.G., Park K.H., Lee J.H., Kim K.H., Chang I.Y., Bae S.C., Kim Y. CCL2 deficient mesenchymal stem cells fail to establish long-lasting contact with T cells and no longer ameliorate lupus symptoms. Sci. Rep. 2017;7:41258. doi: 10.1038/srep41258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali A., Akhter M.A., Haneef K., Khan I., Naeem N., Habib R., Kabir N., Salim A. Dinitrophenol modulates gene expression levels of angiogenic, cell survival and cardiomyogenic factors in bone marrow derived mesenchymal stem cells. Gene. 2015;555:448–457. doi: 10.1016/j.gene.2014.10.045. [DOI] [PubMed] [Google Scholar]
- 30.Sánchez-Luis E., Joaquín-García A., Campos-Laborie F.J., Sánchez-Guijo F., Rivas J.L. Deciphering Master Gene Regulators and Associated Networks of Human Mesenchymal Stromal Cells. Biomolecules. 2020;10:10. doi: 10.3390/biom10040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X., Wang Y., Meng C., Fang N. FMRP regulates endothelial cell proliferation and angiogenesis via the miR-181a-CaM-CaMKII pathway. Cell Biol. Int. 2018;42:1432–1444. doi: 10.1002/cbin.11039. [DOI] [PubMed] [Google Scholar]
- 32.Jiang B.G., Wan Z.H., Huang J., Li L.M., Liu H., Fu S.Y., Yang Y., Zhang J., Yuan S.X., Wang R.Y. Elevated ZC3H15 increases HCC growth and predicts poor survival after surgical resection. Oncotarget. 2016;7:37238–37249. doi: 10.18632/oncotarget.9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y., Ohta T., Mai M. Angiogenesis of AFP producing gastric carcinoma: correlation with frequent liver metastasis and its inhibition by anti-AFP antibody. Oncol. Rep. 2004;11:809–813. [PubMed] [Google Scholar]
- 34.Kikuchi R., Nakamura K., MacLauchlan S., Ngo D.T., Shimizu I., Fuster J.J., Katanasaka Y., Yoshida S., Qiu Y., Yamaguchi T.P. An antiangiogenic isoform of VEGF-A contributes to impaired vascularization in peripheral artery disease. Nat. Med. 2014;20:1464–1471. doi: 10.1038/nm.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haro H., Kato T., Komori H., Osada M., Shinomiya K. Vascular endothelial growth factor (VEGF)-induced angiogenesis in herniated disc resorption. J. Orthop. Res. 2002;20:409–415. doi: 10.1016/S0736-0266(01)00150-4. [DOI] [PubMed] [Google Scholar]
- 36.Robciuc M.R., Kivelä R., Williams I.M., de Boer J.F., van Dijk T.H., Elamaa H., Tigistu-Sahle F., Molotkov D., Leppänen V.M., Käkelä R. VEGFB/VEGFR1-Induced Expansion of Adipose Vasculature Counteracts Obesity and Related Metabolic Complications. Cell Metab. 2016;23:712–724. doi: 10.1016/j.cmet.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y., Linden P., Farnebo J., Cao R., Eriksson A., Kumar V., Qi J.H., Claesson-Welsh L., Alitalo K. Vascular endothelial growth factor C induces angiogenesis in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:14389–14394. doi: 10.1073/pnas.95.24.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rissanen T.T., Markkanen J.E., Gruchala M., Heikura T., Puranen A., Kettunen M.I., Kholová I., Kauppinen R.A., Achen M.G., Stacker S.A. VEGF-D is the strongest angiogenic and lymphangiogenic effector among VEGFs delivered into skeletal muscle via adenoviruses. Circ. Res. 2003;92:1098–1106. doi: 10.1161/01.RES.0000073584.46059.E3. [DOI] [PubMed] [Google Scholar]
- 39.Koblizek T.I., Weiss C., Yancopoulos G.D., Deutsch U., Risau W. Angiopoietin-1 induces sprouting angiogenesis in vitro. Curr. Biol. 1998;8:529–532. doi: 10.1016/s0960-9822(98)70205-2. [DOI] [PubMed] [Google Scholar]
- 40.Rosa A.I., Gonçalves J., Cortes L., Bernardino L., Malva J.O., Agasse F. The angiogenic factor angiopoietin-1 is a proneurogenic peptide on subventricular zone stem/progenitor cells. J. Neurosci. 2010;30:4573–4584. doi: 10.1523/JNEUROSCI.5597-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W., Guo S., Gonzalez-Perez R.R. Leptin pro-angiogenic signature in breast cancer is linked to IL-1 signalling. Br. J. Cancer. 2011;104:128–137. doi: 10.1038/sj.bjc.6606013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toft D.J., Rosenberg S.B., Bergers G., Volpert O., Linzer D.I. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc. Natl. Acad. Sci. USA. 2001;98:13055–13059. doi: 10.1073/pnas.231364798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viñals F., Pouysségur J. Transforming growth factor beta1 (TGF-beta1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF-alpha signaling. Mol. Cell. Biol. 2001;21:7218–7230. doi: 10.1128/MCB.21.21.7218-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaga T., Kawano H., Sakaguchi M., Nakazawa T., Taniyama Y., Morishita R. Hepatocyte growth factor stimulated angiogenesis without inflammation: differential actions between hepatocyte growth factor, vascular endothelial growth factor and basic fibroblast growth factor. Vascul. Pharmacol. 2012;57:3–9. doi: 10.1016/j.vph.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Wang C.Q., Huang Y.W., Wang S.W., Huang Y.L., Tsai C.H., Zhao Y.M., Huang B.F., Xu G.H., Fong Y.C., Tang C.H. Amphiregulin enhances VEGF-A production in human chondrosarcoma cells and promotes angiogenesis by inhibiting miR-206 via FAK/c-Src/PKCδ pathway. Cancer Lett. 2017;385:261–270. doi: 10.1016/j.canlet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Murakami M., Simons M. Fibroblast growth factor regulation of neovascularization. Curr. Opin. Hematol. 2008;15:215–220. doi: 10.1097/MOH.0b013e3282f97d98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta V.B., Besner G.E. HB-EGF promotes angiogenesis in endothelial cells via PI3-kinase and MAPK signaling pathways. Growth Factors. 2007;25:253–263. doi: 10.1080/08977190701773070. [DOI] [PubMed] [Google Scholar]
- 48.Lv S., Cheng G., Zhou Y., Xu G. Thymosin beta4 induces angiogenesis through Notch signaling in endothelial cells. Mol. Cell. Biochem. 2013;381:283–290. doi: 10.1007/s11010-013-1713-8. [DOI] [PubMed] [Google Scholar]
- 49.Xie H., Cui Z., Wang L., Xia Z., Hu Y., Xian L., Li C., Xie L., Crane J., Wan M. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat. Med. 2014;20:1270–1278. doi: 10.1038/nm.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee R.J., Springer M.L., Blanco-Bose W.E., Shaw R., Ursell P.C., Blau H.M. VEGF gene delivery to myocardium: deleterious effects of unregulated expression. Circulation. 2000;102:898–901. doi: 10.1161/01.cir.102.8.898. [DOI] [PubMed] [Google Scholar]
- 51.Kang J., Albadawi H., Patel V.I., Abbruzzese T.A., Yoo J.H., Austen W.G., Jr., Watkins M.T. Apolipoprotein E-/- mice have delayed skeletal muscle healing after hind limb ischemia-reperfusion. J. Vasc. Surg. 2008;48:701–708. doi: 10.1016/j.jvs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Anversa P., Olivetti G., Melissari M., Loud A.V. Stereological measurement of cellular and subcellular hypertrophy and hyperplasia in the papillary muscle of adult rat. J. Mol. Cell. Cardiol. 1980;12:781–795. doi: 10.1016/0022-2828(80)90080-2. [DOI] [PubMed] [Google Scholar]
- 53.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oka T., Maillet M., Watt A.J., Schwartz R.J., Aronow B.J., Duncan S.A., Molkentin J.D. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 55.Desjardins C.A., Naya F.J. The Function of the MEF2 Family of Transcription Factors in Cardiac Development, Cardiogenomics, and Direct Reprogramming. J. Cardiovasc. Dev. Dis. 2016;3:3. doi: 10.3390/jcdd3030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steimle J.D., Moskowitz I.P. TBX5: A Key Regulator of Heart Development. Curr. Top. Dev. Biol. 2017;122:195–221. doi: 10.1016/bs.ctdb.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.George R.M., Firulli A.B. Hand Factors in Cardiac Development. Anat. Rec. (Hoboken) 2019;302:101–107. doi: 10.1002/ar.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VanDusen N.J., Casanovas J., Vincentz J.W., Firulli B.A., Osterwalder M., Lopez-Rios J., Zeller R., Zhou B., Grego-Bessa J., De La Pompa J.L. Hand2 is an essential regulator for two Notch-dependent functions within the embryonic endocardium. Cell Rep. 2014;9:2071–2083. doi: 10.1016/j.celrep.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong X.H., Liu H., Wang S.J., Liang S.W., Wang G.G. Exosomes derived from SDF1-overexpressing mesenchymal stem cells inhibit ischemic myocardial cell apoptosis and promote cardiac endothelial microvascular regeneration in mice with myocardial infarction. J. Cell. Physiol. 2019;234:13878–13893. doi: 10.1002/jcp.28070. [DOI] [PubMed] [Google Scholar]
- 60.Leong Y.Y., Ng W.H., Ellison-Hughes G.M., Tan J.J. Cardiac Stem Cells for Myocardial Regeneration: They Are Not Alone. Front. Cardiovasc. Med. 2017;4:47. doi: 10.3389/fcvm.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lader J., Stachel M., Bu L. Cardiac stem cells for myocardial regeneration: promising but not ready for prime time. Curr. Opin. Biotechnol. 2017;47:30–35. doi: 10.1016/j.copbio.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beltrami A.P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S., Kasahara H., Rota M., Musso E., Urbanek K. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 63.Yoon Y.S., Wecker A., Heyd L., Park J.S., Tkebuchava T., Kusano K., Hanley A., Scadova H., Qin G., Cha D.H. Clonally expanded novel multipotent stem cells from human bone marrow regenerate myocardium after myocardial infarction. J. Clin. Invest. 2005;115:326–338. doi: 10.1172/JCI22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Z., Witman N., Wang W., Li D., Yan B., Deng M., Wang X., Wang H., Zhou G., Liu W. Cell-mediated delivery of VEGF modified mRNA enhances blood vessel regeneration and ameliorates murine critical limb ischemia. J. Control. Release. 2019;310:103–114. doi: 10.1016/j.jconrel.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 65.Ponemone V., Gupta S., Sethi D., Suthar M., Sharma M., Powell R.J., Harris K.L., Jungla N., Arambam P., Kaul U. Safety and Effectiveness of Bone Marrow Cell Concentrate in the Treatment of Chronic Critical Limb Ischemia Utilizing a Rapid Point-of-Care System. Stem Cells Int. 2017;2017:4137626. doi: 10.1155/2017/4137626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm226. W193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw377. W90-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.