Abstract
Insulin receptor substrate 1 (IRS-1) is a critical adapter protein involved in both insulin and insulin-like growth factor (IGF) signaling. Due to the fact that alteration of IRS-1 levels can affect the sensitivity and response to both insulin and IGF-I, we examined the ability of each of these ligands to affect IRS-1 expression. IGF-I (10 nM) stimulation of MCF-7 breast cancer cells caused a transient tyrosine phosphorylation of IRS-1 that was maximal at 15 min and decreased thereafter. The decrease in tyrosine phosphorylation of IRS-1 was paralleled by an apparent decrease in IRS-1 levels. The IGF-mediated decrease in IRS-1 expression was posttranscriptional and due to a decrease in the half-life of the IRS-1 protein. Insulin (10 nM) caused tyrosine phosphorylation of IRS-1 but not degradation, whereas high concentrations of insulin (10 μM) resulted in degradation of IRS-1. IGF-I (10 nM) stimulation resulted in transient IRS-1 phosphorylation and extracellular signal-related kinase (ERK) activation. In contrast, insulin (10 nM) caused sustained IRS-1 phosphorylation and ERK activation. Inhibition of 26S proteasome activity by the use of lactacystin or MG132 completely blocked IGF-mediated degradation of IRS-1. Furthermore, coimmunoprecipitation experiments showed an association between ubiquitin and IRS-1 that was increased by treatment of cells with IGF-I. Finally, IGF-mediated degradation of IRS-1 was blocked by inhibition of phosphatidylinositol 3′-kinase activity but was not affected by inhibition of ERK, suggesting that this may represent a direct negative-feedback mechanism resulting from downstream IRS-1 signaling. We conclude that IGF-I can cause ligand-mediated degradation of IRS-1 via the ubiquitin-mediated 26S proteasome and a phosphatidylinositol 3′-kinase-dependent mechanism and that control of degradation may have profound effects on downstream activation of signaling pathways.
Insulin receptor substrate 1 (IRS-1) was originally identified downstream of the insulin receptor (IR) but can also be phosphorylated in response to insulin-like growth factor I (IGF-I) (57). IRS-1 can also be activated by a number of diverse ligands, including growth hormone, prolactin, oncostatin, interleukin-2 (IL-2), IL-4, IL-7, IL-9, IL-13, IL-15, and α/β interferon (for a review, see reference 66). Following activation, IRS-1 binds a diverse set of downstream signaling molecules including p85, Grb2, Nck/Crk, Syp/Fyn, and SHP2 (66). Through a poorly defined mechanism, IRS-1 can also bind other proteins, including simian virus 40 large T antigen (8), 14-3-3 (27, 39), and αvβ3 integrin (64). The complexity of IRS-1 upstream and downstream signaling reflects an emerging concept of complex cross-talk between extracellular and intracellular signaling pathways and may place IRS molecules in a central position to coordinate multiple signaling pathways (41). The discovery of IRS-2 (58), IRS-3 (31, 50, 55), and IRS-4 (30) has expanded the potential number of divergent signaling pathways.
Although there has been abundant research concerning activation of IRS-1, the mechanisms controlling the duration and strength of the signal remain unclear. Two general mechanisms have been described: regulation of IRS-1 expression and regulation of IRS-1 phosphorylation. IRS-1 expression can be transcriptionally regulated by glucocorticoids (3, 46, 63). We have recently shown that estrogen can induce IRS-1 mRNA and protein expression in breast cancer cell lines and xenografts (32). Increased IRS-1 expression resulted in enhanced tyrosine phosphorylation of IRS-1 in response to IGF-I and increased activation of extracellular signal-related kinase (ERK). Conversely, decreased IRS-1 expression, achieved by antiestrogen treatment, inhibited IGF-I signaling through IRS-1. In a number of cell systems it has been shown that high concentrations of insulin can cause posttranscriptional downregulation of IRS-1 expression (23, 47), a potential mechanism being proteolytic degradation of IRS-1 protein by the calpain pathway (54).
IRS-1 phosphorylation is regulated by receptor tyrosine kinases, protein tyrosine phosphatases, and serine/threonine kinases. Receptor tyrosine kinases phosphorylate IRS-1 on tyrosine residues and activate downstream signaling pathways (66). Following tyrosine phosphorylation, protein tyrosine phosphatases such as SRC homology domain protein tyrosine phophatase (SHP2) are activated and can dephosphorylate IRS-1 (38). In addition, several other phosphatases have been shown to inhibit the action of insulin (2, 18). It has been shown that phosphorylation of the serine or threonine residues of IRS-1, in contrast with dephosphorylation of tyrosines, can inhibit further tyrosine phosphorylation of IRS-1. The kinases involved and the mechanisms of inhibition are poorly understood; however, the serine residues of IRS-1 are phosphorylated by mitogen-activated protein kinase (MAPK) (9), protein kinase C (PKC) (10), Akt/PKB (42), casein kinase II (59), and phosphatidylinositol 3′-kinase (PI3K) (17, 29, 60) following activation of IRS-1 by ligands such as angiotensin II (16) and tumor necrosis factor alpha (22). Okadaic acid, a serine/threonine phosphatase inhibitor, increases phosphorylation of serine residues on IRS-1 and inhibits the action of insulin (25).
In studies aimed at examining factors regulating the action of IRS-1, we have shown that stimulation of IRS-1 tyrosine phosphorylation by IGF-I results in IRS-1 degradation. Low concentrations of insulin (10 nM) that are known to activate only the IR induced sustained IRS-1 activation and downstream ERK phosphorylation. In contrast, high concentrations of insulin (10 μM), which can activate the IGF-I receptor (IGF-IR), were able to degrade IRS-1. IGF-I increased the level of association of IRS-1 with ubiquitin, and 26S proteasome inhibitors blocked degradation of IRS-1. Finally, IGF-I degradation of IRS-1 was blocked by inhibition of PI3K activity, suggesting that this may be a direct negative-feedback mechanism resulting from IRS-1 activation of PI3K. In conclusion, we have revealed a novel mechanism for degradation of IRS-1 that might represent a negative-feedback mechanism important in controlling the activity of this critical signal transduction molecule.
MATERIALS AND METHODS
Materials.
All materials and chemicals were purchased from Sigma (St. Louis, Mo.) unless otherwise noted. IGF-I was purchased from GROPEP (Adelaide, Australia). ICI 182780 was a kind gift from Zeneca Pharmaceuticals (Macclesfield, England). The inhibitors lactacystin, MG132, LY294002, and PD098059 were from Calbiochem (La Jolla, Calif.). All tissue culture reagents were purchased from Life Technologies (Grand Island, N.Y.) unless otherwise stated. [35S]methionine-[35S]cysteine (37 TBq/mmol) was purchased from NEN (Boston, Mass.).
Cell lines.
MCF-7 cells have been maintained in our laboratory for many years (40). Cells were routinely maintained in improved minimal essential medium (IMEM) plus 5% fetal bovine serum (Summit, Ft. Collins, Colo.), 2 mM glutamine, 50 IU of penicillin/ml, and 50 μg of streptomycin/ml. Serum-free medium (SFM) consisted of IMEM plus 10 mM HEPES (pH 7.4), 1 μg of transferrin/ml, 1 μg of fibronectin/ml, 2 mM glutamine, 50 IU of penicillin/ml, 50 μg of streptomycin/ml, and trace elements (Biofluids, Rockville, Md.).
Cell stimulation and lysis.
Cells were plated at a density of 5 × 105 per 6-cm-diameter dish (Becton Dickinson, Lincoln Park, N.J.) and allowed to adhere to the dishes overnight. The next day, the medium was changed to SFM, and 24 h later, the cells were stimulated with various ligands for different periods of time. For experiments using protease inhibitors, cells were first preincubated with the inhibitors (lactacystin [final concentration, 10 μM] and MG132 [final concentration, 50 μM], both in dimethyl sulfoxide [DMSO]) for 30 min prior to stimulation in the presence of an inhibitor. For experiments using PD098059 (25 μM), LY294002 (25 μM), or wortmannin (250 nM), cells were again first preincubated for 30 min with an inhibitor and then stimulated in the presence of the inhibitor as described previously (24). Control cells were incubated with a similar concentration of DMSO alone. After stimulation, cells were washed twice with cold phosphate-buffered saline and then lysed in 150 μl of TNESV buffer, which included fresh protease inhibitors (50 mM Tris-HCl [pH 7.4], 1% NP-40, 3 mM EDTA, 100 mM NaCl, 10 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 20 μg of leupeptin/ml, and 20 μg of aprotinin/ml). Lysates were clarified by centrifugation at 14,000 × g for 15 min at 4°C and stored at −20°C. Protein concentrations were determined by the bicinchoninic acid method in accordance with the manufacturer's (Pierce, Rockford, Ill.) instructions.
Immunoblotting.
Total protein (50 μg) was resuspended in denaturing sample loading buffer (3% dithiothreitol, 0.1 M Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 0.2% bromophenol blue, 20% glycerol), separated by SDS–8% polyacrylamide gel electrophoresis (PAGE), and electrophoretically transferred to a nitrocellulose membrane overnight at 4°C. The remaining steps were all performed at room temperature. The membrane was blocked with 5% milk–TBST (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], 0.05% Tween 20) for 1 h. For antiphosphotyrosine immunoblotting, the membrane was washed six times, for 5 min each time, with TBST and then incubated with a 1:1,000 dilution of a horseradish peroxidase (HRP)-linked primary antibody (RC20; Transduction Laboratories, Lexington, Ky.) in TBST. After the membrane was washed six times (5 min each time) with TBST, bands were visualized by enhanced chemiluminescence (ECL) in accordance with the manufacturer's instructions (Pierce). For phosphorylated-ERK immunoblotting (New England Biolabs, Beverly, Mass.), the membrane was washed six times (5 min each time) with TBST and incubated with a 1:1,000 dilution of primary antibody in TBST. After 1 h, the membrane was again washed six times (5 min each time) with TBST and then incubated with an HRP-linked donkey anti-rabbit antibody (Amersham) at a dilution of 1:1,000 in TBST–5% milk. After the membrane was washed six times (5 min each time) with TBST, bands were visualized by ECL according to the manufacturer's instructions (Pierce). IRS-1 (Upstate Biotechnology, Lake Placid, N.Y. [UBI]) antibodies were used at a final concentration of 0.35 μg/ml, and IRS-2 (UBI) and total MAPK (UBI) antibodies were used at 1 μg/ml. For these blots, the membrane was not washed after the blocking step but was immediately incubated with a 1:1,000 dilution of the primary antibody in TBST–5% milk. After 1 h, the membrane was washed six times (5 min each time) with TBST and then incubated with an HRP-linked donkey anti-rabbit antibody diluted 1:1,000 in TBST–5% milk. After the membrane was washed six times (5 min each time) with TBST, bands were visualized by ECL according to the manufacturer's suggestions (Pierce).
Plasmids and transient transfection.
An expression plasmid for hemagglutinin (HA)-tagged ubiquitin (HA-Ub) was constructed by removing HA-Ub (a 330-bp fragment) from YEp112 (21) by EcoRI and KpnI digestion. This fragment was cloned into pSP72, then excised by EcoRI and XbaI digestion, and subcloned into pcDNA3.1 to generate pcDNA-HAUb. flag–IRS-1 was constructed by first performing genomic PCR on an N-terminal fragment of human IRS-1 from MCF-7 cells, using the primers 5′CAAGTTGAATTCGCCGCCACCATGGACTACAAGGACGACGATGACAAGGCGAGCCCTCCGGAGAGCGAT (EcoRI site, Kozak consensus sequence, flag epitope, and IRS-1) and 3′GCCGACTCTAGACTACTTGCTGGTCAGGCAAAGGCGGTAGAT, which inserted an N-terminal flag epitope. The 1.3-kb fragment was cut with EcoRI (in the 5′ primer) and an internal XhoI. A C-terminal fragment of IRS-1 was digested from IRS-1 cDNA by XhoI and HindIII digestion, the two fragments were ligated together with the XhoI sites, and then the full-length flag–IRS-1 was cloned into EcoRI and HindIII sites in pcDNA3.1. The construct was sequenced to confirm its identity. COS-7 cells were transiently transfected by using Lipofectin in accordance with the manufacturer's instructions. Cells were transfected with 0.5 μg of pcDNA-HAUb plus pflagIRS-1, pcDNA-HAUb alone, pflagIRS-1 alone, or pCDNA3.1 (as a control). After 8 h of transfection, the medium was changed to SFM, and 16 h later, cells transfected with pcDNA-HAUb and pflagIRS-1 were stimulated, or not, with IGF-I (5 nM) for 2 h. All cells were lysed in TNESV buffer, and 500 μg of protein was precleared with 25 μl of a protein A-agarose solution for 30 min at 4°C and then immunoprecipitated with 2 μg of antiflag antibody (Sigma) overnight at 4°C. The next day, 25 μl of protein A-agarose (Pierce) was added, and the suspension was incubated for a further 4 h with rocking at 4°C. The beads were washed three times with TNESV, resuspended in 40 μl of 1× sample loading buffer, and boiled, and polypeptides were separated by SDS–8% PAGE. After transfer of polypeptides to a nitrocellulose membrane, immunoblotting was performed with a 1:1,000 dilution of an antibody against HA (Babco).
Pulse-chase labeling of MCF-7 cells and immunoprecipitation of IRS-1.
MCF-7 cells were plated at a density of 106 per 10-cm-diameter dish, allowed to adhere overnight, and then incubated in SFM for 24 h. Cells were incubated in depletion medium (minimal essential medium) lacking methionine and cysteine but supplemented with 2 mM glutamine) for 1 h and then in labeling medium (depletion medium plus 300 μCi of [35S]methionine-[35S]cysteine/dish) for 1 h. Labeling medium was removed, the dishes were washed five times with chase medium (depletion medium plus 2 mM methionine, 2 mM cysteine, 2 mM glutamine), and then cells were incubated with chase medium with or without IGF-I (10 nM). Cells were lysed in 400 μl of TNESV and then precleared with 25 μl of protein A-agarose for 30 min at 4°C, and IRS-1 was immunoprecipitated with 5 μl of IRS-1 antibodies (UBI) overnight at 4°C. A 25-μl volume of protein A-agarose was added, and after 4 h, the cells were washed five times with TNESV buffer. The beads were resuspended in 40 μl of 1× sample loading buffer, and polypeptides were separated by SDS–6% PAGE. The gel was treated with a fluorography reagent (EnHance; NEN) and dried. Densitometry was performed with a Molecular Dynamics PhosphorImager.
RESULTS
IGF-I stimulation decreases IRS-1 expression in a time- and dose-responsive manner.
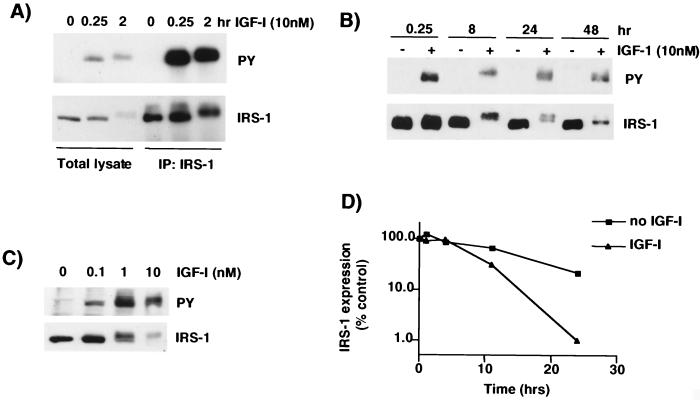
MCF-7 breast cancer cells were stimulated with IGF-I (10 nM), lysed, and immunoblotted for phosphotyrosine-containing proteins. After 15 min of IGF-I stimulation, a single protein of ∼170 kDa (the molecular mass of IRS-1) was readily detectable by immunoblotting with antiphosphotyrosine (Fig. 1A, top panel). A longer incubation with IGF-I (2 h) caused an increase in the molecular mass of this protein and a decrease in phosphorylation of tyrosines. Immunoblotting for IRS-1 revealed a similar migration pattern with a decrease in IRS-1 levels after 2 h of IGF-I stimulation (Fig. 1A, bottom panel). Immunoprecipitation with IRS-1 antibodies followed by immunoblotting with antiphosphotyrosine detected the tyrosine-phosphorylated band, indicating that this is tyrosine-phosphorylated IRS-1. However, upon immunoblotting for IRS-1, we again noted that 2 h of incubation with IGF-I resulted in a shift in the size of IRS-1 and also a significant decrease in IRS-1 expression. Stimulation of cells for 8 h with IGF-I (Fig. 1B) resulted in a dramatic decrease in the electromobility of IRS-1, seen on both IRS-1 and antiphosphotyrosine immunoblots, suggesting that this represents a hyperphosphorylated form. Furthermore, there was a dramatic decrease in the level of IRS-1 protein. A longer incubation with IGF-I (24 h) resulted in a doublet of the hyperphosphorylated form and a less phosphorylated form. At 48 h, most of the IRS-1 migrated at the same molecular mass as IRS-1 under SFM conditions. It should be noted that in SFM, IRS-1 levels did not alter over time course of the experiment, indicating that the decrease in IRS-1 expression was a ligand-mediated event. The apparent decrease in IRS-1 expression was not a result of an inability of the IRS-1 antibody to recognize phosphorylated IRS-1, since a different IRS-1 antibody, which recognizes a different epitope (pleckstrin homology [PH] domain), gave identical results (data not shown). Furthermore, the decrease in IRS-1 was not due to a differential intracellular localization that resulted in a poor extraction by TNESV lysis buffer, since identical results were obtained with 5% SDS lysis buffer (data not shown).
FIG. 1.
IRS-1 is tyrosine phosphorylated and then degraded after IGF-I stimulation. (A) MCF-7 cells were incubated in SFM for 24 h and then stimulated with IGF-I (10 nM) for 15 min and 2 h. Cells were lysed in TNESV buffer, and 300 μg of the resultant protein was used for immunoprecipitation experiments with antibodies to IRS-1. Total cell lysate (lanes 1 to 3 from left) and immunoprecipitated proteins (IP:IRS-1) (lane 4 to 6 from left) were separated by SDS–8% PAGE and immunoblotted with antiphosphotyrosine (PY) or anti-IRS-1 (IRS-1) antibodies. (B) MCF-7 cells were treated as described for panel A, but no immunoprecipitation was performed. (C) MCF-7 cells were incubated with IGF-I (0, 0.1, 1, and 10 nM) for 8 h and lysed, and the resulting lysate was treated as described for panel A. All immunoblots are representative of at least four independent experiments. (D) MCF-7 cells were labeled with [35S]methionine-[35S]cysteine for 1 h, incubated in chase medium with or without IGF-I (5 nM), and then lysed at the indicated time points. Lysates were immunoprecipitated with anti-IRS-1 antibodies and separated by SDS-PAGE, and the gel was then dried. Densitometry was performed with a PhosphorImager. Results are expressed as percentages of the densitometric reading at 0 h. This figure is representative of two independent experiments.
The IGF-I-mediated decrease in IRS-1 expression was directly related to the dose of IGF-I used for stimulation. MCF-7 cells were stimulated with increasing doses of IGF-I for 8 h and then immunoblotted for phosphotyrosine (Fig. 1C, top panel) or IRS-1 (Fig. 1C, lower panel). Increasing concentrations of IGF-I caused a dose-dependent increase in the phosphorylation and the size of IRS-1 and also a dose-dependent decrease in IRS-1 protein levels.
The ability of IGF-I to decrease IRS-1 expression occurred in diverse cell types and was associated with the ability of a cell to phosphorylate IRS-1; IGF-I caused a decrease in IRS-1 expression in two other breast cancer cell lines (ZR-75 and T47D) and in CHO cells but had no effect on IRS-1 expression in HepG2 and MDA-231 cells, which do not phosphorylate IRS-1 (data not shown).
To determine whether the IGF-mediated decrease in IRS-1 expression was a transcriptional effect, we measured steady-state levels of IRS-1 mRNA after exposure to IGF by RNase protection assay. IGF-I had no effect on IRS-1 mRNA expression (data not shown), which is consistent with the inability of insulin (1 μM) to affect IRS-1 mRNA (3). Furthermore, the pattern of IGF-mediated decrease in IRS-1 protein expression was not altered by either actinomycin D (an inhibitor of transcription) at 2 μg/ml or cycloheximide (an inhibitor of translation) at 10 μg/ml (data not shown), suggesting that posttranscriptional mechanisms are responsible for the decreased IRS-1 levels. Therefore, we next examined whether the IGF-I-mediated decrease in IRS-1 was due to a change in the IRS-1 protein's half-life. We measured the half-life of IRS-1 in the presence and in the absence of IGF-I by [35S]methionine-[35S]cysteine pulse-chase labeling of MCF-7 cells followed by IRS-1 immunoprecipitation. In the absence of IGF-I, IRS-1 had a half-life of approximately 15 h (Fig. 1D). When cells were stimulated with IGF-I, IRS-1's half-life decreased to approximately 9 h. This set of experiments indicated that IGF-I caused significant degradation of IRS-1 protein by a posttranscriptional mechanism.
Increased insulin concentrations result in decreased IRS-1 expression.
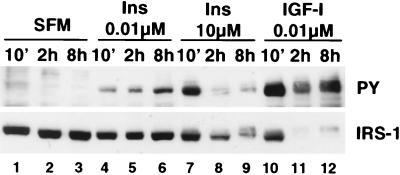
It has previously been shown that insulin at high (micromolar) concentrations causes a decrease in IRS-1 expression (23, 47). We therefore tested the ability of insulin, at low (nanomolar) and at high (micromolar) concentrations, to decrease IRS-1 expression in MCF-7 cells (Fig. 2). Short-term (10-min) stimulation of MCF-7 cells with IGF-I (10 nM) or a nanomolar (0.01 μM) or micromolar (10 μM) concentration of insulin resulted in phosphorylation of IRS-1 tyrosine residues but did not affect IRS-1 expression (compare lanes 1, 4, 7, and 10). Longer incubations (2 and 8 h) with a low concentration of insulin (0.01 μM) resulted in a sustained increase in phosphorylation of IRS-1 (compare lanes 4 to 6 with lanes 1 to 3). At this concentration, insulin had no effect on total IRS-1 expression. In contrast, stimulation of cells with 10 μM insulin, a concentration previously shown to cause a decrease in IRS-1 expression (47), resulted in greater tyrosine phosphorylation of IRS-1 than did a nanomolar concentration of insulin (compare lane 7 with lane 4) and also caused a decrease in IRS-1 expression after 2 and 8 h (lanes 8 and 9). The pattern of phosphorylation and expression of IRS-1 with 10 μM insulin was similar to that seen when cells were treated with IGF-I at 10 nM (lanes 10 to 12).
FIG. 2.
Insulin at high, but not low, concentrations degrades IRS-1. MCF-7 cells were incubated in SFM for 24 h and then stimulated with the indicated concentrations of insulin (Ins) or IGF-I for 10 min (10'), 2 h, or 8 h. Cells were lysed, and 50 μg of lysate was subjected to SDS–8% PAGE and then immunoblotted with antiphosphotyrosine (PY) or anti-IRS-1 (IRS-1) antibodies.
IGF-I and insulin differ in duration of activation of ERK.
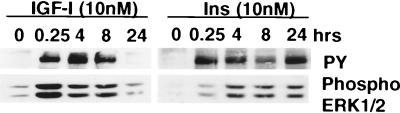
To examine whether the differential length of activation of IRS-1 by insulin and IGF-I affected downstream signaling events, we measured activation of ERK. MCF-7 breast cancer cells were stimulated with IGF-I (10 nM) or insulin (10 nM) for various periods of time. IGF-I stimulation resulted in a time-dependent phosphorylation of IRS-1 tyrosines (Fig. 3). Insulin stimulation resulted in phosphorylation of IRS-1 tyrosine residues, but in contrast to IGF-I, phosphorylation was maintained throughout the length of the stimulation. Immunoblotting for phosphorylated ERK1/2 revealed that IGF-I caused a rapid increase in phosphorylation of ERK after 15 min, after which phosphorylation decreased, whereas insulin caused a sustained activation of ERK over the 24-h period. Immunoblots were probed with antibodies to ERK, and equal levels were observed in all lanes, suggesting that IGF-I does not affect ERK levels in a manner similar to IRS-1 (data not shown).
FIG. 3.
Insulin causes sustained activation of IRS-1 and ERK, while IGF-I causes transient activation. MCF-7 cells were incubated in SFM for 24 h and then stimulated with IGF-I (10 nM) or insulin (Ins; 10 nM) for the indicated periods of time. Cells were lysed, and 50 μg of the resultant protein was immunoblotted with antiphosphotyrosine (PY) or anti-phospho-ERK1/2 (Phospho ERK1/2) antibodies. This figure is representative of three independent experiments.
IGF-I-stimulated degradation of IRS-1 results in a decreased amount of tyrosine-phosphorylated IRS-1 when cells are restimulated with IGF-I.
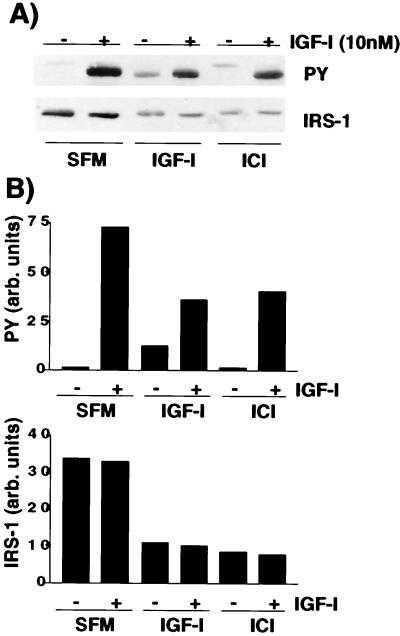
Evidence suggests that phosphorylation of IRS-1 tyrosines is a rate-limiting step in IGF-I-mediated signal transduction. Our group and others have shown that IRS-1 expression can be regulated by the estrogen receptor (32, 48). Estrogen treatment of MCF-7 cells increased IRS-1 expression, and antiestrogen treatment decreased it. The reduced level of IRS-1 expression after antiestrogen treatment resulted in a decreased amount of total tyrosine-phosphorylated IRS-1 following IGF stimulation and reduced downstream signaling through ERK1/2. Therefore, we next tested whether the decrease in IRS-1 expression after IGF treatment would also result in a decreased total amount of tyrosine-phosphorylated IRS-1 if cells were restimulated with IGF-I. As a control, we also decreased IRS-1 expression by the use of the antiestrogen ICI 182780. MCF-7 cells were stimulated with IGF-I (10 nM) or ICI 182780 (1 μM) for 24 h and then were or were not restimulated with IGF-I (10 nM) for 10 min. As expected, compared to treatment with SFM, both IGF-I and ICI 182780 resulted in decreased IRS-1 expression (Fig. 4A, bottom panel). Cells that were incubated in SFM for 24 h and then stimulated with IGF-I for 10 min showed a rapid increase in tyrosine phosphorylation of IRS-1 (Fig. 4A, top panel). Cells that had been prestimulated with IGF-I for 24 h showed weak tyrosine phosphorylation of IRS-1 remaining from that preincubation. IGF-I stimulation now resulted in lower levels of tyrosine-phosphorylated IRS-1 than were found in cells incubated in SFM. However, the reduction in tyrosine-phosphorylated IRS-1 directly paralleled the decrease in total IRS-1 expression (Fig. 4B), suggesting that there had been no change in the ability to phosphorylate IRS-1 but simply a decrease in the total amount of tyrosine-phosphorylated IRS-1. Reduction of IRS-1 expression by ICI 182780 resulted in a reduced amount of tyrosine-phosphorylated IRS-1 compared to cells incubated with SFM and mimicked exactly the effect of IGF-I pretreatment. We have shown previously that this type of reduction affects downstream IGF signaling, such as ERK activation (32).
FIG. 4.
IGF-I-mediated degradation of IRS-1 results in reduced amounts of total tyrosine-phosphorylated IRS-1 following IGF-I stimulation. (A) MCF-7 cells were incubated in SFM for 24 h and then in SFM, or SFM supplemented with IGF-I (5 nM) or ICI 182780 (1 μM) for a further 24 h. Cells were or were not stimulated with IGF-I (10 nM) for 15 min and then lysed, and 50 μg of the resultant protein was separated by SDS–8% PAGE and immunoblotted with antiphosphotyrosine (PY) or anti-IRS-1 (IRS1) antibodies. (B) Densitometric analysis of gels shown in panel A. Gels were scanned and analyzed by using NIH Image 6.0. Values are presented as arbitrary (arb.) densitometric units. The top graph presents values for phosphotyrosine, and the bottom graph shows values for IRS-1.
IRS-1 is degraded by the 26S proteasome in response to IGF-I.
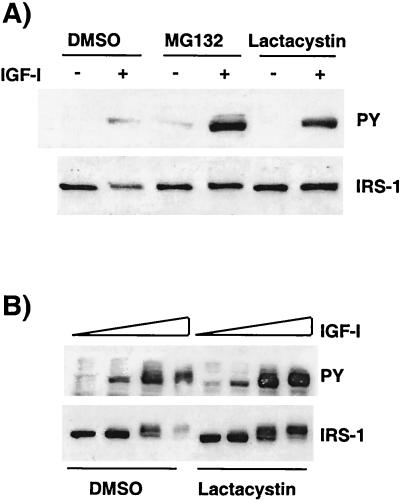
An important mechanism for degradation of a number of critical signaling molecules is the ubiquitin-mediated 26S proteasome activity (20, 43). We used the 26S proteasome inhibitors MG132 and lactacystin (11, 12) to investigate the effect of 26S proteasome activity on IGF-I-mediated degradation of IRS-1. MCF-7 cells were preincubated for 30 min with or without MG132 (50 μM) or lactacystin (10 μM) and then stimulated with IGF-I for 24 h in the absence or presence of the inhibitor. In the absence of the inhibitor, IGF-I caused phosphorylation of IRS-1 tyrosines (Fig. 5A, top panel), which was associated with a decrease in IRS-1 expression (Fig. 5A, bottom panel). In the presence of MG132 or lactacystin, IGF-I treatment resulted in a sustained tyrosine phosphorylation of IRS-1 resulting from a failure to cause degradation of IRS-1. The inhibition of IRS-1 degradation by lactacystin also resulted in an increase in the phosphorylation of IRS-1 tyrosine residues following IGF stimulation. Lactacystin was also able to efficiently block the dose-responsive, IGF-mediated degradation of IRS-1 compared to that evident in DMSO-treated cells (Fig. 5B), and this again resulted in sustained tyrosine phosphorylation of IRS-1.
FIG. 5.
26S proteasome inhibitors block IGF-mediated degradation of IRS-1. (A) MCF-7 cells were incubated in SFM for 24 h; pretreated with DMSO, MG132 (50 μM), or lactacystin (10 μM) for 30 min; and then stimulated with IGF-I (10 nM), or not stimulated, in the presence or absence of inhibitor for a further 24 h. Cells were lysed, and 50 μg of the resultant protein was separated by SDS–8% PAGE and immunoblotted with antiphosphotyrosine (PY) or anti-IRS-1 (IRS-1) antibodies. (B) MCF-7 cells were incubated in SFM for 24 h and then in SFM with or without lactacystin (10 μM) for 30 min. Cells were stimulated with increasing concentrations of IGF-I (0, 0.1, 1, and 10 nM) for 24 h in the presence or absence of lactacystin (10 μM). Cells were lysed, and 50 μg of the resultant protein was separated by SDS–8% PAGE and immunoblotted with antiphosphotyrosine (PY) and anti-IRS-1 (IRS-1) antibodies. This figure is representative of three independent experiments.
IRS-1–ubiquitin association is increased by IGF-I.
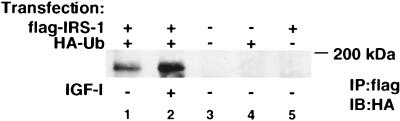
Proteins are targeted for 26S proteasome degradation by covalent attachment of a 76-amino-acid polypeptide termed ubiquitin (43). We next sought to establish whether ubiquitin binds directly to IRS-1. COS-7 cells were transiently transfected with expression plasmids for flag–IRS-1 and HA-Ub. Cells transfected with both expression plasmids were stimulated with 10 nM IGF-I (lane 2) for 2 h; after the cells were lysed, the lysate was immunoprecipitated with antiflag antibodies and immunoblotted with anti-HA antibodies (Fig. 6). Expression of a control plasmid (pcDNA3), flag–IRS-1, or HA-Ub alone resulted in no detectable bands upon HA immunoblotting (lanes 3 to 5). When both flag–IRS-1 and HA-Ub were coexpressed, their association was revealed by flag immunoprecipitation and HA immunoblotting (lane 1), giving a band at the position corresponding to the molecular mass of IRS-1 (∼170 kDa). Furthermore, IGF-I stimulation of cells increased the association of flag–IRS-1 and HA-Ub (lane 2).
FIG. 6.
IRS-1 and ubiquitin associate in vivo. COS-7 cells were transiently transfected with HA-Ub and flag–IRS-1 (lanes 1 and 2), pCDNA3.1 (lane 3), HA-Ub (lane 4), or flag–IRS-1 (lane 5). After transfection, cells were incubated in SFM for 16 h, and then one plate (lane 2) was stimulated with IGF-I (10 nM) for 2 h. Cells were lysed in TNESV, and the lysate was immunoprecipitated with antibodies against the flag epitope (IP:flag) and immunoblotted with antibodies against the HA epitope (IB:HA).
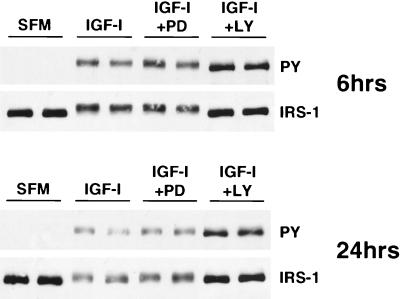
IGF-I-mediated degradation of IRS-1 is blocked by inhibitors of PI3K.
To determine if IGF-I-induced degradation of IRS-1 was a negative-feedback mechanism that resulted from downstream signals that might be elicited through IRS-1, we inhibited two well-characterized pathways activated by IRS-1, the ERK and PI3K pathways. Duplicate plates of MCF-7 cells were preincubated with PD098059 (an ERK inhibitor) or LY294002 (a PI3K inhibitor) for 30 min and then either were or were not stimulated with IGF-I in the presence of the inhibitor for a further 6 or 24 h (Fig. 7). IGF-I treatment (6 h) caused phosphorylation of IRS-1 tyrosines and a decrease in IRS-1 expression. A longer IGF-I treatment (24 h) resulted in a further decrease in IRS-1 phosphorylation that was associated with a decrease in IRS-1 expression. Cotreatment of cells with PD098059 did not affect the ability of IGF to phosphorylate or degrade IRS-1. In contrast, blockade of PI3K with LY294002 completely inhibited IGF-I-mediated IRS-1 degradation at both 6 and 24 h. The result of inhibition of degradation was an increase in tyrosine phosphorylation of IRS-1, similar to the results seen with blockade of the proteasome by lactacystin (Fig. 5). Interestingly, LY blocked the supershift that was caused by IGF-I, which presumably represents the serine phosphorylation of IRS-1 shown by others previously (25). A similar inhibition of IRS-1 degradation was seen with wortmannin (250 nM), another PI3K inhibitor (data not shown).
FIG. 7.
Inhibition of PI3K blocks IGF-mediated degradation of IRS-1. MCF-7 cells were incubated in SFM for 24 h and then incubated with SFM or SEM containing PD90859 (25 μM) or LY294002 (25 μM) for 30 min. Cells were then stimulated with IGF-I for 6 h (top panels) or 24 h (bottom panels) in the presence or absence of inhibitor. Cells were lysed, and 50 μg of the resultant protein was separated by SDS–8% PAGE and immunoblotted with antiphosphotyrosine (PY) and anti-IRS-1 (IRS-1) antibodies. This figure is representative of three independent experiments.
DISCUSSION
IRS-1 is an important downstream signaling molecule for a number of divergent signaling pathways (66). In particular, IRS-1 seems to play a critical role in the proliferative response to insulin and IGF-I. In vitro observations implicating IRS-1 in cell proliferation are substantiated by the fact that at birth, IRS-1 knockout mice (4) are smaller than their wild-type littermates, similar to IGF ligand and IGF receptor knockouts (6, 33). Because most important growth-regulatory genes are under strict regulatory control, we examined the feedback mechanisms controlling IRS-1 action.
Long-term (8- to 48-h) incubation of MCF-7 cells with IGF-I resulted in a decrease in IRS-1 protein expression. We have shown here that this degradation is a posttranscriptional event that can be blocked by inhibitors of the 26S proteasome, implicating ubiquitin activity as a signal for IRS-1 degradation. In confirming this, we have shown that IRS-1 and ubiquitin associate in vivo and that this association is increased by IGF treatment. IRS-1 was degraded in all cell types that exhibited phosphorylation of IRS-1 tyrosine residues in response to IGF-I, but it was not degraded in cells that failed to cause phosphorylation of IRS-1 tyrosines. Furthermore, the level of IRS-1 expression did not change when cells were incubated in SFM alone, and basal IRS-1 expression was not affected by lactacystin, indicating that proteasome degradation of IRS-1 is an IGF-mediated, phosphorylation-dependent event. Insulin-dependent proteasome-mediated degradation of IRS-1 has recently been shown to occur in CHO cells that overexpress the insulin receptor and IRS-1 (56). In these cells, insulin caused degradation of IRS-1, but the degradation was blocked by proteasome inhibitors. Interestingly, insulin did not cause degradation of IRS-2, suggesting that a motif in IRS-1 is responsible for ligand-dependent degradation. Proteasome-mediated degradation of several proteins, including Stat1 (26), c-jun (35), bcl-6 (37), and β-catenin, (1), is regulated by phosphorylation. A recent genetic analysis of yeasts has revealed a potential mechanism for phosphorylation is a signal for ubiquitin degradation; this is termed the F-box hypothesis. In this model, after a protein is phosphorylated, it is recognized by a protein (the F-box receptor) containing a protein recognition domain termed the F box (5, 52). A growing family of proteins contain F-box domains but also have several other protein-protein interaction domains (WD40, LRR, etc.). The F-box receptor can then recruit E1 and E2 ubiquitin-activating and -conjugating enzymes, which leads to polyubiquitination and degradation of the protein. This model is extremely attractive for explaining part of the specificity of ubiquitin degradation and may reveal in part the importance of ubiquitin in such processes as the cell cycle (28).
IRS-1 is a PEST (44)-containing protein with 11 potential repeats rich in proline, serine, and threonine residues (53). Recently, phosphorylation of PEST sequences has been implicated in ubiquitin degradation of a number of cell cycle-regulatory proteins, including cyclin D1 (13), cyclin E (65), and p27 (51). Indeed, it has been proposed that PEST sequences may be accountable in part for the F-box proteolysis pathway (14). However, PEST sequences do not completely identify proteins that will be degraded, since nearly one-third of all proteins contain PEST sequences and, often, deletion of the PEST sequence does not stabilize the protein's half-life.
Several reports have shown that high (micromolar) concentrations of insulin can desensitize cells to further insulin action in animals and in culture. Part of this desensitization may be a result of negative feedback in insulin signaling by degradation of IRS-1 (3, 7, 47). We have shown that in MCF-7 cells, nanomolar concentrations of IGF-I phosphorylate IRS-1 and cause a time- and dose-dependent decrease in IRS-1 expression. The same concentration of insulin weakly phosphorylates IRS-1 and fails to decrease IRS-1 expression. In contrast, insulin at chronically high (micromolar) doses phosphorylates IRS-1 to the same extent as IGF-I (10 nM) and is able to degrade IRS-1. Why is IRS-1 degraded in the presence of chronically high levels of insulin but not with nanomolar concentrations? First, high concentrations of insulin may simply increase insulin receptor-mediated phosphorylation of IRS-1 above a value threshold needed for IRS-1 degradation. Second, it is known that high concentrations of insulin can act through the IGF-IR (15). If the IGF-IR can mediate degradation of IRS-1 by activating different signal transduction pathways to insulin, then insulin acting through the IGF-IR would then be able to degrade IRS-1. Finally, it could be a combination of the two: insulin acts through the IGF-IR, which simply allows increased phosphorylation of IRS-1, which is then degraded. Recent data of Sun et al. (56) indicate that insulin can degrade IRS-1 in CHO cells only when IR is overexpressed. The increased level of IR causes increased phosphorylation of IRS-1 compared to insulin stimulation of cells with endogenous levels of IR and, thus, may again suggest that it is the level of IRS-1 phosphorylation that is the determining factor in its degradation.
The demonstrated inability of insulin at low concentrations to degrade IRS-1 is in contrast to data recently obtained in studies using 3T3-L1 adipocytes (45). In these cells, the scenario is the exact opposite of the situation we observed in MCF-7 cells. In 3T3-L1 adipocytes, insulin at low concentrations can degrade IRS-1. IGF-I at low concentrations fails to degrade IRS-1 and even causes a small transient increase (∼140% of the control value) in IRS-1 expression. Interestingly, IGF-I at high concentrations was able to degrade IRS-1, possibly by acting through the IR. It will be important to compare the levels of activation of IRS-1 by insulin and IGF-I in MCF-7 cells and 3T3-L1 adipocytes to determine if the differences observed are simply a result of a cell-specific ability to phosphorylate IRS-1 or if there are actual receptor-specific signaling pathways. Although the ligands that degrade IRS-1 differ between our cell lines, data from studies of 3T3-L1 adipocytes have also shown that the ligand-mediated degradation of IRS-1 can be specifically blocked with inhibitors of PI3K. Interestingly, calpain activity has been implicated as a mechanism for degradation in 3T3-L1 adipocytes. However, this has been shown mainly in studies using a cell-free system and purified exogenous calpain (53, 54), and more studies are required to determine the potential mechanisms for degradation of IRS-1 in vivo.
The difference in the levels of IRS-1 activation and degradation by insulin and IGF-I may have important physiological consequences. For instance, due to the inability of insulin to degrade IRS-1, no decrease in IRS-1 phosphorylation over time was seen. This was reflected downstream by sustained activation of ERK. In contrast, IGF-I caused a transient phosphorylation of IRS-1 which resulted in a transient activation of ERK. In several different systems the difference between sustained and transient ERK activation has been shown to mediate differential signaling effects (62). The best characterized of these is the ability of nerve growth factor to cause sustained ERK phosphorylation, leading to differentiation in PC12 cells, whereas in the same cell line, epidermal growth factor causes a transient ERK activation that leads to proliferation (34).
Our data indicate that IGF-induced degradation of IRS-1 is mediated by PI3K signaling. This is consistent with data of Sun et al. (56) indicating that insulin-mediated degradation of IRS-1 in CHO cells is blocked by wortmannin. It is possible that the PI3K-mediated degradation of IRS-1 is the result of direct feedback from PI3K activated by IRS-1. Treatment of MCF-7 cells results in a rapid association of the p85 subunit of PI3K with IRS-1 (data not shown). While IGF-IR can also bind and activate PI3K after IGF treatment (61), the majority of PI3K signaling has been shown to occur via insulin receptor substrate molecules (36). It is known that PI3K can phosphorylate IRS-1 on serine residues (17, 29, 60); however, downstream targets of PI3K (e.g., Akt/PKB) are also able to phosphorylate IRS-1 (42). Two recent reports also suggest that PI3K may be involved in negative feedback of IGF signaling. Data from studies using bovine fibroblasts have also shown that pretreatment with insulin desensitizes cells to IGF-mediated proliferation (19). This is associated with decreased phosphorylation of IRS-2 after pretreatment with insulin. PI3K inhibitors can block the insulin pretreatment effect. Additionally, it has recently been shown that degradation of IRS-1 is needed for myoblast differentiation and that the degradation of IRS-1 can be blocked by PI3K inhibitors (49).
In summary, we have shown here that phosphorylation of IRS-1 by IGF-I results in IRS-1 degradation by the 26S proteasome. The degradation of IRS-1 is specifically blocked by two different inhibitors of PI3K but not by an inhibitor of MAPK kinase. This raises the possibility that IRS-1 activation of PI3K results in a direct negative-feedback mechanism that targets IRS-1 for degradation and thus blocks further IGF signaling. This may be an important mechanism for determining the response of a cell to stimuli that can activate IRS-1.
ACKNOWLEDGMENTS
We thank M. Hochstrasser for the HA-Ub construct (Yep112), E. Araki for the human IRS-1 cDNA (pblue-IRS-1), L. R. Dick for technical comments on lactacystin, and J. G. Jackson for helpful comments and suggestions.
This work was supported by Public Health Service (PHS) grants P50CA58183, R01CA74285 (D.Y.), and K01CA77674 (S.O.) and Cancer Center Support grant P30CA54174 from the National Cancer Institute, National Institutes of Health. A.V.L. is a recipient of a Susan G. Komen Breast Cancer Foundation award. This research was supported in part by an award to the University of Texas Health Science Center at San Antonio for the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute (A.V.L.).
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad F, Goldstein B J. Effect of tumor necrosis factor-alpha on the phosphorylation of tyrosine kinase receptors is associated with dynamic alterations in specific protein-tyrosine phosphatases. J Cell Biochem. 1997;64:117–127. [PubMed] [Google Scholar]
- 3.Araki E, Haag III B L, Matsuda K, Shichiri M, Kahn C R. Characterization and regulation of the mouse insulin receptor substrate gene promoter. Mol Endocrinol. 1995;9:1367–1379. doi: 10.1210/mend.9.10.8544845. [DOI] [PubMed] [Google Scholar]
- 4.Araki E, Lipes M A, Patti M, Bruning J C, Haag III B, Johnson R S, Kahn C R. Alternative pathway of insulin-signaling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 5.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 6.Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 7.Cheatham B, Shoelson S E, Yamada K, Goncalves E, Kahn C R. Substitution of the erbB-2 oncoprotein transmembrane domain activates the insulin receptor and modulates the action of insulin and insulin-receptor substrate 1. Proc Natl Acad Sci USA. 1993;90:7336–7340. doi: 10.1073/pnas.90.15.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Ambrosio C, Keller S R, Morrione A, Lienhard G E, Baserga R, Surmacz E. Transforming potential of the insulin receptor substrate 1. Cell Growth Differ. 1995;6:557–562. [PubMed] [Google Scholar]
- 9.De Fea K, Roth R A. Modulation of insulin receptor substrate-1 tyrosine phosphorylation and function by mitogen-activated protein kinase. J Biol Chem. 1997;272:31400–31406. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- 10.De Fea K, Roth R A. Protein kinase C modulation of insulin receptor substrate-1 tyrosine phosphorylation requires serine 612. Biochemistry. 1997;36:12939–12947. doi: 10.1021/bi971157f. [DOI] [PubMed] [Google Scholar]
- 11.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V J, Parent L A, Plamondon L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 12.Dick L R, Cruikshank A A, Grenier L, Melandri F D, Nunes S L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin beta-lactone. J Biol Chem. 1996;271:7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- 13.Diehl J A, Zindy F, Sherr C J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 14.Elledge S J, Harper J W. The role of protein stability in the cell cycle and cancer. Biochim Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 15.Flier J S, Usher P, Moses A C. Monoclonal antibody to the type I insulin-like growth factor (IGF-I) receptor blocks IGF-I receptor-mediated DNA synthesis: clarification of the mitogenic mechanisms of IGF-I and insulin in human skin fibroblasts. Proc Natl Acad Sci USA. 1986;83:664–668. doi: 10.1073/pnas.83.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folli F, Kahn C R, Hansen H, Bouchie J L, Feener E P. Angiotensin II inhibits insulin signaling in aortic smooth muscle cells at multiple levels. A potential role for serine phosphorylation in insulin/angiotensin II crosstalk. J Clin Investig. 1997;100:2158–2169. doi: 10.1172/JCI119752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freund G G, Wittig J G, Mooney R A. The PI3-kinase serine kinase phosphorylates its p85 subunit and IRS-1 in PI3-kinase/IRS-1 complexes. Biochem Biophys Res Commun. 1995;206:272–278. doi: 10.1006/bbrc.1995.1038. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein B J. Regulation of insulin receptor signaling by protein-tyrosine dephosphorylation. Receptor. 1993;3:1–15. [PubMed] [Google Scholar]
- 19.Haddad T C, Conover C A. Insulin and interleukin-4 induce desensitization to the mitogenic effects of insulin-like growth factor-I. Pivotal role for insulin receptor substrate-2. J Biol Chem. 1997;272:19525–19531. doi: 10.1074/jbc.272.31.19525. [DOI] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 21.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotamisligil G S, Peraldi P, Budavari A, Ellis R, White M F, Spiegelman B M. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 23.Inoue G, Cheatham B, Kahn C R. Different pathways of postreceptor desensitization following chronic insulin treatment and in cells overexpressing constitutively active insulin receptors. J Biol Chem. 1996;271:28206–28211. doi: 10.1074/jbc.271.45.28206. [DOI] [PubMed] [Google Scholar]
- 24.Jackson J G, White M F, Yee D. Insulin receptor substrate-1 (IRS-1) is the predominant signaling molecule activated by insulin-like growth factor-I (IGF-I), insulin, and interleukin-4 (IL-4) in estrogen receptor-positive human breast cancer cells. J Biol Chem. 1998;273:9994–10003. doi: 10.1074/jbc.273.16.9994. [DOI] [PubMed] [Google Scholar]
- 25.Kanety H, Feinstein R, Papa M Z, Hemi R, Karasik A. Tumor necrosis factor alpha-induced phosphorylation of insulin receptor substrate-1 (IRS-1). Possible mechanism for suppression of insulin-stimulated tyrosine phosphorylation of IRS-1. J Biol Chem. 1995;270:23780–23784. doi: 10.1074/jbc.270.40.23780. [DOI] [PubMed] [Google Scholar]
- 26.Kim T K, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 27.Kosaki A, Yamada K, Suga J, Otaka A, Kuzuya H. 14-3-3β protein associates with insulin receptor substrate 1 and decreases insulin-stimulated phosphatidylinositol 3′-kinase activity in 3T3L1 adipocytes. J Biol Chem. 1998;273:940–944. doi: 10.1074/jbc.273.2.940. [DOI] [PubMed] [Google Scholar]
- 28.Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 29.Lam K, Carpenter C L, Ruderman N B, Friel J C, Kelly K L. the phosphatidylinositol 3-kinase serine kinase phosphorylates IRS-1. Stimulation by insulin and inhibition by wortmannin. J Biol Chem. 1994;269:20648–20652. [PubMed] [Google Scholar]
- 30.Lavan B E, Fantin V R, Chang E T, Lane W S, Keller S R, Lienhard G E. A novel 160-kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:21403–21407. doi: 10.1074/jbc.272.34.21403. [DOI] [PubMed] [Google Scholar]
- 31.Lavan B E, Lane W S, Lienhard G E. The 60-kDa phosphotyrosine protein in insulin-treated adipocytes is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:11439–11443. doi: 10.1074/jbc.272.17.11439. [DOI] [PubMed] [Google Scholar]
- 32.Lee A V, Jackson J G, Gooch J L, Hilsenbeck S G, Coronado-Heinsohn E, Osborne C K, Yee D. Enhancement of insulin-like growth factor signaling in human breast cancer: estrogen regulation of insulin receptor substrate-1 expression in vitro and in vivo. Mol Endocrinol. 1999;13:787–796. doi: 10.1210/mend.13.5.0274. [DOI] [PubMed] [Google Scholar]
- 33.Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 34.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 35.Musti A M, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 36.Myers M G, Jr, Grammer T C, Wang L M, Sun X J, Pierce J H, Blenis J, White M F. Insulin receptor substrate-1 mediates phosphatidylinositol 3′-kinase and p70S6k signaling during insulin, insulin-like growth factor-1, and interleukin-4 stimulation. J Biol Chem. 1994;269:28783–28789. [PubMed] [Google Scholar]
- 37.Niu H, Ye B H, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogihara T, Isobe T, Ichimura T, Taoka M, Funaki M, Sakoda H, Onishi Y, Inukai K, Anai M, Fukushima Y, Kikuchi M, Yazaki Y, Oka Y, Asano T. 14-3-3 protein binds to insulin receptor substrate-1, one of the binding sites of which is in the phosphotyrosine binding domain. J Biol Chem. 1997;272:25267–25274. doi: 10.1074/jbc.272.40.25267. [DOI] [PubMed] [Google Scholar]
- 40.Osborne C K, Hobbs K, Trent J M. Biological differences among MCF-7 human breast cancer cell lines from different laboratories. Breast Cancer Res Treat. 1987;9:111–121. doi: 10.1007/BF01807363. [DOI] [PubMed] [Google Scholar]
- 41.Pawson T, Scott J P. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 42.Paz K, Hemi R, LeRoith D, Kanety H, Zick Y. The p-tyr binding (PTB) domain of insulin receptor substrate (IRS) proteins serves as a substrate for protein kinase B (PKB). Possible involvement of PKB in insulin resistance induced by hyperinsulinemia. Proc Endocr Soc. 1998;OR17-2:75. [Google Scholar]
- 43.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 44.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 45.Rice K M, Garner C W. IGF-I regulates IRS-1 expression in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 1999;255:614–617. doi: 10.1006/bbrc.1999.0238. [DOI] [PubMed] [Google Scholar]
- 46.Rice K M, Lienhard G E, Garner C W. Regulation of the expression of pp160, a putative insulin receptor signal protein, by insulin, dexamethasone, and 1-methyl-3-isobutylxanthine in 3T3-L1 adipocytes. J Biol Chem. 1992;267:10163–10167. [PubMed] [Google Scholar]
- 47.Rice K M, Turnbow M A, Garner C W. Insulin stimulates the degradation of IRS-1 in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 1993;190:961–967. doi: 10.1006/bbrc.1993.1143. [DOI] [PubMed] [Google Scholar]
- 48.Salerno M, Sisci D, Mauro L, Guvakova M A, Ando S, Surmacz E. Insulin receptor substrate 1 is a target for the pure antiestrogen ICI 182,780 in breast cancer cells. Int J Cancer. 1999;81:299–304. doi: 10.1002/(SICI)1097-0215(19990412)81:2<299::AID-IJC21>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Sarbassov D D, Peterson C A. Insulin receptor substrate-1 and phosphatidylinositol 3-kinase regulate extracellular signal-regulated kinase-dependent and -independent signaling pathways during myogenic differentiation. Mol Endocrinol. 1998;12:1870–1878. doi: 10.1210/mend.12.12.0205. [DOI] [PubMed] [Google Scholar]
- 50.Sciacchitano S, Taylor S I. Cloning, tissue expression, and chromosomal localization of the mouse IRS-3 gene. Endocrinology. 1997;138:4931–4940. doi: 10.1210/endo.138.11.5518. [DOI] [PubMed] [Google Scholar]
- 51.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 52.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 53.Smith L K, Bradshaw M, Croall D E, Garner C W. The insulin receptor substrate (IRS-1) is a PEST protein that is susceptible to calpain degradation in vitro. Biochem Biophys Res Commun. 1993;196:767–772. doi: 10.1006/bbrc.1993.2315. [DOI] [PubMed] [Google Scholar]
- 54.Smith L K, Rice K M, Garner C W. The insulin-induced down-regulation of IRS-1 in 3T3-L1 adipocytes is mediated by a calcium-dependent thiol protease. Mol Cell Endocrinol. 1996;122:81–92. doi: 10.1016/0303-7207(96)03875-0. [DOI] [PubMed] [Google Scholar]
- 55.Smith-Hall J, Pons S, Patti M E, Burks D J, Yenush L, Sun X J, Kahn C R, White M F. The 60 kDa insulin receptor substrate functions like an IRS protein (pp60IRS3) in adipose cells. Biochemistry. 1997;36:8304–8310. doi: 10.1021/bi9630974. [DOI] [PubMed] [Google Scholar]
- 56.Sun X J, Goldberg J L, Qiao L Y, Mitchell J J. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes. 1999;48:1359–1364. doi: 10.2337/diabetes.48.7.1359. [DOI] [PubMed] [Google Scholar]
- 57.Sun X J, Rothenberg P, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill D A, Goldstein B J, White M F. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 58.Sun X J, Wang L M, Zhang Y, Yenush L, Myers M G, Jr, Glasheen E, Lane W S, Pierce J H, White M F. Role of IRS-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 59.Tanasijevic M J, Myers M G, Jr, Thoma R S, Crimmins D L, White M F, Sacks D B. Phosphorylation of the insulin receptor substrate IRS-1 by casein kinase II. J Biol Chem. 1993;268:18157–18166. [PubMed] [Google Scholar]
- 60.Tanti J F, Gremeaux T, Van Obberghen E, Le Marchand-Brustel Y. Insulin receptor substrate 1 is phosphorylated by the serine kinase activity of phosphatidylinositol 3-kinase. Biochem J. 1994;304:17–21. doi: 10.1042/bj3040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tartare-Deckert S, Murdaca J, Sawka-Verhelle D, Holt K H, Pessin J E, Van Obberghen E. Interaction of the molecular weight 85K regulatory subunit of the phosphatidylinositol 3-kinase with the insulin receptor and the insulin-like growth factor-1 (IGF-I) receptor: comparative study using the yeast two-hybrid system. Endocrinology. 1996;137:1019–1024. doi: 10.1210/endo.137.3.8603569. [DOI] [PubMed] [Google Scholar]
- 62.Tombes R M, Auer K L, Mikkelsen R, Valerie K, Wymann M P, Marshall C J, McMahon M, Dent P. The mitogen-activated protein (MAP) kinase cascade can either stimulate or inhibit DNA synthesis in primary cultures of rat hepatocytes depending upon whether its activation is acute/phasic or chronic. Biochem J. 1998;330:1451–1460. doi: 10.1042/bj3301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turnbow M A, Keller S R, Rice K M, Garner C W. Dexamethasone down-regulation of insulin receptor substrate-1 in 3T3-L1 adipocytes. J Biol Chem. 1994;269:2516–2520. [PubMed] [Google Scholar]
- 64.Vuori K, Ruoslahti E. Association of insulin receptor substrate-1 with integrins. Science. 1994;266:1576–1578. doi: 10.1126/science.7527156. [DOI] [PubMed] [Google Scholar]
- 65.Won K A, Reed S I. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 66.Yenush L, White M F. The IRS-signalling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]