Abstract
Breast cancer stem-like cells (BCSCs) play vital roles in tumorigenesis and progression. However, the origin and dynamic changes of BCSCs are still to be elucidated. Using the breast cancer mouse model MMTV-PyMT, we constructed a single-cell atlas of 31,778 cells from four distinct stages of tumor progression (hyperplasia, adenoma/MIN, early carcinoma and late carcinoma), during which malignant transition occurs. We identified that the precise cell type of ERlow epithelial cell lineage gave rise to the tumors, and the differentiation of ERhigh epithelial cell lineage was blocked. Furthermore, we discovered a specific signature with a continuum of gene expression profiles along the tumor progression and significantly correlated with clinical outcomes, and we also found a stem-like cell cluster existed among ERlow epithelial cells. Further clustering on this stem-like cluster showed several sub-clusters indicating heterogeneity of stem-like epithelial cells. Moreover, we distinguished normal and cancer stem-like cells in this stem-like epithelial cell cluster and profiled the molecular portraits from normal stem-like cell to cancer stem-like cells during the malignant transition. Finally, we found the diverse immune cell infiltration displayed immunosuppressive characteristics along tumor progression. We also found the specific expression pattern of cytokines and their corresponding cytokine receptors in BCSCs and immune cells, suggesting the possible cross-talk between BCSCs and the immune cells. These data provide a useful resource for illuminating BCSC heterogeneity and the immune cell remodeling during breast tumor progression, and shed new light on transcriptomic dynamics during the progression at the single-cell level.
Subject terms: Breast cancer, Cancer stem cells
Introduction
Breast cancer is one of the most malignant cancers that seriously threat women’s health and cause casualties [1] and a malignancy with a multistep pathological processes starting with the premalignant atypical ductal hyperplasia (ADH), followed by ductal carcinoma in situ (DCIS) and subsequent malignant invasive ductal carcinoma (IDC) [2]. The full spectrum of distinct cell types and their molecular characteristics during the breast cancer tumorigenesis remain to be well studied, especially at the single-cell level. The mammary tumors developed in MMTV-PyMT breast cancer mice mainly go through four stereotypical stages, including hyperplasia at 4 to 6 weeks of age, adenoma/mammary intraepithelial neoplasia at 8–9 weeks of age, early malignant between 8–12 weeks of age and late-malignant at 13 weeks later, respectively [3]. This progress in mouse model mirrors the pathological procession of human breast cancer patients, and is comparable to human breast diseases classified as benign or in situ proliferative lesions to invasive carcinomas [3].
Breast cancer stem-like cells (BCSCs) are a rare subpopulation of tumor cells characterized with strong tumorigenic capacity. A serial of evidence supported BCSCs as the origin of breast cancer [4–6]. Recent investigations revealed that BCSCs are clinically, molecularly, and biologically heterogeneous [7–9]. However, accumulating evidence has shown that the heterogeneity of BCSCs based on the limited known markers is underestimated, suggesting the existence of more subsets of BCSCs [7]. The single-cell RNA-sequencing technology (scRNA-seq), which emerged in recent years, has played an increasingly important role in biological research [10–12]. Nowadays, scRNA-seq has been widely used in the research of tumor heterogeneity, immune microenvironment, neuroscience, embryonic development, cell differentiation, and others [13,14]. Furthermore, scRNA-seq has proved its power in revealing rare subpopulations [15–17].
Here, we provided the transcriptome analysis of 31,778 single cells including epithelial and immune cells from four different tumor progression stages of MMTV-PyMT breast cancer mouse model. We identified that the precise cell type of ERlow epithelial cell lineage gave rise to the tumors. We also characterized the stem-like cell cluster, and further clustering on this stem-like cell cluster showed several sub-clusters, indicating heterogeneity of stem cells. These results provided evidence that BCSCs are transcriptionally and functionally heterogeneous at the single-cell level.
Results
Single-cell transcriptome charted cell heterogeneity in MMTV-PyMT mouse mammary glands
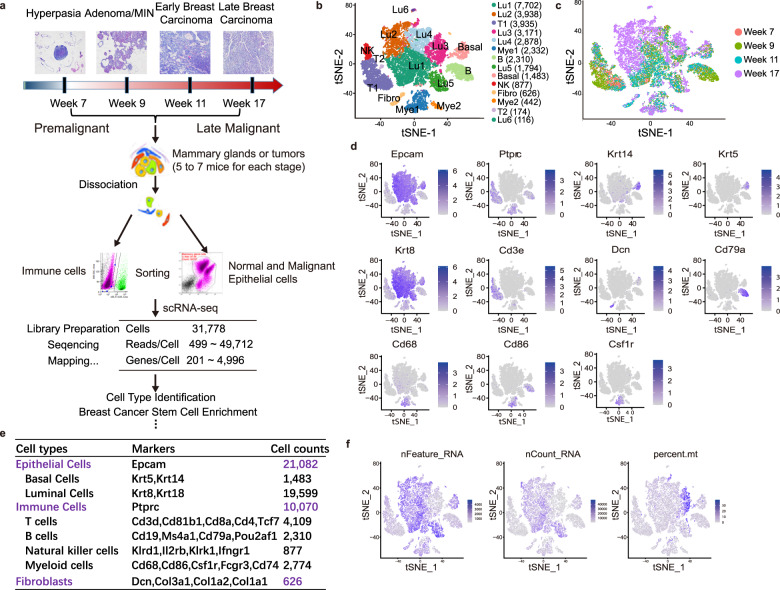
To characterize the single-cell transcriptome dynamics of MMTV-PyMT breast cancer mouse model during tumorigenesis, a total of 31,778 isolated single cells were obtained from mouse mammary glands or tumors, which spanned the cascade from hyperplasia to late breast carcinoma including premalignant, early malignant, and malignant stages. Then we obtained bulk transcriptomics from W07 (Week 7), W09 (Week 9), W11 (Week 11), and W17 (Week 17) mammary glands or tumors of MMTV-PyMT mouse model (Fig. 1a; Fig. S1a).
Fig. 1. Single-cell analysis of mammary glands and tumors from MMTV-PyMT mice.
a A schematic diagram highlighting the workflow including isolation and sequencing of single cells for this study. Single cells were prepared from the 4th pair of mammary glands of MMTV-PyMT mice at different tumor progression stages including hyperplasia, adenoma/mammary intraepithelial neoplasia (MIN, early carcinoma) and malignant tumors (late carcinoma). The transcriptome of single cells was sequenced using the 10x Chromium system. b The t-SNE plot of 31,778 single cells to visualize cell-type clusters based on the expression of known markers, Lu1, 2, 3, 4, 5, and 6: luminal epithelial cell cluster 1, 2, 3, 4, 5, and 6; Basal: basal epithelial cell cluster; Fibro: fibroblast cluster; T1 and 2: T cell cluster 1 and 2; B: B cell cluster; NK: natural killer cell cluster; Mye1 and 2: myeloid cell cluster 1 and 2. c The t-SNE plot of single-cell transcriptomes from the isolated cells at indicated tumor progression stages including week 7, week 9, week 11, and week 17. d The individual gene t-SNE plots showing the expression levels and distribution of representative markers of known cell types, which distinctly separates epithelial cells, immune cells and fibroblasts in the PyMT mouse mammary glands. e A table showed the markers used to annotate the known cell types, including epithelial cells, immune cells and fibroblasts in the PyMT mouse mammary glands. f The number of expressed genes (nFeature_RNA), the distribution of library size (nCount_RNA) and percentage of mitochondrial genes (percent.mt).
A total of 14 clearly separated cell clusters were finally identified (Fig. 1b, c). Based on the expression of known markers, we found that the cells are comprised mainly of clusters of epithelial cells, immune cells, and fibroblasts (Fig. 1d, e). As expected, the epithelial cells were largely basal epithelial cells and luminal epithelial cells, consistent with the cellular characteristics of mammary glands [18, 19]. In total, 19,599 luminal cells were clustered into six separate subsets (Lu1, Lu2, Lu3, Lu4, Lu5, and Lu6) (Fig. 1b). Lu2 and Lu4 were cancer cells, since they emerged almost uniquely in the late carcinoma of week 17, which were consistent with the previous reports that the PyMT mouse model has been characterized as most similar to the luminal B molecular subtype [20] (Fig. 1c). The immune cells comprised subsets of T cells including T1 and T2, B cells and myeloid cells including Mye1 and Mye2 (Fig. 1e). We also noticed that the number of expressed genes (nFeature_RNA), library size (nCount_RNA) and percentage of mitochondrial genes (percent.mt), the three commonly used quality controls in single-cell RNA sequencing, were also clearly distinguished between immune cells and epithelial cells (Fig. 1f). Since the sequencing quality of immune and epithelial cells was different, we determined to filter out low-quality cells by different criteria (Fig. S1b). After filtration, 12,039 epithelial cells and 8,954 immune cells were used for further analysis (Fig. S1c, d).
ERlow luminal epithelial cells gave rise to the tumor cells during breast tumorigenesis
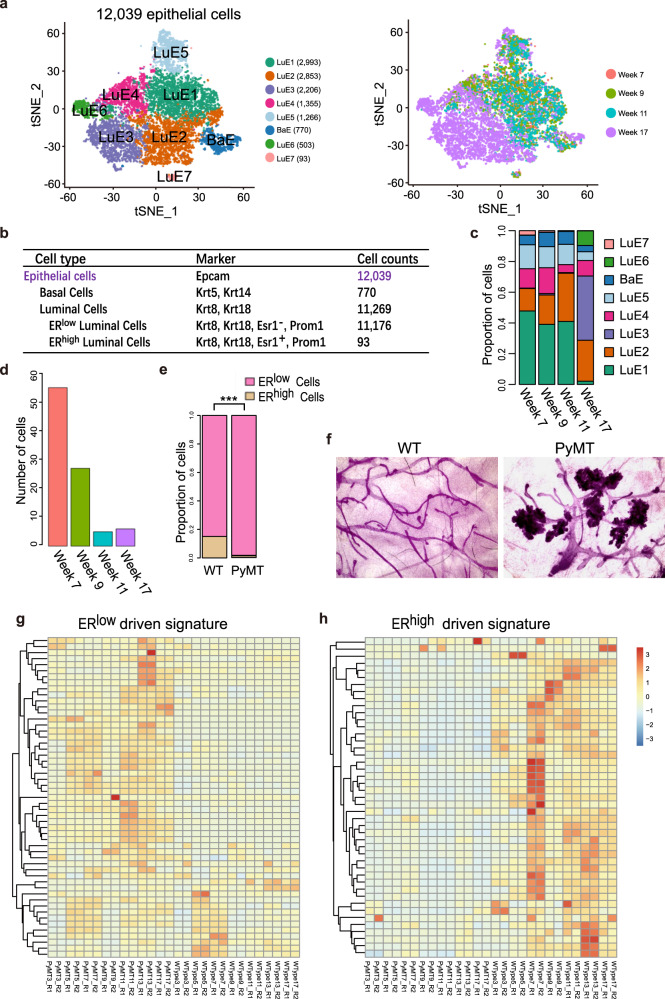
Breast cancer cells originate from epithelium in MMTV-PyMT mouse model, however, which subset of epithelial cells they originate from and the dynamic changes during tumorigenesis are still unknown. Thus, we re-clustered the filtrated 12,039 epithelial cells and obtained 8 clusters mainly comprised 7 luminal cell clusters termed LuE1, 2, 3, 4, 5, 6, and 7 (LuE, Luminal Epithelial cells) and one basal cell cluster termed BaE (BaE, Basal Epithelial cells) (Fig. 2a). 87.9% cells from 17 weeks were clustered together to form cluster LuE2, LuE3, LuE4, and LuE6. On the other hand, 69.7%, 62.3%, and 62.8% cells corresponding from 7, 9, and 11 weeks were clustered together to form cluster LuE1, LuE5, BaE, and LuE7, respectively (Fig. 2a–c). Cells were clearly separated between these two groups of clusters, indicating the unique transcriptional profile for the late carcinoma stage.
Fig. 2. Tumor cells originated from ERlow luminal cells during tumorigenesis.
a The t-SNE plot of 12,039 epithelial cells from the MMTV-PyMT mammary glands colored by cell types (left panel) and stages (right panel), week 7 (hyperplasia), 1,930 cells; week 9 (Adenoma/MIN), 2,886 cells; week 11 (early carcinoma), 2,038 cells; week 17 (late carcinoma), 5,185 cells. LuE1, 2, 3, 4, 5, 6, and 7: luminal epithelial cluster 1, 2, 3, 4, 5, 6, and 7; BaE: basal epithelial cluster. b Summary of cell counts and markers used for the identification of epithelial cell subsets from the mouse mammary glands and tumors. c The proportion of epithelial cell sub-clusters in different stages of tumor progression. d The number of ERhigh epithelial cells across different stages of tumor progression dramatically decreased. e The ratio of ERlow and ERhigh luminal epithelial cells in the mammary glands of MMTV-PyMT mice increased when compared to wild-type mice (WT), which suggests that the normal differentiation of ERhigh cell lineage was blocked, resulting in the accumulation of ERlow cancer cells. f Whole mounts of mammary glands from wild-type (WT) and MMTV-PyMT mice indicated that cancer cells mainly exist in location of alveolar cells. g Gene expression profile of ERhigh luminal signature was upregulated in epithelial cells from WT mice when comparing with epithelial cells from MMTV-PyMT mice. h ERlow luminal signature was highly expressed in MMTV-PyMT mouse epithelial cells relative to WT mice.
Previous studies have shown that luminal progenitor cells can develop into either ERlow or ERhigh luminal progenitor cells, and subsequently differentiate into alveolar and ductal luminal cells [18]. We identified the cluster LuE7 as ERhigh luminal cell cluster by the expression of Esr1 (Fig. 2b; Fig. S2a). We witnessed the percentage of ERhigh luminal cells (LuE7) was decreased along the tumor progression (Fig. 2d) and there was a huge imbalanced percentage distribution of ERlow and ERhigh luminal cells (Fig. 2e). 99.2% luminal cells were belonged to ERlow luminal cells. In order to distinguish the possibility of imbalanced distribution of ERlow and ERhigh luminal cells due to the natural development, we compared the percentage of ERlow cells between MMTV-PyMT and wild-type FVB mice using the published dataset [21]. We identified ERlow and ERhigh luminal cells (Fig. S2b, c) and counted the cell number in luminal epithelial cell clusters (Fig. S2d). ERlow cells were significantly enriched in our data while ERhigh cells were depleted from MMTV-PyMT mouse when comparing to the FVB mouse data (Fig. 2e). Hence, the developmental process from luminal progenitor to ERhigh luminal cells was blocked during tumorigenesis. Since the ERlow subpopulation are the major luminal cells, we proposed that the cancer cells in MMTV-PyMT mouse were mainly ERlow cells and derived from ERlow luminal progenitor cells. It is clear that the tumors are originated from the site of alveolar cells, which are reported as ERlow cells prepared for milk secretion once corresponding conditions and hormone stimulation exist (Fig. 2f). We compared the differentially expressed genes between the ERhigh and ERlow luminal cells and defined the genes upregulated in ERhigh luminal cell population as ERhigh signature and the genes upregulated in ERlow luminal cell population as ERlow signature, respectively. Besides, we included several genes reported as marker genes for ERhigh luminal cell by previous studies into the ERhigh signature such as Esr1, Foxa1, Gata3, Pgr, ect [22] (Table S1). Then we investigated the expression profile of these two signatures in bulk RNA-seq from matched MMTV-PyMT and wild-type FVB mice. Most of ERlow signatures were highly expressed in MMTV-PyMT mammary tissues, while most of ERhigh signatures were highly expressed in FVB counterparts (Fig. 2g, h). It was consistent with the imbalanced percentage distribution of ERlow and ERhigh luminal cells.
Taken together, our data suggested that the specific cell-type ERlow luminal cells gave rise to tumors, and the differentiation of the ERhigh cell lineage was blocked. The accumulation of proliferative ERlow was the fundamental origin of the cancer cells in MMTV-PyMT mouse.
A continuum of gene expression profiles revealed the malignant transition of ERlow luminal cells in the mammary glands
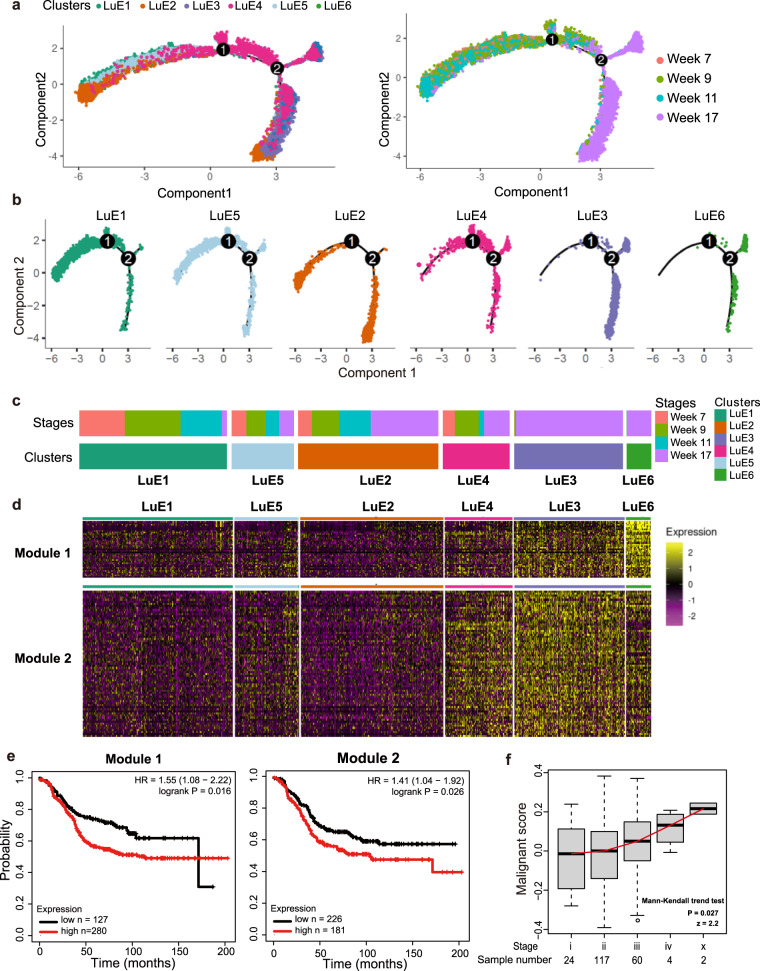
Since we determined that the cancer cells originated from ERlow luminal cells, we then focused on the dynamic changes of these epithelial cells along the transitions from hyperplasia to late carcinoma. To this end, we constructed the single-cell trajectories to trace the ERlow luminal epithelial hierarchy during tumor progression (Fig. 3a). The genes used to construct single-cell trajectories was listed in Table S2. The location of each cell cluster showed a unique pattern on single-cell trajectories. The majority of cells from cluster LuE3 and LuE6 were located on one side of single-cell trajectories, while the cells from cluster LuE1 and LuE5 were mainly located on the other side. Moreover, cells from cluster LuE2 and LuE4 showed an intermediate state (Fig. 3b), suggesting the trajectories of the tumor progression along cluster LuE1, LuE5, LuE2, LuE4, LuE3, and LuE6. Indeed, the cluster of the trajectories was positively correlated with the percentage of cells from week 17, which are mainly cancer cells (Fig. 3c). To gain a more detailed view of gene expression pattern changes over the critical tumor progression and malignant transition, we performed weighted correlation network analysis among the different clusters to explore the specific gene expression pattern and we identified panels of specific signatures on the chronologic tumor progression, which matched the pattern derived from t-SNE plot and single-cell trajectories [23]. The expression of the genes in the modules was gradually increased from left to right along the clusters, composing elevated proportion of cells from week 17 (Fig. 3d). The genes in these two modules were listed in Table S3. Next, we found the significant correlation between genes from these two modules and the clinical outcome of Luminal B breast cancer patients in survival analysis, and patients with these highly expressed genes showed worse prognosis (Fig. 3e). Among these genes, we identified several related to tumor progression as reported in previous studies were also consistent with this pattern (Fig. S3a). Since these genes in both signatures were upregulated along the tumor progression with the similar pattern, we merged these two gene signatures and termed the combined signature as the malignant signature. We also explored the expression profile of malignant signature among different tumor grades in luminal B patient samples from The Cancer Genome Atlas (TCGA) [24]. The results showed that the malignant score was significantly increased in the higher tumor grades (P = 0.027) (Fig. 3f).
Fig. 3. A continuous change of specific gene signature in ERlow luminal cells was correlated with the tumor progression and clinical outcome.
a Pseudo-time trajectory analysis of the six sub-clusters of luminal epithelial cells annotated by clusters (left) and tumor progression stages (right). b Pseudo-time trajectory of cells in each luminal epithelial cell cluster. c The heatmap showed the sample composition of each cluster, indicating that the composition of week 17 was gradually increased along cluster LuE1, LuE5, LuE2, LuE4, LuE3, and LuE6. d Heatmap of expression profile of signature genes in indicated modules identified by WGCNA (Weighted Gene Co-expression Network Analysis). A summary list of genes associated with corresponding WGCNA module were shown in supplementary Table 3. e The Kaplan–Meier relapse free survival curves of patients were grouped by the gene signatures in WGCNA modules. The left corresponded to the module 1 and the right corresponded to the module 2. f Boxplot depicted the distribution of malignant score derived from luminal B patient samples in TCGA. Mann-Kendall trend test was performed using the median value of each stage by “trend” package in R.
Identification of BCSCs in ERlow luminal cells
There is increasing evidence to suggest that diverse solid tumors are hierarchically organized and may be sustained by distinct subpopulations of cancer stem-like cells (CSCs) [25, 26]. Here, we first characterized the heterogeneity of BCSCs at the single-cell level and then characterized the feature of these cells.
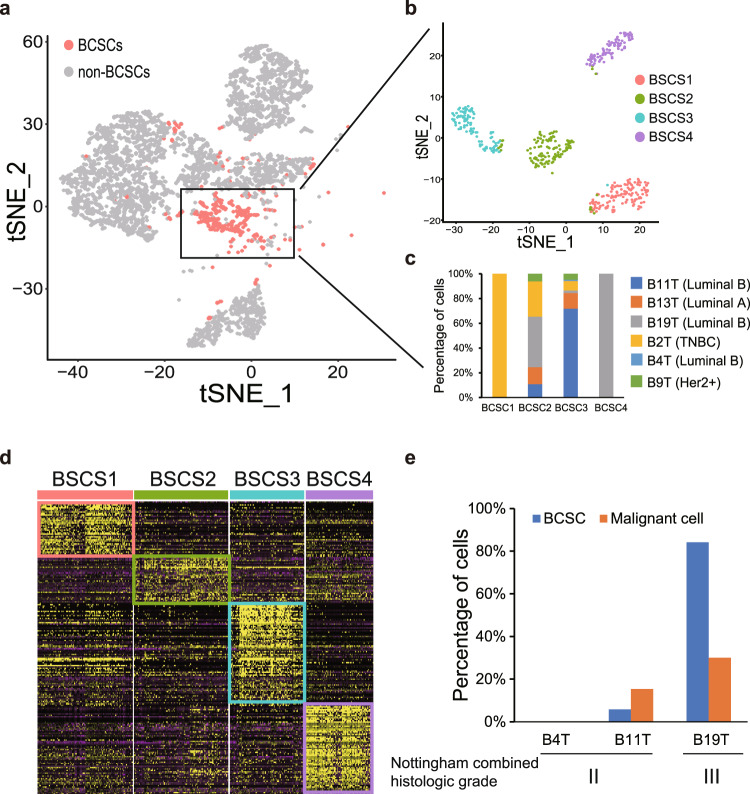
We searched the BCSC-related genes published previously, such as Cd24a, Itgb1, Itga6, Procr, etc in the literature and incorporated these genes into the BCSC signature (Fig. S3b). We scored each ERlow epithelial cell based on the BCSC signature by Gene Set Variation Analysis (GSVA) algorithm [27]. The expression profile of BCSC signature clearly showed a higher expression level on cluster LuE2 (Fig. 4a). Indeed, the score from GSVA algorithm was consistent with the expression profile (Fig. 4b). Since the cluster LuE2 was a mixture of cells from all four tumor progression stages, it is reasonable to assume that cluster LuE2 might include both normal and cancer cells. Next, we investigated the stemness score at different tumor progression stages and found no significant difference among these stages (Fig. S3c, d), suggesting the similarity of stemness in cluster LuE2 among tumor progression stages and the gene expression shared by both normal breast stem-like cells and BCSCs.
Fig. 4. Identification and characterization of cancer stem-like cell cluster in ERlow luminal cells.
a Heatmap of stem cell marker gene expression profile in each cell cluster (up panel) and GSVA score for each cell in each cell cluster (bottom panel). b A bar-plot showed the average GSVA score for each cell cluster. c The t-SNE plot showed the distribution of cancer stem-like cells enriched cluster LuE2 (orange, n = 2,206 cells) within the atlas. d The enlarged t-SNE plot highlighted the cells in cluster LuE2 colored by cancer signature score. e The proportion of BCSCs among cancer cells was defined by the median value of cancer signature score. Cancer cells were defined as the cells with cancer signature score higher than the median value. BCSCs were the cancer cells in LuE2. f Upregulated genes identified from our analysis were confirmed in CD24+CD29+ BCSCs from MMTV-PyMT mice by qPCR, compared to non-CD24+CD29+ BCSCs. The P was calculated by t-test. g The proportion of cancer stem-like and normal stem-like cells defined by the median value of cancer signature score. Cancer stem-like cells were defined as the cells with cancer signature score higher than the median value. Normal stem-like cells were defined as the cells with cancer signature score lower than the median value. h Overlap of the genes between cluster LuE2 marker genes and the differential expressed genes among normal cells and cancer cells in cluster LuE2 were shown by the unsupervised clustering heatmap. i Function enrichment analysis for the genes upregulated in cancer stem-like cells in Fig. 4h. j Function enrichment analysis for the genes upregulated in normal stem-like cells in Fig. 4h.
In order to distinguish between the normal and cancer cells, we extracted the up-/down-regulated genes comparing between bulk RNA-seq of PyMT and wild-type FVB mouse at week 17 as cancer signature genes to identify normal and cancer cells and the cancer signature genes can be found in Table S4. The results clearly showed a spatial pattern of normal and cancer cells on the t-SNE plot (Fig. 4c, d). The percentage of BCSCs among cancer cells was increased along the tumor progression which is consistent with previous studies (Fig. 4e) [28]. In order to confirm our analysis of the BCSC cluster, we identified the upregulated genes in the BCSC cluster compared to the non-BCSC cluster and picked four of these genes for the verification by qPCR. The genes upregulated in the BCSC cluster can be viewed in Table S5. The results showed that these four genes were significantly upregulated in CD24+CD29+ tumor cells from MMTV-PyMT mice, which was compared with non-CD24+CD29+ tumor cells (Fig. 4f). The upregulated genes identified from our analysis were also upregulated in traditionally defined BCSCs, indicating the similarity of these two groups of BCSCs.
Our studies showed that the BCSCs among all stem-like cells were also enriched in the last stage during tumor progression (Fig. 4g). We identified genes showing specific expression pattern between normal and cancer stem-like cells, providing as novel potential biomarkers to mark cancer stem-like cells (Fig. 4h and Table S6). The functional enrichment analysis revealed that these differentially expressed genes were enriched in the various important pathways including TP53, EMT and apoptosis pathways (Fig. 4i, j). In summary, our data revealed both the similarity and dissimilarity of gene expression between normal and cancer stem-like cells.
BCSCs were heterogeneous with different biological function and transcription regulation
It’s well known that tumors are highly heterogeneous [5, 29, 30]. Recent studies reported that the plasticity and heterogeneity are features of CSCs [31–36], but the specific heterogeneous BCSC populations and related markers are still unclear. In order to reveal the heterogeneity of BCSCs, we re-clustered the stem-like cell cluster LuE2 in a more fine-scale into five sub-clusters C1–C5 (Fig. 5a, b). We also checked the composition of cells from each tumor progression stage in the t-SNE plot and found that sub-clusters C1 and C2 existed across week 7, 9, and 11, and were mainly consisted of normal stem-like cells. However, sub-cluster C3, C4 and C5 existed almost mainly at week 17 and were mainly consisted of BCSCs (Fig. 5b, c). There was a clear separation between normal stem-like cells and BCSCs. To illuminate the possible evolution from the normal stem-like cell clusters to BCSC clusters, we also checked the single-cell trajectories and the inferred cell trajectory suggested a branched structure with sub-clusters from week 7, 9, and 11 positioned at the opposite end of the sub-clusters from week 17 (Fig. 5d). Next, we explored marker genes that were uniquely expressed in each sub-cluster (Fig. 5e), and performed functional enrichment analysis by Enrichr [37, 38] and the results showed specific functions were enriched in each sub-cluster, suggesting the functional heterogeneity in BCSCs. For example, genes uniquely expressed in C3 sub-cluster were enriched in cell migration and angiogenesis pathways (Fig. 5f). Then we investigated the transcriptional regulons based on cis-regulatory motif over the different clusters utilizing SCENIC analysis [39] and identified serials of transcription factors (TFs) correlated with each sub-cluster (Fig. 5g). Among these TFs, we found various TFs functioned in cancer/normal stem-like cells such as Sox4, Foxo3 and Myc, indicating their potential regulation on BCSC heterogeneity [40–42].
Fig. 5. The heterogeneity of stem-like cells.
a, b t-SNE plot demonstrated the separation of sub-clusters C1, C2, C3, C4, and C5 in LuE2 cluster colored by sub-clusters (a) and by tumor progression stages (b). c A bar-plot depicted the composition of cells from different tumor progression stages in each sub-cluster. d Pseudo-time analysis showed the single-cell trajectories for LuE2 cluster colored by sub-clusters (top panel) and tumor progression stages (bottom panel). e A heatmap showed the expression profile of marker genes from each sub-cluster. f A heatmap showed the function enrichment for each sub-cluster. Significant enrichment was colored by red. g A heatmap of regulon scores from SCENIC (Single Cell rEgulatory Network Inference and Clustering) analysis. Rows, Individual regulons. Columns, cells organized according to re-clustering of cluster LuE2.
The heterogeneity of BCSCs was confirmed in breast cancer patients
In order to verify the heterogeneity of BCSCs observed in mouse scRNA-seq data, we collected tumor samples from six breast cancer patients which were classified as luminal A, luminal B, TNBC and Her2+ subtypes and performed the scRNA-seq analysis (Table S7). Totally, we obtained 8,990 cells after quality control and performed clustering to define the population structure. 13 clusters were identified including epithelium, immune, stroma, endothelial and other cell clusters (Fig. S4a) [43]. We extracted epithelium cells and scored each epithelial cell cluster based on the BCSC gene signature. The EP8, EP11 and EP12 were identified as BCSC clusters (Fig. S4b, c). The cells from these three clusters were extracted and re-clustered (Fig. 6a), and the t-SNE plot showed the cells were clearly separated into four groups which we named as BCSC1, BCSC2, BCSC3, and BCSC4 (Fig. 6b). Almost all cells in BCSC1 and BCSC4 were from B2T (TNBC) and B19T (Luminal B), respectively. On the other hand, the BCSC2 and BCSC3 were heterogenous clusters comprising the cells from all six patients (Fig. 6c). Furthermore, the cells in each BCSC cluster express a set of specific genes, reflecting the diversity of BCSCs (Fig. 6d).
Fig. 6. The heterogeneity of stem-like cells in breast cancer patient tumors.
a The t-SNE plot highlighted the distribution of cancer stem cells from breast cancer patients. Cancer stem cells were highlighted in color and normal cancer cells were in gray. b Re-clustering of cancer stem cells by Seurat. c Histogram showed the composition of cancer stem cells from each patient. d Heatmap demonstrated the genes specifically expressed in each BCSC cluster. e Histogram showed the percentage of BCSCs and malignant cells in each patient.
Finally, we extracted the epithelium cells from three luminal B patients and evaluated the BCSC and malignant level by scoring based on the BCSC and malignant gene signature (Fig. S4d, e). We selected the clusters with high BCSC and malignant scores and summarized the percentage of cells within the BCSC and malignant clusters. The percentage of BCSC and malignant cells were very low/moderate in B4T/B11T patients. In contrast, both the percentage of BCSCs and malignant cells was high in B19T patient (Fig. 6e). Patient with higher percentage of BCSCs and malignant cells got higher score for histological grade which indicated worse clinical outcome [44].
BCSCs cross-talked with the immune cells through cytokine signals to promote tumor progression
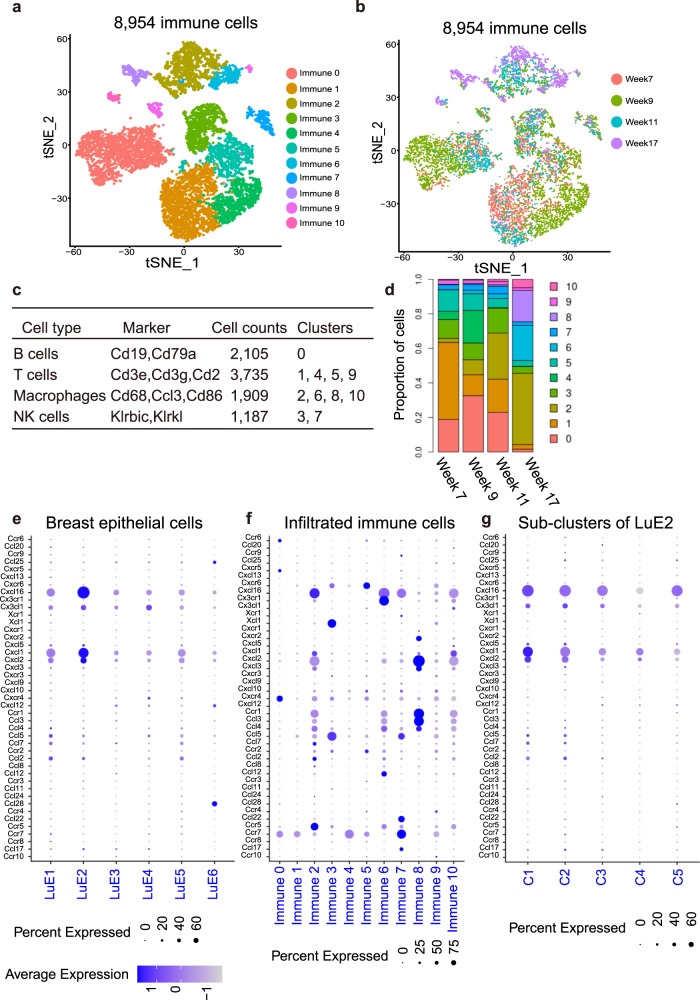
BCSCs are critically regulated by the surrounding microenvironment, especially immune cells [45]. In order to explore the possible cross-talk between the BCSCs and the immune cells, we firstly identified and annotated various immune cell types and the dynamic changes along breast tumor progression in MMTV-PyMT mice. By re-clustering the immune cells, we obtained 11 clusters (Fig. 7a, b), which came from four categories including T cells, B cells, macrophages and natural killer cells (NK cells) (Fig. 7c; Fig S5a–d). We observed that the proportion of the immune cells including B cells, T cells, NK cells, decreased along the tumor progression (Fig. 7d). On the other hand, in clusters 6 and 8, macrophages were almost specifically shown in week 17. To systematically study the interactions between cancer stem-like cells and immune cells, we used the known repository of cytokine and cytokine-receptor interacting pairs that account for the interactions and considered the expression levels of ligands and receptors within each cell type. We found that cancer stem-like cell cluster LuE2 highly expressed Cxcl1, while the immune cells from cluster 8 defined as macrophages specifically highly expressed the corresponding receptor, Cxcr2 (Fig. 7e–f). Furthermore, LuE2 highly expressed Cxcl16 and T cell cluster 5 specifically highly expressed its receptor, Cxcr6 (Fig. 7e, f). Notably, when investigating the expression of the cytokine Cxcl16 and Cxcl1 in LuE2, we found that Cxcl16 showed relatively high expression in C1 (Fig. 7g). The vast majority of cells in C1 were from weeks 7, 9 and 11, indicating a strong immune response in the early stage of tumor development. This result was consistent with significant enrichment of immune functions such as activation of immune response, cytokine-mediated signaling pathway and cytokine production pathways in sub-cluster C1 of LuE2 shown in Fig. 5f. These studies indicated that BCSCs secreted different levels of cytokines among five sub-clusters and functioned through corresponding receptors on immune cells to promote tumor progression.
Fig. 7. The cross-talk between BCSCs and immune cells during tumor progression.
a, b The t-SNE plot showed the distribution of the immune cells from different clusters supervised by Seurat (a) and different tumor progression stages (b) in the atlas. c A table showed the markers used to annotate immune cells and cell accounts of the corresponding cell type. d Dynamic changes of the proportion of the immune cell types spanning from premalignant to late carcinoma. e–g Dot plots showed the expression of paired cytokine and cytokine receptors in ERlow clusters, immune clusters and sub-clusters of LuE2, which indicated that cytokine Cxcl1 and Cxcl16 were highly expressed in sub-clusters of LuE2 and the corresponding receptors Cxcr2 and Cxcr6 were highly expressed in macrophages and T cells.
Discussion
Taking advantage of scRNA-seq, we depicted the dynamic changes of transcriptional profile for epithelial and immune cells isolated from the mammary glands or tumors of MMTV-PyMT mouse model. To our knowledge, this is the first study to define a single-cell atlas of epithelial and immune cells during tumor progression in MMTV-PyMT mouse model.
The traditional method to isolate BCSCs is sorting by FACS based on cell-surface markers, but these BCSCs are usually a mixture of BCSC populations [6, 36]. Taking advantage of scRNA-seq combined with GSVA scoring, we identified the subpopulations of BCSCs and revealed the heterogeneity of BCSCs along the tumor progression. Re-clustering of cells from stem-like cell cluster LuE2 also confirmed the heterogeneity of stem-like cells.
Functional analysis showed that different stem-like cell sub-clusters enriched some unique functions such as the functions related to immune response for these sub-clusters, which may reflect the functional heterogeneity of cancer stem-like cells along tumor progression. These functions were uniquely enriched in the sub-cluster with late developmental stage reflected unique requirement for cancer stem-like cells. It will be helpful to gain insight into the functional requirement for cancer stem-like cells.
Evidence showed that BCSCs contribute to educate and reconstitute the immune microenvironment [46]. In our study, we found that the cancer stem-like cell cluster LuE2 secreted the specific cytokine Cxcl16 and Cxcl1. The cytokines can function through affecting and cross-talking with the corresponding immune cells. On the other hand, increasing evidence have demonstrated the role of immune microenvironment in the generation and maintenance of BCSCs [47]. Here, we revealed the dynamic changes of BCSC populations, where the proportion of immune cell types T, B, and NK cells decreased and the proportion of macrophages increased, which provided benefits for survival of the BCSCs.
Our findings provide unparalleled insight into the cellular heterogeneity of breast cancer with different types of premalignant lesions and malignant lesions, which may be helpful for identifying markers for cancer prevention and facilitate our understanding of breast cancer pathogenesis. Furthermore, our findings on the cross-talk of BCSCs and immune cells provide thoughts for the combination of immunotherapy and cancer stem cell targeted therapy for precise medicine.
Methods and materials
Breast cancer patient tumor tissues
All the breast cancer patient tumor tissues were obtained from Shanghai Cancer Hospital affiliated with Fudan University. An informed consent was obtained from all the involved patients, and the study was approved by the institution’s ethics committee (Fudan University Shanghai Cancer Center Institutional Review Board, 050432-4-1212B) (Shanghai, China).
Mice and tissue collections
MMTV-PyMT and wild-type FVB mice were housed in standard animal cages under specific pathogen-free conditions in the Department of Laboratory Animal Science of Fudan University. Animal experiments were approved according to the experimental animal guidelines of the Care and Use of Laboratory Animals of Fudan University and approved by the Fudan University Shanghai Cancer Center Institutional Review Board (JS-082).
Mammary glands or tumors from 7-week old, 9-week old, 11-week old and 17-week old MMTV-PyMT FVB mice were excised, dissected and minced into small pieces and then resuspended with collagenase-hyaluronidase digestion reagent (Catalog #07912, STEMCELL Technologies, USA). Mammary glands or tumors for each stage were from 5 to 7 mice (seven mice for the first and second stages, and five mice for the third and fourth stages). The number of mice used in each experimental group was determined by power analysis and on the basis of prior experience with animal models. No mouse was excluded from the analyses and No randomization of mice was needed in this study. This study included a lot of complicated experimental design, the researchers were limited, and the feasibility of blinding was poor, thus blinding was not efficiently applied.
Tissue pieces were digested for approximately 1 hr at 37 °C and shaken once every 15 min. Cell aggregates were removed by filtering cell suspension with 40 μm filter. Cell suspensions were centrifuged at 1200 rpm for 5 m and resuspended for subsequent experiments.
Fluorescent-activated cell sorting (FACS)
Dissociated cells from the mammary glands or tumors of MMTV-PyMT mice were suspended in FACS buffer containing anti-mouse cell lineage antibody cocktails: CD45 (1:50, 555483, BD), CD31 (1:50, 555446, BD), CD140b (1:50, 558821, BD), anti-CD24 (1:50, 138506, BioLegend, USA) and anti-CD29 (1:80, 102226, BioLegend). A MoFlo Astrios instrument (Beckman Coulter, Brea, USA) was used for sorting. Data acquisition and analysis were performed using Summit software.
Dissociated single cells were separately sorted by fluorescence-activated cell sorting (FACS) based on specific cell-surface markers. We used antibodies against endothelial marker CD31, b1-integrin CD29, heat-stable antigen CD24, hematopoietic marker CD140b and CD45 antigens to gate on the CD31−CD140b−CD45− (Lineage−, Lin−) epithelial cell population including both normal and tumor epithelial cells and the CD31−CD140b−CD45+ immune cell population. Then, we defined four distinct Lin- epithelial cell subpopulations based on the expression of CD29 and CD24. The rare CD29-CD24- population was excluded from the Lin- subpopulation since it was reported as the possible stromal population [19].
Library preparation and sequencing for mouse sample
Single-cell sequencing was constructed using the 10x Genomics Chromium platform for droplet-enabled scRNA-seq according to the manufacturer’s instructions. Library generation was performed following the Chromium Single Cell 3′ Reagents Kits version 2 user guide in order to capture 5000 cells to 10000 cells/chip position (CG00052 Rev B). All the remaining procedures including the library construction were performed according to the standard manufacturer’s protocol. Each library was sequenced on the Illumina HiSeq 4000 platform to achieve an average of 48,488 reads per cell.
The raw data was processed by the Cell Ranger Single-Cell Software Suite (release 2.0), including using “cellranger mkfastq” to demultiplexes raw base call files into fastq-format files and then using “cellranger count” to perform reads alignment, filtering, barcode counting, and UMI counting. The reads were aligned to the mm10 reference genome using a pre-built annotation package downloaded from the 10X Genomics website. The output from different lanes was eventually aggregated using “cellranger aggr” with default parameters.
Library preparation and sequencing for human sample
The fresh tumor tissue was stored in the GEXSCOPETM Tissue Preservation Solution (Singleron) and transported to the Singleron lab on ice as soon as possible. The specimens were washed with Hanks Balanced Salt Solution (HBSS) for 3 times and minced into 1–2 mm pieces. Then the tissue pieces were digested with 2 ml GEXSCOPETM Tissue Dissociation Solution (Singleron) at 37 °C for 15 min in 15 ml centrifuge tube with sustained agitation. After digestion, using 40-micron sterile strainers to filter the samples and centrifuging the samples at 1000 rpm for 5 m. Then the supernatant was discarded, and the sediment was resuspended in 1 ml PBS (HyClone). To remove the red blood cells, 2 mL GEXSCOPETM red blood cell lysis buffer (Singleron) was added at 25 °C for 10 m. The solution was then centrifuged at 500×g for 5 min and suspended in PBS. The sample was stained with trypan blue (Sigma) and microscopically evaluated.
Single-cell suspensions with 1×105 cells/mL in concentration in PBS (HyClone) were prepared. Single-cell suspensions were then loaded onto microfluidic devices and scRNA-seq libraries were constructed according to Singleron GEXSCOPER protocol by GEXSCOPER Single-Cell RNA Library Kit (Singleron Biotechnologies) [48]. Individual libraries were diluted to 4 nM and pooled for sequencing. Pools were sequenced on Illumina HiSeq X with 150 bp paired end reads.
Raw reads were processed to generate gene expression profiles using an internal pipeline. Briefly, after filtering read one without poly T tails, cell barcode and UMI was extracted. Adapters and poly A tails were trimmed (fastp V1) before aligning read two to GRCh38 with ensemble version 92 gene annotation (fastp 2.5.3a and featureCounts 1.6.2) [49]. Reads with the same cell barcode, UMI and gene were grouped together to calculate the number of UMIs per gene per cell. The UMI count tables of each cellular barcode were used for further analysis.
Single-cell RNA-Seq data processing
Here, we applied Seurat package to normalize and scale the single-cell gene expression matrix [50, 51]. It was first normalized by “NormalizeData” function with setting normalization method as “LogNormalize”. The uninteresting variations were removed by implementing by “ScaleData” function. Finally, the corrected expression matrix was used as an input for further analysis.
Cell filtration for mouse scRNA-seq data
Due to the different QC for Immune and breast cells, we adopted two different criteria. The quality of cells was assessed based on three metrics step by step: [1] The number of total UMI counts per cell (library size); [2] The number of detected genes per cell; [3] The proportion of mitochondrial gene counts. After identification of epithelial cells and immune cells in Fig. 1b, Low-quality epithelial cells were filtered if the quality of the cell does not meet the following standards: “total counts: > 5,000; number of genes: >2,000; the proportion of mitochondrial gene counts: <8%”. The criterion for the immune cell is “total counts: >1000; number of genes: >500; the proportion of mitochondrial gene counts: <8%”.
Cell filtration for human scRNA-seq data
Low-quality cells were filtered if the quality of the cells did not meet the following criteria: “number of genes: >400; number of genes: <7000; the proportion of mitochondrial gene counts: <20%”.
Dimension reduction, cell clustering, and annotation
We selected the top 3000 largest variable genes as highly variable genes (HVGs) and performed the subsequent analysis such as PCA clustering, WGCNA network analysis and construction of single-cell trajectories based on this set of HVGs. The expression profile of HVGs was centered and scaled values. It was implemented by “FindVariableGenes” function with default parameters. We then used the “RunPCA” function to perform the principal component analysis (PCA). The number of significant principal components was determined by “jackstraw” function. The analysis identified 50 significant principal components to supply to the “RunPCA” function. We then utilized the “FindClusters” function to conduct the cell clustering with resolution setted as 0.4. We annotated cell clusters based on the expression of curated known cell markers on t-SNE plot.
Differential expression analysis
Differential gene expression analysis was performed by the “FindMarkers” function. The statistical method to identify differentially expressed genes was based on Wilcox rank sum test. The “FindMarkers” function was run with default parameters.
Survival analysis and enrichment analysis
The survival analysis was performed on KMplot (http://kmplot.com/analysis/index.php?p=service&cancer=breast) [52]. We supplied multiple gene names to the KMplot by using “use mean expression of selected genes” option. We restricted our analysis in Luminal B type patients. The enrichment analysis was performed on MsigDB (https://www.gsea-msigdb.org/gsea/msigdb) with default parameters [53, 54]. Gene expression profile of breast cancer patient was downloaded from the TCGA website (https://portal.gdc.cancer.gov/). Raw reads counts were extracted from files with the suffix “htseq.counts”. The trimmed mean of M-values (TMM) normalized expression value was generated by “edgeR” package [55]. The clinical information was also downloaded from the TCGA website. The information of cancer subtype was retrieved from TCGA previous study [24].
Cancer signature scoring
The signature genes were the genes differentially expressed between MMTV-PyMT and control FVB mouse at week 17 in bulk RNA-seq. The edgeR package in R was applied to identify differentially expressed genes [55]. Genes with FDR < 0.05 and expression fold change >1.5 (upregulated genes) or <0.67 (down-regulated genes) were defined as differentially expressed genes. Cancer signature scoring was calculated by subtracting the mean expression of the down-regulated genes from the mean expression of the upregulated genes [14, 43]. We defined cancer and normal cells by the median score based on cancer signature genes. Cells with score lower than the median were defined as normal cells and cells with score higher than the median as cancer cells.
Gene set variation analysis (GSVA)
Pathway analyses were predominantly performed on the gene sets described in the Molecular Signatures Database (MSigDB) and exported using the GSEABase package (version 1.36.0). To reduce pathway overlaps and redundancies, genes associated with multiple gene sets were trimmed from these gene sets and thus retained genes are only associated with one gene set by following previous study [56]. Most gene sets retained >70% of the associated genes. Next, to assign stemness of cancer stem-like cell estimates to individual cells. We applied GSVA using standard settings, as implemented in the GSVA package (version 1.22.4). Differences in pathway activities scored per cell by GSVA between the different clusters.
Reconstruction of differentiation trajectories using Monocle
Using the R package Monocle (version 2.8.0), differentiation hierarchies within different clusters were reconstructed [57]. Cell fate decisions and differentiation trajectories were reconstructed with the Monocle 2 package, which utilized reverse graph embedding based on a user-defined gene list to generate a pseudo-time plot that could account for both branched and linear differentiation processes.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2020YFA0112300, 2016YFA0101202), NSFC grants (81930075, 81772799, 81773155), “Ten Thousand Plan” - National High-Level Talents Special Support Plan WR-YK5202101, the National Key Research and Development Program of China (2018YFA0507501), Program for Outstanding Medical Academic Leader in Shanghai (2019LJ04), Program of Shanghai Academic/Technology Research Leader (20XD1400700), Shenzhen Science and Technology Innovation Commission Project, Shenzhen Municipal Government of China (KQTD20170810160226082), the Fudan University Research Foundation (IDH1340042), the Research Foundation of the Fudan University Shanghai Cancer Center (YJRC1603).
Author contributions
S.L.L. designed and supervised the study and wrote the manuscripts. G.J.J. and J.C.L.T. analyzed and interpreted the scRNA-seq data and wrote the manuscripts. J.C.L.T. performed the RT-PCR verification for BCSC. L.Z. collected and prepared the mouse samples for scRNA-seq. M.X.D. assisted the experiments. Z.X.C. wrote the manuscript. J.F. assisted the scRNA-seq. L.X.Z. and X.W.B. provided helpful suggestions. All authors have reviewed the manuscript and approved the final version.
Data availability
The scRNA-seq data for MMTV-PyMT mouse has been deposited in the NCBI under the accession code PRJNA762594. The bulk RNA-seq data for MMTV-PyMT and wild-type control mouse has been deposited in the NCBI under the accession code PRJNA761912. The scRNA-seq data for breast cancer patients have been deposited in the NCBI under the accession code PRJNA764023.
Competing interests
The authors declare no competing interests
Footnotes
Edited by Massimiliano Agostini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guojuan Jiang, Juchuanli Tu, Lei Zhou.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04261-y.
References
- 1.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomark Prev. 2017;26:444–57.. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 2.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–9. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Turashvili G, Brogi E. Tumor heterogeneity in breast cancer. Front Med. 2017;4:227.. doi: 10.3389/fmed.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicha MS. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res. 2008;10:105. doi: 10.1186/bcr1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–60. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–30. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 12.Suva ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–308. e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–13.. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, Bao F, Dai Q, Wu LF, Altschuler SJ. Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nat Methods. 2019;16:311–4. doi: 10.1038/s41592-019-0353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paik DT, Cho S, Tian L, Chang HY, Wu JC. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020;17:457–73.. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Zhang Q, Song J, Ruan Q, Ruan W, Chen Y, et al. A highly sensitive, accurate, and automated single-cell RNA sequencing platform with digital microfluidics. Anal Chem. 2020;92:8599–606.. doi: 10.1021/acs.analchem.0c01613. [DOI] [PubMed] [Google Scholar]
- 18.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–58. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu NY, Nolan E, Lindeman GJ, Visvader JE. Stem cells and the differentiation hierarchy in mammary gland development. Physiol Rev. 2020;100:489–523. doi: 10.1152/physrev.00040.2018. [DOI] [PubMed] [Google Scholar]
- 20.Hollern DP, Andrechek ER. A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast Cancer Res. 2014;16:R59.. doi: 10.1186/bcr3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong M, Thennavan A, Urrutia E, Li Y, Perou CM, Zou F, et al. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Briefings Bioinform. 2020;416:27.. doi: 10.1093/bib/bbz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang G, Wang X, Sheng D, Zhou L, Liu Y, Xu C, et al. Cooperativity of co-factor NR2F2 with pioneer factors GATA3, FOXA1 in promoting ERalpha function. Theranostics. 2019;9:6501–16. doi: 10.7150/thno.34874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer cell. 2018;33:690–705. e9. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak K. Heterogeneity in breast cancer. J Clin Investig. 2011;121:3786–8. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–38. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 27.Hnzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–52. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2017;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 30.Januškeviciene I, Petrikaite V. Heterogeneity of breast cancer: the importance of interaction between different tumor cell populations. Life Sci. 2019;239:117009. doi: 10.1016/j.lfs.2019.117009. [DOI] [PubMed] [Google Scholar]
- 31.Luo M, Brooks M, Wicha MS. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des. 2015;21:1301–10. doi: 10.2174/1381612821666141211120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magee Jeffrey A, Piskounova E, Morrison Sean J. Cancer Stem Cells: Impact, Heterogeneity, and Uncertainty. Cancer Cell. 2012;21:283–96.. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannello F. Understanding breast cancer stem cell heterogeneity: time to move on to a new research paradigm. BMC Med. 2013;11:169. doi: 10.1186/1741-7015-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–87. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41.. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang A, Chen L, Li C, Zhu Y. Heterogeneity in cancer stem cells. Cancer Lett. 2015;357:63–8. doi: 10.1016/j.canlet.2014.11.040. [DOI] [PubMed] [Google Scholar]
- 37.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–6. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, et al. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966–83. doi: 10.1038/s41418-019-0389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30:833–44. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu R, Choi HS, Zhen X, Kim SL, Kim JH, Ko YC, et al. Betavulgarin isolated from sugar beet (Beta vulgaris) suppresses breast cancer stem cells through Stat3 signaling. Molecules. 2020;25:2999.. doi: 10.3390/molecules25132999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakha EA, El-Sayed ME, Lee AH, Elston CW, Grainge MJ, Hodi Z, et al. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26:3153–8. doi: 10.1200/JCO.2007.15.5986. [DOI] [PubMed] [Google Scholar]
- 45.Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast. Cancer Cell. 2019;177:1330–45 e18. doi: 10.1016/j.cell.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen OW, Nielsen HL, Gudjonsson T, Villadsen R, Rank F, Niebuhr E, et al. Epithelial to mesenchymal transition in human breast cancer can provide a nonmalignant stroma. Am J Pathol. 2003;162:391–402. doi: 10.1016/S0002-9440(10)63834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the NICHE in regulating stemness of tumor cells? cell stem cell. 2015;16:225–38. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dura B, Choi JY, Zhang K, Damsky W, Thakral D, Bosenberg M, et al. scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3’ mRNA profiling. Nucleic Acids Res. 2019;47:e16. doi: 10.1093/nar/gky1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 50.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20.. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi M, Wang Y, Tang W, Cui X, Wu H, Tang Y, et al. Identification of TP53 mutation associated-immunotype and prediction of survival in patients with hepatocellular carcinoma. Ann Transl Med. 2020;8:321. doi: 10.21037/atm.2020.02.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data for MMTV-PyMT mouse has been deposited in the NCBI under the accession code PRJNA762594. The bulk RNA-seq data for MMTV-PyMT and wild-type control mouse has been deposited in the NCBI under the accession code PRJNA761912. The scRNA-seq data for breast cancer patients have been deposited in the NCBI under the accession code PRJNA764023.