Abstract
Preclinical studies on gene delivery into mouse lymphocytes are often hampered by insufficient activity of lentiviral (LV) and adeno-associated vectors (AAVs) as well as missing tools for cell type selectivity when considering in vivo gene therapy. Here, we selected designed ankyrin repeat proteins (DARPins) binding to murine CD8. The top-performing DARPin was displayed as targeting ligand on both vector systems. When used on engineered measles virus (MV) glycoproteins, the resulting mCD8-LV transduced CD8+ mouse lymphocytes with near-absolute (>99%) selectivity. Despite its lower functional titer, mCD8-LV achieved 4-fold higher gene delivery to CD8+ cells than conventional VSV-LV when added to whole mouse blood. Addition of mCD8-LV encoding a chimeric antigen receptor (CAR) specific for mouse CD19 to splenocytes resulted in elimination of B lymphocytes and lymphoma cells. For display on AAV, the DARPin was inserted into the GH2-GH3 loop of the AAV2 capsid protein VP1, resulting in a DARPin-targeted AAV we termed DART-AAV. Stocks of mCD8-AAV contained similar genome copies as AAV2 but were >20-fold more active in gene delivery in mouse splenocytes, while exhibiting >99% specificity for CD8+ cells. These results suggest that receptor targeting can overcome blocks in transduction of mouse splenocytes.
Keywords: splenocytes, gene therapy, viral vectors, chimeric antigen receptor, CAR, AAV capsid engineering, cancer immunotherapy
Graphical abstract

Mouse models are crucial for translating viral vectors into clinical gene therapy and cancer immunotherapy. For gene delivery to mouse T lymphocytes, Buchholz and colleagues addressed lentiviral and AAV vectors to murine CD8, a marker on cytotoxic T cells, which resulted in strong gene transfer with high target-cell selectivity.
Introduction
This is a pivotal time for gene therapy research. Even as new milestones are achieved, old and new setbacks caution against optimism in favor of thorough and robust preclinical work. Approved gene therapy products demonstrate the real-world efficacy of gene therapy using viral vectors. Autologous chimeric antigen receptor (CAR) T cell products achieve response rates not seen for previous benchmark therapies in patients with advanced hematological cancer,1,2 and in vivo delivery of the SMN1 gene with vectors based on adeno-associated virus 9 (AAV9) improves the quality of life of children with spinal muscular atrophy.3 In addition to these and other approved gene therapy medicinal products, many clinical trials are ongoing worldwide that promise benefit for patients suffering from severe diseases.4, 5, 6 However, adverse events such as in the insertional oncogenesis caused upon retroviral-vector-mediated gene transfer into hematopoietic stem cells of patients with X-linked severe combined immunodeficiencies7,8 or the recent fatalities in clinical trials for AAV8-based gene therapy demand diligent preclinical research.9
To this end, animal models are required that satisfy high standards of representativeness. Humanized mouse models are crucial for the demonstration of in vivo activity and biodistribution of viral vectors. Still, some important aspects require (confirmatory) studies in syngeneic mouse models. Especially in cancer immunotherapy, syngeneic mouse models—which rely on a fully active murine immune system and compatibility with the chosen tumor type—can be essential for safety and proof-of-concept demonstration to enter into early clinical trials. For example, proof-of-concept data for the now-approved CAR T cell product axicabtagene ciloleucel were obtained in a syngeneic mouse model.10 CAR T cells are based on synthetic signaling molecules constructed from domains of several immune receptors that enable T cell receptor (TCR)-independent activation of T cells upon encounter of tumor markers.11,12 In all approved CAR T cell therapies, patient T cells are armed with CARs ex vivo—most commonly using retro- or lentiviral vectors—and then re-infused into the patient. While preclinical CAR T cell studies are often restricted to immunodeficient mouse models transplanted with human CAR T cells, additional studies in syngeneic mouse models can provide important information about CAR T cell interactions with the immune system, especially when studying the tumor microenvironment.13,14
However, use of syngeneic mouse models, especially when genetically manipulating T lymphocytes, can be challenging for LV and AAV vectors due to incompatibilities with murine cell biology. Therefore, there is a need for vectors mediating efficient gene delivery into murine cells. Hypothesizing that efficient receptor recognition and cell entry can at least partially overcome this problem, targeting LV and AAV vectors to surface markers of mouse T lymphocytes appears as a straightforward strategy.
For both vector types, receptor targeting relies on surface engineering, which essentially includes two modifications: the elimination of natural receptor binding and the display of targeting ligands, such as designed ankyrin repeat proteins (DARPins), exhibiting high affinity for the target receptor.15 We have established this approach for AAV2 by displaying a Her2/neu-specific DARPin at the N terminus of the capsid protein VP2 and eliminating binding to heparan-sulfate proteoglycan (HSPG) through two point mutations.16 Later, this approach was extended to other surface markers.17, 18, 19 Based on work with randomized reporter insertion, the GH2/3 loop of the AAV2 capsid has been suggested as an alternative insertion site, potentially offering more exposed surface display of targeting ligands.20 Recently, retargeting of AAVs by insertion of nanobodies into the GH2/3 loop was demonstrated.21
Receptor-targeted LVs (RT-LVs) are enveloped particles pseudotyped with engineered envelope glycoproteins of the paramyxoviridae measles virus (MV) or Nipah virus. The attachment protein, which in the case of MV is the hemagglutinin H, is fused to the targeting ligand and blinded for its natural receptors. At this time, more targeted LVs than AAVs have been described, including RT-LVs directed against surface markers on human lymphocytes such as CD4, CD8, and CD3.22,23 All these RT-LVs have been shown to selectively deliver CARs to the targeted lymphocyte subtypes not only in vitro but even in vivo, enabling the generation of CAR T cells upon a single vector injection into humanized mice.24, 25, 26
Here, we demonstrate an adaptable workflow that resulted in the generation of mouse CD8-targeted LV and AAV vectors. Murine CD8 (mCD8), which only has negligible identity to its human counterpart (<30% query coverage and <70% identity), was chosen as target receptor due to its function as distinguished marker of cytotoxic T cells, co-enabling recognition of antigens presented on major histocompatibility complex (MHC) class I by the T cell receptor. Our data demonstrate high gene transfer activity into murine CD8+ T lymphocytes by both mCD8-LV and mCD8-AAV, concurrent with high selectivity for the target cells.
Results
To generate DARPins specific for murine CD8, a heterodimer of CD8α and CD8β was generated by fusing each chain to human IgG Fc domains, which were modified to associate as knob-hole pairs27 (Figure 1A). After purification, the protein was immobilized for selection of DARPin libraries by ribosome display (Figure 1B). Three selection rounds were performed, followed by one round of selection in solution, which included a counterselection with the CD8αα-Fc homodimer. After selection, crude lysates of DARPin-expressing E. coli were assessed by ELISA for binding to the CD8αβ-Fc heterodimer and the CD8αα-Fc homodimer. Signals at least one order of magnitude above background were observed for 14 DARPins. Most bound both CD8 dimers, indicating recognition of the α chain. Four DARPins (MSF9, MSG10, MSF11, and MSB12) apparently recognized the β-chain, since signals were observed only for the CD8αβ-Fc heterodimer (Figure 1C). Next, binding of crude lysate-derived DARPins- to mouse splenocytes was determined. Costaining with antibodies distinguished between cells expressing the CD8αβ heterodimer or the CD8αα homodimer. Four DARPins (MSD9, MSE10, MSH11, and MSC12) were found to be the top binders (Figure 1D). All of them were CD8α binders, since no DARPin was found to bind to CD8β+ cells only. Since MSE10 turned out to be most efficient in binding to cell-exposed CD8 also on cell lines (Figure S1), this DARPin was selected for display on vector particles.
Figure 1.
Selection of DARPins for murine CD8
(A) To-scale approximation of the structure of CD8αβ-Fc generated from structures PDB: 2ATP, 4NQT. Knob mutations are indicated in red, hole mutations in blue. CH, constant heavy domain of Fc; Avi, Avi-tag. (B) Workflow of ribosome display for DARPin selection. An mRNA library was generated from the DNA DARPin library (1) and then translated in a combined reaction that yields polypeptide bound to ribosomes arrested on the mRNA (2). These ternary complexes were then subjected to pre-selection before being bound to immobilized CD8αβ-Fc and washed stringently (3). After washing, ternary complexes were eluted, RNA was reverse-transcribed and amplified by reverse transcriptase PCR, and adapters were ligated before another round of screening was initiated (4a). For further characterization, the PCR product was cloned into expression plasmids (4b). The inlay shows a ribbon representation of the DARPin structure, including N- and C-terminal caps (NC and CC) as well as internal repeats (IR), generated from structure PDB: 4J8Y. (C) Screening results for the indicated output DARPins. Crude lysates from E. coli expressing the DARPins selected by ribosome display were tested for binding to CD8αβ-Fc (light green bars), CD8αα-Fc (dark green bars), or huFc (gray bars) by ELISAs. DARPin binding was detected colorimetrically with HRP-coupled anti-HA antibodies. Dotted line indicates background set at maximum signal observed for binding to huFc. Bars represent data from n = 1 run. Orange triangles indicate DARPins MSD9, MSE10, MSH11, and MSC12. (D) Screening results for binding of DARPins to murine splenocytes. Crude lysates from E. coli expressing the DARPins were used to decorate murine C57BL/6 splenocytes. DARPins were detected via their HA tags using anti-HA-PE antibodies. In parallel, cells were stained either for CD8α or CD8β, respectively. Results were then gated for CD8α+ cells (light green bars), CD8β+ cells (dark green bars), and CD8-negative cells (gray bars). Bars represent data from n = 1 mouse. Orange triangles indicate top-performing DARPins MSD9, MSE10, MSH11, and MSC12. See also Figure S1.
Building on previous work, the C terminus of receptor-blinded, truncated hemagglutinin (HΔ18) protein from MV was fused genetically to the N terminus of MSE10 via a G4S linker (Figure 2A). Pseudotyped LV stocks were purified and concentrated from cell culture supernatant by sucrose cushion centrifugation. Characterization of eleven stocks of mCD8-LV and nine stocks of VSV-LV (pseudotyped with the G protein of vesicular stomatitis virus) by nanoparticle tracking analysis revealed a consistently smaller size of VSV-LV than mCD8-LV, with mean particle sizes of 118.0 nm (95% confidence interval [CI]: 114.6–121.3 nm) for VSV-LV and 130.9 nm (95% CI: 127.2–134.6 nm) for mCD8-LV (Figures 2B and 2C). This is in line with previous data describing that RT-LVs pseudotyped with modified paramyxovirus glycoproteins are larger than VSV-LVs.28 Particle counts of the LV stocks were usually slightly higher for VSV-LV than for mCD8-LV after 150-fold or 333-fold concentration (Figure 2D). Both vector stocks were produced using identical amounts of packaging plasmid. The difference in particle counts between LVs pseudotyped with VSV-G and paramyxovirus glycoproteins has been previously observed and may arise from the vesicle-forming activity of VSV-G protein29 and/or the use of two versus three plasmids with a cytomegalovirus (CMV) promoter during production, which may have resulted in sequestration of transcription factors.
Figure 2.
Displaying MSE10 on LV particles mediates selective gene transfer
(A) To-scale approximation of vector mCD8-LV structure. MV H protein is blinded for its natural receptors by point mutations and fused at its C terminus with DARPin MSE10 directed against mCD8. The inlay illustrates the envelope protein complex. MV HΔ18 is shown as a dimer, MV FΔ30 as a trimer. The illustration was created using structures PDB: 5YZD, 2ZB5, and cellPACK recipe HIV-1_0.1.6. (B–D) Characterization of mCD8-LV stocks by nanoparticle tracking analysis. Particle size distribution and concentration were determined using a NanoSight NS300 analyzer. VSV-LV served as a reference. (B) Example histograms from one measurement. Size modes ± standard error of VSV-LV (dashed line, blue bands) and mCD8-LV (solid line, red bands) are indicated. Bands represent standard error of n = 4 sequential technical replicates. (C) Particle sizes and (D) particle concentrations of 9 VSV-LV stocks and 11 mCD8-LV stocks from 5 measurement sessions, respectively. Each symbol represents one vector stock. Solid lines in (C) are arithmetic means. Error bars represent 95% confidence intervals (CIs). White symbols in (D) indicate 150-fold concentration, colored symbols 333-fold concentration. (E–G) Titration of gene transfer activity on murine Pan T cells. Splenic Pan T cells from BALB/c mice were thawed. 4 × 104 cells/well stimulated with αmCD3/αmCD28-beads and recombinant human IL7 and IL15 (rhIL7+15) were transduced with serial dilutions of GFP-transferring VSV-LV or mCD8-LV by spinfection. Cells were analyzed by flow cytometry and qPCR 5 days post spinfection. Symbols represent data from n = 3 mice. (E) Percentage of GFP+ viable singlet cells. Transducing titers for VSV-LV (blue triangles) and mCD8-LV (red circles) stocks are shown. (F) Geometric mean fluorescence intensities (MFIs) of GFP+ viable singlet cells at the indicated particle doses. (G) Representative flow cytometry plots showing GFP versus CD8 signals of viable singlet cells from one mouse. Vector dose was 1 × 1010 particles/well. (H) Numbers of integrated vector copies per cell (VCNs) for VSV-LV (blue triangles) and mCD8-LV (red circles) stocks. DNA was extracted from total cells, without prior sorting. (I and J) Titration of VSV-LV and mCD8-LV stocks equipped with promoter variants. 4 × 104 Pan T cells per well stimulated with αmCD3/αmCD28-beads and recombinant human IL7 and IL15 (rhIL7+15) were transduced with the indicated amounts of GFP-transferring VSV-LV (blue) or mCD8-LV (red) stocks carrying GFP under control of either SFFV (●), MCMV (▪), EF1a (▴), or PGK (▾) promoter. Cells were analyzed by flow cytometry 5 days post spinfection. (I) Percentage of GFP+ viable singlet cells. Symbols represent means of data from n = 3 mice. Red symbols indicate mCD8-LV, blue symbols VSV-LV. (J) MFI of GFP+ viable singlet cells. See also Figure S2.
For gene transfer experiments, Pan T cells from BALB/c mice were activated using αmCD3/αmCD28-beads and recombinant human IL7 and IL15 (rhIL7+15). The cells were incubated with vector doses ranging from 2.5 × 105 to 3.2 particles/cell of VSV-LV or mCD8-LV carrying gfp as reporter gene. Cultures were analyzed for the presence of GFP by flow cytometry and for vector integration by quantitative PCR (qPCR) 5 days later. Remarkably, mCD8-LV was very active in gene delivery, its transducing titer being within one order of magnitude of VSV-LV (Figures 2E and 2F). In contrast to the latter, however, gene delivery was highly selective for CD8+ splenocytes, resulting in a decrease in CD8 intensity, which was likely due to masking of the epitope for antibody staining (Figure 2G). The highest dose (i.e., 2.5 × 105 particles/cell) resulted in approximately 60% GFP+ cells for VSV-LV and 40% for mCD8-LV. Notably, mCD8-LV reached a plateau of 40% GFP+ cells already with 2 × 109 particles/well. The earlier plateau and steeper increase in MFI (mean fluorescence intensity) of cells treated with mCD8-LV is consistent with mCD8-LV only transducing a subpopulation of target cells (i.e., CD8+ cells). Transducing titers were 1.07 × 109 transducing units (TU) per mL for VSV-LV and 1.17 × 108 TU/mL for mCD8-LV (Figures 2E and 2F). This difference was well in line with the vector copy numbers (VCNs), which were approximately one order of magnitude higher for cells transduced with VSV-LV compared to cells incubated with mCD8-LV for all particle counts (range: 5.0- to 11.5-fold) (Figure 2H). At 2 × 109 particles/well, mean VCNs were 30.5 for VSV-LV-transduced cells and 2.9 for mCD8-LV-transduced cells. Gene transfer into primary mouse T lymphocytes was stable at least for a monitoring period of 2 weeks, and there was no detectable impact on viable cell counts relative to the untransduced control with both VSV-LV and mCD8-LV (Figure S2).
To assess the influence of the chosen promoter, Pan T cells were next transduced with defined amounts of particles carrying the GFP reporter under the control of promoters from the spleen focus-forming virus (SFFV), the human elongation factor 1a (EF1a), phosphoglycerate kinase (PGK), or the murine CMV (MCMV) immediate early promoter. We observed no relevant difference in GFP expression, indicating that all four promoters are equally suitable for gene expression in the mouse system at least for short-term cultivation (Figures 2I and 2J). Accordingly, the SFFV promoter was used for all further work.
Having determined the basic properties of mCD8-LV, we next set out to assess the gene transfer activity of mCD8-LV in whole mouse blood. Blood from BALB/c mice was incubated with 2 × 1010 particles/150 μL (i.e., approximately 5 × 104 particles/leukocyte) in the presence of rhIL7. The cultures were agitated on a shaker at 180 rpm at 37°C for 6 h to simulate vector binding in circulating blood. After allowing for vector binding, erythrocytes were cleared, and the remaining cells were activated with αmCD3/αmCD28-beads and cultivated for 5 days in the presence of rhIL7+15 before cells were analyzed by flow cytometry and qPCR (Figure 3A). Both LVs transduced over 5% of total cells (Figures 3B and 3C). After treatment with mCD8-LV, over 20% of CD8+ cells were GFP+, while only around 5% GFP-positivity among CD8+ cells was achieved with VSV-LV. For VSV-LV, only 25.5% of GFP+ cells generated were CD8+ (95% CI: 22.4%–28.6%), while for mCD8-LV, 99.2% of the GFP+ cells were CD8+ (95% CI: 97.0%–101.4%) (Figure 3E). The transduction rate on CD8+ cells was more than 5-fold higher with mCD8-LV than with VSV-LV, reaching up to 27.5% of viable CD8+ cells. As observed on Pan T cells, genomic integration was lower for mCD8-LV than for VSV-LV (Figure 3F).
Figure 3.
mCD8-LVs specifically transduce CD8+ cells in murine whole blood
Blood from three BALB/c mice collected in lithium heparin tubes was transferred to 48-well plates and mixed with rhIL7 before VSV-LVs or mCD8-LVs were added at 2 × 1010 particles/well. Blood was incubated at 37°C for 6 h at 180 rpm. Subsequently, erythrocytes were removed by centrifugation over a Histopaque cushion. Purified cell preparations were then mixed with αmCD3/αmCD28-beads and rhIL7+15 and cultured for 5 days before analysis by flow cytometry and qPCR. (A) Workflow. (B) Representative flow cytometry plots showing GFP versus CD8 signals of viable singlet cells from one mouse. (C) Percentage of GFP+ viable singlet cells, determined by flow cytometry. (D) Percentage of GFP+ cells among viable singlet CD8+ cells. (E) Percentage of CD8+ cells among GFP+ cells. p value is from paired t test. (F) Vector copies/cell. DNA was extracted from total cells, without prior sorting. Symbols represent data from n = 3 mice, bars represent means, error bars represent 95% CIs.
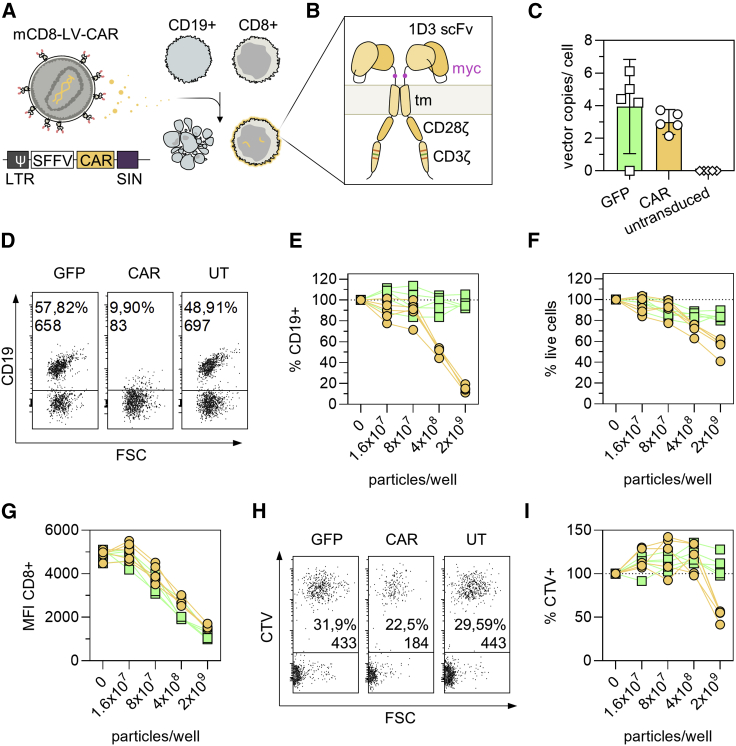
Next, we devised an experiment to demonstrate the use of mCD8-LV for transfer of a CAR gene (Figure 4). This CAR covers a scFv specific for murine CD19 as well as murine CD3ζ stimulatory and CD28 costimulatory domains10 (Figure 4A). Whole splenocytes (containing endogenous CD19+ cells) from BALB/c mice were cultivated without full activation, only in the presence of IL7 either alone or as 10:1 coculture with CD19+ A20 cells, a murine lymphoma cell line derived from BALB/c (Figure 4B). Cultures were transduced with mCD8-LVCAR or the corresponding GFP-transferring mCD8-LVGFP as control and analyzed by flow cytometry and qPCR 2 days later. At this time point and in the absence of activation, CAR and GFP levels were low, yet clearly detectable over background (Figure S3). VCNs were in the expected range of 1–3 copies/cell (Figure 4C). In line with this, a strong decrease in the frequency of CD19+ splenocytes was detectable in splenocyte-only cultures that had received mCD8-LVCAR. Addition of 2 × 109 particles of mCD8-LVCAR decreased the numbers of CD19+ cells to less than 20% of the content of the controls (Figure 4D). This effect was dose dependent, becoming apparent at 8 × 107 particles, and was most pronounced at 2 × 109 particles (Figure 4E). Additionally, the total number of viable cells in the culture decreased to approximately 60% of that of the untreated control (Figure 4F). This confirms that loss of the CD19 signal was due not simply to masking but indeed to target cell killing. Underlining the vectors’ specificity, the CD8 signal decreased with increasing vector dose likely caused by (partial) masking of the epitope of the CD8 antibody by the bound vector particles (Figure 4G). We also detected specific killing of A20 cells in cocultures with splenocytes. In routine cell culture, A20 cells proliferated aggressively, usually requiring 1:15 subculturing every 2 days. Yet, the highest dose of mCD8-LVCAR (2 × 109 particles/well) caused substantial reduction of A20 cells to about 50% of the control culture (Figures 4H and 4I).
Figure 4.
CAR gene delivery with mCD8-LV
4 × 104 BALB/c splenocytes were cultivated alone or in co-culture with 4 × 103 A20 tumor cells stained with Cell Trace Violet (CTV). Cultures were treated with defined particle amounts of mCD8-LV encoding GFP or a chimeric antigen receptor (CAR) directed against mCD19. After spinfection, cells were cultivated for 2 days in the presence of rhIL7 before analysis by flow cytometry and qPCR. (A) Workflow. (B) CAR structure. The CAR used in this experiment was previously published. It employs a single-chain variable fragment (scFv) of the ID3 monoclonal antibody against mouse CD19. It is a second-generation CAR with a CD3ζ stimulatory and CD28ζ costimulatory domain. The first and third immunoreceptor tyrosine-based activation motifs (ITAMs) were ablated by mutation to decrease T cell apoptosis and increase survival in vivo.10 A myc tag (purple) between scFv and transmembrane domain (tm) facilitates detection of the CAR. (C) Similar VCNs were obtained for both vector stocks. Symbols represent data of lymphocytes from n = 5 mice. Bars represent means. Error bars represent 95% CIs. DNA was extracted from total cells, without prior sorting. (D–G) Killing of CD19+ B lymphocytes in splenocyte-only cultures. (D) Representative flow cytometry plots from one mouse showing CD19 signals of viable singlet cells after no treatment or transduction with 2 × 109 particles/well. Percent CD19 positivity and CD19+ cell counts are reported. (E) Percentage of CD19+ cells relative to untransduced control. (F) Percentage of viable cells relative to untransduced control. (G) Geometric MFI of CD8+ cells. (E–G) Symbols represent data of lymphocytes from n = 5 mice treated with mCD8-LVGFP (green squares) or mCD8-LVCAR (yellow circles). (H and I) Killing of A20 target cells in cocultures. Flow cytometry data, obtained from viable singlet cell populations are reported. (H) Representative flow cytometry plots from one mouse after no treatment or transduction with 2 × 109 particles/well. CTV+ cells are A20 tumor cells. (I) Percentage of CTV+ (A20) cells relative to untransduced control. Symbols represent data from n = 5 mice treated with mCD8-LVGFP (green squares) or mCD8-LVCAR (yellow circles). See also Figures S3 and S4.
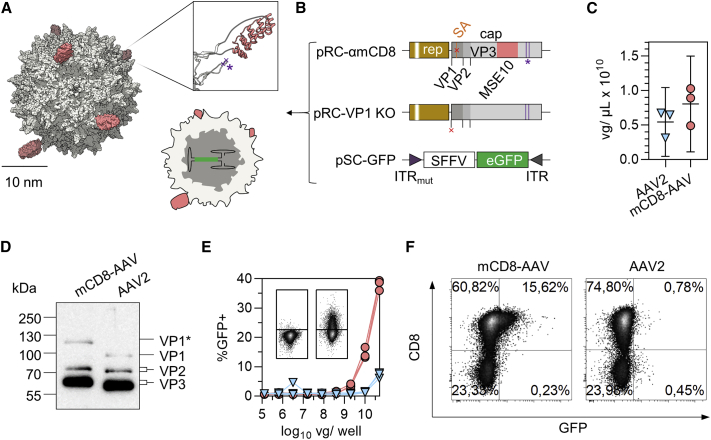
Having established that MSE10-LV mediates efficient and selective gene transfer of reporter and therapeutic genes into murine CD8+ T lymphocytes, we next displayed this DARPin on the surface of AAV2 vector particles (Figure 5A). The coding sequence of MSE10 was inserted into the GH2/3 loop of VP1 and the production of VP2 and VP3 was prevented by deletion of the splice acceptor site downstream of the VP1 translation initiation site resulting in pRC-mCD8. In pRC-VP1KO, the VP1 start codon was deleted, thus encoding VP2 and VP3 only (Figure 5B). In both plasmids, the natural affinity for HSPG was ablated by point mutations as demonstrated before.16 The corresponding mCD8-AAV particles were produced by packaging the GFP reporter followed by purification via an iodixanol gradient. Vector genome (vg) counts in mCD8-AAV stocks and AAV2 stocks were similar (Figure 5C). Western blot analysis of purified particle stocks revealed the presence of all three VP proteins exhibiting the expected large shift in electrophoretic mobility for MSE10-VP1 as well as slight shifts due to the point mutations in VP2 and VP3 (Figure 5D). Yet, performance of the vectors on primary cells was markedly different. At the same doses of vg copies, mCD8-AAV generated 10-fold more GFP+ T cells than AAV2, reaching 38.1% transduced cells at the highest dose, while exhibiting strong preference for CD8+ cells (Figures 5E and 5F).
Figure 5.
Generation and characterization of mCD8-AAV
(A) Structure of mCD8-AAV. To-scale approximation of electron density surfaces of AAV2 capsid (gray) decorated with DARPins (red). Cartoon shows cross-section of particle with self-complementary (sc) genome. Inlay shows ribbon representation of DARPin inserted in the GH2-GH3 loop of AAV2 VP1. Purple asterisk and crosses indicate locations of point mutations, which ablate HSPG binding (R585A, R588A). Panel was created using structures PDB: 1LP3, 4J8Y. (B) Generation of mCD8-AAV. Vectors were produced by transfection of HEK293T cells with the indicated plasmids. The genetic structure of cap (gray) is visualized. Start codons for VP1, VP2, and VP3 are indicated by black ticks. Point mutations ablating natural affinity for HSPG (R585A, R588A; HSPGmut) are indicated by purple lines and asterisk, the splice acceptor site downstream of VP1 (SA) in orange, deletions of SAS or start codon with red crosses. The inverted terminal repeats (ITRs) are indicated. (C) Vector genome (vg) titers. Symbols represent technical replicates from three extractions and PCRs. Error bars represent 95% CIs. (D) Detection of MSE10-VP1 (labeled with VP1∗) by western blotting. 2 × 1010 genome copies of vector stock were lysed in urea buffer and run on SDS-PAGE. After blotting, AAV2 capsid was visualized using the B1 monoclonal antibody. Blot was exposed for 7 s. Contrast was adapted, retaining relative pixel intensities. (E and F) Pan T cells (4 × 104 cells/well) from BALB/c mice fully activated with αmCD3/αmCD28-beads and rhIL7+15 were transduced by spinfection with AAV2 (blue triangles) or mCD8-AAV (red circles) at the indicated doses of vg/well. Cultures were analyzed 5 days post transduction by flow cytometry. (E) Graph summarizes the percentage of GFP+ viable singlet cells. Symbols represent data from n = 3 mice. Representative flow cytometry plots in inlay from one mouse show GFP signals versus FSC in wells treated with 1.28 × 105 vg/well (left) and 5 × 1010 vg/well (right) mCD8-AAV. (F) Representative flow cytometry plots show percentages of GFP+ versus CD8+ cells from one mouse.
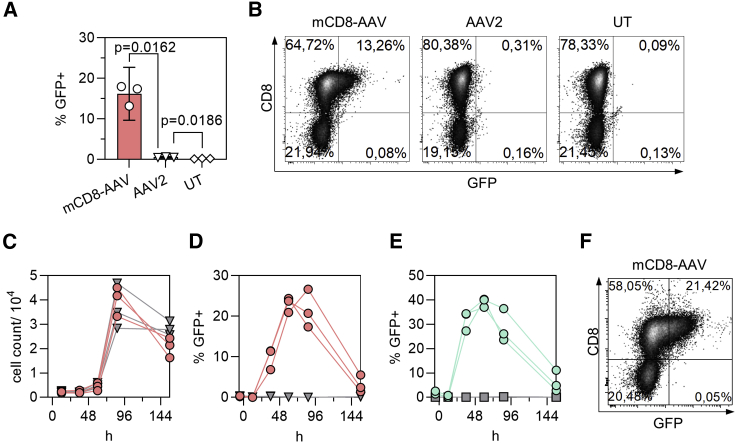
On whole splenocytes, which contain high amounts of off-target cells, especially B lymphocytes, mCD8-AAV was not only highly efficient in gene transfer but with >99% specificity also highly selective for CD8+ cells (Figures 6A and 6B). With AAV2, in contrast, the fraction of GFP+ cells was 26-fold lower (0.57% versus 16.2%) (Figure 6A). As expected for a non-integrating vector, the rate of GFP+ cells peaked between 2 and 4 days after vector particles had been added to the cells and then decreased over time (Figures 6C–6F). Notably, over the whole monitoring period, only CD8+ cells appeared GFP+ (Figures 6E and 6F), and there was no negative impact on cell viability (Figure 6C).
Figure 6.
Displaying MSE10 on AAV2 mediates selective transient transduction of murine splenocytes
Whole splenocytes from BALB/c mice were activated, transduced with 1 × 1010 vg/well (2.5 × 105 vg/cell) (A and B) or with 5 × 1010 vg/well (i.e., 1.25 × 106 vg/cell) (C–F). (A) Percentage of GFP+ viable singlet cells 5 days after start of incubation with mCD8-AAV, AAV2, or no treatment (UT). Symbols represent data from n = 3 mice. Bars represent means, error bars represent 95% CIs. Multiplicity-adjusted p values are from repeated-measures one-way ANOVA with Tukey’s multiple comparisons test. (B) Representative flow cytometry plots from one mouse 5 days after start of incubation with mCD8-AAV, AAV2, or no treatment (UT). (C–F) Transduction kinetics on primary cells. Symbols represent data from n = 3 mice. (C) Cell count over time after incubation with vector (red circles) or without (gray triangles). (D) GFP+ cells among viable singlets after incubation with vector (red circles) or without (gray triangles). (E) Specificity of gene transfer. GFP+ cells among CD8+ cells (cyan circles) or CD8− cells (gray squares) after treatment with mCD8-AAV. (F) Representative flow cytometry plot from one mouse at 86 h post vector addition.
Discussion
This paper describes LV and AAV vectors using mouse CD8 as attachment receptor. Key to this achievement was the identification of mCD8-specific DARPins. As versatile high-quality binders, they are instrumental for the rational engineering workflow resulting in receptor-targeted vectors. DARPins have favorable physicochemical properties, among them high stability, low immunogenicity, and the independence from post-translational modification like cysteine bridges or glycosylation, which turned out to be especially beneficial for the modification of the AAV capsid.15,30 We have previously reported that ribosome display can consistently and conveniently deliver such DARPins.31,32 Here, we adapted the DARPin selection approach by offering mCD8α and mCD8β subunits heterodimerized via knob-in-hole IgG Fc domains as bait in the selection process. Notably, with the exception of one candidate, all selected DARPins recognized the CD8α chain. This was curious, since a counterselection in solution had been implemented with the mCD8αα homodimer. However, when we previously selected human CD8 binders, the vast majority of obtained DARPins were specific for the α chain.32 This suggests that instead of counterselecting, the added mCD8αα rather caused off-rate selection, enriching for DARPins binding mCD8α with a low off-rate. Yet, with DARPins MSG10 and MSF11, candidate binders for the mouse CD8β chain were obtained, which are now available for further studies.
We are aware that DARPin MSE10 recognizing mCD8α mediates binding to all types of CD8+ cells, including—besides T lymphocytes—CD8+ dendritic and natural killer cells as well as γδ T cells. However, in this proof-of-concept study, we decided on MSE10, owing to the fact that its high binding efficiency offered the highest likelihood to mediate efficient transduction of mouse CD8+ lymphocytes upon vector engineering.
Compatibility of LVs with mouse lymphocytes is a well-known concern in the gene therapy field.33 Human immunodeficiency virus 1 (HIV-1) cannot productively infect murine cells, as there are several blocks in the infection cycle.34 Apart from the incompatibility of mouse CD4 and CCR5/CXCR4 with the HIV-1 fusion machinery, pre- and post-integrational blocks have been identified. These include blocks to nuclear import35 and chromosomal integration as well as post-integration blocks to Gag expression, processing, and release.36 In light of this, it was surprising that we achieved highly efficient gene delivery into mouse lymphocytes with our approach. However, one principal difference between infection with HIV-1 and transduction with LVs is the number of particles to which a single cell is exposed. During HIV-1 infection, each cell is exposed to far fewer particles than in LV transduction. During transduction, tens to hundreds of thousands of pseudotyped particles per cell may be applied, potentially outcompeting the activities of restriction factors. This holds true also for mCD8-LV, which was produced with particle numbers well in the range of those reported for VSV-LV or other RT-LVs such as hCD8-LV.28 It is also important to mention that, although we applied relatively high particle numbers, no relevant cytotoxicity was observed. Still, we cannot rule out any influence on T cell phenotype.
Promoter choice can be a critical issue, especially when switching from the human system to mouse cells. Previously, the EF1a or PGK promoters have been suggested for mouse lymphocytes instead of the SFFV promoter.37 Alternatively, use of the immediate early MCMV promoter has been recommended.38 Normalizing vector input to defined particle counts, we found no relevant difference between these promoters in transducing titers for both VSV-LV and mCD8-LV. Although the choice of promoter is apparently not relevant for short-term experiments, using a promoter other than SFFV may be beneficial for longer-term in vivo experiments, as inactivation of the SFFV promoter by methylation in mouse cells has previously been reported.39
Another factor critically influencing successful transduction is the activation status of the lymphocytes. While LVs—in contrast to retroviral vectors—are able to transduce non-dividing cells,40 cytokines are thought to contribute crucially to reverse transcription of the vector transgene in immune cells. The restriction factor SAMHD1, which is active against HIV in human and mouse lymphocytes, ensures low levels of dNTPs in non-dividing cells, restricting reverse transcription of LV payloads. Stimulation with IL7 is known to induce inactivating phosphorylation of SAMHD1, thus facilitating reverse transcription.41,42 In addition to cytokines, prolonged activation via T cell-receptor-agonistic antibodies or superantigens was shown to improve reverse transcription and chromosomal integration of mouse lymphocytes by HIV-136 and transduction of mouse lymphocytes with VSV-LV.37 Accordingly, mCD8-LV achieved highly specific transduction of CD8+ lymphocytes in whole blood of BALB/c mice, even though only IL7 was supplied initially. T cell activation was then performed to keep the cells alive in culture and enhance reporter gene expression for easy detection. Because of this, we cannot completely rule out that some transduction occurred after activation through particles sticking to the T cells. Yet, transduction under these conditions was likely facilitated by the inactivation of SAMHD1 by IL7 together with high particle numbers accumulating on CD8+ cells.
While the gene delivery activity of mCD8-LV on murine lymphocytes was below that of its non-targeted counterpart, VSV-LV, mCD8-targeting with AAV increased gene delivery rates by more than one order of magnitude compared to non-targeted AAV2. This was surprising, since AAV vectors are known to be rather inefficient on lymphocytes. Only recently, in vivo transduction of mouse lymphocytes with serotype-8-derived vectors was described.43 Our data suggest that efficient binding to T cells via CD8 enables efficient gene delivery with AAV2 vectors. This stresses the crucial role of high-affinity targeting ligands displayed on the particle surface. While future work needs to address the precise mechanism of cell entry of mCD8-AAV, previous work with a glutamate-receptor-targeted AAV, displaying DARPins on the N terminus of VP2, proved that entry of DARPin-targeted AAVs requires expression of the AAV receptor AAVR.19,44 Irrespective of mechanism, the unprecedentedly high gene delivery activity demonstrated for mCD8-AAV together with the flexibility of the targeting approach (in which exchanging the DARPin enables transduction via other receptors of interest) prompted us to call these vectors DART-AAVs (DARPin-targeted). CAR T cell generation is a potential application for both mCD8-targeted vectors. Here, we demonstrate killing of endogenous B cells and syngeneic murine tumor cells upon ex vivo CD19-CAR delivery with mCD8-LV. Notably, CAR-mediated killing was achieved without full T cell activation, stimulating only with IL7. This setting, in which T cells are in a non-activated mode, models in vivo CAR T cell generation, where vectors are injected systemically and CAR T cells develop in the organism.24
The availability of mCD8-LV now enables syngeneic mouse models for in vivo CAR T cell generation in the context of B cell lymphoma. While the integrative nature of LVs enables stable gene expression in rapidly dividing cells (such as activated CAR T cells), there is only limited control over the pharmacokinetics of transduction after in vivo administration. AAVs possess a favorable biosafety level compared to LVs (level 1 versus 2) and a proven safety profile for in vivo gene delivery. This may make them superior for clinical translation not only over LVs but also over CD8-targeted nanocarriers, which were recently shown to deliver CAR mRNA in tumor mouse models and for which only limited safety data exist so far.45 Yet, also for AAV-mediated CAR gene delivery, comprehensive preclinical research will be required. The high potency of mCD8-AAV on mouse lymphocytes facilitates studies in fully immunocompetent animals, thus allowing for a detailed characterization of the elicited immune responses against vector particles and CARs. Finally, it should be noted that the potential applications of the vectors described here go far beyond CAR therapy. By offering precise spatiotemporal control over transgene expression in mouse lymphocytes, either stably or transiently, they can become a valuable tool for basic and applied research.
Materials and methods
Plasmids and molecular cloning
CD8-Fc expression constructs were generated using the cloning strategy previously described for the generation of DARPins against human CD8.32 In brief, coding sequences for murine CD8α residues 21–181 (UniProt: P01731) and murine CD8β residues 21–161 (UniProt: P10300) were fused to that of the constant region of human immunoglobulin G1 (IgG1-Fc) in conjunction with a glycine-serine linker and an Avi tag (G4S-Avi). The plasmid pMP71-mCD8a includes the coding sequence of CD8a (kindly provided by Wolfgang Uckert, Berlin, Germany). The coding sequence of CD8β was synthesized de novo (GeneArt; Thermo Fisher Scientific, Regensburg, Germany). The IgG1-Fc domain followed by G4S-Avi is encoded in the mammalian expression vector phuFc-Avi.31 To generate CD8αα-Fc, the extracellular CD8α coding region (excluding disulfide residues responsible for dimerization) was PCR-amplified using primers 1 and 2 (Table S1), purified, and cloned into phuFc-Avi via digestion with NcoI/NotI to obtain pCD8α-huFc-Avi. The extracellular CD8β coding sequence (excluding disulfide residues responsible for dimerization) was cloned into phuFc-Avi harboring an additional XbaI restriction site downstream of the Avi tag (phuFc-AviX) via digestion with NcoI/NotI to obtain pCD8β-huFc-Avi. For generation of CD8αβ-Fc heterodimers, a strategy described by Merchant et al.27 and relying on knobs-into-holes mutations in the constant region of human IgG1 harboring a C-terminal Avi-tag (Fc) was used and performed as described before.32 The CD8α domain of pCD8α-huFc-Avi was cloned into phuFc-Aviknob via NcoI/NotI restriction sites to generate CD8α-Fcknob, and the CD8β domain of pCD8β-huFc-Avi was cloned into phuFc-Avihole via NcoI/NotI restriction sites to generate CD8β-Fchole. To generate MV HΔ18 envelope plasmids, the coding sequence for the MSE10-DARPin (Table S3) was inserted into pHnse-Δ18mut-L346 by SfiI/NotI restriction cloning, generating pHnse-Δ18mut-L3-MSE10. To generate LV transfer vector plasmids encoding GFP under various promoters, promoter sequences from PGK, MCMV, and EF1α (Table S2) were amplified by PCR with primers 3 and 4 (PGK), 5 and 6 (MCMV), or 7 and 8 (EF1a) from pGL3-PGK (kindly provided by Arne Auste), pMCMV3 (Addgene: 85711), and pTN-CD20.CAR (kindly provided by Michael Hudecek, Würzburg, Germany), respectively. Amplified promoters were cloned into the transfer plasmid via EcoRI/BamHI, resulting in plasmids pPGK-GFP-W, pMCMV-GFP-W, and pEF1a-GFP-W. To generate plasmids encoding mCD8-targeted AAV2 capsid proteins, MSE10 DARPin (Table S3) was amplified by PCR from lentiviral envelope plasmid using primers 9 and 10. Amplified DARPin was cloned into an intermediate plasmid from which an insert containing the DARPin and the mutations knocking out the HSPG binding site (R585>A, R588>A) was cloned into pRC_RR_VP1_r1c3 (Addgene: 65724) by BsiWI/XcmI, generating pRC-mCD8.
Cell culture and media
LentiX-293T cells (Takara Bio, Kusatsu, Japan) were cultured in Dulbecco’s modified Eagle’s medium high glucose (Sigma-Aldrich, St. Louis, MO, USA; D6546) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, F7524, lot BCCB7222) and 2 mM L-glutamine (Sigma-Aldrich, G7513) (i.e., DMEM complete). HEK293T cells were subcultured twice a week at ratios between 1:8 and 1:10 using 0.25% trypsin in 1 mM EDTA-PBS without Ca2+ or Mg2+. A20 cells (ATCC, Manassas, VA, USA; TIB-208) were cultured in RPMI 1640 (Sigma-Aldrich, R0883-500ML) supplemented with 10% FBS, 2 mM L-glutamine, and 50 μM β-mercaptoethanol (Carl Roth, Karlsruhe, Germany). A20 cells were subcultured three times a week at ratios between 1:15 and 1:20. Primary murine splenocytes were kept in T cell medium (TCM) (i.e., RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, 1× NEAA, 1× sodium pyruvate, 25 mM HEPES) (Sigma-Aldrich, H3537-500ML), penicillin-streptomycin, and 50 μM β-mercaptoethanol.
Production of LV
We produced RT-LVs following our established protocol for the generation of LVs pseudotyped with paramyxoviral glycoproteins.28 In brief, LentiX-293T cells (Takara Bio) were transfected with the pHnse-Δ18mut-L3-MSE10 and pCG-FΔ3047 envelope plasmids, the transfer plasmid, and the packaging plasmid pCMVd8.9 (Addgene: 2221) in a 7.7:23:87:83 ratio. For VSV-LV production, VSVG envelope (pMD2.G, Addgene: 12259), transfer, and packaging plasmid were transfected in a ratio of 35:100:65. Supernatants harvested 48 h after transfection were clarified by 0.45 μm filtration. Filtrates were either treated with Pierce Universal Nuclease (Thermo Fisher Scientific, Waltham, MA, USA, 88702) at 50 U/mL for 30 min at 37°C, or vector stocks were treated with 3 U DNase I (Invitrogen, Waltham, MA, USA, AM2222) per 40 μL for 30 min at 37°C and quenched with a final concentration of 5 mM EDTA directly prior to transduction. Particles were purified and concentrated 150- to 333-fold into PBS from the clarified supernatant by overnight centrifugation through a 20% sucrose cushion at 4,500 × g and 4°C. Concentrated stocks were aliquoted and stored at −80°C.
Production of AAVs
AAV2 and mCD8-AAV particles were generated by transient transfection of HEK293T cells. 24 h prior to transfection, 1.8 × 107 HEK293T cells were seeded per 14 cm dish. On the day of transfection, the cell culture medium was replaced by 12 mL DMEM (Sigma-Aldrich, D6546) with 15% fetal calf serum (FCS) and 2 mM L-glutamine. For the transfection mix, 30 μg of total DNA per dish was mixed in 2 mL DMEM without additives, vortexed, and added to 1.9 mL DMEM supplemented with 120 μL 18 mM polyethylenimine (PEI) solution. For production of AAV2, helper plasmids pXX6-80,48 packaging plasmid pRC22 (Addgene: 104963), and the self-complementary transfer vector pscAAV-SFFV-GFP16 were mixed in a ratio of 60:20:20. For production of mCD8-AAV, pXX6-80, and pscAAV-SFFV-GFP, the complementary capsid plasmid pRC-VP1-KO and pRC-mCD8 were mixed in a ratio of 15:5:5:5. The mixture was vortexed vigorously and incubated for 20 min at room temperature. A total of 4 mL of transfection mix per dish was added to the HEK293T cells. Medium was changed after 5 h to 18 mL DMEM supplemented with 10% FCS and 2 mM L-glutamine. 48 h after transfection, cells were scraped off, pelleted (1,800 × g, 30 min, 4°C) and lysed using Tris-HCl/NaCl (pH 8.5). Four freeze-and-thaw cycles in liquid nitrogen were conducted followed by a Benzonase treatment (50 U/mL cell lysate; Sigma-Aldrich, E1014-25KU) for 30 min at 37°C. Lysate was clarified (3,700 × g, 20 min, 4°C) before loading onto an iodixanol gradient.49 The gradient was centrifuged for 2 h at 290,000 × g in a 70Ti rotor (Beckman Coulter, Brea, CA, USA), and AAV particles were harvested from the 40% iodixanol layer. Stocks were aliquoted and stored at −80°C.
Isolation of murine splenocytes and splenic Pan T cells
Spleens from male or female adult CAC BALB/cAnNCrl or C57BL/6 mice were transferred to 5 mL of ice-cold TCM (see Cell culture and media). Next, cell suspensions were generated by grinding the spleens through 70 μm cell strainers (Thermo Fisher Scientific, 352350) using the back sides of plungers of 5 mL syringes. Strainers were rinsed several times with TCM from the respective well, using 1 mL per rinse. The cell suspension was homogenized by pipetting with a 1 mL micropipette, then transferred to 15 mL Falcon tubes and washed with 8 mL of TCM (300 × g, 4°C, 5 min). Red blood cell lysis was performed by resuspending cell pellets in 5 mL 1× Pharm Lyse (Becton Dickinson [BD], Franklin Lakes, NJ, USA, 555899) ammonium chloride-based lysis buffer and incubating for 6 min at room temperature. Lysis was stopped by two consecutive washes with 8 and 13 mL of TCM, respectively. Cell pellets were then resuspended in 5 mL of TCM, and cell count and viability were determined using the LUNA-FL Automated Fluorescence Cell Counter (Logos Biosystems, Annandale, VA, USA, L20001), LUNA Cell Counting Slides (Logos Biosystems, L12001), and acridine orange/propidium iodide dye (Logos Biosystems, F23001). Splenocytes were frozen by pelleting (300 × g, 5 min) followed by resuspension in FBS (Sigma-Aldrich, F7524) containing 10% DMSO (Sigma-Aldrich, D8418-100ML) to a final concentration of 1 × 106–1 × 107 cells/mL and transferred to −80°C in isopropanol-filled freezing vessels. Cells were transferred to liquid nitrogen vapor phase storage 1 to 7 d later. Splenic Pan T cells were isolated from splenocytes via the Mouse Pan T Cell Isolation Kit II (Miltenyi Biotec, Bergisch Gladbach, Germany, Ref 130-095-130), using MS Columns (Miltenyi Biotec, 130-042-201), following the manufacturer’s protocol.
Transduction of splenocytes and Pan T cells
BALB/c splenocytes were activated and transduced on the day of thawing (or isolation). 4 × 104 cells/well (50 μL per well) were mixed in TCM with recombinant human (rh) IL7 (Miltenyi Biotec, 130-095-361), rhIL15 (Miltenyi Biotec, 130-095-764), and 4 × 104 αmCD3/αmCD28-beads/well (Invitrogen, 11456D) and seeded into wells of 96-well plates. Vector stocks were diluted in TCM, and 50 μL/well of vector-containing supernatant was added to the cells, resulting in final cytokine concentrations of 25 U/mL for rhIL7 and 50 U/mL for rhIL15. For LV transduction, cells were spinfected (1.5 h, 800 × g, 32°C). AAV-transduced cells were incubated for 5–6 h at 37°C. Afterward, 100 μL/well of TCM containing 25 U/mL of rhIL7 and 50 U/mL of rhIL15 was added.
Transduction of whole blood
Blood from CAC BALB/cAnNCrl mice was collected in lithium heparin tubes, and 150 μL/well was transferred into wells of 48-well plates. Recombinant human IL7 was added to each well to a final concentration of 25 U/mL together with 2 × 1010 LV particles per well (or an equivalent volume of Dulbecco’s phosphate-buffered saline [DPBS, Lonza, Basel, Switzerland]). Blood was incubated on an orbital shaker (INFORS HT, Bottmingen, Switzerland, Celltron, throw 25 mm) at 180 rpm at 37°C, 5% CO2, 90% relative humidity for 6 h. Afterward, erythrocytes were eliminated by Histopaque centrifugation. Samples were diluted with 1 mL each of room-temperature DPBS and layered onto 1 mL of room temperature Histopaque 1083 (Sigma-Aldrich, 10831-100ML) in 15 mL tubes. Tubes were centrifuged at 400 × g, room temperature, for 30 min. After centrifugation, all material above the Histopaque plug was collected (as indicated in Figure 3). The material was washed once with 10 mL of PBS (300 × g, 10 min, room temperature). Pellets were resuspended in 1 mL of DPBS, and cell numbers were determined using the Luna Fl. Cells were activated using rhIL7+15 as well as αmCD3/αmCD28-beads (as described in Transduction of splenocytes and Pan T cells) and seeded into 96-well plates at approximately 3 × 104 cells/well (200 μL per well). Cells were incubated at 37°C, 5% CO2, 90% relative humidity for 5 days prior to analysis.
Recombinant protein expression, purification, and ribosome display
Target protein expression, protein purification, and ribosome display were performed as previously described for selection of DARPins against human CD8.32 In brief, CD8αα and CD8αβ target proteins for ribosome display were produced by transient transfection of the CD8 constructs in the presence or absence of the biotinylation constructs pDisplaysBirA and pDisplay-BirAER into HEK293T cells. Proteins from the cell culture supernatant were purified via Protein A-Sepharose and size exclusion chromatography performed by using a Superdex 200 HighLoad 16/600 column (GE Healthcare, Chicago, IL, USA) in a high-pressure liquid chromatography (ÄKTAxpress, GE Healthcare) system. DARPins were selected from the S-N3C library, a DARPin library with reduced hydrophobicity.50 In order to enrich binders of high affinity, three rounds of on-plate ribosomal display were conducted. The fourth round was carried out in solution to enable counterselection with CD8αα-Fc proteins. After the last round of selection, DARPin-encoding DNA fragments were analyzed for CD8 binding by single clone analysis.
ELISA
To screen the output of the DARPin selection for binding to CD8, an ELISA with recombinant CD8-FcBiotin protein was conducted as previously described.31 In brief, 96-well Maxisorb plates (Thermo Fisher Scientific) were coated with 100 mL 20 mM Neutravidin solution in TBS for 1 h at room temperature. After washing, unspecific binding was blocked with TBS-T supplemented with 0.5% BSA for 1 h at room temperature. To immobilize the target protein, wells were coated with 10–20 nM of biotinylated CD8αα-Fc, CD8αβ-Fc, or Fc-only protein solution in blocking buffer. After washing, wells were incubated with diluted DARPin crude lysate preparations for 1 h at room temperature. To quantify the amount of bound DARPin proteins, wells were subsequently incubated with anti-HA antibody (1:5,000, Abcam, Cambridge, UK, clone 16B12, ab130275) and a horseradish peroxidase (HRP)-conjugated rabbit anti-mouse antibody (1:1,000; P0260, Agilent, Santa Clara, CA, USA) for 1 h at room temperature each. The plates were washed three times with TBS-T before and after each antibody incubation step. The bound antibodies were detected using SureBlue TMB substrate (SeraCare, Milford, MA, USA) and terminated after 5–10 min using 1N H2SO4. The reaction product was quantified in a microtiter plate reader at 450 nm.
Nanoparticle tracking
Particle numbers were determined by nanoparticle tracking analysis using the NanoSight NS300 system (Malvern Instruments, Malvern, UK) and detection via a green laser. For this, LV stocks were diluted in 0.2 μm-filtered DPBS (Lonza, BE17-512F) to achieve 30–60 particles/frame. A continuous flow protocol was used. Size mode and concentration data are from technical quadruplicates. For each vector stock, four 60 s videos were recorded and analyzed sequentially.
Western blotting
AAV2 stocks were incubated in urea buffer (200 mM Tris/HCl [pH 8.0], 5% SDS, 8 M urea, 0.1 mM EDTA, 2.5% DTT, 0.03% bromophenol blue) at 95°C for 10 min and loaded on 10% SDS-PAGE gels. After electrophoretic separation, proteins were blotted on nitrocellulose membranes (Amersham, Amersham, UK, 10600004). Membranes were incubated with anti-AAV VP1/VP2/VP3 rabbit polyclonal antibody at a dilution of 1:50 (Progen, Heidelberg, Germany, 61084) overnight at 4°C followed by an incubation with the polyclonal rabbit anti-mouse immunoglobulin HRP conjugate at a dilution 1:2,000 (Agilent, P0260) for 90 min at room temperature. After application of Chemiluminescent Peroxidase Substrate (Sigma-Aldrich, CPS160-1KT), luminescent signals were detected on the chemiluminescence reader MicroChemi (DNR Bio-Imaging Systems, Neve Yamin, Israel). All antibodies were diluted in TBS-T (50 mM Tris/HCl, 150 mM NaCl, 0.1% Tween-20 [pH 7.4]) containing 2% powdered milk.
Immunostaining and flow cytometry
Immunostaining for flow cytometry was performed in the 96-tube format in 1.4 mL round-bottom tubes (Micronic, Lelystad, the Netherlands, MP32022). Wash buffers were used cold (4°C). For washes, cells were centrifuged at 300 × g, 4°C for 5 min. After centrifugation, supernatant was removed using a multichannel aspirator with a spacer, leaving 100–150 μL of dead volume. After removal of supernatant, pellets were resuspended by 1–3 s of vortexing. To determine immune phenotype and transgene positivity, cell suspensions were washed twice with 400 μL of wash buffer (2% FCS, 0.1% sodium azide, 1 mM EDTA in PBS) per wash per tube. Subsequently, 1 μL of mouse FcR blocking reagent (Miltenyi Biotec, 130-092-575) in 20 μL of wash buffer was added to each tube to block unspecific FcR-mediated binding of antibodies. Cells were vortexed and incubated for 10 min at room temperature before antibody cocktails in wash buffer were added to the cells at 20 μL/tube. Cells were immunostained using αmCD19-VioBlue (2 μL/tube; Miltenyi Biotec, 130-118-463), αmCD3-BV421 (1 μL/tube; Becton Dickinson, 130-118-463), αc-myc-fluorescein isothiocyanate (FITC) (0.5 μL/tube; Miltenyi Biotec, 130-116-485), αmCD8a-PerCP-Cy5.5 (1 μL/tube; Becton Dickinson, 561109), and αmCD4-APC (1 μL/tube; Becton Dickinson PharMingen, 561091) antibodies. Appropriate isotype controls were used to inform gating decisions. To assess cell viability, Fixable Viability Dye eFluor780 (Thermo Fisher Scientific, 65-0865-14) was added to antibody mixes at 0.12 μL/tube. After 3–5 s of vortexing, cells were incubated at 4°C for 30–45 min. Cells were then washed twice before cell pellets were resuspended by vortexing in 200 μL of 0.5% formaldehyde in PBS. Samples were kept in the dark at 4°C until measurement. Flow cytometry was performed on a MACSQuant Analyzer 10 (Miltenyi Biotec), and data were analyzed using FCS Express 6 (De Novo Software).
For the killing assay, A20 cells were stained with Cell Trace Violet (Invitrogen, C34557) prior to co-culture setup following the manufacturer’s instructions. Cells were then pelleted, resuspended in 2 mL of TCM, and counted for culture setup. See Figure S4 for the gating strategy employed to analyze the killing assay.
To assess binding of DARPin candidates to primary cells, 1–2 × 105 splenocytes from one C57BL/6 mouse were incubated with DARPin crude lysate for 60 min at 4°C. Subsequently, cells were washed and stained for viability via eFluor 450 (Affymetrix, Santa Clara, CA, USA, 65-0863-14), the DARPins’ HA tags by αHA-PE (Miltenyi Biotec, 130-120-717), and CD8a or CD8b by αmCD8a-APC (Miltenyi Biotec, 130-102-540) or αmCD8b-APC (BioLegend, San Diego, CA, USA, 140410).
LV vector copy number qPCR
Genomic DNA (gDNA) was extracted in the 96-well format using DNeasy 96 Blood & Tissue Kit (QIAGEN, Hilden, Germany, 69581) following the manufacturer’s instructions. gDNA was eluted into AE buffer. To determine lentiviral integrations per cell (VCN), quantitative real-time PCR was performed using primers 11 and 12 against woodchuck hepatitis virus post-transcriptional response element (WPRE) and primers 13 and 14 against murine albumin Alb. Twenty microliter reactions containing 8 μL of template and 0.5 μM primers were set up using QuantiTect SYBR Green PCR Kit (QIAGEN, 204143) and subsequently run on a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). A standard for WPRE was generated by serial dilution of pSEW-GFP lentiviral transfer plasmid51 in AE buffer. An Alb standard was generated by extracting gDNA from 1 × 106–1 × 102 cells of the murine T cell line 58m8ab (kindly provided by Wolfgang Uckert, Berlin, Germany).
AAV genome copy qPCR
Ten microliters of purified vector stock was used for DNA extraction using the DNeasy Blood & Tissue kit (QIAGEN, 69506). Genomic AAV vector titers were determined via inverted terminal repeat (ITR)-specific qPCR using primers 15, 16, and probe 17 with the LightCycler 480 Probes Master mix (Roche, 04707494001) on a LightCycler 480 Real-Time PCR System (Roche). The transfer cassette from pssAAV-hSyn-eYFP transfer plasmid,19 isolated by PvoII-XmnI digest and gel purification, served as a standard.
Protein structure visualizations
Protein structures were approximated using the indicated PDB structures and UCSF Chimera 1.13.1. Spacing of unstructured domains was approximated using α-helical peptides of appropriate length.
Statistics and graphing
All statistical analyses were carried out in GraphPad Prism version 8.4.2. Flow cytometry signals were assumed to be normally distributed. Accordingly, tests were performed assuming normal distribution. Data generated from cells of the same mouse were handled as matched data. Differences of population means were quantified by repeated-measures one-way ANOVA (or paired t test, depending on number of groups to be compared) and Tukey’s multiple comparisons test. Selected multiplicity-adjusted p values are reported.
Acknowledgments
The authors would like to thank Markus Seeger (Institute of Medical Microbiology, University of Zurich) for providing the DARPin library as well as Gundula Braun and Manuela Gallet (Paul-Ehrlich-Institut) for excellent technical assistance. This work was supported by a grant from the Federal Ministry of Health (03292364) to C.J.B. K.B. and D.G. appreciate funding through the German Center for Infection Research (DZIF, BMBF; TTU-HIV 08.415).
Author contributions
Conceptualization: C.J.B., J.H., A.M.F., and A.M.; Methodology: A.M., A.M.F., K.B., and D.M.G.; Investigation: A.M., D.M.G., A.M.F., and M.M.; Formal analysis: A.M.; Resources: K.B., D.G., D.M.G.; Writing – original draft: A.M. and C.J.B.; Revision: A.M. and C.J.B.; Visualization: A.M.; Supervision: J.H. and C.J.B.; Funding acquisition: C.J.B.
Declaration of interests
C.J.B. is listed as inventor on patents covering T cell targeted lentiviral vectors. D.G. is a co-founder, shareholder, and Chief Scientific Officer of AaviGen GmbH. All the other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.09.014.
Supplemental information
References
- 1.Hartmann J., Schüßler-Lenz M., Bondanza A., Buchholz C.J. Clinical development of CAR T cells-challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med. 2017;9:1183–1197. doi: 10.15252/emmm.201607485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elsallab M., Levine B.L., Wayne A.S., Abou-El-Enein M. CAR T-cell product performance in haematological malignancies before and after marketing authorisation. Lancet Oncol. 2020;21:e104–e116. doi: 10.1016/S1470-2045(19)30729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Medicines Agency . 2020. Assessment report Zolgensma: International non-proprietary name: onasemnogene abeparvovec Procedure No. EMEA/H/C/004750/0000. [Google Scholar]
- 4.Naldini L. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol. Med. 2019;11:e9958. doi: 10.15252/emmm.201809958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendell J.R., Al-Zaidy S.A., Rodino-Klapac L.R., Goodspeed K., Gray S.J., Kay C.N., Boye S.L., Boye S.E., George L.A., Salabarria S. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu D., Schieferecke A.J., Lopez P.A., Schaffer D.V. Adeno-Associated Virus Vector for Central Nervous System Gene Therapy. Trends Mol. Med. 2021;27:524–537. doi: 10.1016/j.molmed.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 8.Fischer A., Hacein-Bey-Abina S. Gene therapy for severe combined immunodeficiencies and beyond. J. Exp. Med. 2020;217:e20190607. doi: 10.1084/jem.20190607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(2020). High-dose AAV gene therapy deaths. Nat. Biotechnol. 38, 910. [DOI] [PubMed]
- 10.Kochenderfer J.N., Yu Z., Frasheri D., Restifo N.P., Rosenberg S.A. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood. 2010;116:3875–3886. doi: 10.1182/blood-2010-01-265041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.June C.H., Sadelain M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michels A., Hartmann J., Buchholz C.J. Chimäre Antigenrezeptoren (CARs) in der Onkologie: eine Übersicht zu klinischer Anwendung und neuen Entwicklungen. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2020;63:1331–1340. doi: 10.1007/s00103-020-03222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kueberuwa G., Zheng W., Kalaitsidou M., Gilham D.E., Hawkins R.E. A Syngeneic Mouse B-Cell Lymphoma Model for Pre-Clinical Evaluation of CD19 CAR T Cells. J. Vis. Exp. 2018;(140):58492. doi: 10.3791/58492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alizadeh D., Wong R.A., Gholamin S., Maker M., Aftabizadeh M., Yang X., Pecoraro J.R., Jeppson J.D., Wang D., Aguilar B. IFNγ Is Critical for CAR T Cell-Mediated Myeloid Activation and Induction of Endogenous Immunity. Cancer Discov. 2021;11:2248–2265. doi: 10.1158/2159-8290.CD-20-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchholz C.J., Friedel T., Büning H. Surface-Engineered Viral Vectors for Selective and Cell Type-Specific Gene Delivery. Trends Biotechnol. 2015;33:777–790. doi: 10.1016/j.tibtech.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Münch R.C., Janicki H., Völker I., Rasbach A., Hallek M., Büning H., Buchholz C.J. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol. Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münch R.C., Muth A., Muik A., Friedel T., Schmatz J., Dreier B., Trkola A., Plückthun A., Büning H., Buchholz C.J. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 2015;6:6246. doi: 10.1038/ncomms7246. [DOI] [PubMed] [Google Scholar]
- 18.Muik A., Reul J., Friedel T., Muth A., Hartmann K.P., Schneider I.C., Münch R.C., Buchholz C.J. Covalent coupling of high-affinity ligands to the surface of viral vector particles by protein trans-splicing mediates cell type-specific gene transfer. Biomaterials. 2017;144:84–94. doi: 10.1016/j.biomaterials.2017.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann J., Thalheimer F.B., Höpfner F., Kerzel T., Khodosevich K., García-González D., Monyer H., Diester I., Büning H., Carette J.E. GluA4-Targeted AAV Vectors Deliver Genes Selectively to Interneurons while Relying on the AAV Receptor for Entry. Mol. Ther. Methods Clin. Dev. 2019;14:252–260. doi: 10.1016/j.omtm.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Judd J., Wei F., Nguyen P.Q., Tartaglia L.J., Agbandje-McKenna M., Silberg J.J., Suh J. Random Insertion of mCherry Into VP3 Domain of Adeno-associated Virus Yields Fluorescent Capsids With no Loss of Infectivity. Mol. Ther. Nucleic Acids. 2012;1:e54. doi: 10.1038/mtna.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichhoff A.M., Börner K., Albrecht B., Schäfer W., Baum N., Haag F., Körbelin J., Trepel M., Braren I., Grimm D. Nanobody-Enhanced Targeting of AAV Gene Therapy Vectors. Mol. Ther. Methods Clin. Dev. 2019;15:211–220. doi: 10.1016/j.omtm.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank A.M., Buchholz C.J. Surface-engineered lentiviral vectors for selective gene transfer into subtypes of lymphocytes. Mol. Ther. Methods Clin. Dev. 2018;12:19–31. doi: 10.1016/j.omtm.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank A.M., Braun A.H., Scheib L., Agarwal S., Schneider I.C., Fusil F., Perian S., Sahin U., Thalheimer F.B., Verhoeyen E., Buchholz C.J. Combining T-cell-specific activation and in vivo gene delivery through CD3-targeted lentiviral vectors. Blood Adv. 2020;4:5702–5715. doi: 10.1182/bloodadvances.2020002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer A., Thalheimer F.B., Hartmann S., Frank A.M., Bender R.R., Danisch S., Costa C., Wels W.S., Modlich U., Stripecke R. In vivo generation of human CD19-CAR T cells results in B-cell depletion and signs of cytokine release syndrome. EMBO Mol. Med. 2018;10:e9158. doi: 10.15252/emmm.201809158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamali A., Kapitza L., Schaser T., Johnston I.C.D., Buchholz C.J., Hartmann J. Highly Efficient and Selective CAR-Gene Transfer Using CD4- and CD8-Targeted Lentiviral Vectors. Mol. Ther. Methods Clin. Dev. 2019;13:371–379. doi: 10.1016/j.omtm.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal S., Hanauer J.D.S., Frank A.M., Riechert V., Thalheimer F.B., Buchholz C.J. In vivo generation of CAR T cells selectively in human CD4+ lymphocytes. Mol. Ther. 2020;28:1783–1794. doi: 10.1016/j.ymthe.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merchant A.M., Zhu Z., Yuan J.Q., Goddard A., Adams C.W., Presta L.G., Carter P. An efficient route to human bispecific IgG. Nat. Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 28.Weidner T., Agarwal S., Perian S., Fusil F., Braun G., Hartmann J., Verhoeyen E., Buchholz C.J. Genetic in vivo engineering of human T lymphocytes in mouse models. Nat. Protoc. 2021;16:3210–3240. doi: 10.1038/s41596-021-00510-8. [DOI] [PubMed] [Google Scholar]
- 29.Mangeot P.-E., Dollet S., Girard M., Ciancia C., Joly S., Peschanski M., Lotteau V. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 2011;19:1656–1666. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plückthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 31.Hartmann J., Münch R.C., Freiling R.-T., Schneider I.C., Dreier B., Samukange W., Koch J., Seeger M.A., Plückthun A., Buchholz C.J. A Library-Based Screening Strategy for the Identification of DARPins as Ligands for Receptor-Targeted AAV and Lentiviral Vectors. Mol. Ther. Methods Clin. Dev. 2018;10:128–143. doi: 10.1016/j.omtm.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank A.M., Weidner T., Brynza J., Uckert W., Buchholz C.J., Hartmann J. CD8-Specific Designed Ankyrin Repeat Proteins Improve Selective Gene Delivery into Human and Primate T Lymphocytes. Hum. Gene Ther. 2020;31:679–691. doi: 10.1089/hum.2019.248. [DOI] [PubMed] [Google Scholar]
- 33.Kerkar S.P., Sanchez-Perez L., Yang S., Borman Z.A., Muranski P., Ji Y., Chinnasamy D., Kaiser A.D.M., Hinrichs C.S., Klebanoff C.A. Genetic engineering of murine CD8+ and CD4+ T cells for preclinical adoptive immunotherapy studies. J. Immunother. 2011;34:343–352. doi: 10.1097/CJI.0b013e3182187600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair S., Rein A. Antiretroviral restriction factors in mice. Virus Res. 2014;193:130–134. doi: 10.1016/j.virusres.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsurutani N., Yasuda J., Yamamoto N., Choi B.-I., Kadoki M., Iwakura Y. Nuclear import of the preintegration complex is blocked upon infection by human immunodeficiency virus type 1 in mouse cells. J. Virol. 2007;81:677–688. doi: 10.1128/JVI.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J.X., Diehl G.E., Littman D.R. Relief of preintegration inhibition and characterization of additional blocks for HIV replication in primary mouse T cells. PLoS ONE. 2008;3:e2035. doi: 10.1371/journal.pone.0002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilham D.E., Lie-A-Ling M., Taylor N., Hawkins R.E. Cytokine stimulation and the choice of promoter are critical factors for the efficient transduction of mouse T cells with HIV-1 vectors. J. Gene Med. 2010;12:129–136. doi: 10.1002/jgm.1421. [DOI] [PubMed] [Google Scholar]
- 38.Addison C.L., Hitt M., Kunsken D., Graham F.L. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 1997;78:1653–1661. doi: 10.1099/0022-1317-78-7-1653. [DOI] [PubMed] [Google Scholar]
- 39.Herbst F., Ball C.R., Tuorto F., Nowrouzi A., Wang W., Zavidij O., Dieter S.M., Fessler S., van der Hoeven F., Kloz U. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol. Ther. 2012;20:1014–1021. doi: 10.1038/mt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 41.Behrendt R., Schumann T., Gerbaulet A., Nguyen L.A., Schubert N., Alexopoulou D., Berka U., Lienenklaus S., Peschke K., Gibbert K. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coiras M., Bermejo M., Descours B., Mateos E., García-Pérez J., López-Huertas M.-R., Lederman M.M., Benkirane M., Alcamí J. IL-7 Induces SAMHD1 Phosphorylation in CD4+ T Lymphocytes, Improving Early Steps of HIV-1 Life Cycle. Cell Rep. 2016;14:2100–2107. doi: 10.1016/j.celrep.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breuer C.B., Hanlon K.S., Natasan J.-S., Volak A., Meliani A., Mingozzi F., Kleinstiver B.P., Moon J.J., Maguire C.A. In vivo engineering of lymphocytes after systemic exosome-associated AAV delivery. Sci. Rep. 2020;10:4544. doi: 10.1038/s41598-020-61518-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., Carette J.E. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parayath N.N., Stephan S.B., Koehne A.L., Nelson P.S., Stephan M.T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 2020;11:6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anliker B., Abel T., Kneissl S., Hlavaty J., Caputi A., Brynza J., Schneider I.C., Münch R.C., Petznek H., Kontermann R.E. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat. Methods. 2010;7:929–935. doi: 10.1038/nmeth.1514. [DOI] [PubMed] [Google Scholar]
- 47.Funke S., Schneider I.C., Glaser S., Mühlebach M.D., Moritz T., Cattaneo R., Cichutek K., Buchholz C.J. Pseudotyping lentiviral vectors with the wild-type measles virus glycoproteins improves titer and selectivity. Gene Ther. 2009;16:700–705. doi: 10.1038/gt.2009.11. [DOI] [PubMed] [Google Scholar]
- 48.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reul J., Muik A., Buchholz C.J. Ligand Coupling to the AAV Capsid for Cell-Specific Gene Transfer. Methods Mol. Biol. 2019;1950:35–50. doi: 10.1007/978-1-4939-9139-6_3. [DOI] [PubMed] [Google Scholar]
- 50.Seeger M.A., Zbinden R., Flütsch A., Gutte P.G.M., Engeler S., Roschitzki-Voser H., Grütter M.G. Design, construction, and characterization of a second-generation DARP in library with reduced hydrophobicity. Protein Sci. 2013;22:1239–1257. doi: 10.1002/pro.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demaison C., Parsley K., Brouns G., Scherr M., Battmer K., Kinnon C., Grez M., Thrasher A.J. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.