Abstract
Simian foamy viruses (SFVs) are highly prevalent in a variety of nonhuman primate species ranging from prosimians to apes. SFVs possess a broad host range, and human infections can occur by cross-species transfer (W. Heneine et al., Nat. Med. 4:403–407, 1998). Retrovirus screening of potential sources of infection, such as laboratory research animals and simian-derived biological products, could minimize human exposure to SFVs by reducing the risk of potential retrovirus infection in humans. We describe a variety of sensitive assays for SFV isolation and detection which were developed with a prototype strain of SFV serotype 2. The Mus dunni cell line (M. R. Lander and S. K. Chattopadhyay, J. Virol. 52:695–698, 1984) was found to be highly sensitive for SFV production on the basis of various general and specific retrovirus detection assays such as reverse transcriptase assay, transmission electron microscopy, immunofluorescence assay, and Western blotting. A highly sensitive PCR assay was developed on the basis of the sequences in primary SFV isolates obtained from pig-tailed macaques (Macaca nemestrina) and rhesus macaques (Macaca mulatta). Analysis of naturally occurring SFV infection in macaques indicated that analysis by a combination of assays, including both highly sensitive, specific assays and less sensitive, broadly reactive assays, is important for evaluation of retrovirus infection.
Foamy viruses (FVs) occur in several animal species including nonhuman primates (2, 21, 30); however, there is no direct evidence that FVs are indigenous to humans (3, 17, 40). Simian FVs (SFVs) possess a broad cell tropism (20, 21, 29, 46), tissue tropism (1, 13, 15, 21, 22, 33, 34, 43, 45), and species tropism (2, 19, 21, 23). In fact, cross-species infection of humans has been reported in handlers of SFV-infected primates (10, 19, 21, 32, 40, 41). In one case SFV was isolated from an infected individual 20 years postexposure (41). In vitro studies have also shown that SFVs can persist in a latent state (6, 11) and can be reactivated to produce infectious virus (39). Although there is no pathogenesis directly associated with SFVs (47), the long-term consequences of SFV infection in humans may not yet be known. Various animal models demonstrate that activation and increased replication of retroviruses can result in acute or slow diseases (44). Therefore, the long-term presence of a latent, potentially inducible, infectious retrovirus in humans can raise some public health safety concerns.
To minimize the risk of cross-species infection of humans with SFVs, sensitive detection assays can be used to identify FV-infected animals and to analyze simian-derived biological products for SFV contamination. This is consistent with the current testing strategy outlined for retrovirus testing of cell substrates for live, attenuated viral vaccines (25). Extensive and rigorous retrovirus testing is particularly important in the case of xenotransplantation since the recipients of animal tissues or organs would be immunosuppressed and thus potentially more susceptible to virus infections. In this paper we describe a variety of sensitive detection assays that may be used to investigate SFV infections in animals and humans and to analyze monkey-derived biological products for retrovirus contamination.
MATERIALS AND METHODS
Cells and viruses.
The Mus dunni cell line (wild mouse fibroblast ATCC CRL-2017 [28]) was kindly provided by Janet Hartley (National Institute of Allergy and Infectious Diseases, National Institutes of Health). The following cell lines were obtained from the American Type Culture Collection (ATCC; Rockville, Md.): Vero (African green monkey kidney; ATCC CCL-81), Cf2Th (canine thymus; ATCC CRL-1430), A204 (human rhabdomyosarcoma; ATCC HTB-82), and A549 (human lung carcinoma; ATCC CCL-185) cells. HeLa (human epithelioid carcinoma) cells were obtained from the AIDS Research and Reference Reagent Program (Richard Axel, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health). Primary chicken embryo fibroblasts (CEFs) were prepared from 10-day-old embryos (SPAFAS, Inc., Preston, Conn.). Cells were maintained in a 75-cm2 flask in 13 ml of Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (heat inactivated; Gibco, Grand Island, N.Y.), 2 mM l-glutamine, 250 U of penicillin per ml, and 250 μg of streptomycin per ml (designated complete medium).
The M. dunni, Cf2Th, Vero, and HeLa cell lines were infected in parallel at 75% confluence with 0.1 ml (281 50% tissue culture infective doses [TCID50s]) of freshly reconstituted SFV serotype 2 (SFV-2; (102.75 TCID50s per 0.2 ml in MRC-5 cells; 22 days; ATCC catalog no. VR 277, FV-34, lot 4D, 91-10). The infection was set up in duplicate in complete medium containing Polybrene (4 μg per ml); one uninfected flask was set up as a negative control for each of the cell lines. The cells were split when they reached confluence (every 3 to 4 days) or were fed fresh medium. Filtered supernatants (0.45-μm-pore-size test tube top filter units; Corning, Cambridge, Mass.) were collected, aliquoted, and saved at −80°C for reverse transcriptase (RT) analysis. The cultures were regularly monitored for cytopathic effect (CPE). CPEs that ranged from 1+ to 4+, which were equivalent to about 25 to 95% cell death, were noted. The cultures were propagated until extensive cell lysis (4+ CPE [≥90%]) occurred or until there was extensive accumulation of floating dead cells in the supernatant.
A204, A549, and CEF cells were infected with 0.2 ml (103.35 TCID50s) of a new lot of SFV-2 which contained a higher virus titer (103.5 TCID50s per 0.5 ml in A72 cells; 7 days; ATCC; lot 5W, 95-12). The infection and propagation of the cells were done under the conditions described above. The cultures were monitored for CPE, and filtered supernatants were collected for RT analysis.
To assess the contribution of cellular polymerases to the RT activity in samples that were collected at times of extensive cell lysis, confluent uninfected cultures were lysed by four freeze-thaw cycles of the flask containing 13 ml of complete medium, and the samples were subsequently handled similarly to the infected cultures.
The sensitivity of SFV detection in infected macaques was determined by cocultivation of monkey peripheral blood mononuclear cells (PBMCs) with M. dunni cells. The animals were maintained in accordance with the Guide for the Care and Use of Laboratory Animals (31) under a protocol approved by the Center for Biologics Evaluation (CBER) Animal Care and Use Committee. A pig-tailed macaque (Macaca nemestrina), designated animal Mn97, had previously been identified as seropositive on the basis of immunofluorescence assay (IFA; Microbiological Associates Inc., Rockville, Md.). PBMCs from Mn97 were prepared by the Ficoll-Hypaque procedure from heparinized blood, aliquoted, and cryopreserved. Unstimulated PBMCs (5 × 106) were cocultured with 2 × 106 trypsinized M. dunni cells in 20 ml of complete Dulbecco’s modified Eagle medium containing Polybrene (4 μg/ml). The medium was changed on the next day and was replaced with medium without Polybrene. The cultures were further propagated and monitored for CPE, and filtered supernatants were collected as described above until extensive cell lysis occurred. Generally, at each medium change, prior to filtration, the PBMCs in the supernatant were pelleted by centrifugation at 1,500 rpm for 10 min (Beckman GS-6KR centrifuge with a GH-3.8 horizontal rotor; Beckman, Columbia, Md.) and added back to the M. dunni cells. To determine the lowest number of PBMCs from Mn97 from which virus could be recovered, cells were serially diluted and cocultured with 7.5 × 105 M. dunni cells initially in a 25-cm2 flask in 4 ml of complete medium in the presence of Polybrene. The cells were subsequently transferred to a 75-cm2 flask. Two 10-fold dilution series of PBMCs (1.8 × 106 to 0.018 cells and 4.5 × 105 to 0.045 cells) were tested in independent cocultivation experiments. The cultures were handled as described above, and supernatants were collected until termination of the cultures due to extensive cell lysis.
RT assay.
RT assay was performed with 10 μl of sample and 50 μl of RT cocktail for 2 h in a 37°C water bath. The RT cocktail consisted of 50 mM Tris-HCl (pH 8.3), 60 mM NaCl, 20 mM dithiothreitol, 0.05% Nonidet P-40, 0.6 mM MnCl2, 10 μg of poly(A) per ml, and 5 μg of pdT12-18 per ml. One microliter of [α-32P]dTTP (0.5 μCi; >400 Ci/mmol; Amersham Corp., Arlington Heights, Ill.) was added per ml of cocktail just prior to use. Five microliters of the reaction mixture was spotted in duplicate onto DE81 filter paper (Whatman) and air dried. Unbound 32P was removed by four 5-min washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and two 1-min washes in 95% ethanol. The filters were dried and exposed to X-ray film overnight at −80°C. Subsequently, the radioactivity in the spots was counted in a liquid scintillation counter. All the samples from the different cultures were tested in the same RT experiment to avoid any differences in the results due to assay variability. Two independent RT experiments, each spotted in duplicate, were performed with each infected cell culture and the uninfected control flask. The results (means ± standard deviations [SDs]) of the two experiments were calculated.
Transmission electron microscopy (TEM).
Uninfected and SFV-2-infected cells (with CPEs ranging from about 1+ to 3+) were pelleted and fixed for 2 to 3 h with 2% glutaraldehyde–2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.3). The samples were stored in phosphate-buffered saline (PBS) with 4% sucrose at 4°C. The cells were subsequently postfixed with 2% osmium tetroxide, dehydrated with graded alcohols, and embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and were examined for virus particles with a Zeiss EM 912 Omega electron microscope.
IFA.
Infected cells were plated onto coverslips. At the appearance of a 1+ to 2+ CPE, the cells were fixed with ice-cold acetone-methanol (1:1). Fixed cells were incubated in a humidified chamber on a rocker for 30 min at 37°C with a 1:40 dilution (in PBS [pH 7.4]) of plasma from animal Mn97. The cells were then incubated with fluorescein isothiocyanate-conjugated anti-monkey serum (1:160 dilution; Sigma Chemical Co., St. Louis, Mo.) and observed for staining with a ×60 objective in an Olympus fluorescence microscope. Uninfected cells (negative controls) were treated in parallel.
Western blot analysis.
Uninfected and SFV-2-infected cells (at a CPE of about 2+) were harvested by scraping and washed with Dulbecco’s PBS (pH 7.4). The cell lysates were prepared in buffer consisting of 50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 100 mM sodium chloride, 0.2% deoxycholate, and 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Behring Diagnostics, La Jolla, Calif.). The protein concentration was determined with a protein assay dye (Bio-Rad, Hercules, Calif.). Five micrograms of protein was heat denatured and analyzed on an 8 to 16% Tris-glycine-polyacrylamide gel (Novex, San Diego, Calif.), the gel was run for 3 h at 90 V in 1× Tris-glycine running buffer (24.8 mM Tris, 192 mM glycine, 0.1% sodium dodecyl sulfate), and the protein was transferred to a nitrocellulose filter in 24.8 mM Tris–192 mM glycine–20% methanol. After protein transfer onto the nitrocellulose filter at 30 V for 1.5 h, the filter was blocked overnight in PBS (pH 7.3)–0.05% Tween–5% nonfat dried milk (designated PBST+5%) at room temperature and was then incubated with a 1:100 dilution of plasma from animal Mn97. The filter was incubated for 2 h at room temperature and then overnight at 4°C on a rocker. The filter was washed four times in PBST+5% and was then incubated for 2 h at room temperature with a 1:500 dilution of horseradish peroxidase-conjugated goat anti-monkey immunoglobulin G (Cappel Research Products, Durham, N.C.) in PBST+5%. The filter was then washed (whole molecule; six times for 5 min each in PBS-Tween, and the protein bands were visualized by chemiluminescence with the Supersignal CL-HRP-substrate system (Pierce, Rockford, Ill.). The substrate was added to the filter generally for 3 to 5 min, and the filter was then blotted with paper to remove excess substrate and then exposed to BioMax MR film (Kodak, Rochester, N.Y.) for various times ranging from 5 s to 2 min.
PCR assay.
DNA was prepared from monkey PBMCs and from M. dunni cells infected with prototype strains of SFV-1 and SFV-2 (SFV-1 strain FV-21 [catalog no. VR-276; ATCC]; SFV-2 strain FV-34 [catalog no. VR-277; ATCC]). Set A primers were synthesized on the basis of the published sequence of SFV-1 (27, 29). The outer primer pair of set A consisted of forward primer 1 (5′-GGAATGCAGTGGGTATAGAG-3′) and reverse primer 2 (5′-CCTGATATCAATTGTGGTGG-3′), and the inner primer pair consisted of forward primer 3 (5′-CAGTGAATTCCAGAATCTCTTC-3′) and reverse primer 4 (5′-TATCCTTAGGAACTAACACCT-3′). Set B primers were synthesized on the basis of the highly conserved sequences identified in the long terminal repeats (LTRs) of SFVs, which had been isolated with primer set A from naturally infected rhesus and pig-tailed macaques (42). The outer primer pair of set B consisted of forward primer 3 (5′-CAGTGAATTCCAGAATCTCTTC-3′) and reverse primer 5 (5′-CACTTATCCCACTAGATGGTTC-3′), and the inner primer pair consisted of forward primer 6 (5′-CCAGAATCTCTTCATACTAACTA-3′) and reverse primer 7 (5′-GATGGTTCCCTAAGCAAGGC-3′). The PCR conditions for both the outer and inner primer pairs of primer sets A and B were the same. The reaction was carried out with a 100-μl reaction mixture with 5 U of Taq DNA polymerase according to the manufacturer’s instructions (Boehringer Mannheim, Indianapolis, Ind.). For the first amplification, 35 cycles were done at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Ten microliters of the reaction mixture was reamplified under the same PCR conditions. The sensitivity of PCR detection by the outer primer pair of set B was determined by using a panel of DNAs which were created by spiking different copy numbers of cloned SFV-2 LTR DNA in 105 cell equivalents of M. dunni DNA. The SFV-2 LTR fragment was amplified with the outer primer pair of set A and was cloned into vector DNA. PCR-amplified DNA products were analyzed on agarose gels and were visualized by staining with ethidium bromide. PCR primers, which were used to detect the human β-actin gene (Clontech, Palo Alto, Calif.) as a control for the presence of DNA in the sample, amplified an 838-bp DNA fragment. The PCR mixture without DNA was used as the negative control.
RESULTS
SFV replication in different cell lines.
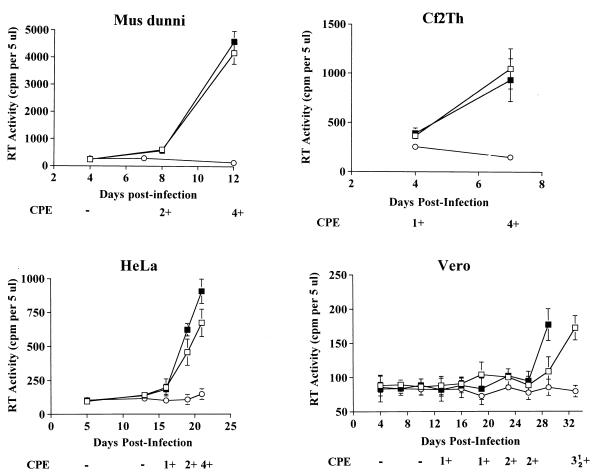
Both SFV-1 and SFV-2 replicated efficiently in M. dunni cells. The kinetics of SFV-2 replication were analyzed in the Cf2Th, M. dunni, HeLa, and Vero cell lines, and the results are shown in Fig. 1. In general, RT activity increased over time postinfection (p.i.) and peaked when the maximum CPE (4+) was reached, at which time the culture was terminated. The highest RT activity was seen in M. dunni cells, in which a CPE was first noted on day 7 p.i.; on day 8 the cells reached a CPE of 2+ and progressed rapidly to a CPE of 4+ at the end of passage 3 (p3), on day 12. The peak RT activity at the time of termination of the culture was about 16-fold above that at the initial time point. Although SFV was most abundantly produced in M. dunni cells, Cf2Th cells were the most sensitive to the CPE, which was first seen at day 4 and which progressed rapidly, with the culture being terminated due to extensive cell death at the end of p2 on day 7. However, the RT activity at this time was about fivefold lower than the peak RT activity in M. dunni cells. In the case of HeLa cells, the peak RT activity was similar to that seen in Cf2Th cells; however, the kinetics of virus replication and CPE progression were delayed. In the HeLa cells, a CPE was first noted on day 16, which was at the end of p4, and progressed until day 21, when the culture was terminated (end of p6). In this case, the peak RT level reached at the time of termination of the culture was about fourfold above the background level. SFV-2 replication was very slow in Vero cells: a CPE was first noted at p5 on day 13 and the CPE progressed slowly to 2+ on day 23. The duplicate cultures in the two infected flasks were terminated on day 29 and day 33, respectively, at the end of p10 when a 3.5+ CPE was seen. Low-level RT activity was detected in the infected Vero cell cultures at the termination of the cultures. Although this level of RT activity was above that for the negative control culture supernatant, it was not clear whether the RT activity was associated with virus or cellular enzymes which were released in the supernatant due to the cell lysis caused by SFV. Thus, the RT activity in the supernatants of infected Vero cells was compared with that in an uninfected cell lysate (data not shown). The results indicated a low level RT activity in both samples. Therefore, SFV infection of Vero cells was confirmed by other detection assays (described below).
FIG. 1.
Kinetics of SFV-2 virus replication in different cell lines. SFV-2 virus replication was monitored at various time points p.i. of M. dunni cells, Cf2Th cells, HeLa cells, and Vero cells. Filtered supernatant was assayed in two independent RT assays. Each sample was spotted in duplicate. The means ± SDs (error bars) for each infected culture (□, ■) and the uninoculated control cells (○) are shown. The progression of cytopathogenicity is indicated, ranging from no CPE (−) to about 90% CPE (4+).
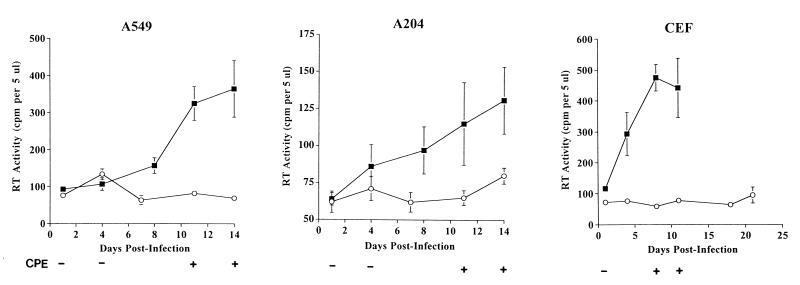
To further assess the replication of SFV in human cells, A204 and A549 cell lines were infected with SFV by using an eightfold greater concentration of infectious virus than that used in the studies described above. The results, shown in Fig. 2, indicate low levels of RT activity in both cell lines on day 14 p.i. To investigate the broad host range of SFV-2, CEF cells were infected and the kinetics of replication of SFV were analyzed (Fig. 2). Although the cells were highly sensitive to the CPE and extensive cell lysis was seen on day 11 p.i., the level of RT activity was low. Infection of chicken cells with FVs has previously been shown for SFV-1 (38) and baboon FV serotype 10 (36).
FIG. 2.
Kinetics of SFV-2 replication in human and chicken cells. A204, A549, and CEF cells were inoculated with SFV-2 at an eightfold greater concentration than that used in the experiment described in the legend to Fig. 1. RT activity was detected in filtered supernatants of SFV-2 infected cells (■). Parallel uninfected cells were the negative control (○). The CPE at various time points is indicated as negative (−) or positive (+). The infection was done in a single experiment; the RT data are the means ± SDs (error bars) for two independent RT assays in which each sample was spotted in duplicate.
IFA analysis.
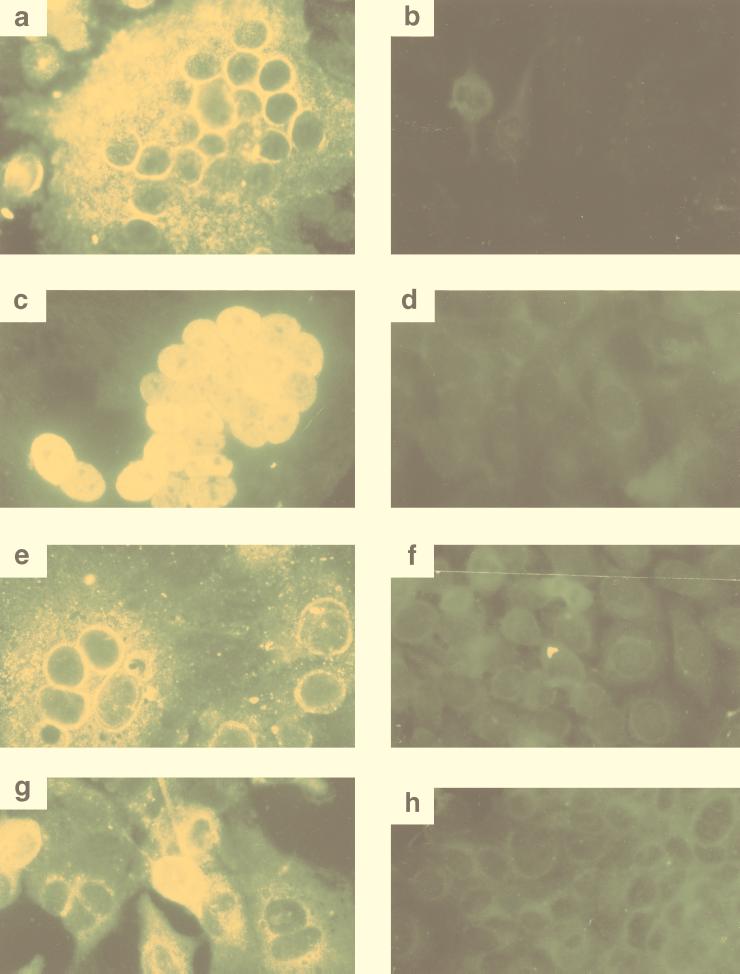
SFV-2 infection in the different cell lines was further analyzed by IFA with plasma from animal Mn97. Intense staining and multinucleated syncytia were seen in the infected cells but not in the uninfected cells (Fig. 3), indicating high levels of viral protein expression. The intensity and cellular localization of the stained signal varied in the different infected cell lines: intense granular cytoplasmic and perinuclear staining was seen in M. dunni cells (Fig. 3a) and HeLa cells (Fig. 3e), while little staining was seen in the nucleus. In Vero cells (Fig. 3g) cytoplasmic and nuclear staining was seen; however, the staining was mostly perinuclear. In the case of the Cf2Th cells (Fig. 3c), intense nuclear staining and diffuse cytoplasmic staining were seen. The differences in the cellular locations of the signals may be due to asynchronous infection of the cells in the culture. Differential expression of viral proteins in the cytoplasm versus the nucleus has previously been shown to be dependent on the time p.i. in the case of SFV-1 (16) and human foamy virus (37).
FIG. 3.
Immunofluorescence of SFV-2 infected cells. IFAs of uninfected and virus-infected M. dunni (a and b, respectively), Cf2Th (c and d, respectively), HeLa (e and f, respectively) and Vero (g and h, respectively) cells were done with plasma from animal Mn97 as described in Materials and Methods. Multinucleated syncytia as well as singly stained cells were seen in virus-infected cells, whereas no signal was seen in the uninfected cultures. Intense cytoplasmic and perinuclear staining was seen in infected M. dunni, HeLa, and Vero cells (a, e, and g, respectively). In Cf2Th cells, intense nuclear and diffuse cytoplasmic staining were seen (c).
TEM analysis.
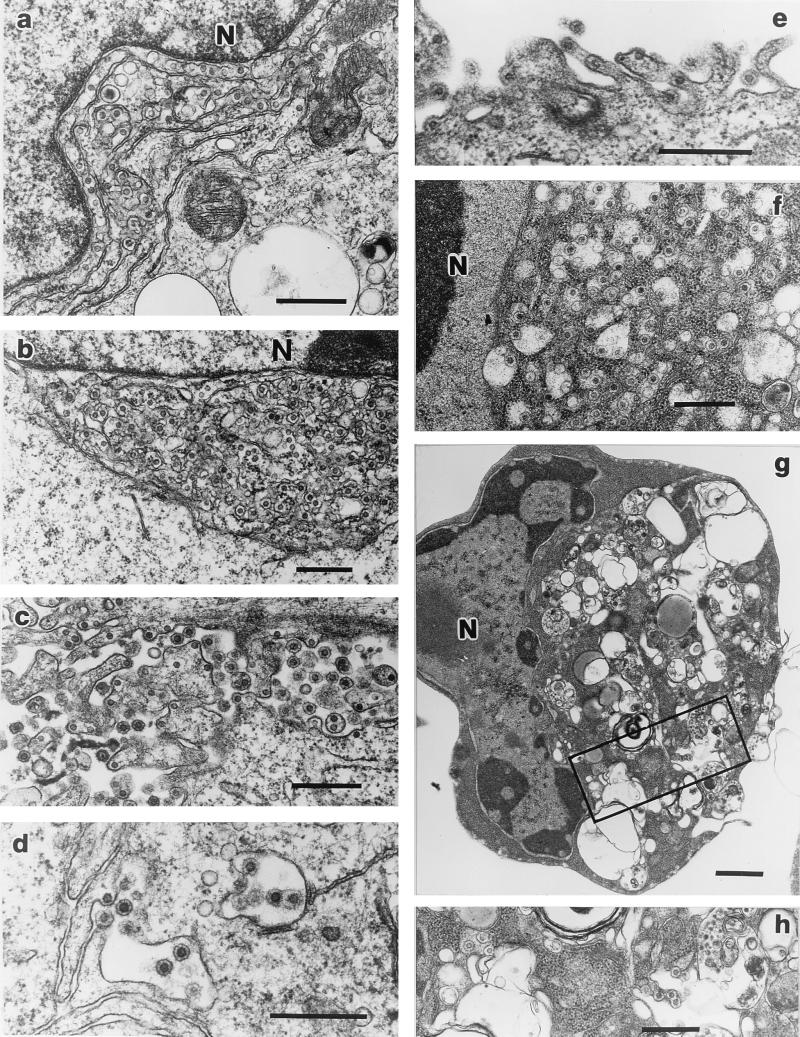
SFV-2 infection of the different cell lines was further confirmed by TEM. Analysis of normal and infected cells indicated particles characteristic of SFV in the infected cells; however, the amount and location of virus produced varied in the different cell lines. In addition, analysis of cells at different stages of the CPE (1+ to 3+) indicated an increase in the number of virus particles with progression of the CPE. In general, the levels of extracellular virus production directly correlated with the RT activity. Abundant intracellular and extracellular virus was seen in M. dunni cells (Fig. 4a to c). In the case of the M. dunni cells, large numbers of virus cores were associated with parallel arrays of rough endoplasmic reticulum (ER) membranes surrounding the nucleus (Fig. 4a). In some cases very enlarged structures consisting of extensively developed membranes and viral cores were seen adjacent to the nucleus (Fig. 4b). Numerous SFV particles with a characteristic spiked envelope were seen budding from the plasma membrane and extracellularly (Fig. 4c). In contrast to M. dunni, few infected Vero cells were observed, and these had very few intracellular or extracellular particles (Fig. 4d). In the case of SFV-infected Cf2Th cells, little budding occurred at the plasma membrane, and therefore, few extracellular particles were seen (Fig. 4e). However, in these cells there was abundant intracellular accumulation of enveloped virus in dilated ER membranes (Fig. 4f). SFV-infected HeLa cells produced few extracellular particles, like the Cf2Th and Vero cells; however, they were different from the other infected cells, including M. dunni, in that they were highly vacuolated and contained enveloped particles (Fig. 4g and h).
FIG. 4.
TEM of SFV-2-infected cells. SFV particles were seen intracellularly and budding from the plasma membrane. Infected M. dunni cells (a to c) had viral cores associated with extensively duplicated ER membranes, which were adjacent to the nucleus (a and b). Abundant mature particles were seen budding extracellularly from the plasma membrane (c). Infected Vero cells had few mature particles, which bud from the plasma membranes (d). In the case of infected Cf2Th cells (e and f) apoptotic cells were seen with few extracellular particles (e) and abundant accumulation of enveloped particles intracytoplasmically in dilated cisternae of the ER (f). In infected HeLa cells, enveloped particles were seen in the vacuolated cytoplasm of apoptotic cells (g and h). Few particles were seen to be budding from the plasma membrane. N, nucleus; the boxed region in panel g is magnified in panel h to show virus particles. Bars, 0.5 μm in panels a to f and h and 1 μm in panel g.
SFV-2 infection of HeLa cells and Cf2Th cells resulted in apoptosis. Cellular changes indicative of apoptosis were seen, including chromatin condensation and nuclear segmentation (Fig. 4f and 4g) (24). These results indicate that SFV can have two mechanisms of cell death: lysis and apoptosis. The difference between the nucleus of an apoptotic cell and that of a nonapoptotic cell can be seen by comparing the chromatin distributions in the nuclear regions (designated N) of Cf2Th and HeLa cells (Fig. 4f and g, respectively) with that in the nuclear region of M. dunni cells (Fig. 4a).
Western blot analysis.
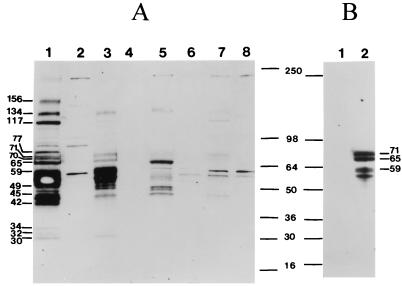
To analyze the efficiency of virus production in the different cell lines, similar amounts of protein lysates from infected and uninfected cells were analyzed by Western blotting (Fig. 5). Numerous virus-specific proteins were highly expressed in the SFV-2-infected M. dunni cells (Fig. 5A, lane 1). Some of the proteins were comparable in size to those previously reported for the SFV-1 and SFV-3 Gag, Pol, and Env proteins, e.g., 134 kDa for the Pol precursor (35), 77 kDa for the RT (80 kDa) (8), 117 kDa for the Env precursor (27), 71 kDa for the Env glycoprotein (70 kDa) (7, 9) and 70 kDa for the Gag precursor (69 kDa) (27). Small amounts of the gag-encoded major core protein (30 kDa) (8, 9) were detected, indicating inefficient processing of the precursor proteins. A novel large protein (156 kDa) was detected in SFV-infected M. dunni cells. However, specific SFV reagents were not available to characterize the different proteins. Proteins of similar sizes were detected in Cf2Th, Vero, and HeLa cells (Fig. 5A, lanes 3, 5, and 7, respectively). Some of the bands were seen only when larger amounts of the lysates were analyzed (data not shown). In general, protein expression in the different cell lines correlated with the levels of virus production seen by the other detection assays (i.e., M. dunni > Cf2Th > HeLa > Vero); however, the number and amount of proteins detected in the different cell lines depended upon the stage of infection at the time of preparation of the cell lysate. This was evidenced by the increased expression of the 71- and 65-kDa proteins in Cf2Th cells when the cells were harvested when the CPE was 3+ (Fig. 5B, lane 2) compared with that when the cells were harvested when the CPE was 2+ (Fig. 5A, lane 3). Western blot analysis of virus concentrated from filtered supernatant of SFV-2-infected M. dunni cells by pelleting through a sucrose cushion produced a protein profile identical to that for the infected cell lysate, thus indicating the presence of both extracellular and intracellular particles in the virus preparation. The small amount of the 30-kDa protein, which corresponds to the major viral capsid protein, in the virus preparation indicated a low yield of mature virions (data not shown).
FIG. 5.
Western blot analysis of SFV-2-infected cells. Five micrograms of protein from lysates of the following SFV-2-infected cells (at a CPE of 2+) were immunoblotted with SFV-positive monkey plasma (A): M. dunni (lane 1), Cf2Th (lane 3), Vero (lane 5), and HeLa (lane 7). Uninfected cell lysates were the negative controls, as follows: M. dunni (lane 2), Cf2Th (lane 4), HeLa (lane 6), and Vero (lane 8) cells. In addition, Cf2Th cells were analyzed when the CPE was 3+ (B). Lane 1, uninfected cells; lane 2, infected cells. The sizes of some of the prominently visible SFV-2 proteins in M. dunni are indicated. The molecular masses were calculated from standard markers (SeeBlue; Novex, San Diego, Calif.) and are indicated in kilodaltons. The filter shown in panel A was incubated in substrate for 3 min and remained at room temperature for about 30 min before autoradiography. It was then exposed to X-ray film for 1 min. The filter shown in panel B was incubated in substrate for 10 s and immediately exposed to X-ray film for 10 s.
Isolation of SFV from pig-tailed macaque PBMCs by using the M. dunni cell line.
The efficiency of SFV isolation in M. dunni cells was further assessed in coculture studies with PBMCs from animal Mn97. Initial cocultivation studies with unstimulated PBMCs and M. dunni cells indicated that SFV could be detected by the CPE at p3 on day 12. This was confirmed by the presence of RT activity in a sample collected at p4 on day 14. Subsequently, various dilutions of PBMCs from Mn97 were cocultivated with M. dunni cells to estimate the sensitivity of isolation of SFV from an infected monkey. The results indicated that virus could be recovered from at least 4.5 × 105 cells on day 21 at p6 after coculture (Table 1). FVs were not detected with 1.8 × 105 cells at day 25 (p8) or other, lower dilutions, including 4.5 × 104 cells, which remained negative even at p11 (39 days). On the basis of these results the sensitivity of virus detection in M. dunni cells is at least 1 infected cell in 4.5 × 105 cells at p6.
TABLE 1.
Detection of SFV-infected PBMCs from animal Mn97 by coculture with M. dunni cells
| No. of PBMCsa | CPE | RT activityb | Day p.i. | Passage no. |
|---|---|---|---|---|
| 4.5 × 106 | 2+ | + | 21 | 6 |
| 1.8 × 106 | 2+ | + | 18 | 5 |
| 4.5 × 105 | 1+ | − | 21 | 6 |
| 2+ | + | 28 | 8 | |
| 1.8 × 105 | − | − | 25 | 8 |
| 4.5 × 104 | − | − | 39 | 11 |
| 1.8 × 104 | − | − | 25 | 8 |
Two independent dilution series are indicated.
The RT activity was detected (+) or not detected (−) in the sample at the indicated time point or a previous one. Similar results were obtained in two independent RT assays.
Development of sensitive PCR primers for detection of SFV in macaques.
To study natural SFV infection and virus transmission in macaques, 28 monkeys (16 Macaca nemestrina and 12 Macaca mulatta macaques) were analyzed by the various detection assays described above (26). SFV infection was initially determined by screening monkey plasma for viral antibodies by IFA, and the results were further evaluated by PCR analysis and virus isolation with monkey PBMCs. The IFA and PCR results for selected animals are shown in Table 2. Initially, PCR primer set A was developed on the basis of known SFV-1 sequences and was used in the analysis. As seen in Table 2, there was a correlation between the IFA data and the PCR data for four positive and two negative animals. However, MmG2K and MmJ4G were positive by IFA and negative by PCR. To address this discordance in the results, PCR primer set B was developed on the basis of highly conserved sequences present in the LTRs of viral DNAs of primary SFV isolates of different macaque species. As shown in Table 2, this set of primers was able to detect SFV sequences in MmG2K and MmJ4G, which were negative in tests with primer set A. Thus, there was a perfect correlation between the IFA and PCR data when the more sensitive primer set B was used. In addition, primer set B could also detect SFVs in Macaca nemestrina as well as SFV-1 from Macaca cyclopsis. Thus, primer set B was found to be broadly reactive for the detection of SFVs in different macaque species.
TABLE 2.
Analysis of SFV infection in macaquesa
| Monkeyb | IFA result | PCR result
|
|
|---|---|---|---|
| Primer set A | Primer set B | ||
| Mn97 | + | + | |
| Mn72 | + | + | |
| MmK3A | + | + | |
| MmK3T | + | + | |
| MmG2K | + | − | + |
| MmJ4G | + | − | + |
| Mn32 | − | − | − |
| MmA2N | − | − | − |
Data are presented for samples collected in 1996.
Mn, Macaca nemestrina; Mm, Macaca mulatta.
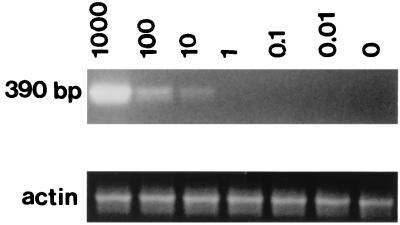
The sensitivity of PCR detection was determined by evaluating the detection of SFV sequences in a background of uninfected cellular DNA by using SFV-2 as a prototype virus. A 390-bp DNA fragment was amplified with the outer primer pair of primer set B. The results indicated detection of 10 viral copies in 105 cell equivalents of uninfected M. dunni DNA (Fig. 6). The specificity of the primers was demonstrated by the absence of amplified fragments in DNAs prepared from uninfected monkey or human PBMCs (data not shown).
FIG. 6.
Detection of SFV by PCR. The sensitivity of PCR detection with set B outer primer pair was determined by amplification of M. dunni DNA (105 cell equivalents) containing different numbers of copies of cloned SFV-2 DNA. The number of copies of cloned DNA and the size of the PCR-amplified fragment are indicated. Primers used for detection of the human β-actin gene, which was used as a control for the presence of DNA in each sample, amplified the fragments of the expected sizes.
DISCUSSION
The broad host range and high prevalence of SFVs in nonhuman primates can result in cross-species transmission of SFVs from infected animals to humans (10, 21, 32, 39, 41, 45). Although there is no evidence of SFV-induced disease in animals or humans (47), the virus can remain in a latent, inducible state in vitro (6, 11) and persist long term in vivo (41). In fact, Schweizer et al. (41) were successful in isolating infectious SFV, after prolonged coculture, from stimulated PBMCs of an infected individual who had had a persistent and clinically latent infection for about 20 years (41). Since retroviruses can cause acute or slow diseases and since transient immune suppression following FV infection has been shown in rabbits (40), the presence of an infectious, albeit latent, retrovirus such as SFV in humans raises concerns regarding public health safety. Therefore, to minimize the potential risk of cross-species transfer of retroviruses to humans, sensitive detection assays should be used to screen potential sources of infection such as nonhuman primates and simian-derived biological products. In this paper we have described various assays that may be used to detect SFV in infected animals or humans and in monkey-derived biological products. We have used some of the assays to analyze naturally occurring SFV infections in Macaca mulatta and Macaca nemestrin monkeys. The results indicated that rigorous testing is necessary for retrovirus detection, such as use of a combination of selected general and specific assays for the detection of known as well as novel viruses. Furthermore, the most sensitive assays which can identify the presence of low-level or latent retroviruses should be used: for example, assays that use the M. dunni cell line, which is highly sensitive for SFV isolation, and the set B PCR primers, which can detect SFVs in different macaque species.
Analysis of different cell lines for susceptibility to SFV-2 infection identified M. dunni cells as the most sensitive cell line for virus production. In addition, the comparative infectivity studies indicated that SFV-2 replicated at low levels in human cells and poorly in monkey cells compared with the levels of replication in cells of other species, e.g., mouse and dog cells. The inefficient viral replication in primate cells may reflect the inability of the virus to induce pathogenesis in this species. The initial low level of virus replication could produce immune responses in vivo, such as high levels of neutralizing antibodies (43), which might result in a low-level virus burden and chronic infection without clinical symptoms. The persistence of a retrovirus in the absence of disease induction in its natural host has also been seen in the case of the simian immunodeficiency virus (SIV) (4). However, upon cross-species transfer, SIV can replicate efficiently, resulting in AIDS in Asian macaques. In general, increased viremia can result in retrovirus-induced diseases. For example, murine amphotropic, replication-competent retrovirus was found to be nonpathogenic in healthy macaques (12) but induced T-cell lymphomas in highly immunosuppressed animals (14). Furthermore, live, attenuated SIV that contained deletions in nef, vpr, and negative regulatory element (NRE) regions protected adult monkeys against infection with pathogenic virus (48), whereas the same virus produced AIDS in infant monkeys (5). Thus, to fully assess whether SFV may be pathogenic in humans, studies with animals need to be done to evaluate SFV replication with regard to the host immune status and age as well as the virus dose and route of exposure.
ACKNOWLEDGMENTS
We thank T. Bryan and M. Lundquist for technical assistance, A. Thompson for preparation of the CEF cells, P. Snoy and R. Olsen for veterinary services, and K. Peden and H. Golding for comments on the manuscript.
REFERENCES
- 1.Agrba V Z, Kokosa L V, Cuvirov G N, Bierwolf D, Widmaier R, Graffi A, Lapin B A, Sangulia I A, Rudolph M. Isolation of foamy virus type II out of haematopoietic cells from baboons with haemagoblastoses and from healthy animals. Arch Geschwulstforsch. 1978;48:97–111. [PubMed] [Google Scholar]
- 2.Aguzzi A. The foamy virus family: molecular biology, epidemiology and neuropathology. Biochim Biophys Acta. 1993;1155:1–24. doi: 10.1016/0304-419x(93)90019-9. [DOI] [PubMed] [Google Scholar]
- 3.Ali M, Taylor G P, Pitman R J, Parker D, Rethwilm A, Cheingsong-Popov R, Weber J N, Bieniasz P D, Bradley J, McClure M O. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res Hum Retroviruses. 1996;12:1473–1483. doi: 10.1089/aid.1996.12.1473. [DOI] [PubMed] [Google Scholar]
- 4.Allan J S. Pathogenic properties of simian immunodeficiency viruses in nonhuman primates. In: Koff W, et al., editors. Annual review of AIDS research. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1991. pp. 191–206. [Google Scholar]
- 5.Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 6.Baker E F. Latent simian foamy virus. S Afr Med J. 1989;76:451–452. [PubMed] [Google Scholar]
- 7.Benzair A B, Rhodes-Feuillette A, Lasneret J, Emanoil-Ravier R, Peries J. Purification and characterization of the major envelope glycoprotein of simian foamy virus type I. J Gen Virol. 1985;66:1449–1455. doi: 10.1099/0022-1317-66-7-1449. [DOI] [PubMed] [Google Scholar]
- 8.Benzair A B, Rhodes-Feuillette A, Lasneret J, Emanoil-Ravier R, Peries J. Purification and characterization of simian foamy virus type I structural core polypeptides. Arch Virol. 1986;87:87–96. doi: 10.1007/BF01310545. [DOI] [PubMed] [Google Scholar]
- 9.Cavalieri F, Rhodes-Feuillette A, Benzair A B, Emanoil-Ravicovitch R, Peries J. Biochemical characterization of simian foamy virus type I. Arch Virol. 1981;68:197–202. doi: 10.1007/BF01314572. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Nonhuman primate spumavirus infections among persons with occupational exposure in United States, 1996. Morbid Mortal Weekly Rep. 1997;46:129–131. [PubMed] [Google Scholar]
- 11.Clarke J K, Samuels J, Dermott E, Gay F W. Carrier cultures of simian foamy virus. J Virol. 1970;5:624–631. doi: 10.1128/jvi.5.5.624-631.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornetta K, Morgan R A, Gillio A, Sturm S, Baltrucki L, O’Reilly R, Anderson W F. No retroviremia or pathology in long-term follow-up of monkeys exposed to a murine amphotropic retrovirus. Hum Gene Ther. 1991;2:215–219. doi: 10.1089/hum.1991.2.3-215. [DOI] [PubMed] [Google Scholar]
- 13.DiGiacomo R F, Hooks H J, Sulima M P, Gibbs C J, Gajudesek C. Pelvic endometriosis and simian foamy virus infection in a pigtailed macaque. J Am Vet Med Assoc. 1977;171:859–861. [PubMed] [Google Scholar]
- 14.Donahue R E, Kessler S W, Bodine D, McDonagh K, Dunbar C, Goodman S, Agricola B, Byrne E, Raffeld M, Moen R, Bacher R J, Zsebo K M, Nienhuis A W. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176:1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman M D, Dunnick N R, Barry D W, Parkman P D. Isolation of foamy virus from rhesus, African green and cynomolgus monkey leukocytes. J Med Primatol. 1975;4:287–295. doi: 10.1159/000459871. [DOI] [PubMed] [Google Scholar]
- 16.Fleming W A, Clarke J K. Fluorescence assay of foamy virus. J Gen Virol. 1970;6:277–284. doi: 10.1099/0022-1317-6-2-277. [DOI] [PubMed] [Google Scholar]
- 17.Goepfert P A, Ritter G D, Jr, Peng X, Gbakima A A, Zhang Y, Mulligan M M. Analysis of West African hunters for foamy virus infections. AIDS Res Hum Retroviruses. 1996;12:1725–1730. doi: 10.1089/aid.1996.12.1725. [DOI] [PubMed] [Google Scholar]
- 18.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri S, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T M. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 19.Hooks J J, Detrick-Hooks B. Simian foamy virus-induced immunosuppression in rabbits. J Gen Virol. 1979;44:383–390. doi: 10.1099/0022-1317-44-2-383. [DOI] [PubMed] [Google Scholar]
- 20.Hooks J J, Detrick-Hooks B. Spumavirinae: foamy virus group infections. Comparative aspects and diagnosis. In: Kurstak E, Kurstak C, editors. Comparative diagnosis of viral disease. New York, N.Y: Academic Press, Inc.; 1981. pp. 599–618. [Google Scholar]
- 21.Hooks J J, Gibbs J., Jr The foamy viruses. Bacteriol Rev. 1975;39:169–185. doi: 10.1128/br.39.3.169-185.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston P B. A second immunologic type of simian foamy virus: monkey throat infections and unmasking by both types. J Infect Dis. 1961;109:1–9. doi: 10.1093/infdis/109.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Johnston P B. Taxonomic features of seven serotypes of simian and ape foamy viruses. Infect Immun. 1971;3:793–799. doi: 10.1128/iai.3.6.793-799.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr J F R, Harmon B V. Definition and incidence of apoptosis: a historical perspective. In: Tomei L D, Cope F O, editors. Apoptosis: the molecular basis of cell death. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 5–50. [Google Scholar]
- 25.Khan A S. Retrovirus screening of vaccine cell substrates. Dev Biol Stand. 1996;88:155–160. [PubMed] [Google Scholar]
- 26. Khan, A. S., et al. Unpublished data.
- 27.Kupiec J-J, Kay A, Hayat M, Ravier R, Peries J, Galibert F. Sequence analysis of the simian foamy virus type 1 genome. Gene. 1991;101:185–194. doi: 10.1016/0378-1119(91)90410-d. [DOI] [PubMed] [Google Scholar]
- 28.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergia A, Leung N J, Blackwell J. Cell tropism of the simian foamy virus type 1 (SFV-1) J Med Primatol. 1996;25:2–7. doi: 10.1111/j.1600-0684.1996.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 30.Morris J A, Saglam M, Bozeman F M. Recovery of a new syncytium virus from a cottontail rabbit. J Infect Dis. 1965;115:495–499. doi: 10.1093/infdis/115.5.495. [DOI] [PubMed] [Google Scholar]
- 31.National Research Council. Guide for the care and use of laboratory animals. Washington, D.C: National Academy Press; 1996. [Google Scholar]
- 32.Neumann-Haefelin D, Rethwilm A, Bauer G, Gudat F, zur Hausen H. Characterization of a foamy virus isolated from Cercopithecus aethiops lymphoblastoid cells. Med Microbiol Immunol. 1983;172:75–86. doi: 10.1007/BF02124508. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien T C, Albrecht P, Schumacher H P, Tauraso N M. Isolation of foamy virus types 1 and 2 from primary rhesus monkey brain cultures. Proc Soc Exp Biol Med. 1971;137:1318–1323. doi: 10.3181/00379727-137-35780. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien T C, Albrecht P, Hannah J, Tauraso N, Robbins B, Trimmer R. Foamy virus serotypes 1 and 2 in rhesus monkey tissues. Arch Gesamte Virusforsch. 1972;38:216–224. doi: 10.1007/BF01249672. [DOI] [PubMed] [Google Scholar]
- 35.Renne R, Friedl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3) Virology. 1992;186:597–608. doi: 10.1016/0042-6822(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes-Feuillette A, Saal F, Lasneret J, Dubouch P, Peries J. Isolation and characterization of a new simian foamy virus serotype from lymphocytes of a Papio cynocephalus baboon. J Med Primatol. 1979;8:308–320. doi: 10.1159/000460216. [DOI] [PubMed] [Google Scholar]
- 37.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnitzer T J. Phenotypic mixing between two primate oncoviruses. J Gen Virol. 1979;42:199–206. doi: 10.1099/0022-1317-42-1-199. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer M, Fleps U, Jackle A, Renne R, Turek R, Neumann-Haefelin D. Simian foamy virus type 3 (SFV-3) in latently infected Vero cells: reactivation by demethylation of proviral DNA. Virology. 1993;192:663–666. doi: 10.1006/viro.1993.1084. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer K-O, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. Markers of foamy virus infections in monkeys, apes, and accidently infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res Hum Retroviruses. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer M, Falcone V, Gange J, Turek R, Neumann-Haefelin D. Simian foamy virus isolated from an accidentally infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shahabuddin, M., et al. Unpublished data.
- 43.Swack N S, Hsiung G D. Pathogenesis of simian foamy virus infection in natural and experimental hosts. Infect Immun. 1975;12:470–474. doi: 10.1128/iai.12.3.470-474.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teich N, Wyke J, Kaplan P. Pathogenesis of retrovirus-induced disease. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses: molecular biology of tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. pp. 187–248. [Google Scholar]
- 45.von Laer D, Neumann-Haefelin D, Heeney J L, Schweizer M. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology. 1996;221:240–244. doi: 10.1006/viro.1996.0371. [DOI] [PubMed] [Google Scholar]
- 46.Voss G, Nick S, Stahl-Hennig C, Ritter K, Hunsmann G. Generation of macaque B lymphoblastoid cell lines with simian Epstein-Barr-like viruses: transformation procedure, characterization of cell lines and occurrence of simian foamy virus. J Virol Methods. 1992;39:185–195. doi: 10.1016/0166-0934(92)90137-3. [DOI] [PubMed] [Google Scholar]
- 47.Weiss R A. A virus in search of a disease. Nature (London) 1988;333:497–498. doi: 10.1038/333497a0. [DOI] [PubMed] [Google Scholar]
- 48.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]