Graphical abstract

Keywords: Tumor mutational burden (TMB), Molecular alterations, Differential expression, Signaling pathway, Immunology, Survival
Abstract
Whether tumor mutational burden (TMB) is related to improved survival outcomes or the promotion of immunotherapy in various malignant tumors remains controversial, and we lack a comprehensive understanding of TMB across cancers. Based on the data obtained from The Cancer Genome Atlas (TCGA), we conducted a multiomics analysis of TMB across 21 cancer types to identify characteristics related to TMB and determine the mechanism as it relates to prognosis, gene expression, gene mutation and signaling pathways. In our study, TMB was found to have a significant relationship with prognosis for 21 tumors, and the relationship was different in different tumors. TMB may also be related to different outcomes for patients with different tumor subtypes. TMB was confirmed to be correlated with clinical information, such as age and sex. Mutations in GATA3 and MAP3K1 in beast invasive carcinoma (BRCA), TCF7L2 in colon adenocarcinoma (COAD), NFE2L2 in esophageal carcinoma (ESCA), CIC and IDH1 in brain lower grade glioma (LGG), CDH1 in stomach adenocarcinoma (STAD), and TP53 in uterine corpus endometrial carcinoma (UCEC) were demonstrated to be correlated with lower TMB. Moreover, we identified differentially expressed genes (DEGs) and differentially methylated regions (DMRs) according to different TMB levels in 21 cancers. We also investigated the correlation between enrichment of signaling pathways, immune cell infiltration and TMB. In conclusion, we identified multiomic characteristics related to the TMB in 21 tumors, providing support for a comprehensive understanding of the role of TMB in different tumors.
1. Introduction
Currently, immunotherapy is used to treat many cancer types, such as lung squamous cell carcinoma (LUSC), bladder urothelial carcinoma (BLCA) and melanoma [1], [2], [3]. Compared with those receiving traditional treatment, patients receiving immune checkpoint inhibitor (ICI) treatment may achieve long-term survival. However, ICIs are not effective in all cancer patients [4], [5], [6], and most cancer patients treated with ICIs do not benefit. It is vital to determine precise biomarkers to predict the efficacy of immunotherapy.
Cancer is a genetic disease, and neoplastic transformation results from the accumulation of somatic mutations in the DNA of affected cells [7]. TMB is defined as the total number of somatic mutations per megabase (Mb) in the exon region, excluding synonymous mutations [8]. Whole-exome sequencing (WES) is considered the gold standard for detecting TMB [9]. In some tumors, the TMB has been shown to be an independent predictor of ICI success and can identify patients who may benefit from immunotherapy [7], [10], [11]. Generally, patients with high TMB obtain better results from immunotherapy [12], [13]. TMB is considered to be a predictive biomarker of tumor behavior and the immune response [14], [15]. TMB is also regarded as a prognostic biomarker [16], [17]. Studies have shown that TMB is related to clinical characteristics [18], [19] and immune cell infiltration [20], [21] in some tumor patients. TMB is also affected by gene mutations and the expression of mismatch repair genes (MMRs) [22], [23]. A recent report concluded that high TMB favors the prognosis of patients receiving immunotherapy and that it is not evident in all cancers [24]. Obviously, it is important to conduct further research on TMB.
Currently, most studies on TMB have focused on the analysis of the genome in individual cancers, such as melanoma and bladder cancer [7]. Few studies have directed attention to the influence and role of TMB across cancers. Therefore, there is an urgent need to conduct a comprehensive and exhaustive analysis of the function of TMB in different tumors.
To identify multiomic characteristics correlated with TMB and explore the related mechanisms in multiple cancers, we calculated the TMB of samples and classified the samples into high-TMB group and low-TMB group. The Kaplan-Meier method and Cox risk regression were utilized to study the relationship between TMB and overall survival (OS) and progression-free survival (PFS). Wilcoxon rank-sum test and linear regression analysis were performed to analyze the relationship between TMB and other clinical characteristics. We identified gene mutations related to TMB via the Wilcoxon rank-sum test and proportion test, and the correlation between the expression of MMRs and TMB was evaluated by the Spearman method. DEGs were selected by the edgeR package of R, and DMRs in promoter regions were identified by Wilcoxon rank-sum test. Considering the findings, we confirmed genes whose differential expression was related to differential DNA methylation using the Spearman method. Then, we screened the signaling pathways significantly enriched in the high- and low-TMB groups using GSEA software [25], [26]. Considering the TIMER database [27], [28], [29] and the ImmuCellAI tool [30], we measured the relationship between the TMB and the abundance of immune cell infiltration. We sought to explain the relationship between TMB and prognosis in terms of gene mutation and immune cell infiltration. Taken together, our study may help provide a comprehensive understanding of the role and influence of TMB in 21 tumors and help predict the prognosis of patients with different cancers.
2. Methods
2.1. Patient cohorts
There are data of 33 cancers available in UCSC-Xena database (https://xenabrowser.net/datapages/) using a unified process, including adrenocortical carcinoma (ACC), BLCA, BRAD, Cervical Cancer (CESC), Cholangiocarcinoma (CHOL), COAD, Large B-cell Lymphoma (DLBC), ESCA, Glioblastoma (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), Acute Myeloid Leukemia (LAML), LGG, liver hepatocellular carcinoma (LIHC), Lung Adenocarcinoma (LUAD), LUSC, Mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), Prostate Cancer (PRAD), Rectal Cancer (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), STAD, Testicular Cancer (TGCT), thyroid carcinoma (THCA), thymoma (THYM), UCEC, Uterine Carcinosarcoma (UCS) and Ocular melanomas (UVM). Survival data, clinical information and mRNA expression data of 33 cancers in TCGA datasets were downloaded from the UCSC-Xena database, PFS information was obtained from the study of Liu, J et al. [31]. The tumor subtype data was obtained from the study of Sanchez-Vega et al. [32]. DNA methylation data was retrieved from Firehose case datasets (http://gdac.broadinstitute.org). Somatic mutation data were retrieved using the TCGAbiolinks R package [33]. Levels of immune cell infiltration for TCGA 33 tumors were retrieved from TIMER2.0 (http://timer.cistrome.org/) [27], [28], [29] and ImmuCellAI (http://bioinfo.life.hust.edu.cn/ImmuCellAI/) [30]. Since the data in this study were obtained from public databases, no moral conflict was encountered.
2.2. Calculation of TMB
TMB refers to the number of somatic mutations per 1 million bases in the exon region, excluding intron variants and synonymous mutations [8]. Currently, WES is considered the gold standard for detecting TMB [9]. Using next-generation sequencing (NGS) techniques, WES can be performed to detect somatic mutations in the entire exome (approximately 30 to 50 Mb of a coding sequence) [34]. We used 40 Mb as the estimate of exome size. Therefore, the TMB was equal to the number of somatic mutations after removing synonymous mutations and intron variants divided by 40.
2.3. Analysis of the relationship between TMB and prognosis and other clinical characteristics
To investigate the correlation between TMB and prognosis of 33 cancer types, we categorized the survival data into low- and high-TMB groups according to TMB level, and ultimately selected the optimal cut-point in survival analysis. Kaplan-Meier method and Cox regression analysis was used to evaluate the correlation between TMB and OS, which was defined as the time from diagnosis to the date of death. TMB was found to be significant related to the OS of patients diagnosed with 21 of the 33 cancers assessed, and subsequent analysis was carried out related to these 21 tumors: ACC, BLCA, BRCA, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LGG, LIHC, LUSC, OV, PAAD, PCPG, SARC, SKCM, STAD, THCA, THYM and UCEC. According to histological and molecular characteristics, the 21 cancers were categorized into 32 tumor subtypes [32]. We calculated the differences in TMB in different cancer subtypes by Wilcoxon rank-sum test and studied the relationship between TMB and OS for different subtypes of 21 tumors. The same method was used to detect the relationship between TMB and PFS, in our study, PFS referred to the events included only deaths with tumor, excluding deaths from other reasons [31].
In order to verify our results, we then conducted a similar analysis on the relationship between TMB and OS and PFS that the survival data were grouped into low- (top 1/3 by TMB) and high-TMB groups (bottom 1/3). A P value < 0.05 in the log-rank test was considered to be significant.
Then, we investigated other clinical information associated with TMB. Wilcoxon rank-sum test was conducted to study the association between sex and TMB, and a linear regression analysis was performed to study the association between age, race, tumor grade, tumor stage, smoking status and TMB. A P value < 0.05 was considered to be significant.
2.4. Gene mutation and expression of MMRs
To avoid potential bias caused by ultramutated samples, we eliminated samples with a mutation frequency > 1,000 [35]. Driver gene mutations can drive tumorigenesis [36], and 375 driver genes have been reported [37]. Therefore, we paid attention to highly mutated driver genes in each tumor (≥5% mutation frequency) due to their potential biological significance and detectability in the analysis [35].
The somatic mutation data of 21 cancers were classified into mutant and wild groups by driver genes, and we identified the driver genes whose mutations related to TMB by Wilcoxon rank-sum test. Then, we grouped the data into high- (bottom 1/3) and low-TMB groups (top 1/3 by TMB), and a proportion test was performed to compare the difference in driver gene mutation frequency between the two groups. In addition, to execute research on the cause of ultramutations, we compared the proportion of ultramutated samples in the mutant group and wild group for each driver gene by proportion test. A total of 29 MMRs had been reported in articles [38], [39], [40], [41], [42], [43], [44], and correlation coefficients of the expression of the MMRs and TMB were calculated by the Spearman method. A P value < 0.05 was considered to be significant.
2.5. Assessment of gene expression and DNA methylation
To identify DEGs and DMRs, the mRNA expression data and DNA methylation data were divided into high- and low-TMB groups (top/bottom tertiles by TMB) respectively. The DEGs in the two groups with |log2(FC)| >1 and false discovery rate (FDR) < 0.01 were identified by the edgeR package in R. The DMRs in promoter regions (hereafter, DMRs refers to DMRs in promoter regions) were identified by Wilcoxon rank-sum test with criteria set to FDR < 0.05 and |difference| > 0.1. Gene expression is regulated by epigenetic alterations, and the most common alteration is DNA methylation [45]. To identify the differential expression of genes related to abnormal DNA methylation, genes with both differential expression and methylation were selected to analyze the correlation between the level of gene expression and DNA methylation by the Spearman method. DNA methylation sites in which one probe corresponds to multiple genes were removed, and a rho value < 0 and a P value < 0.05 indicated that the DNA methylation may have some effect on the expression of genes [46]. Results indicating sites with minimum rho values were retained. In addition, the DMRs were annotated according to chromosome by “IlluminaHumanMethylation450kanno.ilmn12.hg19” package of R.
2.6. Signaling pathways and immune cell infiltration analysis
Gene set enrichment analysis (GSEA) was carried out with GSEA_4.0.2 software [25], [26] using TMB level (top/bottom tertiles by TMB) as the phenotype, and “h.all.v7.2.symbols.gmt gene sets” was selected as the reference gene set, which was retrieved from the MSigDB database (http://software.broadinstitute.org/gsea/msigdb/). Only pathways with NOM P value < 0.05 were considered to be significantly enriched. A similar analysis using optimal cut-point by TMB was conducted to confirm the results.
The results of the CIBERSORT algorithm [47] was considered with the TIMER database [27], [28], [29] were chosen to explore the difference of the abundance of infiltrated immune cells between two groups by Wilcoxon rank-sum test, both top/bottom tertiles (1/3) and optimal cut-point were used for high/low TMB groups in the analysis. Then the similar analysis was performed on the immune cell infiltration data of TCGA tumors obtained from the ImmuCellAI tool [30].
2.7. Statistical analysis
All statistical analyses were completed with R 4.0 software (https://www.r-project.org/). Unless noted otherwise, a P value < 0.05 was considered to be significant.
3. Results
3.1. Driver gene mutations and mismatch repair gene expression related to TMB
Our follow-up analysis focused on the 21 cancer types for which OS was related to TMB. In our study, most driver genes had a higher TMB in the mutant group (Fig. 1A), except for MAP3K1 (Difference = -0.29, P value = 0.01) and GATA3 (Difference = -0.23, P value < 0.01) in BRCA, TCF7L2 (Difference = -0.45, P value = 0.05) in COAD, NFE2L2 (Difference = -0.65, P value = 0.02) in ESCA, CIC (Difference = -0.05, P value = 0.02) and IDH1 (Difference = -0.25, P value < 0.01) in LGG, CDH1 (Difference = -0.96, P value < 0.01) in STAD, and TP53 (Difference = -0.36, P value < 0.01) in UCEC, in which mutations were ultimately determined to be correlated with lower TMB (Fig. 1B-G). There was no significant difference in TMB between the patients with mutant driver genes and the wild type in KIRC and other 3 cancers, including PCPG, THCA and THYM, which showed lower TMB (Fig. 1A, S2A). The results suggest that most driver gene mutations are related to higher TMB in 19 cancers.
Fig. 1.
The molecular alterations related to TMB in 21 cancer types. (A) Wilcoxon rank-sum test was used to identify driver gene mutations related to TMB in each tumor type, the difference refers to difference between the median level of TMB in the mutant and wild groups. Only genes with significantly results in at least three cancers are shown in the heat map (P value < 0.05). (B-G) Driver genes that are related to a lower TMB. (H-J) Driver genes that influence ultramutation. (K) Spearman correlation between the expression of mismatch repair genes and TMB.
Moreover, we found that, among the 21 cancer types, cancers with a higher TMB such as COAD, SKCM, STAD and UCEC exhibited high frequency of ultramutations, while cancers with a lower TMB such as KICH, KIRP, PCPG and THCA samples exhibited no ultramutations (Table S1B). A greater proportion of ultramutated samples were found in the wild groups of APC, KRAS and TP53 in COAD and TP53 in STAD and UCEC than in the corresponding mutant groups (Fig. 1H-J). Then, we performed linear regression analysis to correct for single mutations and simultaneous mutations of the three driver genes in COAD, and it was found that mutation of the remaining gene has a significant negative linear relationship with the formation of ultramutations when the other two gene mutations were corrected (Table S1B).
The loss of function due to MMRs has been shown to lead to irreparable DNA replication errors [48], and the expression of MMRs has been found to be related to TMB levels [49]. The Spearman method was applied to calculate the correlation between the expression of MMRs and TMB. The results showed that in most tumors, such as LGG and SKCM, the high expression of most MMRs is associated with a high TMB. While high expression of MLH1 and HFM1 was associated with a low TMB in several tumors, which was confirmed in a previous study [50]. With regard to THYM and THCA, the high expression of most MMRs was associated with lower TMB (Fig. 1K), like the expression of HMGB1 had a strong correlation with TMB in THYM with a rho = -0.64 and P value = 6.31E-15 (Table S1C). Taken together, these results indicate that the correlation between TMB and driver gene mutations and expression of MMRs is different in different cancers, and most driver gene mutations and high expression of MMRs have a positive relationship with TMB in most cancer types.
3.2. Differentially expressed genes and differentially methylated regions related to TMB
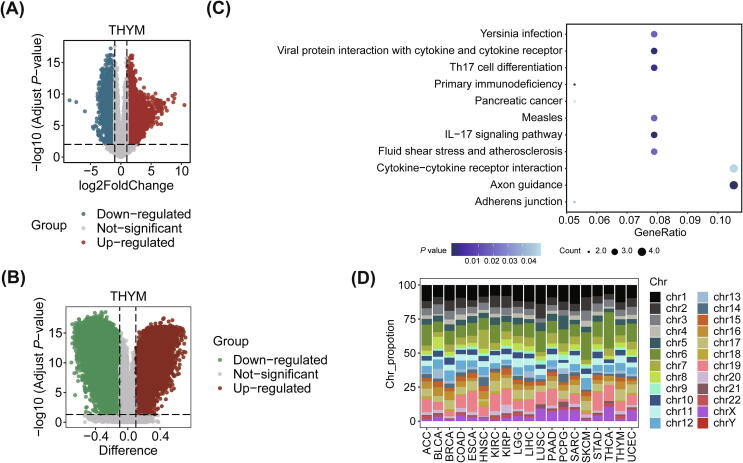
We identified DEGs (Table S2A) and DMRs (Table S2B) in the high- and low-TMB groups (top/bottom tertiles by TMB) in 21 cancers. THYM showed the most DEGs (6244 genes) and DMRs (3676 regions) (Fig. 2A, B), and the proportion of differential expression of gene related to abnormal methylation was also the highest in the DEGs (proportion = 1.25%, Table S3). A KEGG analysis was conducted on genes whose differential expression was related to DNA methylation in THYM (78 genes, Table S2D) using clusterProfiler package of R [51], and a P value < 0.05 was considered to be significant, there were 4 of 38 genes were enriched in Axon guidance (P value = 0.01), Cytokine − cytokine receptor interaction (P value = 0.05), respectively (Fig. 2C). In the analysis about DMRs, we found that KICH exhibited no DMRs, and there were only 4 DMRs located on chromosome 1, chromosome 5, chromosome 6 and chromosome 10 in OV (Table S2C). Most of the DMRs in other cancers were located on chromosome 1, chromosome 6 and chromosome 19 (Fig. 2D). For example, there were about 13.85% of DMRs locating on chromosome 1 in LUSC, 15.15% of DMRs locating on chromosome 19 in ESCA, 26.67% of DMRs locating on chromosome 6 in THCA (Table S2C). Overall, differential gene expression was widely observed between high and low TMB groups in different tumors, but differential methylation of promoter regions occurred less frequently.
Fig. 2.
The DEGs and DMRs correlated to TMB. (A) Volcano map showing the DEGs with FDR < 0.01 and |log2(FC)| > 1 in THYM. The upregulated and downregulated genes are marked in red and blue, respectively. (B) Volcano map showing the DMRs in the promoter regions with FDR < 0.05 and |difference| > 0.1 in THYM. The difference refers to the difference between the median values of DNA methylation in the high-(bottom 1/3) and low-TMB groups (top 1/3 by TMB). (C) Bubble plot showing pathway enrichment based on the KEGG results of genes with differential expression related to DNA methylation (P value < 0.05). (D) Percentage stacked histogram showing the distribution of DMRs in promoter regions in chromosomes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Signaling pathway and immune cell infiltration associated with TMB
To better understanding the impact of TMB on cell pathways and the microenvironment, we using both the top/bottom tertiles method and the optimal cut-point method which are decided by survival analysis to compare the differences of these tumor characters.
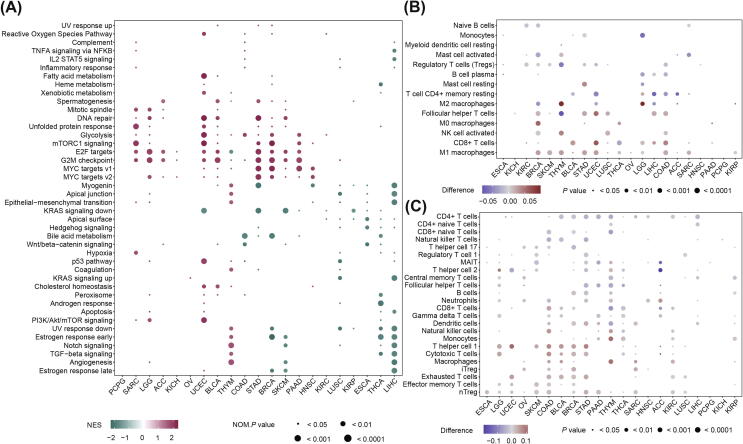
For signaling pathways, 243 results were significantly enriched in analysis with top/bottom tertiles method (Table S4A), 216 results were enriched in analysis with optimal cut-point method (Table S4B), 160 (65.84%) of which were also enriched in the analysis used top/bottom tertiles method, and the results of these two methods were mostly consistent (Table S4C). In the SARC, LGG, ACC and KICH, significant pathways were enriched in the high-TMB group, such as G2M checkpoint and E2F targets (Fig. 3A, S1A). While in the KIRP, ESCA, THCA and LIHC, all of significant pathways were enriched in the low-TMB group (Fig. 3A, S1A). For other tumors, the pattern of signaling pathway enrichment between the two groups was more complex (Fig. 3A, S1A). In 5 cancers including SARC, LGG, UCEC, STAD and BRCA, high TMB was positively associated with DNA repair, unfolded protein response and G2M checkpoint pathways, which were tumor suppressor pathways, and positively related to carcinogenic pathways including Glycolysis, mTORC1 signaling pathways and E2F targets (Fig. 3A, S1A). Notably, E2F targets, G2M checkpoint and MYC targets V1 were enriched in the low-TMB group of THYM (Fig. 3A, S1A). The results showed that the enrichment of signaling pathways according to TMB levels is different in different tumors.
Fig. 3.
The relationship between TMB and signaling pathways and infiltration of immune cells among cancer types. (A) The signaling pathways enriched in the high- (bottom 1/3) and low-TMB groups (top 1/3 by TMB). Only pathways with significant results in at least three cancers were shown in the heatmap. (B) Wilcoxon rank-sum test was conducted to identify the immune cell infiltration correlated to TMB based on the CIBERSORT algorithm, the difference refers to difference between the median abundance of immune cell infiltration in high- (bottom 1/3) and low-TMB groups (top 1/3 by TMB). (C) Wilcoxon rank-sum test was conducted to identify the immune cell infiltration correlated to TMB based on the ImmuCellAI, the difference refers to difference between the median abundance of immune cell infiltration in high- (bottom 1/3) and low-TMB groups (top 1/3 by TMB).
We chose the results of the CIBERSORT algorithm and ImmuCellAI tool to study the relationship between TMB and immune cell infiltration. Using these two tools, 76.74% and 72.16% of the results of the optimal cut-point method, respectively, are also significant in the results of the top/bottom tertiles method. The heatmaps showed that in most tumors, macrophages, effector memory T cells (Tem), T helper cell 1 (Th1), regulatory T cells (nTreg, iTreg) and other infiltrating immune cells were increased in patients with a high TMB, while the abundance of infiltrating CD4 + T cells, CD4 + naive T cells, naive B cells, CD8 + naive T cells and other immune cells was decreased (Fig. 3B-C, S1B-C). For PCPG, the infiltration of mucosal associated invariant T cells (MAIT) was related to low TMB with a difference = -0.03 and P value = 0.02 (Fig. 3C, S1C). We can conclude that the relationship between the TMB and immune cell infiltration is varied according to different cancer types.
3.4. Correlation between TMB and patient prognosis and other clinical factors
TMB of each sample was calculated to study the distribution of TMB in 33 cancers, and we found that the highest levels of TMB in SKCM followed by LUSC and other epithelial cancers, while THCA, leukemias and pediatric tumors, such as PCPG, LAML and THYM exhibited a lower TMB (Fig. S2A).
Then, Kaplan-Meier analysis and optimal cut-point were used to identify the correlation between TMB and OS of 33 cancers. It turned out that high TMB was associated with the poor prognosis of patients with ACC, BRCA, COAD, ESCA, HNSC, KICH, KIRC, LGG, LIHC, PAAD, PCPG, SARC, THCA or THYM, while the opposite was true for patients with STAD (Fig. 4A), BLCA, KIRP, LUSC, OV, SKCM and UCEC (Fig. S2B). And when using top/bottom tertiles by TMB to analyze the relationship between OS and TMB in 33 cancers, high TMB was still related to the poor outcome of patients with ACC, ESCA, HNSC, KIRC and LGG, and the longer survival for patients with BLCA, OV, STAD and UCEC (Fig. S2C). This suggests that the relationship between TMB and OS may vary according to cancer types.
Fig. 4.
Kaplan–Meier analysis of OS and PFS according to the TMB in STAD and clinical information correlated to TMB among cancer types. The survival curves show the relationship between TMB and OS (A) and PFS (B) in patients with STAD. (C) Heat map showing the linear correlation between age, race, tumor stage, tumor grade, smoking status and TMB. (D) Box plots showing the difference in TMB between males and females with different cancers.
Among 21 cancers, 8 types of tumors including BRCA, COAD, ESCA, HNSC, LGG, SARC, STAD and UCEC were further stratified into 32 subtypes [32]. For example, BRCA was stratified into 5 molecular subtypes (Fig. S2D): luminal A (LumA), luminal B (LumB), basal-like (Basal), HER2-enriched (Her2) and normal-like (Normal). Through an analysis of TMB levels of the 32 molecular subtypes of these 8 tumors, we found that TMB levels differed in different subtypes of each tumor, like in BRCA, Her2 subtype was associated with a higher TMB, LumA subtype was associated with a lower TMB (Fig. S2D). Kaplan-Meier analysis was conducted to identify the association between TMB and OS of 32 subtypes of 8 tumor, and we found that OS of 15 kinds of subtypes in 8 cancers had significant relationship with TMB (optimal cut-point), OS of 4 kinds of subtypes in 5 cancers (COAD, ESCA, LGG, SARC and STAD) still had the consistent relationship with TMB when using top/bottom tertiles to perform a similar analysis (Fig. S2E, F). For example, in COAD, patients with chromosomal instability (CIN) subtype tended to have a poor survival outcome in high-TMB group (optimal cut-point method: HR = 2.56, P value < 0.01. top/bottom tertiles method: HR = 2.02, P value = 0.03) (Fig. S2E, F). Even for the same cancer, the relationship between TMB and prognosis may be different between different subtypes, such as in BRCA, patients with basal subtype tended to have a better survival outcome in high-TMB group (HR = 0.39, P value = 0.04), while patients with HER2 subtype tended to have a better survival outcome in low-TMB group (HR = 4.79, P value = 0.02) (Fig. S2E). This suggests that the relationship between TMB and OS may vary according to molecular subtypes of tumor.
The similar analysis was utilized to investigate the association between TMB and PFS of 21 tumors. It turned out that PFS of patients with 16 cancers was correlated with TMB (optimal cut-point), except for BRCA, COAD, KIRP, LUSC and THYM (Fig. 4B, S2G). Moreover, the results were in accordance with those in STAD (Fig. 4A, B) and other 15 tumors (Fig. S2B, G). There were still 9 cancers including ACC, BLCA, ESCA, KIRC, LGG, OV, STAD, THCA and UCEC in which PFS was related to TMB when using top/bottom tertiles (Fig. S2H).
Compared with top/bottom tertiles, the optimal cut-point refers to the result with the minimum P value, which increases the number of statistically significant cancers in the survival analysis but doesn’t change the relationship between prognosis and TMB. In general, TMB has divergent relationship with prognosis in different cancer types and can be used as a prognostic marker in pan-cancer, which was reported in the research of Wu, H et al. [52].
Through an analysis of association between TMB and other clinical information, statistically significant differences were observed in BLCA, BRCA and other 16 cancer types. We identified that male patients or patients with high grade BLCA, older patients with BRCA, older patients with SKCM and COAD patients with low stage BLCA, the patients in this study had a higher TMB (Fig. 4C, D). We also found that race in BLCA had a strong correlation with TMB (Estimation = 0.96, P value < 0.01), and white people tended to have a higher TMB (Fig. 4C). Male patients with LIHC, KIRP, PAAD or THCA tended to have high levels of TMB, while for those with STAD, the opposite was found (Fig. 4D). Thus, the correlation between clinical factors and TMB of different cancer types is heterogeneous.
4. Discussion
In some tumors, TMB has a predictive effect on the efficacy of immunotherapy and the prognosis of cancer patients [7], [17], it is related to the clinical characteristics [18], gene expression [53], DNA methylation [54], enrichment of signaling pathway [53], immune cell infiltration [20], gene mutations and the expression of MMRs [22], [23]. To date, few studies have been conducted to enable a comprehensive pan-cancer analysis of TMB. Through a multiomics analysis of TCGA datasets on 21 types of cancer in which the OS was shown to be related to TMB (Fig. S2B), we revealed different roles of TMB in each tumor and analyzed the possible reasons for its relationship with prognosis.
By studying driver gene mutations associated with TMB, what impressed us was mutations of GATA3 and MAP3K1 in BRCA, TCF7L2 in COAD, NFE2L2 in ESCA, CIC and IDH1 in LGG, CDH1 in STAD, and TP53 in UCEC tended to result in lower TMB (Fig. 1A-G). As for PCPG, THCA and THYM, the reason why no driver gene mutation was significantly associated with TMB may be related to their lower TMB level. When investigating the occurrence of ultramutations, we found that mutations in APC, KRAS and TP53 inhibit the occurrence of ultramutations in COAD, and TP53 mutation had the same effect on ultramutation in STAD and UCEC (Fig. 1H-J). Further analysis is needed to determine the specific reasons for these outcomes and the related mechanisms. In addition, high expression of most MMRs were related to a lower TMB in THCA and THYM, which may explain the low TMB levels in them (Fig. 1K).
The analysis of DEGs and DMRs driven by TMB showed that THYM samples had the most DEGs and DMRs (Table S3), and the proportion of DEGs related to abnormal DNA methylation was also the largest in DEGs (proportion = 1.25%, Table S3).
Our survival results showed that TMB has different relationship with prognosis of patients with different tumors (Fig. S2B, C, E-H), and in several cancers such as BLCA, STAD and UCEC, the relationship between TMB and prognosis was shown in the previous researches [21], [55], [56], [57], [58]. In our study, the relationship between TMB and clinical information may indicate that white, male patients or patients with high-grade in BLCA, older patients with SKCM, female patients with THCA, PAAD, LIHC or STAD, male patients with KIRP and patients with COAD in a later stage (Fig. 4C, D) may be more likely to have a better prognosis according to their relationship between TMB and prognosis. This finding suggested that TMB can help us predict which kind of patients may have a better prognosis.
In addition to differences in TMB levels, amount of immune infiltration, signaling pathways, molecular subtypes, differences in age and gender in the high/low groups might also affect survival. Combined with our analysis and existing reports, we found that the correlation between TMB and survival outcomes of patients may be explained by the immune microenvironment and gene mutations.
The reason that a high TMB level was negatively correlated with overall survival of patients with BRCA, COAD, KICH, KIRC, LGG or LIHC may be related to a high TMB being conducive to the infiltration of Tregs in KIRC, CD8 + T cells and macrophages in LGG (Fig. 3B-C, S2B-C). In the previous study, increased infiltration of Tregs is linked to poor outcomes in patients with kidney cancer [59], the proportion of immune cells (CD8 + T cells, neutrophils and macrophages) is negatively correlated with the OS of LGG patients [60]. For patients with BRCA, an increased proportion of Tregs [61] and TP53 mutations have been positively correlated with TMB and poor outcomes [18], [62]. In this study, we found that TMB was higher in the TP53 mutant group (Table S1A) and that the infiltration level of Tregs was positively correlated with TMB in BRCA (Fig. 3B-C, S2B-C). Patients with COAD and a high TMB have been previously shown to exhibit abundant immune cell infiltration and poor prognosis [63]. We surmised that poor prognosis related to high TMB is related to the MUC4 mutation because the MUC4 mutation frequency was higher in the high TMB group (Table S1A) and has been previously shown to be related to poor prognosis [64]. In combination with the aforementioned findings, we attributed the poor prognosis of KICH patients with a high TMB to the higher frequency of TP53 mutations (Table S1A). In patients with LIHC, we found that the CTNNB1 mutation group had a higher TMB (Table S1A), which was significantly correlated with a better prognosis in LIHC [65]. The TMB of the TP53 mutation group was also higher (Table S1A), which has previously been reported to be related to a higher TMB and worse outcome in patients with hepatocellular carcinoma [66].
The reason why high TMB is beneficial to the outcomes of patients with BLCA, OV, SKCM, STAD or UCEC may be that high TMB can increase the abundance of CD8 + T cells in BLCA patients [55], induce the activation of antitumor immune cells in OV patients [67], increase the proportion of M1 macrophages in SKCM patients and decrease the level of Tregs in STAD patients (Fig. 3B), and increase level of T cell CD8 + and M1 macrophages in patients with UCEC (Fig. 3B-C, S2B-C). As previously reported, patients with BLCA and high levels of CD8 + T cells may have a better prognosis [55]. Low levels of macrophages have been shown to be a risk factor for SKCM [20]. Both Tregs and M2 macrophages protect tumor cells from detection and elimination by the immune system, leading to undesirable results for patients with STAD [68], [69]. The reduction in B cells and CD8 + T cells has been significantly associated with a poor prognosis of UCEC [57].
In addition, we speculated that high TMB in ACC, ESCA, HNSC, PAAD, PCPG, SARC, THCA, or THYM is related to poor prognosis because it is negatively correlated with relatively larger fractions of antitumor immune cells, such as T cells and B cells, or positively correlated with the infiltration of immune cells that protect tumors, such as Tregs (Fig. 3B-C, S2B-C). For KIRP and LUSC, patients with and a high TMB may have a better outcome (Fig. S2B), and further study is needed to explore the reasons for these outcomes.
We also found that not all tumors with a high TMB are related to increased CD8 + T cell infiltration (Fig. 3 B-C, S1B-C) or good prognosis (Fig. S2B), which was reported in a recently published article [24].
There are limitations of our research. The data studied are all from the TCGA database; therefore, our analysis lacks data about other races, such as Asians. The clinical information, such as tumor stages and grades, was insufficient for the analysis of some cancer samples. Our optimal cut-point can stratify patients according to prognosis, but cannot stratify patients for immunotherapy efficacy. At present, there is no unified standard for the cut-off value of TMB-H. According to a report, TMB thresholds for effective prediction of immunotherapy response in various malignancies are not well established, the relationship between immunotherapy response and the level of TMB still needs specific research. The TMB cut-off for immunotherapy also varies according to cancer types [7]. Moreover, our study may need to be updated according to new discoveries in the future.
5. Conclusions
Taken together, our study presents a systematic analysis of tumor mutational burden and its related factors across cancers and highlights the potential role of TMB in different cancers. And TMB can be used as a prognostic marker in pan-cancer. Our research further supports a comprehensive understanding of the role of TMB across cancers and has important clinical implications.
Funding
This work was supported in part by grants from the Natural Science Foundation of Anhui Province (2008085QH378 to K.D.), Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to Z.L.) and Tip-Top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program (2019TQ05Y351 to Z.L.).
CRediT authorship contribution statement
Lin Li: Conceptualization, Data curation, Investigation, Visualization, Writing – original draft. Long Bai: Data curation, Investigation, Visualization. Huan Lin: Data curation, Investigation, Visualization. Lin Dong: Conceptualization, Investigation, Methodology. Rumeng Zhang: Conceptualization, Investigation, Methodology. Xiao Cheng: Conceptualization, Investigation, Methodology. Zexian Liu: Conceptualization, Methodology, Supervision, Funding acquisition. Yi Ouyang: Supervision, Methodology. Keshuo Ding: Conceptualization, Methodology, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.013.
Contributor Information
Yi Ouyang, Email: ouyangy@sysucc.org.cn.
Keshuo Ding, Email: dingks@ahmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Brahmer J., Reckamp K.L., Baas P., Crinò L., Eberhardt W.E.E., Poddubskaya E. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. New Eng J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powles T., Eder J.P., Fine G.D., Braiteh F.S., Loriot Y., Cruz C. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 3.Robert C., Ribas A., Wolchok J.D., Hodi F.S., Hamid O., Kefford R. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet (London, England) 2014;384(9948):1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 4.Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167(2):397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonpavde G., Dranitsaris G., Necchi A. Improving the Cost Efficiency of PD-1/PD-L1 Inhibitors for Advanced Urothelial Carcinoma: A Major Role for Precision Medicine? Eur Urol. 2018;74(1):63–65. doi: 10.1016/j.eururo.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Science translational medicine 2016;8(328):328rv4. [DOI] [PMC free article] [PubMed]
- 7.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Annal Oncol Off J Eur. Soc. Med Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meléndez B., Van Campenhout C., Rorive S., Remmelink M., Salmon I., D’Haene N. Methods of measurement for tumor mutational burden in tumor tissue. Trans Lung Canc Res. 2018;7(5):661–667. doi: 10.21037/tlcr.2018.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fancello L., Gandini S., Pelicci P.G., Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immuno Ther Cancer. 2019;7(1):183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY) 2015;348(6230):124-128. [DOI] [PMC free article] [PubMed]
- 11.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (New York, NY) 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellmann M.D., Ciuleanu T.-E., Pluzanski A., Lee J.S., Otterson G.A., Audigier-Valette C. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. New Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samstein R.M., Lee C.-H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman A.M., Kato S., Bazhenova L., Patel S.P., Frampton G.M., Miller V. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birkbak N.J., Kochupurakkal B., Izarzugaza J.M.G., Eklund A.C., Li Y., Liu J. Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS ONE. 2013;8(11):e80023. doi: 10.1371/journal.pone.0080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clinic Oncol Off J Am Soc Clinic Oncol 2019;37(14):1217-1227 [DOI] [PMC free article] [PubMed]
- 17.Danilova L., Wang H., Sunshine J., Kaunitz G.J., Cottrell T.R., Xu H. Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. PNAS. 2016;113(48):E7769–E7777. doi: 10.1073/pnas.1607836113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Lin A., Li Y., Ding W., Meng H., Luo P. Age and Mutations as Predictors of the Response to Immunotherapy in Head and Neck Squamous Cell Cancer. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.60896910.3389/fcell.2020.608969.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draaisma K., Wijnenga M.M.J., Weenink B., Gao Y.a., Smid M., Robe P. PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathologica Communications. 2015;3(1) doi: 10.1186/s40478-015-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J., Wu X., Yu J., Zhu Y., Cang S. Prognostic Role of Tumor Mutation Burden Combined With Immune Infiltrates in Skin Cutaneous Melanoma Based on Multi-Omics Analysis. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.570654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Zhang X., Li J., Ma X., Feng F., Liu L. ADRB1 was identified as a potential biomarker for breast cancer by the co-analysis of tumor mutational burden and immune infiltration. Aging. 2021;13(1):351–363. doi: 10.18632/aging.104204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1) doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endris V., Buchhalter I., Allgäuer M., Rempel E., Lier A., Volckmar A.-L. Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer. 2019 doi: 10.1002/ijc.32002. [DOI] [PubMed] [Google Scholar]
- 24.McGrail D.J., Pilié P.G., Rashid N.U., Voorwerk L., Slagter M., Kok M. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Annal Oncol Off J Eur Soc. Med Oncol. 2021;32(5):661–672. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic acids research 2020;48(W1):W509-w14. [DOI] [PMC free article] [PubMed]
- 28.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B.o., Severson E., Pignon J.-C., Zhao H., Li T., Novak J. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Advanced science (Weinheim, Baden-Wurttemberg, Germany) 2020;7(7):1902880. [DOI] [PMC free article] [PubMed]
- 31.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173(2):400-16.e11. [DOI] [PMC free article] [PubMed]
- 32.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173(2):321-37.e10. [DOI] [PMC free article] [PubMed]
- 33.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic acids research 2016;44(8):e71. [DOI] [PMC free article] [PubMed]
- 34.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Y., Liu L., Chen H.u., Wang Y., Xu Y., Mao H. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell. 2016;29(5):711–722. doi: 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Jiménez F., Muiños F., Sentís I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 37.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh P., Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129(7-8):391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xavier A., Olsen M.F., Lavik L.A., Johansen J., Singh A.K., Sjursen W. Comprehensive mismatch repair gene panel identifies variants in patients with Lynch-like syndrome. Mol Genet Genomic Med. 2019;7(8) doi: 10.1002/mgg3.v7.810.1002/mgg3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravind L., Walker D.R., Koonin E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27(5):1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell reports 2018;23(1):239-54.e6. [DOI] [PMC free article] [PubMed]
- 42.Jiricny J. The multifaceted mismatch-repair system. Nature reviews Molecular cell biology 2006;7(5):335-346. [DOI] [PubMed]
- 43.Li G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 44.Marti T.M., Kunz C., Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol. 2002;191(1):28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 45.Irizarry R.A., Ladd-Acosta C., Wen B.o., Wu Z., Montano C., Onyango P. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 47.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Li X, Liang H, Wei L, Cui Q, Yao M, et al. A new mutL homolog 1 c.1896+5G>A germline mutation detected in a Lynch syndrome-associated lung and gastric double primary cancer patient. Molecular genetics & genomic medicine 2019;7(8):e787. [DOI] [PMC free article] [PubMed]
- 49.Ling B.o., Ye G., Zhao Q., Jiang Y., Liang L., Tang Q. Identification of an Immunologic Signature of Lung Adenocarcinomas Based on Genome-Wide Immune Expression Profiles. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.60370110.3389/fmolb.2020.603701.s00110.3389/fmolb.2020.603701.s00210.3389/fmolb.2020.603701.s00310.3389/fmolb.2020.603701.s00410.3389/fmolb.2020.603701.s00510.3389/fmolb.2020.603701.s006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Indraccolo S., Lombardi G., Fassan M., Pasqualini L., Giunco S., Marcato R. Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clinic Canc Res Off J Am Assoc Canc Res. 2019;25(6):1828–1837. doi: 10.1158/1078-0432.CCR-18-1892. [DOI] [PubMed] [Google Scholar]
- 51.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu HX, Wang ZX, Zhao Q, Chen DL, He MM, Yang LP, et al. Tumor mutational and indel burden: a systematic pan-cancer evaluation as prognostic biomarkers. Annals of translational medicine 2019;7(22):640. [DOI] [PMC free article] [PubMed]
- 53.Kang K., Xie F., Mao J., Bai Y.i., Wang X. Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.57314110.3389/fonc.2020.573141.s00110.3389/fonc.2020.573141.s00210.3389/fonc.2020.573141.s00310.3389/fonc.2020.573141.s004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai L., Bai H., Duan J., Wang Z., Gao S., Wang D.i. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J ImmunoTher Cancer. 2019;7(1) doi: 10.1186/s40425-019-0660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv J., Zhu Y., Ji A., Zhang Q., Liao G. Mining TCGA database for tumor mutation burden and their clinical significance in bladder cancer. Biosci Rep. 2020;40(4) doi: 10.1042/BSR20194337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F., Wei X.L., Wang F.H., Xu N., Shen L., Dai G.H. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Annal Oncol Off J Eur Soc Med Oncol. 2019;30(9):1479–1486. doi: 10.1093/annonc/mdz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao L., Fu X., Han X., Yu Y., Ye Y., Gao J. Tumor mutation burden in connection with immune-related survival in uterine corpus endometrial carcinoma. Canc Int. 2021;21(1):80. doi: 10.1186/s12935-021-01774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan S., Gao X., Qin Q., Li H., Yuan Z., Zhao S. Association between tumor mutation burden and immune infiltration in ovarian cancer. Int Immunopharmacol. 2020;89:107126. doi: 10.1016/j.intimp.2020.107126. [DOI] [PubMed] [Google Scholar]
- 59.Zhu G., Pei L., Yin H., Lin F., Li X., Zhu X. Profiles of tumor-infiltrating immune cells in renal cell carcinoma and their clinical implications. Oncol Lett. 2019 doi: 10.3892/ol10.3892/ol.2019.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin W., Jiang X., Tan J., Xin Z., Zhou Q., Zhan C. Development and Validation of a Tumor Mutation Burden-Related Immune Prognostic Model for Lower-Grade Glioma. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West N.R., Kost S.E., Martin S.D., Milne K., deLeeuw R.J., Nelson B.H. Tumour-infiltrating FOXP3(+) lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br J Cancer. 2013;108(1):155–162. doi: 10.1038/bjc.2012.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyu H., Li M., Jiang Z., Liu Z., Wang X. Correlate the TP53 Mutation and the HRAS Mutation with Immune Signatures in Head and Neck Squamous Cell Cancer. Comput Struct Biotechnol J. 2019;17:1020–1030. doi: 10.1016/j.csbj.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z., Xie X., Wang X., Zhang X., Li W., Sun T. Correlations Between Tumor Mutation Burden and Immunocyte Infiltration and Their Prognostic Value in Colon Cancer. Front Genet. 2021;12 doi: 10.3389/fgene.2021.62342410.3389/fgene.2021.623424.s00110.3389/fgene.2021.623424.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng L., Li Y., Gu H., Xiang L., Xiong Y., Wang R. Mucin 4 mutation is associated with tumor mutation burden and promotes antitumor immunity in colon cancer patients. Aging. 2021;13(6):9043–9055. doi: 10.18632/aging.202756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mo Z., Wang Y., Cao Z., Li P., Zhang S. An Integrative Analysis Reveals the Underlying Association Between CTNNB1 Mutation and Immunotherapy in Hepatocellular Carcinoma. Front Oncol. 2020;10:853. doi: 10.3389/fonc.2020.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., Yan K., He X., Zhu H., Song J., Chen S. LRP1B or TP53 mutations are associated with higher tumor mutational burden and worse survival in hepatocellular carcinoma. J Cancer. 2021;12(1):217–223. doi: 10.7150/jca.48983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Perrone G, Ruffini PA, Catalano V, Spino C, Santini D, Muretto P, et al. Intratumoural FOXP3-positive regulatory T cells are associated with adverse prognosis in radically resected gastric cancer. European journal of cancer (Oxford, England : 1990) 2008;44(13):1875-1882. [DOI] [PubMed]
- 69.Huang X., Pan Y., Ma J., Kang Z., Xu X., Zhu Y. Prognostic significance of the infiltration of CD163(+) macrophages combined with CD66b(+) neutrophils in gastric cancer. Cancer Med. 2018;7(5):1731–1741. doi: 10.1002/cam4.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.