Summary
The crosstalk between intestinal epithelial cells (IECs) and Th17-polarized CD4+ T cells is critical for mucosal homeostasis, with HIV-1 causing significant alterations in people living with HIV (PLWH) despite antiretroviral therapy (ART). In a model of IEC and T cell co-cultures, we investigated the effects of IL-17A, the Th17 hallmark cytokine, on IEC ability to promote de novo HIV infection and viral reservoir reactivation. Our results demonstrate that IL-17A acts in synergy with TNF to boost IEC production of CCL20, a Th17-attractant chemokine, and promote HIV trans-infection of CD4+ T cells and viral outgrowth from reservoir cells of ART-treated PLWH. Importantly, the Illumina RNA-sequencing revealed an IL-17A-mediated pro-inflammatory and pro-viral molecular signature, including a decreased expression of type I interferon (IFN-I)-induced HIV restriction factors. These findings point to the deleterious features of IL-17A and raise awareness for caution when designing therapies aimed at restoring the paucity of mucosal Th17 cells in ART-treated PLWH.
Subject areas: Immunology, Immune response, Virology
Graphical abstract

Highlights
-
•
IL-17A acts in synergy with TNF to enhance CCL20 production in IEC exposed to HIV
-
•
IL-17A/TNF-activated IEC efficiently promote HIV trans-infection of CD4+ T cells
-
•
IL-17A reprograms IEC to boost HIV outgrowth from CD4+ T cells of ART-treated PLWH
-
•
IL-17A decreases the expression of IFN-I-induced HIV restriction factors in IEC
Immunology; Immune response; Virology
Introduction
The implementation of viral-suppressive antiretroviral therapy (ART) transformed the HIV-1 epidemics into a manageable chronic viral infection (Barre-Sinoussi et al., 2013; Ghosn et al., 2018). However, ART is unable to cure infection in people living with HIV (PLWH) (Cohn et al., 2020; Lederman et al., 2016; Margolis et al., 2020; Sengupta and Siliciano, 2018) and to reverse the state of chronic immune activation that leads to premature aging and non-AIDS co-morbidities (Clifford and Ances, 2013; El-Far and Tremblay, 2018; Gabuzda et al., 2020). This justifies the need for the identification of new therapeutic interventions toward an HIV cure (Deeks et al., 2016).
The gut-associated lymphoid tissues (GALTs) play a unique role during primary HIV/SIV transmission but also during chronic infections (Brenchley et al., 2006; Grossman et al., 2006). Although it is well-established that GALT-infiltrating CD4+ T cells are rapidly targeted by infection and massively depleted during the early acute phases of HIV/SIV infections (Brenchley et al., 2004; Mehandru et al., 2004; Veazey, 2019), intestinal epithelial cells (IECs) are the first cells to interact with viral particles during rectal transmission (Gonzalez et al., 2019). Indeed, rectal IEC monolayers facilitates HIV/SIV acquisition and dissemination (Gonzalez et al., 2019; Real et al., 2018; Ribeiro Dos Santos et al., 2011), in part via transcytosis, a process involving virion capture, intracellular trafficking, and trans-infection of subjacent target cells (e.g., CD4+ T cells) in the absence of integrative infection in IECs (Alfsen et al., 2005; Bomsel, 1997). In addition to transcytosis, the capacity of IECs to support productive HIV infection has been studied on multiple models, including cell lines and primary cells. Studies on IEC lines demonstrated that latent and productive infection occurs depending on the use of low and high HIV titers, respectively (Bouhlal et al., 2002; De Mareuil et al., 1995; Fantini et al., 1991a, 1991b, 1992b; Faure et al., 1994; Gauthier and Tremblay, 2010; Kagnoff and Roebuck, 1999; Omary et al., 1991). In contrast to IEC lines, primary IECs were reported to be permissive to integrative infection when exposed to laboratory-adapted HIV strains but not primary isolates (Chenine et al., 1998; Meng et al., 2002; Ribeiro Dos Santos et al., 2011). Finally, primary IECs were documented to selectively transfer CCR5-tropic HIV strains to subjacent CD4+ T cells (Margolis and Shattock, 2006; Meng et al., 2002) via the formation of virological synapses (Real et al., 2018). Thus, the interplay between IECs and CD4+ T cells plays a key role in HIV/SIV mucosal transmission (Gonzalez et al., 2019; Margolis and Shattock, 2006).
Among CD4+ T cells preferentially infiltrating the GALT, CCR6+ Th17-polarized cells represent the first targets of HIV/SIV infection during acute phases and contribute to viral reservoir persistence in ART-treated PLWH (Anderson et al., 2020; Gosselin et al., 2017; Haase, 2011; Kelley et al., 2017; Masson et al., 2015; Planas et al., 2019; Shanmugasundaram et al., 2014; Stieh et al., 2016; Wacleche et al., 2016, 2017). At homeostasis, Th17 cells maintain the integrity of the intestinal physical/biological barrier, in part by producing the lineage-specific cytokine IL-17A under the control of the transcription factor RORγt (Dong, 2008; Kelley et al., 2017; Yang et al., 2008). IL-17A acts on IECs to strengthen the expression of tight junction proteins and to induce the secretion of antimicrobial peptides and the production of chemokines specifically attracting Th17 cells (e.g., CCL20, CCL25) (Hirota et al., 2007; Liang et al., 2006; Sano et al., 2015; Wang et al., 2009; Yamazaki et al., 2008). It is noteworthy that IL-17A acts in synergy with other cytokines, such as TNF (Friedrich et al., 2014; Guo et al., 2014; Lee et al., 2008a), and represents an “alarmin” that promotes antibacterial immunity at mucosal sites (Minegishi and Karasuyama, 2009). In line with these observations, studies in a model of Rhesus macaques exposed to Salmonella showed an increased expression of IL-17A and TNF, as well as other IL-17A-regulated genes in the gut (Raffatellu et al., 2008). Similar conclusions regarding an enrichment of gene signatures associated with Th17 cells were reported in the rectal mucosa of HIV-negative men who have sex with men (MSM) that engaged in condomless receptive anal intercourse (CRAI), along with an increase of rectal CD4+ T cells that produced greater levels of IL-17A compared with HIV-negative men who had never engaged in anal intercourse (Kelley et al., 2017). HIV-negative MSM who engaged in CRAI also shown a microbiota enriched for the Prevotellaceae family. Finally, our most recent study indicates the expression of IL-17A mRNA in colon biopsies of ART-treated PLWH (Gabriel et al., 2021). These lines of evidence support the production of IL-17 and TNF in vivo, at the mucosal level, after exposure to different components of the microbiota.
The depletion of Th17 cells, together with their functional alterations observed in the blood and GALT of PLWH are not completely restored with ART (Brenchley et al., 2008; Schuetz et al., 2014). The paucity of Th17 cells is caused by multiple mechanisms, including infection per se and an altered Th17 trafficking into mucosal sites (Loiseau et al., 2016; Mavigner et al., 2012; Planas et al., 2019; Wacleche et al., 2017). The depletion of Th17 cells is the main cause of gut barrier dysfunction leading to microbial translocation and chronic immune activation in ART-treated PLWH (Mudd and Brenchley, 2016; Somsouk et al., 2015; Tincati et al., 2016). In this context, the restoration of Th17-mediated mucosal immunity in the GALT represents an important research priority for HIV remission (Planas et al., 2019; Wacleche et al., 2017). For example, the administration of IL-21, a cytokine essential for Th17 survival, increased the frequency of Th17 cells in SIV-infected rhesus macaques and induced the expression of genes related to antimicrobial and anti-inflammatory functions (Micci et al., 2015; Pallikkuth et al., 2013). This intervention also reduced the residual plasma viremia and decreased the size of SIV-DNA reservoirs in ART-treated SIV-infected animals (Micci et al., 2015; Pallikkuth et al., 2013). Taking into consideration the importance of improving Th17-mediated immunity in HIV-infected ART-treated PLWH, it is highly relevant to evaluate the impact of IL-17A on HIV reservoir establishment and persistence at mucosal sites.
In this manuscript, we used the HT-29 IEC line and primary CD4+ T cells from uninfected individuals, as well as T cells from untreated and ART-treated PLWH, to investigate the effects of IL-17A on the interplay between IECs and CD4+ T cells, with relevance for early HIV transmission events, as well as viral reservoir dynamics during ART-treated chronic infection. Our results support a model in which IL-17A, despite its beneficial effects on mucosal homeostasis, acts on IECs to promote HIV dissemination and viral outgrowth from reservoir cells by compromising the type I IFN-mediated anti-viral immunity at mucosal sites. These findings raise awareness for caution when designing therapies aimed at restoring the paucity of mucosal Th17 cells in ART-treated PLWH.
Results
IL-17A acts in synergy with TNF to boost CCL20 production by IECs in the presence of HIV-1
The chemokine CCL20 attracts CCR6+ Th17 cells at mucosal sites (Wang et al., 2009; Yamazaki et al., 2008), with CCL20 expression in IECs being induced by the synergic action of IL-17A and TNF (Friedrich et al., 2014). HIV/SIV infections were previously associated with an altered CCL20 expression in IECs (Loiseau et al., 2016; Mavigner et al., 2012; Raffatellu et al., 2008), a deficit likely contributing to an impaired recruitment of Th17 cells into the GALT. This prompted our initial hypothesis that HIV exposure alters the capacity of IECs to secrete CCL20. To address this hypothesis, HT-29 IECs were activated with IL-17A or IL-17F in the presence or the absence of TNF, and exposed to two different CCR5-tropic HIV strains, HIVTHRO (a transmitted founder (T/F) molecular clone (Ochsenbauer et al., 2012) and HIVNL4.3BaL [a molecular clone containing the env- NL4.3 backbone, with an CCR5-tropic env from an HIV strain isolated from the bronchoalveolar lavage (BaL) of an individual with chronic infection (Bobardt et al., 2008). We used CCR5-tropic HIV considering the essential role of such strains in HIV transmission (Parrish et al., 2012). It is noteworthy that HT-29 cells are documented to support low level integrative infection (Gauthier and Tremblay, 2010), as a consequence of galactosyl ceramide (GalCer)-mediated HIV entry (Delezay et al., 1997; Fantini et al., 1993), despite the fact that HT-29 IECs express low to undetectable levels of the main HIV receptor CD4 and co-receptors CCR5 and CXCR4 (Dwinell et al., 1999; Jordan et al., 1999) (data not shown). In the absence of HIV, IL-17A acted in synergy with TNF to induce the production of CCL20 (Figure 1A, left panel). The production of IL-8/CXCL8, a chemokine ligand of CXCR1-2, mainly mediating the migration of neutrophils (Murphy, 1997), was induced by TNF, but was not influenced by IL-17A (Figure 1A, right panel). In contrast to IL-17A, IL-17F did not affect the production of CCL20 or IL-8 (data not shown). In contrast to our initial hypothesis, exposure to HIVTHRO did not decrease, but rather significantly increases CCL20 production by IECs in response to TNF alone, as well as TNF + IL-17A (Figure 1B, left panel), with no major impact on IL-8 production (Figure 1B, right panel). A similar effect was observed when cells were exposed to HIVNL4.3BaL (data not shown). Levels of CCL20 produced by cytokine-activated and HIV-exposed HT-29 IECs remained stable in the presence of the integrase inhibitor Raltegravir or the gp120/CD4-mediated entry inhibitor BMS-806 (Figure S1). This suggests that CCL20 production was not dependent on HIV entry and integration in IECs. Overt CCL20 production may contribute to the abundant recruitment of CCR6+ Th17 cells at mucosal sites during the initial steps of HIV transmission.
Figure 1.
IL-17A acts in synergy with TNF to promote CCL20 production by IEC in the presence or the absence of HIV-1
(A) Intestinal epithelial cells (IEC; HT29 cell line) were cultured in 48 well-plates until 80% confluence (150,000 cells/well). IEC were activated with TNF (10 ng/mL) and/or IL-17A (10 ng/mL) for 24 h.
(B) IEC cells were cultured in 48 well-plates wells until 80% confluence and activated with TNF (10 ng/mL) and/or IL-17A (10 ng/mL) for 3h and then exposed to T/F THRO (HIVTHRO) HIV strain (50 ng/well) for 24 h. Levels of CCL20 (A and B, left panels) and IL-8 (A and B, right panels) were quantified in cell culture supernatants by ELISA 24 h post-stimulation with cytokines (A) and/or HIV exposure (B). Paired t Test evaluated the statistical differences. p values are indicated on the graphs. Results are mean ± SD from three independent experiments performed with HT-29 IEC.
IL-17A-activated IECs efficiently transmit HIV infection to CD4+ T cells
The ability of IECs to capture and disseminate HIV to subjacent T cells is well-documented (Alfsen et al., 2005; Bomsel, 1997; Margolis and Shattock, 2006; Meng et al., 2002; Real et al., 2018). Cytokines such as TNF and IFN-γ were reported to facilitate HIV transmission from IECs to CD4+ T cells (Fantini et al., 1992a; Yasen et al., 2017). To investigate the effects of IL-17A on IEC-mediated HIV transmission, cytokine-activated IECs were exposed to HIVTHRO and co-cultured with CD3/CD28-activated CD4+ memory T cells from uninfected individuals, as depicted in Figure 2A. The highest levels of HIVTHRO replication were observed when CD4+ T cells were co-cultured with HIV-exposed TNF + IL-17A-activated (IECTNF+IL-17A) compared with inactivated (IECMedium) and TNF-activated IEC (IECTNF), as measured by ELISA (Figures 2B and 2C). Similar results were obtained when HIV-p24+ T -cells were quantified by flow cytometry, indicative that IECTNF+IL-17A highly efficiently transmit HIV infection to T cells (data not shown).
Figure 2.
IL-17A-activated IEC efficiently transmit HIV infection to CD4+ T cells
(A) Shown is the experimental flow chart. Briefly, IEC were cultured in 48 well-plates until they reached 80% confluence (150,000 cells/well). IEC were then, activated with TNF (10 ng/mL) and/or IL-17A (10 ng/mL) for 24 h before being exposed to HIVTHRO for 3 h. Unbound HIV was removed by extensive washing and cells were further cultured for 24 h. In parallel, memory CD4+ T cells isolated from PBMCs of HIV-uninfected individuals were activated with CD3/CD28 Abs for 3 days. Cytokine-activated HIV-exposed IEC cells were co-cultured with CD3/CD28-activated memory CD4+ T cells (500,000 cells/well) and media supplemented with IL-2 (5 ng/mL) was refreshed every 3 days for 9 days.
(B and C) Levels of HIV-p24 in cell culture supernatant were quantified by ELISA at days 3, 6, and 9. Shown are the mean ± SD of HIV-p24 levels between the different IEC treatment from one representative donor at days 3, 6, and 9 (B) and from n = 6 different individuals at day 9 post co-culture (C).
(D) Levels of CCL20 in cell culture supernatants were quantified at day 3 post-coculture (D). For Figures 2C and 2D, each symbol represents results generated with memory CD4+ T cells from a different individual (mean values of ELISA triplicate measurements). The bars represent the median value of all individuals. Friedman test with Dunn's post-test evaluated the statistical differences. p values are indicated on the graphs (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001).
Robust HIV replication in T cells co-cultured with HIV-exposed IECTNF+IL-17A coincided with the highest levels of CCL20 (Figure 2D), a chemokine reported to promote HIV latency (Cameron et al., 2010), but also involved in HIV resistance through the induction of APOBEC3G (Lafferty et al., 2010). To determine whether CCL20 modulates HIV replication, CD3/CD28-activated CD4+ T cells were infected with HIV in vitro and cultured in the presence or the absence of recombinant human CCL20 (Figure 2A). Results in Figures S2B–S2E demonstrate that CCL20 does not directly influence HIV replication in CD4+ T cells.
To determine whether cytokine-activated IEC differ in their ability to harbor HIV after exposure, integrated HIV-DNA, as well as early (RU5) and late (Gag) reverse transcripts were quantified 24 hours post-HIV exposure (Figure S3A), using published protocols (Cattin et al., 2019). Integrated HIV-DNA together with early and late reverse transcripts were increased in IECTNF+IL-17A and IECIL-17A (Figure S3B). In addition, cell-associated HIV-p24 levels, quantified in cell lysates by crude HIV-p24 ELISA 48 hours post-HIV exposure (Figure S3A), were increased in both IECTNF and IECTNF+IL-17A compared with IECMedium (Figure S3C).
Thus, HIV-exposed IECTNF+IL-17A favor HIV transmission/replication to/in CD4+ T cells by mechanisms involving an augmented level of HIV uptake and an increased integrative infection in an environment rich in CCL20. The finding that recombinant CCL20 exerts no direct effects on HIV replication in vitro suggests that in vivo, this chemokine rather contributes to the recruitment of CCR6+ cells, including Th17 cells, at mucosal sites, thus facilitating their infection.
IL-17A-activated IECs promote HIV replication in HIV-infected memory CD4+ T cells
HIV reservoirs persist in colon-infiltrating CCR6+CD4+ T cells of ART-treated PLWH (Anderson et al., 2020; Gosselin et al., 2017; Haase, 2011; Kelley et al., 2017; Masson et al., 2015; Planas et al., 2019; Shanmugasundaram et al., 2014; Stieh et al., 2016; Wacleche et al., 2016, 2017). To further explore the effect of IL-17A on the interplay between IECs and CD4+ T cells during HIV infection, we studied the ability of cytokine-activated IECs to promote viral replication in memory CD4+ T cells infected with HIVTHRO in vitro (Figure 3A). HIV-infected T cells were identified by flow cytometry as CD3+CD4lowHIV-p24+ at day 9 post-infection (Figures 3B and 3C), with CD4 downregulation being indicative of a productive infection (Richard et al., 2018). Also, HIV-p24 levels were measured in cell-culture supernatants by ELISA (Figure 3D). For HIVTHRO-infected CD4+ T cells, the frequency of CD3+CD4lowHIV-p24+ T cells was similar in all experimental conditions (Figure 3B), HIV-p24 levels in cell-culture supernatants, indicative of productive viral replication, were the highest when HIVTHRO-infected T cells were co-cultured with IL-17A-activated IECs (IECIL-17A) compared to IECTNF (Figure 3D). The viability of cells at day 9 post-coculture was around 80%, without major differences between conditions (Figure 3E).
Figure 3.
IL-17A-activated IEC promote HIV replication/outgrowth in HIV-infected memory CD4+ T cells
(A) Shown is the experimental flow chart. Briefly, IEC were activated with TNF and/or IL-17A as described in Figure 1A. In parallel, memory CD4+ T cells of HIV-uninfected individuals were isolated and activated as in Figure 2. After activation, memory CD4+ T cells of HIV-uninfected individuals were infected with HIVTHROin vitro. Cytokine-activated IEC (150,000 cells/well) were co-cultured with in vitro infected memory CD4+ T cells (500,000 cells/well). Media containing IL-2 (5 ng/mL) was refreshed every 3 days for 9 days.
(B and C) The intracellular expression of HIV-p24 in memory CD4+ T cells was quantified by flow cytometry upon staining with fluorochrome-conjugated CD3, CD4 and HIV-p24 Abs. Shown are dot plots presenting the frequency of CD4lowHIV-p24+ T cells across the different experimental conditions for one representative donor (B) and the statistical analysis of results obtained with HIVTHRO-infected memory CD4+ T cells of n = 4 different individuals at day 9 post co-culture (C).
(D) Levels of HIV-p24 in cell culture supernatant were quantified by ELISA at days 3, 6, 9, and 12 post-culture (D). Shown is the statistical analysis of results obtained with HIVTHRO-infected memory CD4+ T cells of n = 4 different individuals at day 9 post co-culture.
(E) Cell viability was quantified by flow cytometry upon staining with Live/Dead Fixable Aqua Dead Cell Stain Kit. Shown is the statistical analysis of the percentage of live cells obtained with HIVTHRO-infected memory CD4+ T cells of n = 4 different individuals at day 9 post co-culture. Each symbol represents results generated with memory CD4+ T cells from a different individual (mean values of ELISA triplicate measurements). The bars represent the median value of all individuals. Friedman test with Dunn's post-test evaluated the statistical differences. p values are indicated on the graphs (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001).
To determine whether cytokines used to activate IECs may act directly on T cells to modulate HIV replication, CD3/CD28-activated memory CD4+ T cells of HIV-uninfected individuals were infected with HIVTHRO in vitro and cultured in the presence or the absence of IL-17A and/or TNF. The addition of these cytokines had no effect on HIV replication measured by flow cytometry and ELISA in T cells cultured alone (Figure S4), indicating that IL-17A acts via IEC to exert its pro-viral effects in T cells.
IL-17A acts on IECs to decrease Tetherin/BST2 expression
In an effort to identify molecular mechanisms used by IL-17A to promote HIV dissemination and replication, in the context of the IEC:T cell interplay, we first evaluated the expression of BST2, an HIV restriction factor counteracted by HIV Nef and Vpu, with BST2 documented to play a crucial role in HIV release and cell-to-cell transmission (Casartelli et al., 2010; Neil et al., 2008). We used flow cytometry to quantify first the surface expression of BST2 on IECs upon exposure to cytokines and cultured alone (as in Figures 1A and 1B), as well as on memory CD4+ T cells infected with HIVTHRO in vitro (as in Figure 3A).
The expression of BST2 was significantly decreased in IECTNF+IL-17A compared with IECMedium, with no significant differences between IECMedium and IECTNF or IECIL-17A (Figures 4A and 4B). Considering the possibility that covert productive HIV infection occurs in HT-29 cells (Chenine et al., 1998), a decreased BST2 expression may facilitate the release of virions produced by HIV-infected IECTNF+IL-17A, compared with IECMedium for an efficient transmission to T cells, as observed in Figure 2.These results suggest the increased dissemination potential of IECTNF+IL-17A compared with IECTNF(Figure 2), despite similar HIV capture capacity (Figure S3C).
Figure 4.
IL-17A acts on IEC to decrease Tetherin/BST2 expression
(A and B) IEC were activated as described in Figure 1. BST2 expression on IEC was quantified by flow cytometry on day 3 post-activation. Shown are histograms of BST2 expression on IEC (A), the frequency of BST2+ IEC (B, left panel) and MFI of BST2 expression (B, right panel) (mean ± SD of triplicate wells) of one experiment performed with HT-29 cells; similar results were obtained in a second experiment.
(C–E) Cytokine-activated IEC were co-cultured with memory CD4+ T cells activated via CD3/CD28 and infected with HIVTHROin vitro. The expression of BST2 in memory CD4+ T cells was quantified by flow cytometry using fluorochrome-conjugated CD3, CD4, HIV-p24 and BST-2 Abs. Shown are histograms of BST2 expression (C) on total T cells (CD3+ cells, C, upper panel) and CD4lowHIV-p24+ T cells (C, lower panel). Shown is statistical analysis of MFI BST2 expression on total T cells (D, left panel) and CD4lowHIV-p24+ T cells (D, right panel) in n = 4 different individuals at day 9 post co-culture. The bars represent the median value of all individuals. Friedman test with Dunn's post-test addressed the statistical differences. p values are indicated on the graphs (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001). (E) Correlation between the HIV-p24 levels in cell culture supernatant and the MFI of BST2 expression on total T cells (E, left panel) and CD4lowHIV-p24+ T cells (E, right panel) were calculated using. Spearman correlation (SC, p and r values) and linear regression (LR, p and r2 values) models for all individuals and experimental conditions tested. Each symbol represents results generated with T cells from a different individual.
The expression of BST2 was also significantly decreased on total T cells and CD4lowHIV-p24+ T cells in co-cultures performed with memory CD4+ T cells infected with HIVTHRO in vitro and IECIL-17A compared with IECTNF (Figures 4C and 4D). Noteworthy, the BST2 expression on total T cells and CD4lowHIV-p24+ T cells negatively correlated with levels of productive HIV replication in cell-culture supernatants (Figure 4E).
In addition to CD4 downregulation, another way to identify productively HIV-infected T cells is based on their downregulation of BST2 (Van Damme et al., 2008). Consistent with HIV replication results in Figure 3D, the highest frequency of BST2lowHIV-p24+ cells was observed in the presence of IECIL-17A compared with IECMedian (Figure S5A top panels and 5B).
Together, these results point to the BST2 downregulation on IEC, as well as CD4+ T cells as a mechanism facilitating HIV infection in IEC: T cell co-cultures upon IL-17A exposure in the presence or the absence of TNF.
IL-17A counteracts the TNF-mediated type I IFN expression
To further investigate mechanisms underlying the pro-viral effects promoted by IECTNF+IL-17A and IECIL-17A, bioactive type I IFN levels were quantified in cell culture supernatants at day 3 of co-culture between HIVTHRO-infected T cells with cytokine-activated IEC. The highest levels of type I IFN were detected in IECTNF:T cell co-cultures, while the lowest levels were detected in T cells co-cultured with IECIL-17A and IECMedium (Figure 5A). Interestingly, IL-17A decreased type I IFN levels in co-cultures of T cells with IECTNF+IL-17A compared with IECTNF (paired t test p = 0.0065, Figure 5A). It is noteworthy that type I IFN levels positively correlated with the MFI expression of BST2, a known IFN stimulated gene (ISG) that limits HIV release from infected cells (Liberatore and Bieniasz, 2011), on T cells from all experimental conditions at day 3 post-co-culture (Figure 5B). Furthermore, a co-culture between HIVTHRO-infected T cells and cytokine-activated IECs in the presence of soluble IFN alpha/beta receptor (B18R), which prevents IFN signaling, was performed as depicted in Figure 6A. The presence of B18R in co-cultures of T cells with IECTNF counteracted the effect of TNF and increased HIV replication to a similar extent of what is observed in co-cultures of T cells with IECTNF+IL-17A (Figure 6B). In addition to BST2, the other ISG ISG15 (Freitas et al., 2020) and IFIT1 (Fensterl and Sen, 2015) were upregulated in co-cultures with IECTNF (Figure 6C), and this upregulation was abrogated when T cells were cultured with IECTNF+IL-17A as observed in Figures 4C and 4D. B18R blocked the upregulation of ISG in co-cultures of T cells with IECTNF as compared with the ones with IECTNF+IL-17A. These results reveal that the proviral features of IL-17A are mediated, at least in part, by its capacity to downregulate type I IFN production and responses induced by TNF.
Figure 5.
IL-17A-activated IEC decrease type I interferon levels in positively relationship with BST2 expression on HIV-infected T cells
Cytokine-activated IEC were co-cultured with HIVTHRO-infected memory CD4+ T cells, as described in Figure 3. Bioactive human Type I interferons (IFN) levels were quantified using the reporter cell line HEK-Blue IFN-α/β in cell culture supernatants collected at days 3 post-co-culture.
(A) Shown are type I IFN levels in co-cultures performed with HIVTHRO-infected memory CD4+ T-cells from n = 4 different individuals and the bars represent the median value of all individuals Friedman test with Dunn's post-test evaluated the statistical differences. p values are indicated on the graphs (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001).
(B) The correlation between type I IFN levels with the MFI of BST2 expression on total T-cells (B) was evaluated using SC and LR models in all donors and experiments conditions. Each symbol represents results generated with T cells from a different individual.
Figure 6.
IL-17A acts on IEC to modulate HIV dissemination to T cells and the expression of IFN-stimulated genes
(A) Shown is the experimental flow chart (A). Briefly, IEC were activated with TNF and/or IL-17A, as in Figure 1A. In parallel, memory CD4+ T-cells of HIV-uninfected individuals were isolated and activated, as in Figure 2. After activation, the memory CD4+ T-cells was infected with HIVTHRO. Cytokine-activated IEC (150,000 cells/well) were then co-cultured with in vitro infected memory CD4+ T cells (500,000 cells/well) for 9 days. Media containing IL-2 (5 ng/mL) with or without B18R (100 ng/mL) was refreshed every 3 days.
(B) The intracellular expression of HIV-p24 in memory CD4+ T cells was quantified by flow cytometry upon staining with fluorochrome-conjugated CD3, CD4 and HIV-p24 Abs and levels of HIV-p24 in cell culture supernatants were quantified by ELISA. Shown is the statistical analysis of results obtained with HIVTHRO-infected memory CD4+ T cells of n = 5 different individuals at day 9 post co-culture by flow cytometry (left panel) and ELISA (right panel). (C) The expression of HIV-p24 in memory CD4+ T cells was quantified by flow cytometry upon staining with fluorochrome-conjugated BST2 and the expression of ISG15 and IFIT1 were quantified by RT-PCR. Shown is the statistical analysis of results obtained with cells of n = 5 different individuals at day 3 post co-culture. Each symbol represents results generated with memory CD4+ T cells from a different individual. Paired t Test evaluated the statistical differences among DMSO and B18R. p values are indicated on the graphs.
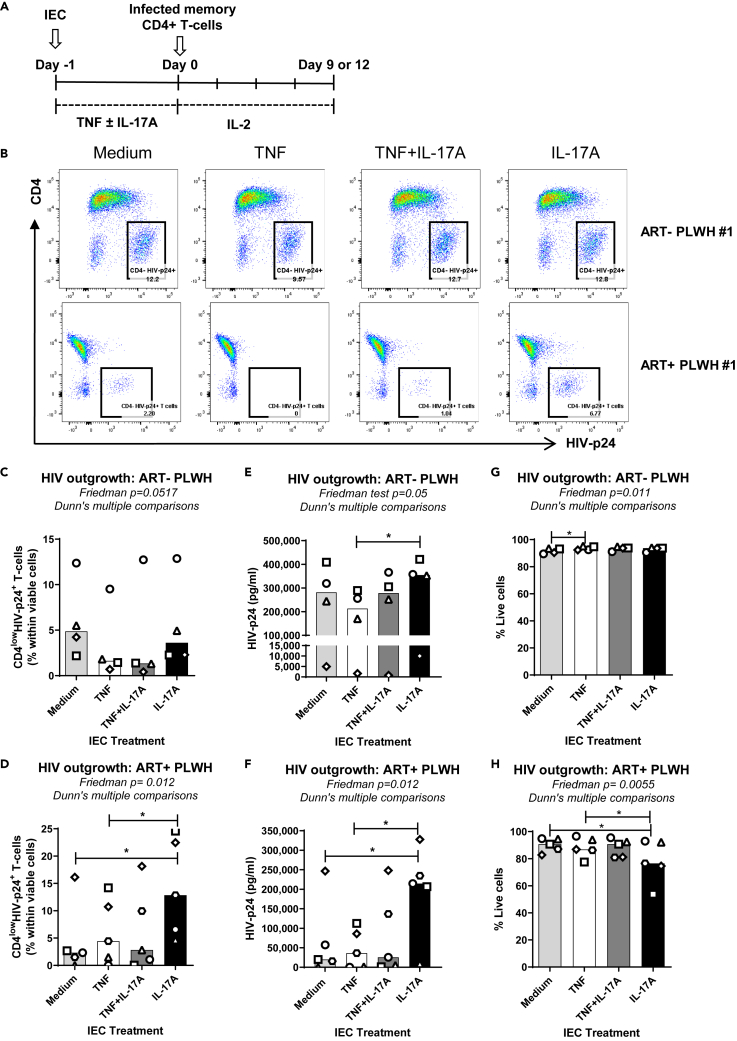
IL-17A activated IEC promote HIV outgrowth in memory CD4+ T cells of PLWH
To further explore the effect of IL-17A on the interplay between IECs and CD4+ T cells during chronic HIV infection, we studied the ability of cytokine-activated IEC to promote viral outgrowth from memory CD4+ T cells of both suppressed ART-treated PLWH (ART + PLWH) and untreated PLWH (ART- PLWH) (Figure 7A), with detectable plasma viremia (Table 1). Similar results to memory CD4+ T-cells infected with HIVTHRO in vitro were obtained with T cells of ART- PLWH (Figure 7B top panels and C), with an increased viral outgrowth measured by ELISA observed in the presence of IECIL-17A versus IECTNF (Figure 7E). Remarkably, when co-cultures were performed with T cells of ART + PLWH, the highest levels of HIV outgrowth, as measured by both FACS and ELISA, were achieved in the presence of IECIL-17A (Figure 7B bottom panels, D and F). An increased frequency of BST2lowHIV-p24+ cells was observed when memory CD4+ T cells from ART- and ART + PLWH were co-cultured with IECIL-17A in comparison with IECTNF (Figure S5A middle and bottoms panels, C and D). Additionally, a decrease in BST2 expression on total T cells and/or CD4lowHIV-p24+ T cells was also noticed for memory CD4+ T cells isolated from ART- (Figures S6A and S6B), as well as ART + PLWH (Figures S6D and S6E) upon co-culture with IECIL-17A compared to IECTNF, with a negative correlation between BST2 expression and levels of HIV outgrowth (Figures S6C and S6F). The cell viability decreased in co-cultures of IECIL-17A with T cells of ART- PLWH in line with a higher HIV outgrowth (Figure 7H). In conclusion, these results show that the activation of IEC with IL-17A promotes viral outgrowth from CD4+ T cells isolated from ART- and especially ART + PLWH.
Figure 7.
IL-17A-activated IEC promote HIV outgrowth in HIV-infected memory CD4+ T cells
(A) Shown is the experimental flow chart. Briefly, IEC were activated with TNF and/or IL-17A, as in Figure 1A. In parallel, memory CD4+ T cells of untreated (ART-) and ART-treated (ART+) PLWH were isolated and activated as in Figure 2. Cytokine-activated IEC (150,000 cells/well) were co-cultured with either memory CD4+ T cells isolated from ART- PLWH (500,000 cells/well) or ART + PLWH (1,000,000 cells/well). Media containing IL-2 (5 ng/mL) was refreshed every 3 days for 9 to 12 days.
(B–D) The intracellular expression of HIV-p24 in memory CD4+ T cells was quantified by flow cytometry upon staining with fluorochrome-conjugated CD3, CD4 and HIV-p24 Abs. Shown are dot plots presenting the frequency of CD4lowHIV-p24+ T cells across the different experimental conditions for one representative donor of each group (B) and the statistical analysis of results obtained with memory CD4+ T cells of n = 4 different ART- PLWH at day 9 post co-culture (C), and memory CD4+ T cells of n = 5 different ART + PLWH at day 12 post co-culture (D).
(E–F) Levels of HIV-p24 in cell-culture supernatant were quantified by ELISA at days 3, 6, 9, and 12 post-culture. Shown is the statistical analysis of results obtained with memory CD4+ T cells of n = 4 different ART- PLWH at day 9 post co-culture (E), and memory CD4+ T cells of n = 5 different ART + PLWH at day 12 post co-culture (F).
(G and H) Cell viability was quantified by flow cytometry upon staining with Live/Dead Fixable Aqua Dead Cell Stain Kit by flow cytometry. Shown is the statistical analysis of the percentage of live cells obtained in co-cultures with ART- (G) and ART + PLWH (H). Each symbol represents results (mean values of ELISA triplicate measurements) generated with memory CD4+ T cells from a different individual. The bars represent the median value of all individuals. Friedman test with Dunn's post-test evaluated the statistical differences. p values are indicated on the graphs (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001).
Table 1.
Clinical parameters of untreated and ART-treated PLWH study participants
| ID | Sex | Agea | CD4 countsb | CD4:CD8 Ratio | Viral loadc | Time since infectiond | ART | Time on ARTd |
|---|---|---|---|---|---|---|---|---|
| ART+ #1 | M | 31 | 824 | 0.9 | <40 | 58 | Atripla | 44 |
| ART+ #2 | F | 28 | 535 | 1.8 | <50 | 177 | Viracept Truvada |
151 |
| ART+ #3 | M | 50 | 569 | 1.2 | <50 | 111 | Darunavir Raltegravir |
101 |
| ART+ #4 | M | 21 | 796 | 2.0 | <40 | 8 | Stribild | 4 |
| ART+ #5 | M | 50 | 827 | 2.3 | <40 | 140 | Complera | 30 |
| ART+ #6 | M | 26 | 331 | NA | <40 | 86 | NA | 24 |
| ART+ #7 | M | 42 | 795 | 1.1 | <40 | 46 | Tivicay/Edurant Viread |
33 |
| ART+ #8 | M | 55 | 963 | 1.5 | <40 | 124 | Prevista Kivexa Norvir |
107 |
| ART- #1 | M | 40 | 1,068 | 1.2 | 22,812 | 2 | None | NA |
| ART- #2 | M | 47 | 529 | 1.2 | 3,189 | 110 | None | NA |
| ART- #3 | M | 39 | 946 | 0.7 | 8,809 | 11 | None | NA |
| ART- #4 | M | 24 | 316 | 0.5 | 9,496 | 55 | None | NA |
N.A, information not available; M, male; F, female.
Years.
Cells/μl.
HIV-RNA copies/ml plasma.
Months.
Transcriptional reprogramming associated with IL-17A-mediated pro-viral features
To further explore in an unbiased manner mechanism underlying the pro-viral features of IL-17A, the Illumina RNA sequencing technology was used on RNA extracted from cytokine-activated IEC cultured alone (Figures 8A and 8B; Data S1, S2) and co-cultured with CD4+ T cells isolated from ART + PLWH (Figures 8C and 8D; Data S3, S4), using culture/co-culture protocols described in Figures 1 and 3, respectively.
Figure 8.
IL-17A modulates anti-viral responses in IEC cultured alone or co-cultured with memory CD4+ T cells of ART + PLWH
RNA sequencing was performed with RNA extracted from IEC activated with TNF and/or IL-17A as described in Figure 1A and from cytokine-activated IEC (150,000 cells/well) co-cultured with CD3/CD28-activated memory CD4+ T cells of n = 3 different ART + PLWH individuals (1 000,000 cells/well) for 3 days as described in Figure 3A.
(A and C) Hierarchical clustering was performed on the scaled fold-changes to identify eight clusters of transcriptional signatures grouped in three major clusters (I, II, and III) associated to cytokine-activated IEC (A) and cytokine-activated IEC co-cultured with memory CD4+ T cells (C).
(B and D) Gene set enrichment analysis was performed on each gene cluster using the Gene Ontology: Biological Processes database. The most significant differentially expressed pathways were within cluster 3 for IEC (B) and cluster 1 for cytokine-activated IEC co-cultured with memory CD4+ T cells of ART + PLWH individuals (D). Only pathways with FDR<0.01 are depicted. Nodes (pathways) are color coded with significance.
For cytokine-activated IEC cultured alone, large sets of differentially expressed genes were identified using a fold-change (FC) cutoff of 1.3. We found 6,639, 5,429, and 3,983 transcripts upregulated, along with 5,297, 3,925, and 6,437 transcripts downregulated in IECTNF, IECIL-17A+TNF, and IECIL-17A compared with IECMedium, respectively (Data S1). To gain insights into gene sets modulated by IL-17A and TNF alone or in combination, we used a hierarchical clustering based on the scaled FC of each condition compared to medium. We identified 3 main clusters (MCs) characterized by specific patterns of response in IECTNF, IECIL-17A+TNF, and IECIL-17A, that were subdivided into 8 distinct transcriptional signatures with slight variations within each of the 3 MC (Figure 8A; Data S1). Clusters 2, 5, 7, and 8 (MC I) included gene sets expressed at higher levels in IECIL-17A+TNF compared with IECTNF or IECIL-17A (Figure 8A), thus reflecting transcriptional changes in the expression of different effector molecules prompted by the synergic action of IL-17A and TNF (e.g., the cytokines IL-4 and IL-1β, and the chemokines CCL20, CXCL2, and CXCL3; Data S1). Clusters 4 and 6 (MC II) included gene sets with higher expression levels in IECIL-17A compared with IECTNF or IECIL-17A+TNF (Figure 8A) (e.g., genes involved in the monocarboxylic and organic acid transport: SLC16A14, PPARD, and SLC10A5; Data S1). Finally, clusters 1 and 3 (MC III) included gene sets with higher expression levels in IECTNF compared with IECIL-17A+TNF (Figure 8A), thus revealing transcripts downregulated by IL-17A in TNF-activated IEC (e.g., the cytokine IL-32 and the interferon stimulated genes IFITM1, IFIT1, ISG15, IFI6, and OAS2; Data S1).
We further used a gene set enrichment analysis (GSEA), focusing on the Gene Ontology: Biological Processes database. Each of the gene modules was compared to a reference background consisting of all transcripts measured (28,083). The graphical representation of enriched GO terms that reached statistical significance (false discovery rate, FDR, <5%) was generated using ClueGO on Cytoscape software. Only clusters 1, 3, and 4 were enriched in GO terms with statistical significance and a functional trend (Data S2). Cluster 3 was associated with immune response processes such as “Defense response to virus” and “Response to Type I Interferon” (Figure 8B). Other gene sets were upregulated in IECTNF but downregulated with the addition of IL-17 in IECIL-17A+TNF; noteworthy, some of those genes were also downregulated in IECIL-17A (Figures S7A and S7B). For example, enriched pathways in IECTNF versus IECIL-17A+TNF and/or IECIL-17A included genes with a demonstrated activity against HIV, such as the viral sensor MB21D1 (Gao et al., 2013) and the restriction factors APOBEC3G, IFI16, IFITM1, SAMHD1, IFIT1, IFITM3, ISG15, MX2, RSAD2 and TRIM22 (Colomer-Lluch et al., 2018; Hotter et al., 2019; Hotter and Kirchhoff, 2018; Nasr et al., 2012, 2017) (Figures S7A and S7B). While cluster 3 was associated with an upregulation of viral response associated transcripts, genes associated with the GO term “Defense response to virus” were also included in Cluster 1; those genes were downregulated in IECIL-17A+TNF compared to IECTNF and they included the viral sensor TMEM173 (Trotard et al., 2016), and the HIV restriction factors BST2, APOBEC3D, and IFITM2 (Neil et al., 2007; Qian et al., 2015; Sato et al., 2014) (Figure S8C).
A similar analysis was performed in cytokine-activated IEC co-cultured with CD4+ T cells isolated from ART + PLWH (Data S3). This analysis allowed us to identify similar patterns of gene expression also grouped in 3 MC and categorized in 8 clusters (Figure 7C, Data S3). Only Cluster 1, which mainly included genes that were upregulated in co-cultures performed with IECTNF and downregulated in co-cultures performed with IECTNF+IL-17A and IECIL-17A, showed statistical significance for the immune response process “Defense to virus” and “Response to Type I Interferon” (Figure 7D; Data S4). Among cluster 1 genes, some were known HIV restriction factors such as APOBEC3F, BST2, HERC5, IFIT1, IFITM1, IFITM2, ISG15, MX2, RSAD2, TRIM22, and TRIM5 (Figures S8A and S8B).
These results demonstrate that IL-17A, in the presence or the absence of TNF, abrogates the anti-viral type I IFN-mediated responses in IEC cultured alone and co-cultured with CD4+ T cells carrying HIV reservoirs in ART + PLWH. This abrogation of anti-viral responses highly likely explain the pro-viral features of IL-17A.
IL-17 acts on primary human IEC to modulates CCL20 production and ISGs expression
To extend our results obtained on the HT-29 cells in a more physiological setting, we performed experiments with commercially available primary human IEC (phIEC) isolated from one HIV-uninfected individual. We confirm here that TNF acts in synergy with IL-17A in promoting CCL20 production in phIEC (Figure 9A). Furthermore, the expression of the ISGs ISG15 and IFIT1 mRNA was increased in phIECTNF and decreased in phIECTNF+IL-17 and phIECIL-17 compared to phIECMedium (Figures 9B and 9C). These results support a model in which IL-17A can increase the production of pro-inflammatory chemokines such as CCL20, while interfering with antiviral mechanisms such as those mediated by ISGs in vivo.
Figure 9.

Effect of IL-17 on primary human intestinal epithelial cells
Primary human Primary human intestinal epithelial cells (phIEC) were cultured in 48 well-plates for 5 days until 90% confluence (150,000 cells/well). phIEC were activated with TNF (10 ng/mL) and/or IL-17A (10 ng/mL) for 24 h.
(A–C) IEC cells were cultured in 48 well-plates wells until 80% confluence and activated with TNF (10 ng/mL) and/or IL-17A (10 ng/mL) for 24h. Levels of CCL20 were quantified in cell-culture supernatants by ELISA (mean ± SD values of experimental triplicates) (A) and ISG15 and IFIT1 mRNA expression were quantified by RT-PCR (mean value of RT-PCR duplicate measurements) (B and C).
Discussion
In this study, we provide evidence supporting the deleterious role of the Th17 hallmark cytokine IL-17A in the context of HIV-1 infection by its capacity to reprogram the pro-inflammatory and pro-viral features of IEC. Briefly, we demonstrate that IL-17A in the presence of TNF stimulation and/or HIV exposure, increases the capacity of IEC to produce CCL20, a chemokine that recruits CCR6+ Th17 cells into mucosal sites (Wang et al., 2009; Yamazaki et al., 2008). We revealed the capacity of IL-17A to enhance HIV trans-infection from IEC to CD4+ T cells and to promote HIV outgrowth from CD4+ T cells of ART-treated PLWH. By using an RNA-Sequencing approach, we identified pro-inflammatory and anti-viral gene sets modulated by IL-17A in IEC alone and in IEC:T cell co-cultures, with the decrease in type I IFN production/responses (i.e., BST2, IFIT1, ISG15) representing a common pattern of IL-17A exposure. In this context, the dual role played by IL-17A in controlling mucosal immunity versus HIV infection should be considered when seeking to implement new therapeutic interventions aimed at the restoration of mucosal Th17 cells in ART-treated PLWH.
By its capacity to target both IEC and Th17 cells, HIV causes profound alterations of the GALT physical and biological barrier functions (Mudd and Brenchley, 2016; Tincati et al., 2016). Such alterations are observed during the very early acute phases of infection and are not resolved by viral-suppressive ART, even with early ART initiation (Ananworanich et al., 2012; Deleage et al., 2016; Epple et al., 2009; George et al., 2005; Hensley-McBain et al., 2018; Schuetz et al., 2014). Nevertheless, recent studies reported IEC alterations even before the depletion of Th17 cells occurs in the intestinal mucosa of SIV-infected macaques (Hensley-McBain et al., 2018), thus emphasizing the key role played by IEC in HIV/SIV pathogenesis.
It is well established that Th17 cells, via their production of IL-17A, represent essential players in the defense against fungal (e.g., Candida albicans) and bacterial (e.g., Staphylococcus aureus) pathogens at mucosal and skin barrier surfaces (Henry et al., 2010; Li et al., 2018). Nevertheless, IL-17A-producing Th17 cells play a pathogenic role in the context of auto-immunity (e.g., psoriasis and ankylosing spondylitis) (Burkett and Kuchroo, 2016) and viral infections, such as the 2009 influenza A (H1N1), Respiratory Syncytial Virus (RSV) (Li et al., 2012; Niwa et al., 2018; Ryzhakov et al., 2011), and the new SARS-CoV-2 infection (De Biasi et al., 2020; Wiche Salinas et al., 2020). However, whether IL-17A plays a deleterious role during HIV/SIV remains unknown. The purpose of the current study was to investigate the pro-viral features of IL-17A in the context of IEC and CD4+ interactions.
Previous reports demonstrated that IL-17A acts in synergy with TNF to promote CCL20 production by IEC (Friedrich et al., 2014; Lee et al., 2008b). This synergy is mechanistically explained by the TNF-dependent upregulation of IL-17 receptors on IEC, as well as the stabilization of messenger RNA (Herjan et al., 2018). Here, we confirmed these findings in HT-29 IEC and further demonstrated that IL-17A but not IL-17F increases the TNF-mediated CCL20 production in IEC. The chemokine CCL20 binds on its receptor CCR6 and regulates trafficking of CCR6+ cells, including dendritic cells (DC) and Th17 cells (Wang et al., 2009; Yamazaki et al., 2008). Noteworthy, studies by the group of A.T. Haase demonstrated that glycerol monolaurate, an antimicrobial compound, inhibits the production of CCL20 and prevented SIV infection of rhesus macaques by interfering with the trafficking on CCR6+ DC into the female genital tract (Li et al., 2009; Shang et al., 2017). Thus, the CCL20/CCR6 axis plays a key role in immune cell trafficking into mucosal sites, as well as in the distal dissemination of HIV/SIV infection from the portal sites of viral entry. A deficit in CCL20 expression was reported in the intestine of PLWH and SIV-infected macaques (Loiseau et al., 2016; McGary et al., 2017; Raffatellu et al., 2008), and was suggested to represent one mechanism explaining the paucity of mucosal Th17 cells during HIV/SIV infection. In contrast to our initial hypothesis, we found that HIV exposure in vitro does not reduce, but rather increases CCL20 production by IEC in response to TNF + IL-17A. In line with these findings, HIV/SIV challenge in vivo was reported to induce the secretion of multiple chemokines promoting the migration of CD4+ T cells that are preferential infection targets (e.g., CCR6+ T cells) (Li et al., 2009; McGary et al., 2017; Nazli et al., 2010; Sankapal et al., 2016; Shang et al., 2017; Zhou et al., 2018). Thus, during the initial steps of HIV/SIV infection, IL-17A may contribute to the recruitment of CCR6+ Th17 cells at mucosal sites. In contrast, during the chronic phase, when HIV/SIV-infected Th17 cells are massively depleted (Brenchley et al., 2008; Loiseau et al., 2016), an alteration in CCL20 expression at the intestinal level may be the consequence of an altered IL-17A production.
Previous studies demonstrated that CCL20 acts on T -cells to increase their resistance to infection via the induction of the HIV restriction factor APOBEC3G (Lafferty et al., 2017). Other studies reported on the capacity of CCL20 to promote HIV latency (Cameron et al., 2010). In our experimental settings, we found that high levels of HIV replication coincided with the highest levels of CCL20 production in IEC:T cell co-cultures, and exogenous CCL20 does not interfere with HIV replication in memory CD4+ T cells. Therefore, the effects of CCL20 on HIV replication remain controversial.
IEC are the first cells to interact with HIV/SIV during the initial phases of viral transmission. HIV capture, transcytosis, and release from EC are positively and negatively regulated by multiple host factors (Gonzalez et al., 2019). In non-rectal EC, HIV is internalized and retained in vacuoles or multi-vesicular bodies (Yasen et al., 2017). TNF can modulate the retention of HIV into female genital tract EC and can increase their capacity to release HIV to infect CD4+ T cells (Yasen et al., 2017). IEC release exosomes that contain HIV restriction factors and miRNAs after stimulation with TLR-3 ligand. Those factors restrict HIV replication in macrophages infected in vitro (Guo et al., 2018). We observed that IL-17A, in combination with TNF, increased the capacity of IECs to capture and transmit HIV to CD4+ T cells. These results attribute to IL-17A a direct role in promoting HIV dissemination from the portal sites of entry. This idea is in line with results emerging from population studies where individuals with increased levels of IL-17A or IL-17A-induced chemokines exhibited an increased risk for HIV acquisition (Arnold et al., 2016; Chege et al., 2012; Gosmann et al., 2017). Of note, it is established that HIV-Tat protein induces the production of IL-17A on CD4+ T cells (Cafaro et al., 2016; Johnson et al., 2013). The latter finding, in addition to the facts that Th17 cells have a superior permissiveness to HIV infection (Gosselin et al., 2010; Monteiro et al., 2011) and are located at portal sites of HIV entry (Kelley et al., 2017; Masson et al., 2015; Stieh et al., 2016), highlight the relevance of evaluating the Th17 responses and their correlation to protection versus susceptibility to HIV acquisition in vaccination trials (Perreau et al., 2008; Stephenson et al., 2020).
One important finding of our study is that IL-17A-stimulated IECs exhibit an increased capacity to promote/facilitate viral replication when co-cultured with primary CD4+ T cells infected with HIVTHRO in vitro or CD4+ T cells from viremic and ART-treated PLWH. Although solid evidence supports that Th17 cells are depleted during HIV/SIV infections (Brenchley et al., 2008; Schuetz et al., 2014), IL-17A can be produced by other cell types of the innate and adaptive immune system, including neutrophils (Hu et al., 2017) and NKT cells (Campillo-Gimenez et al., 2010), which massively infiltrate mucosal sites during HIV/SIV infections (Somsouk et al., 2015). Whether IL-17A-activated IECs may contribute to HIV reservoir reactivation in ART-treated PLWH, thus leading to a state of chronic immune activation in the GALT, remains an important question to address in future studies. This knowledge is highly relevant in the context of therapies aimed to Th17 cell restoration in the GALT. Noteworthy, the administration of IL-21 proved to be beneficial in reestablishing the frequency of Th17 cells and in decreasing the size of viral reservoir during ART in the gut of SIV-infected rhesus macaques (Micci et al., 2015). This is in line with the fact that IL-21, in addition to its effect on Th17 cell survival (Planas et al., 2019; Wacleche et al., 2017), it also exhibits anti-viral activities (Adoro et al., 2015) that may counteract the pro-viral activity of IL-17A. The pro-viral activity of IL-17A may also be neutralized in the presence of ART. However, ART does not target HIV transcription and therefore IL-17A may still facilitate HIV provirus reactivation, as in a “shock and kill strategy”. Noteworthy, studies reported that the administration of the TLR-9 agonist MGN1703 decreased the size of the gut HIV-reservoir and correlated with the intestinal expression of IL-17R (Krarup et al., 2018). Further studies should investigate better how IL-17A impacts HIV reactivation in the gut in ART-treated PLWH.
In search of molecular mechanisms underlying the pro-viral features of IL-17A, we performed RNA sequencing and used bioinformatics tools for data mining. The GO enrichment analysis indicated that IL-17A dampens the response to type I IFN in IECs cultured alone and in the presence of CD4+ T cells from ART-treated PLWH. Genes included in the GO term response to type I IFN were essentially linked to HIV sensing and restriction and were upregulated by TNF but downregulated by IL-17A. One of the transcripts downregulated by IL-17A was BST2, an HIV restriction factor limiting the release of de novo produced virions (Neil et al., 2008). In line with this, we found the lowest levels of BST2 expression on IECs stimulated with TNF + IL-17A versus TNF or IL-17A alone. These results may explain the superior ability of IECs activated with TNF + IL-17A to transmit HIV compared with IECs activated with either TNF or IL-17A alone. Previous studies demonstrated covert productive HIV replication in HT-29 cells (Fantini et al., 1991b). Therefore, levels of BST-2 expression on IECs may determine their ability to transmit HIV infection to subjacent CD4+ T cells. We also found that HIVTHRO-infected CD4+ T cells expressed the lowest levels of BST-2 when co-cultured with IECs activated with IL-17A compared with TNF, in line with the highest levels of HIV replication in IECIL-17A:T cell co-cultures. Consistent with the fact that BST-2 expression is regulated by type I IFN (Van Damme et al., 2008), we observed a positive correlation between BST-2 expression and type I IFN levels in cell culture supernatants. Our results also provide evidence that IL-17A promote a pro-inflammatory transcriptional program, while impairing the capacity of IEC to sense HIV via pathogen recognition receptors. Indeed, the RNA sequencing results revealed that IL-17A exposure of IEC results in a decrease in TLR3 and RIG-I and an increase in the expression of multiple pro-inflammatory cytokines/chemokines, as well as a decrease in A20, a negative regulator inflammation (Chitre et al., 2018).
Previous studies demonstrated that early type I IFN supplementation can prevent the establishment of SIV infection, while a delay in the type I IFN production lead to an accelerated disease progression (Sandler et al., 2014). Accordingly, our results raise the hypothesis that increased IL-17A levels/responses prior to infection could dampen or delay the type I IFN responses, thus facilitating HIV/SIV acquisition. Indeed, in a model of SIV infection, mucosal type I IFN responses were detected at late time points post-infection and coincided with vanished IL-17A responses (Hensley-McBain et al., 2018). Further studies targeting the IL-17A pathway blockade to boost type I IFN responses during early HIV/SIV infection or before exposure should test this hypothesis.
In conclusion, our results provide evidence supporting the deleterious role of IL-17A during acute and chronic HIV infection by its capacity to transcriptionally reprogram IEC for an efficient HIV capture and transmission but also an increased capacity to promote replication/reactivation in subjacent infected CD4+ T cells. Our results point to IL-17A as a potential therapeutic target to limit HIV acquisition at mucosal sites and reduce the deleterious effects of HIV reactivation in ART-treated PLWH. The successful use of IL-17A Abs for the treatment of specific autoimmune conditions (e.g., psoriasis) (Burkett and Kuchroo, 2016) opens the path for testing such Abs for HIV cure/remission.
Limitations of the study
We acknowledge that one major limitation of our study is the absence of experiments with polarized IEC. We indeed performed our co-culture experiments on plastic grown IEC which does not reflect the physiological interplay between T cells and IEC. In vivo, T cells have access to the serosal basolateral side of the epithelial cell barrier whereas in our study T cells had access to the apical side. A polarized distribution of chemokine receptors or polarized secretion of cell products in IECs could affect the interpretation of our experiments and the application of our conclusions to an in vivo scenario. Future experiments should validate our results performing co-cultures of polarized human primary IECs with T cells, as reported previously (Real et al., 2018).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified NA/LE Mouse Anti-Human CD3 (Clone UCHT1) | BD | Cat#555329; RRID: AB_395736 |
| Purified NA/LE Mouse Anti-Human CD28 (Clone CD28.2) | BD | Cat#555725; RRID: AB_396068 |
| HIV-1 core (p24) antigen-FITC (Clone KC57) | Beckman Coulter | Cat#6604665; RRID: AB_1575987 |
| Mouse anti-human CD3 Alexa 700 (Clone UCHT1) | BD | Cat# 557943; RRID: AB_396952 |
| Mouse anti-human CD3 Pacific Blue (Clone UCHT1) | BD | Cat#558117; RRID: AB_397038 |
| Mouse anti-human CD317 BV421 (BST2, Clone Y129) | BD | Cat#566381; RRID: AB_2744363 |
| Mouse anti-human CD4 PerCP-Cy5.5 (Clone RPA-T4) | Biolegend | Cat#300530; RRID: AB_893322 |
| Rabbit polyclonal anti-BST2 | Provided by Dr. Éric A. Cohen (IRCM, Montreal, QC, Canada (Dube et al., PlosPathogens, 2010) | |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (405 nm excitation) | Invitrogen | Cat#L34957 |
| Goat polyclonal Secondary Antibody to Rabbit IgG - Fc Alexa Fluor 594 | Abcam | Cat#ab150092; RRID:AB_2893495 |
| Bacterial and virus strains | ||
| HIV-1 THRO plasmid (pTHRO.c/2626), subtype B | NIH AIDS Reagent Program (Contribution of Dr. John Kappes and Dr. Christina Ochsenbauer) | Cat#11745 |
| NL4.3BaL HIV plasmid | From Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA) | N/A |
| Biological samples | ||
| Leukaphereses of ART treated and untreated people living with HIV | Recruited at the Montreal Chest Institute, McGill University Health Centre and Centre Hospitalier de l’Université de Montréal with the help of Dr Jean-Pierre Routy’s group | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| rhIL-2 | R&D Systems | Cat#202-IL-050 |
| rhIL-17A | R&D Systems | Cat#7955-IL-025 |
| rhTNF-α | R&D Systems | Cat#210-TA-020 |
| Recombinant Viral B18R Protein | R&D Systems | Cat#8185-BR-025 |
| rhCCL20 | R&D Systems | Cat#360-MP-025 |
| Critical commercial assays | ||
| Memory CD4+ T Cell Isolation Kit, human | Miltenyi | Cat#130-091-893 |
| Fixation/Permeabilization Solution Kit (Cytofix/Cytoperm) | BD | Cat#554714 |
| p24 ELISA | Homemade. Hybridome provided by Dr. Michel J. Tremblay (Bounou et al., J Virol., 2002) | |
| Human CCL20/MIP-3 alpha DuoSet ELISA | R&D Systems | Cat#DY360-05 |
| Human IL-8/CXCL8 DuoSet ELISA | R&D Systems | Cat#DY208-05 |
| SmGM- 2 Smooth Muscle Cell Growth Medium -2 BulletKit | Lonza | Cat#CC-3182 |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat #204245 |
| AllPrep DNA/RNA/miRNA Universal kit | Qiagen | Cat#80224 |
| Rneasy Plus Mini Kit | Qiagen | Cat#74136 |
| Deposited data | ||
| RNA-Sequencing data set | This paper | GEO: GSE147045 |
| Gene Ontology: Biological Processes database | Broad Institute | https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=BP |
| Homo Sapiens database GRCh 37 version75 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
| Experimental models: cell lines | ||
| HT-29 cell line | ATCC | Cat#HTB-38; RRID:CVCL_0320 |
| HEK-Blue™ IFN-α/β Cells | InvivoGen | Cat#hkb-ifnab |
| Human Intestinal Epithelial Cells (inEpC) | Lonza | Cat#CC-2931, Lot 0000751353 |
| ACH-2 cell line | NIH HIV Reagent Program | Cat#ARP-349; RRID:CVCL_0138 |
| Oligonucleotides | ||
| See Table S1: Oligonucleotides | ||
| Software and algorithms | ||
| Kallisto software version 0.44.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| R, pheatmap package | The R Foundation | https://CRAN.R-project.org/package=pheatmap |
| FlowJo version 10 | BD | https://www.flowjo.com/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ |
| Cytoscape 3.7.2 | National Resource for Network Biology | https://cytoscape.org/ |
| ClueGO | National Resource for Network Biology | https://apps.cytoscape.org/apps/cluego |
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Petronela Ancuta (petronela.ancuta@umontreal.ca).
Material availability
This Study did not generate new unique reagents.
Experimental model and subject details
Human subjects
HIV-uninfected individuals (HIV-), virally suppressed ART-treated PLWH (ART+PLWH) and untreated PLWH (HIV+PLWH) (Table 1), were recruited at the Montreal Chest Institute, McGill University Health Centre and Centre Hospitalier de l’Université de Montréal (CHUM, Montreal, QC, Canada). Peripheral blood mononuclear cells (PBMC) (109–1010 cells) were collected by leukapheresis and frozen until use, as previously described (Boulassel et al., 2003). Plasma viral load in ART-treated PLWH was measured using the Amplicor HIV-1 monitor ultrasensitive method (Roche).
The collection of leukapheresis from HIV-uninfected individuals, untreated ART- PLWH and ART+ treated PLWH was conducted in compliance with the principles included in the Declaration of Helsinki. This study received approval from the Institutional Review Board of the McGill University Health Centre and the CHUM-Research Centre, Montreal, Quebec, Canada. Written informed consents were obtained from all study participants.
Epithelial cell line
The female human colorectal adenocarcinoma intestinal epithelial cell (IEC) line HT-29 (ATCC, Manassas, VA, USA) was seed in T75 culture flask (Corning, Tewksbury, MA, USA) with McCoy's 5A medium (ATCC, Manassas, VA, USA) containing 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (P/S; original solution at 10,000 U/ml) (Gibco, Carlsbad, CA, USA). Cells were split every 4-6 days when reaching 90-95% confluence using Trypsin-EDTA (Gibco, Carlsbad, CA, USA). No cell authentication was performed on the HT-29 cell line.
HEK293-type I IFN reporter cell line
Bioactive human Type I interferons (IFN) were quantified using the Female Human HEK293- Type I IFN reporter cell line provided by Dr. Eric Cohen accordingly to the protocol described by the original provider (InvivoGen, San Diego, CA, USA). Briefly, cells were cultured with DMEM media (Gibco, Carlsbad, CA, USA) containing 10% FBS, 1% P/S, Blasticidin (InvivoGen, San Diego, CA, USA) and Zeocin (InvivoGen, San Diego, CA, USA). Cells were harvest every 4-6 days after reaching 90-95% confluence using Versene (Gibco, Grand Island, NY, USA). Experiments were performed with cells at passages 3-7. No cell authentication was performed on the HT-29 cell line.
Primary intestinal epithelial cells
Primary human IEC were purchased from Lonza (lot 0000751353; unknown sex as reported by the certificate of analysis) and seed in culture following the instructions of the provider. Prior to primary IEC culture, Rat-tail type 1 collagen was used to cover the surface of 48 well plates at a concentration of 30 ug/ml and incubated for 30 min at 37°C. Then, collagen was aspirated, and wells were washed with PBS twice. Primary IEC were cultured at density of ∼100,000 cell/well with SmBM smooth muscle cell basal medium (SmBM-2™, Lonza USA) and SmGM™-2 Single Quots™ (Lonza, USA) formulates which contains insulin, hFGF-B, hEGF-B, FBS and gentamicin/amphotericin-B. Cells were incubated at 33°C with CO2 5% for 5 days to reach 90% confluence and be used for subsequent experiments.
Method details
Cytokines, chemokines and proteins
Human recombinant TNF-α, IL-17A, IL-2, CCL20 and B18R Protein (R&D Systems) were used at concentrations indicated in the Results or Figure legends.
Primary CD4+ T-cell isolation and culture
Memory CD4+ T-cells were isolated from PBMCs of HIV-uninfected and ART-treated PLWH by negative selection using magnetic beads (Miltenyi Biotec, Auburn, CA, USA), as we previously described (Planas et al., 2017). T-cells (2x106 cells/ml per 48 well-plate well) were stimulated with immobilized CD3 and soluble CD28 Abs (1 μg/mL) for 3 days prior to infection and/or co-culture experiments with IEC. T-cells were cultured with RPMI1640 media (Gibco, Grand Island, NY, USA) containing 10% FBS and 1% P/S. After infection and/or co-culture with IEC, T-cells were maintained in culture in the presence of IL-2 (5 ng/ml; R&D Systems, Minneapolis, MN, USA) for up to 9 days, with 50% media being refreshed every 3 days.
HIV-1 exposure and infection in vitro
The molecular clones of CCR5-tropic HIV-1 strain used in this study were: transmitted Founder (T/F) THRO and the recombinant NL4.3BAL viruses. The T/F THRO HIV plasmid was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pTHRO.c/2626 (cat# 11745) from Dr. John Kappes and Dr. Christina Ochsenbauer. The NL4.3BaL HIV plasmid was a gift from Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA). HIV-1 plasmid was amplified by MiniPrep and MaxiPrep and viral stocks were produced and titrated, as we described it previously (Gosselin et al., 2010; Monteiro et al., 2011). HT29 IEC cultured in 48 well-plates (150,000 cells/well) until 80% confluence were exposed to HIV (50 ng HIV-p24/150,000 cells) for 3 hours. Unbound virions were removed by extensive washing. In a set of experiments, cytokine-activated HIV-exposed HT-29 IEC were cultured either alone or co-cultured with CD3/CD28-activated memory CD4+ T-cells from HIV-uninfected individuals. In another set of experiments, memory CD4+ T-cells from HIV-uninfected individuals were infected with HIV (20 ng HIV-p24/106 cells) and then co-cultured with cytokine-activated HT-29 IEC. Viral replication was measured by flow cytometry intracellular HIV-p24 staining and HIV-p24 ELISA in cell-culture supernatant, as previously described (Gosselin et al., 2017; Zhyvoloup et al., 2017). Briefly, the IEC:T cell co-cultures were harvested at day 9 post infection (for HIV infection in vitro) or day 9-12 post culture (for VOA). Cells were stained on the surface with fluorochrome-conjugated CD3, CD4, and BST2 Abs and intracellularly with anti-HIV-p24 Abs. The positivity gates were established using the fluorescence minus one (FMO) strategy. Acquisition was performed using the BD LSRFortessa flow cytometer (BD Bioscience, USA). In parallel, cell culture supernatants collected every 3 days were used for HIV-p24 ELISA.
Flow cytometry staining and analysis
The following fluorochrome-conjugated Abs were used for flow cytometry analysis: HIV-p24 FITC (KC57) (Beckman Coulter, Fullerton, CA, USA), CD3 Pacific blue (UCHT1), CD3 Alexa Fluor 700 (UCHT1), BST2 BV421 (Y129) (BD Biosciences, San Diego, CA, USA) and CD4 PerCP/Cy5.5 (RPA-T4) (Biolegend, San Diego, CA, USA). BST2 staining was also performed with homemade primary rabbit polyclonal BST2 Abs (provided by Dr. Éric Cohen) and secondary goat anti-rabbit IgG conjugated with Alexa Fluor 594 (Abcam, Cambridge, UK). Live/Dead Fixable Aqua Dead Cell Stain Kit (Vivid, Life Technologies, Burlington, Ontario, CA) was used to exclude dead cells. Cells were analyzed with the BD-LSRII cytometer and BD-Diva (BD Biosciences) and FlowJo version 10 (Tree Star, Inc., Ashland Oregon, USA). The positivity gates were placed using fluorescence minus one (FMO) strategy (Roederer, 2002).
ELISA
HIV-p24 levels in cell culture supernatant or cell lysates were quantified using a homemade sandwich ELISA, as described previously (Bounou et al., 2002; Cattin et al., 2019; Gosselin et al., 2017). Prior to the ELISA, virions in cell supernatants and cells were lysed using a homemade buffer solution (PBS 1X, Tween 20 0.05%, Triton X-100 2.5%, Trypan Blue 1% and Thimerosal 0.02% in deionized water) for 1h at 37°C. Levels of CCL20 and IL-8 were measured in the cell culture supernatant, according to the manufacturer’s protocols (R&D Systems, Minneapolis, MN, USA).
HIV-DNA quantification
Integrated HIV-DNA, as well as early (RU5) and late (Gag) reverse transcripts were quantified in cell lysates by nested real-time PCR using specific primers (Table S1), as we previously described (Gosselin et al., 2010; Monteiro et al., 2011; Planas et al., 2017; Cattin et al., 2019). Briefly, integrated HIV-DNA, as well as early (RU5) reverse transcripts and total HIV-DNA (which detects both late reverse transcripts and integrated HIV-DNA by targeting the Gag gene) were quantified in cell lysates by nested real-time PCR using primers purchased at IDT. The CD3 gene was used to normalize the number of HIV-DNA copies per cell number (two CD3 copies per cell) (Planas et al., 2017; Cattin et al., 2019).
Real-time RT-PCR
Total RNA was extracted from the primary intestinal epithelial cells with the RNeasy Plus Mini kit (Qiagen). IFIT1 and ISG15 gene expression was evaluated by One step SYBR green RT-PCR (Qiagen) using a LightCycler 480. IFIT1 primers were purchased from Qiagen (QuantiTect Primer Assay). ISG15 primers were purchased from IDT with the following sequence Forward 5′-ACTCATCTTTGCCAGTACAGGAG-3′, Reverse: 5′-CAGCATCTTCACCGTCAGGTC-3′ (Table S1). The amplification cycles used for ISG15 RT-PCR was reverse transcription at 50°C for 30 min, 15 min at 95°C and then 35 cycles each at 94°C for 10 sec, 56°C for 10 sec, and 72°C for 10 sec. The relative expression of the target genes mentioned above were normalized relative to the internal control 28S rRNA. The sequence of primers used were the 28S rRNA forward primer 5′-CGAGATTCCTGTCCCCACTA-3′ and 28S rRNA reverse primer 5′-GGGGCCACCTCCTTATTCTA-3′ (purchased from IDT) (Table S1). Each reaction was performed in triplicates. Melting curve analysis was used to determine the uniformity of the thermal dissociation profile for each product amplified by real-time RT-PCR.
Illumina RNA sequencing and analysis
Total RNA was extracted from cytokine-activated IEC and cytokine-activated IEC co-cultured with CD4+ memory T-cells using RNeasy Plus mini kit (Qiagen, Germantown, Maryland, USA). Genome-wide transcriptional profiling was performed by Genome Québec (Montreal, Québec, Canada) using the Illumina RNA-Sequencing model NovaSeq S4 6000 PE100. Briefly, the paired-end sequencing reads were aligned to coding and non-coding transcripts from Homo Sapiens database GRCh 37 version75 and quantified with the Kallisto software version 0.44.0. The entire RNA-Sequencing data set and the technical information requested by Minimum Information About a Microarray Experiment (MIAME) are available at the Gene Expression Omnibus database under accession GSE147045. Log2-counts per million (log CPM) transformed transcript-level data of IECTNF, IECTNF+IL-17A and IECIL-17A alone or co-cultured with memory CD4+ T-cells of ART + PLWH was compared respectively with IEC Medium alone or co-cultured with memory CD4+ T-cells of ART + PLWH, to calculate the fold change (Data S6).
Gene set enrichment analysis
ClueGO application on the software Cytoscape 3.7.2 and a Fisher exact-t-Test was applied to find significant enrichments and produce networks of enriched pathways. The latest Gene Ontology: Biological Processes database was used, as available at https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=BP. To stablish modules of genes with similar responses to cytokine-activations, hierarchical clustering was performed on the scaled fold-changes with the pheatmap package in R. Only genes with an absolute fold-change greater than 1.3 in at least one of the cytokine-activation conditions were considered, therefore focusing mainly on larger transcriptional changes. Two different resolutions of clustering were utilized, yielding either 3 main clusters or 8 clusters. Each of the gene modules was compared to a reference background consisting of all genes measured (28,083 genes). The R scripts used for analysis are available on Data S5. The matrix database with fold change gene information used to run this analysis is available on Data S6.
Quantification and statistical analysis
Statistical analyses were performed with GraphPad Prism 10 software (GraphPad Software, Inc., La Joya, CA, USA). Paired t Test were performed to assess the statistical difference between two conditions for the same donor and p values were added on the graph. Friedman along with Dunn's multiple comparisons test evaluated the statistical differences between more than two conditions (∗, p < 0.1; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001). Spearman Correlation and Linear regression models were applied to explore the relationship between two quantitative variables. p values of ≤0.05 were considered statistically significant. More details of the statistical analysis are included in Figure legends.
Acknowledgments
This work was supported in part by funds from the Canadian Institutes of Health Research to PA (#MOP-114957; #TCO125276; IBC-154053), National Institutes of Health (NIH) to C.T. and P.A. (R01AG054324), as well as infrastructure funding from the Canadian Foundation for Innovation (CFI) to P.A. and C.T. Core facilities and human cohorts were supported by the Fondation du CHUM and the Fonds de recherche du Québec – Santé (FRQ-S) HIV/AIDS and Infectious Diseases Network. TWS was supported by Doctoral awards from the Université de Montréal and the FRQ-S. The authors thank Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA) for providing the HIV NL4.3BAL molecular clone. Dr. Dominique Gauchat and Philippe St Onge (Flow Cytometry Core Facility, CHUM-Research Center, Montréal, QC, Canada) for expert technical support with polychromatic flow cytometry sorting; Olfa Debbeche (NLC3 Core Facility CHUM-Research Center, Montréal, QC, Canada); Mario Legault for his help with ethical approvals and informed consents; and Josée Girouard, Angie Massicotte, and Cynthia Dion, for their key contribution to study participant recruitment and access to blood samples and clinical information from HIV-infected and uninfected participants. The authors address a special thanks to all study participants for their key contribution to this work.
Author contributions
T.R.W.S. contributed to the study design, performed all experiments, analyzed results, prepared figures, and wrote the manuscript. J.-P.G. and O.T. performed RNA-Seq analysis. O.T. contributed with figures from RNA-seq analysis. H.T. generated preliminary results for Figure 1A. A.G., Y.Z., L.R.M., E.M.G., and D.V. performed experiments. J.-P.R. and C.T. allowed access to biological samples and study participants clinical information. M.E.-F., N.C., and A.L.L. contributed to study design and provided protocols for IEC culture and HIV quantification. M.G.B. and E.A.C. kindly provided reagents and expertise for BST2 experiments. P.A. designed the study, contributed to data analysis and figure preparation, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103225.
Supplemental information
Data and code availability
-
•
RNA-Sequencing data sets have been deposited in the Gene Expression Omnibus (GEO) database: GSE147045 and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
-
•
All original code has been included in Data S5 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adoro S., Cubillos-Ruiz J.R., Chen X., Deruaz M., Vrbanac V.D., Song M., Park S., Murooka T.T., Dudek T.E., Luster A.D. IL-21 induces antiviral microRNA-29 in CD4 T cells to limit HIV-1 infection. Nat. Commun. 2015;6:7562. doi: 10.1038/ncomms8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfsen A., Yu H., Magerus-Chatinet A., Schmitt A., Bomsel M. HIV-1-infected blood mononuclear cells form an integrin- and agrin-dependent viral synapse to induce efficient HIV-1 transcytosis across epithelial cell monolayer. Mol. Biol. Cell. 2005;16:4267–4279. doi: 10.1091/mbc.e05-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananworanich J., Schuetz A., Vandergeeten C., Sereti I., de Souza M., Rerknimitr R., Dewar R., Marovich M., van Griensven F., Sekaly R. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS ONE. 2012;7:e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.L., Khoury G., Fromentin R., Solomon A., Chomont N., Sinclair E., Milush J.M., Hartogensis W., Bacchetti P., Roche M. Human immunodeficiency virus (HIV)-Infected CCR6+ rectal CD4+ T cells and HIV persistence on antiretroviral therapy. J. Infect. Dis. 2020;221:744–755. doi: 10.1093/infdis/jiz509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K.B., Burgener A., Birse K., Romas L., Dunphy L.J., Shahabi K., Abou M., Westmacott G.R., McCorrister S., Kwatampora J. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9:194–205. doi: 10.1038/mi.2015.51. [DOI] [PubMed] [Google Scholar]
- Barre-Sinoussi F., Ross A.L., Delfraissy J.F. Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- Bobardt M.D., Cheng G., de Witte L., Selvarajah S., Chatterji U., Sanders-Beer B.E., Geijtenbeek T.B., Chisari F.V., Gallay P.A. Hepatitis C virus NS5A anchor peptide disrupts human immunodeficiency virus. Proc. Natl. Acad. Sci. U S A. 2008;105:5525–5530. doi: 10.1073/pnas.0801388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel M. Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat. Med. 1997;3:42–47. doi: 10.1038/nm0197-42. [DOI] [PubMed] [Google Scholar]
- Bouhlal H., Chomont N., Haeffner-Cavaillon N., Kazatchkine M.D., Belec L., Hocini H. Opsonization of HIV-1 by semen complement enhances infection of human epithelial cells. J. Immunol. 2002;169:3301–3306. doi: 10.4049/jimmunol.169.6.3301. [DOI] [PubMed] [Google Scholar]
- Boulassel M.R., Spurll G., Rouleau D., Tremblay C., Edwardes M., Sekaly R.P., Lalonde R., Routy J.P. Changes in immunological and virological parameters in HIV-1 infected subjects following leukapheresis. J. Clin. Apher. 2003;18:55–60. doi: 10.1002/jca.10051. [DOI] [PubMed] [Google Scholar]
- Bounou S., Leclerc J.E., Tremblay M.J. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol. 2002;76:1004–1014. doi: 10.1128/jvi.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Paiardini M., Knox K.S., Asher A.I., Cervasi B., Asher T.E., Scheinberg P., Price D.A., Hage C.A., Kholi L.M. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Price D.A., Douek D.C. HIV disease: fallout from a mucosal catastrophe? Nat. Immunol. 2006;7:235–239. doi: 10.1038/ni1316. [DOI] [PubMed] [Google Scholar]
- Brenchley J.M., Schacker T.W., Ruff L.E., Price D.A., Taylor J.H., Beilman G.J., Nguyen P.L., Khoruts A., Larson M., Haase A.T., Douek D.C. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett P.R., Kuchroo V.K. IL-17 blockade in psoriasis. Cell. 2016;167:1669. doi: 10.1016/j.cell.2016.11.044. [DOI] [PubMed] [Google Scholar]
- Cafaro A., Piccaro G., Altavilla G., Gigantino V., Matarese G., Olivieri E., Ferrantelli F., Ensoli B., Palma C. HIV-1 Tat protein vaccination in mice infected with Mycobacterium tuberculosis is safe, immunogenic and reduces bacterial lung pathology. BMC Infect Dis. 2016;16:442. doi: 10.1186/s12879-016-1724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P.U., Saleh S., Sallmann G., Solomon A., Wightman F., Evans V.A., Boucher G., Haddad E.K., Sekaly R.P., Harman A.N. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc. Natl. Acad. Sci. U S A. 2010;107:16934–16939. doi: 10.1073/pnas.1002894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo-Gimenez L., Cumont M.C., Fay M., Kared H., Monceaux V., Diop O., Muller-Trutwin M., Hurtrel B., Levy Y., Zaunders J. AIDS progression is associated with the emergence of IL-17-producing cells early after simian immunodeficiency virus infection. J. Immunol. 2010;184:984–992. doi: 10.4049/jimmunol.0902316. [DOI] [PubMed] [Google Scholar]
- Casartelli N., Sourisseau M., Feldmann J., Guivel-Benhassine F., Mallet A., Marcelin A.G., Guatelli J., Schwartz O. Tetherin restricts productive HIV-1 cell-to-cell transmission. Plos Pathog. 2010;6:e1000955. doi: 10.1371/journal.ppat.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A., Wiche Salinas T.R., Gosselin A., Planas D., Shacklett B., Cohen E.A., Ghali M.P., Routy J.P., Ancuta P. HIV-1 is rarely detected in blood and colon myeloid cells during viral-suppressive antiretroviral therapy. AIDS (London, England) 2019;33:1293–1306. doi: 10.1097/QAD.0000000000002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chege D., Chai Y., Huibner S., Kain T., Wachihi C., Kimani M., Barasa S., McKinnon L.R., Muriuki F.K., Kariri A. Blunted IL17/IL22 and pro-inflammatory cytokine responses in the genital tract and blood of HIV-exposed, seronegative female sex workers in Kenya. PLoS ONE. 2012;7:e43670. doi: 10.1371/journal.pone.0043670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenine A.L., Matouskova E., Sanchez G., Reischig J., Pavlikova L., LeContel C., Chermann J.C., Hirsch I. Primary intestinal epithelial cells can be infected with laboratory-adapted strain HIV type 1 NDK but not with clinical primary isolates. AIDS Res. Hum. retroviruses. 1998;14:1235–1238. doi: 10.1089/aid.1998.14.1235. [DOI] [PubMed] [Google Scholar]
- Chitre A.S., Kattah M.G., Rosli Y.Y., Pao M., Deswal M., Deeks S.G., Hunt P.W., Abdel-Mohsen M., Montaner L.J., Kim C.C. A20 upregulation during treated HIV disease is associated with intestinal epithelial cell recovery and function. Plos Pathog. 2018;14:e1006806. doi: 10.1371/journal.ppat.1006806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford D.B., Ances B.M. HIV-associated neurocognitive disorder. Lancet Infect Dis. 2013;13:976–986. doi: 10.1016/S1473-3099(13)70269-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L.B., Chomont N., Deeks S.G. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe. 2020;27:519–530. doi: 10.1016/j.chom.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M., Ruiz A., Moris A., Prado J.G. Restriction factors: from intrinsic viral restriction to shaping cellular immunity against HIV-1. Front Immunol. 2018;9:2876. doi: 10.3389/fimmu.2018.02876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mareuil J., Guettari N., Bolmont C., Salaun D., Baillon J.G., Hostomsky Z., Hirsch I. Restriction of HIV-1 replication in intestinal cells is genetically controlled by the gag-pol region of the HIV-1 genome. Virology. 1995;207:160–167. doi: 10.1006/viro.1995.1062. [DOI] [PubMed] [Google Scholar]
- Deeks S.G., Lewin S.R., Ross A.L., Ananworanich J., Benkirane M., Cannon P., Chomont N., Douek D., Lifson J.D., Lo Y.R. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat. Med. 2016;22:839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage C., Schuetz A., Alvord W.G., Johnston L., Hao X.P., Morcock D.R., Rerknimitr R., Fletcher J.L., Puttamaswin S., Phanuphak N. Impact of early cART in the gut during acute HIV infection. JCI Insight. 2016;1 doi: 10.1172/jci.insight.87065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delezay O., Koch N., Yahi N., Hammache D., Tourres C., Tamalet C., Fantini J. Co-expression of CXCR4/fusin and galactosylceramide in the human intestinal epithelial cell line HT-29. AIDS (London, England) 1997;11:1311–1318. doi: 10.1097/00002030-199711000-00004. [DOI] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Dwinell M.B., Eckmann L., Leopard J.D., Varki N.M., Kagnoff M.F. Chemokine receptor expression by human intestinal epithelial cells. Gastroenterology. 1999;117:359–367. doi: 10.1053/gast.1999.0029900359. [DOI] [PubMed] [Google Scholar]
- El-Far M., Tremblay C.L. Gut microbial diversity in HIV infection post combined antiretroviral therapy: a key target for prevention of cardiovascular disease. Curr. Opin. HIV AIDS. 2018;13:38–44. doi: 10.1097/COH.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple H.J., Schneider T., Troeger H., Kunkel D., Allers K., Moos V., Amasheh M., Loddenkemper C., Fromm M., Zeitz M., Schulzke J.D. Impairment of the intestinal barrier is evident in untreated but absent in suppressively treated HIV-infected patients. Gut. 2009;58:220–227. doi: 10.1136/gut.2008.150425. [DOI] [PubMed] [Google Scholar]
- Fantini J., Baghdiguian S., Yahi N., Chermann J.C. Selected human immunodeficiency virus replicates preferentially through the basolateral surface of differentiated human colon epithelial cells. Virology. 1991;185:904–907. doi: 10.1016/0042-6822(91)90570-2. [DOI] [PubMed] [Google Scholar]
- Fantini J., Bolmont C., Yahi N. Tumor necrosis factor-alpha stimulates both apical and basal production of HIV in polarized human intestinal HT29 cells. Immunol. Lett. 1992;34:85–90. doi: 10.1016/0165-2478(92)90031-i. [DOI] [PubMed] [Google Scholar]
- Fantini J., Cook D.G., Nathanson N., Spitalnik S.L., Gonzalez-Scarano F. Infection of colonic epithelial cell lines by type 1 human immunodeficiency virus is associated with cell surface expression of galactosylceramide, a potential alternative gp120 receptor. Proc. Natl. Acad. Sci. U S A. 1993;90:2700–2704. doi: 10.1073/pnas.90.7.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J., Yahi N., Baghdiguian S., Chermann J.C. Human colon epithelial cells productively infected with human immunodeficiency virus show impaired differentiation and altered secretion. J. Virol. 1992;66:580–585. doi: 10.1128/jvi.66.1.580-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J., Yahi N., Chermann J.C. Human immunodeficiency virus can infect the apical and basolateral surfaces of human colonic epithelial cells. Proc. Natl. Acad. Sci. U S A. 1991;88:9297–9301. doi: 10.1073/pnas.88.20.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E., Yahi N., Zider A., Cavard C., Champion S., Fantini J. Physical contact with lymphocytes is required for reactivation of dormant HIV-1 in colonic epithelial cells: involvement of the HIV-1 LTR. Virus Res. 1994;34:1–13. doi: 10.1016/0168-1702(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Fensterl V., Sen G.C. Interferon-induced Ifit proteins: their role in viral pathogenesis. J. Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B.T., Scholte F.E.M., Bergeron E., Pegan S.D. How ISG15 combats viral infection. Virus Res. 2020;286:198036. doi: 10.1016/j.virusres.2020.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M., Diegelmann J., Beigel F., Brand S. IL-17A alone weakly affects the transcriptome of intestinal epithelial cells but strongly modulates the TNF-alpha-induced expression of inflammatory mediators and inflammatory bowel disease susceptibility genes. Inflamm. Bowel Dis. 2014;20:1502–1515. doi: 10.1097/MIB.0000000000000121. [DOI] [PubMed] [Google Scholar]
- Gabriel E.M., Salinas T.R.W., Gosselin A., Larouche-Anctil E., Durand M., Landay A.L., El-Far M., Tremblay C.L., Routy J.P., Ancuta P. Overt IL-32 isoform expression at intestinal level during HIV-1 infection is negatively regulated by IL-17A. AIDS (London, England) 2021 doi: 10.1097/QAD.0000000000002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda D., Jamieson B.D., Collman R.G., Lederman M.M., Burdo T.H., Deeks S.G., Dittmer D.P., Fox H.S., Funderburg N.T., Pahwa S.G. Pathogenesis of aging and age-related comorbidities in people with HIV: highlights from the HIV ACTION workshop. Pathog. Immun. 2020;5:143–174. doi: 10.20411/pai.v5i1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N., Sun L., Chen Z.J. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier S., Tremblay M.J. Interleukin-4 inhibits an early phase in the HIV-1 life cycle in the human colorectal cell line HT-29. Clin. Immunol. (Orlando, Fla.) 2010;135:146–155. doi: 10.1016/j.clim.2009.12.001. [DOI] [PubMed] [Google Scholar]
- George M.D., Reay E., Sankaran S., Dandekar S. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J. Virol. 2005;79:2709–2719. doi: 10.1128/JVI.79.5.2709-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn J., Taiwo B., Seedat S., Autran B., Katlama C. Hiv. Lancet. 2018;392:685–697. doi: 10.1016/S0140-6736(18)31311-4. [DOI] [PubMed] [Google Scholar]
- Gonzalez S.M., Aguilar-Jimenez W., Su R.C., Rugeles M.T. Mucosa: key interactions determining sexual transmission of the HIV infection. Front Immunol. 2019;10:144. doi: 10.3389/fimmu.2019.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmann C., Anahtar M.N., Handley S.A., Farcasanu M., Abu-Ali G., Bowman B.A., Padavattan N., Desai C., Droit L., Moodley A. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in Young South African women. Immunity. 2017;46:29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin A., Monteiro P., Chomont N., Diaz-Griffero F., Said E.A., Fonseca S., Wacleche V., El-Far M., Boulassel M.R., Routy J.P. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J. Immunol. 2010;184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin A., Wiche Salinas T.R., Planas D., Wacleche V.S., Zhang Y., Fromentin R., Chomont N., Cohen E.A., Shacklett B., Mehraj V. HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS (London, England) 2017;31:35–48. doi: 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman Z., Meier-Schellersheim M., Paul W.E., Picker L.J. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat. Med. 2006;12:289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- Guo L., Xu X.Q., Zhou L., Zhou R.H., Wang X., Li J.L., Liu J.B., Liu H., Zhang B., Ho W.Z. Human intestinal epithelial cells release antiviral factors that inhibit HIV infection of macrophages. Front Immunol. 2018;9:247. doi: 10.3389/fimmu.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Jiang X., Xiao Y., Zhou T., Guo Y., Wang R., Zhao Z., Xiao H., Hou C., Ma L. IL-17A signaling in colonic epithelial cells inhibits pro-inflammatory cytokine production by enhancing the activity of ERK and PI3K. PLoS ONE. 2014;9:e89714. doi: 10.1371/journal.pone.0089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A.T. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu. Rev. Med. 2011;62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- Henry T., Kirimanjeswara G.S., Ruby T., Jones J.W., Peng K., Perret M., Ho L., Sauer J.D., Iwakura Y., Metzger D.W., Monack D.M. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J. Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley-McBain T., Berard A.R., Manuzak J.A., Miller C.J., Zevin A.S., Polacino P., Gile J., Agricola B., Cameron M., Hu S.L. Intestinal damage precedes mucosal immune dysfunction in SIV infection. Mucosal Immunol. 2018;11:1429–1440. doi: 10.1038/s41385-018-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herjan T., Hong L., Bubenik J., Bulek K., Qian W., Liu C., Li X., Chen X., Yang H., Ouyang S. IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat. Immunol. 2018;19:354–365. doi: 10.1038/s41590-018-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Yoshitomi H., Hashimoto M., Maeda S., Teradaira S., Sugimoto N., Yamaguchi T., Nomura T., Ito H., Nakamura T. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 2007;204:2803–2812. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotter D., Bosso M., Jonsson K.L., Krapp C., Sturzel C.M., Das A., Littwitz-Salomon E., Berkhout B., Russ A., Wittmann S. IFI16 targets the transcription factor Sp1 to suppress HIV-1 transcription and latency reactivation. Cell Host Microbe. 2019;25:858–872.e813. doi: 10.1016/j.chom.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotter D., Kirchhoff F. Interferons and beyond: induction of antiretroviral restriction factors. J. Leukoc. Biol. 2018;103:465–477. doi: 10.1002/JLB.3MR0717-307R. [DOI] [PubMed] [Google Scholar]
- Hu S., He W., Du X., Yang J., Wen Q., Zhong X.P., Ma L. IL-17 production of neutrophils enhances antibacteria ability but promotes arthritis development during Mycobacterium tuberculosis infection. EBioMedicine. 2017;23:88–99. doi: 10.1016/j.ebiom.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.P., Patel K., Johnson K.R., Maric D., Calabresi P.A., Hasbun R., Nath A. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc. Natl. Acad. Sci. U S A. 2013;110:13588–13593. doi: 10.1073/pnas.1308673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan N.J., Kolios G., Abbot S.E., Sinai M.A., Thompson D.A., Petraki K., Westwick J. Expression of functional CXCR4 chemokine receptors on human colonic epithelial cells. J. Clin. Invest. 1999;104:1061–1069. doi: 10.1172/JCI6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagnoff M.F., Roebuck K.A. Human immunodeficiency virus type 1 (HIV-1) infection and expression in intestinal epithelial cells: role of protein kinase A and C pathways in HIV-1 transcription. J. Infect. Dis. 1999;179:S444–S447. doi: 10.1086/314801. [DOI] [PubMed] [Google Scholar]
- Kelley C.F., Kraft C.S., de Man T.J., Duphare C., Lee H.W., Yang J., Easley K.A., Tharp G.K., Mulligan M.J., Sullivan P.S. The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 2017;10:996–1007. doi: 10.1038/mi.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krarup A.R., Abdel-Mohsen M., Schleimann M.H., Vibholm L., Engen P.A., Dige A., Wittig B., Schmidt M., Green S.J., Naqib A. The TLR9 agonist MGN1703 triggers a potent type I interferon response in the sigmoid colon. Mucosal Immunol. 2018;11:449–461. doi: 10.1038/mi.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty M.K., Sun L., Christensen-Quick A., Lu W., Garzino-Demo A. Human beta defensin 2 selectively inhibits HIV-1 in highly permissive CCR6(+)CD4(+) T cells. Viruses. 2017;9 doi: 10.3390/v9050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty M.K., Sun L., DeMasi L., Lu W., Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman M.M., Cannon P.M., Currier J.S., June C.H., Kiem H.P., Kuritzkes D.R., Lewin S.R., Margolis D.M., McCune J.M., Mellors J.W. A cure for HIV infection: "not in my lifetime" or "just around the corner"? Pathog. Immun. 2016;1:154–164. doi: 10.20411/pai.v1i1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Wang P., Kattah M.G., Youssef S., Steinman L., DeFea K., Straus D.S. Differential regulation of chemokines by IL-17 in colonic epithelial cells. J. Immunol. 2008;181:6536–6545. doi: 10.4049/jimmunol.181.9.6536. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Wang P., Kattah M.G., Youssef S., Steinman L., DeFea K., Straus D.S. Differential regulation of chemokines by IL-17 in colonic epithelial cells. J. Immunol. 2008;181:6536–6545. doi: 10.4049/jimmunol.181.9.6536. [DOI] [PubMed] [Google Scholar]
- Li C., Yang P., Sun Y., Li T., Wang C., Wang Z., Zou Z., Yan Y., Wang W., Wang C. IL-17 response mediates acute lung injury induced by the 2009 pandemic influenza A (H1N1) virus. Cell Res. 2012;22:528–538. doi: 10.1038/cr.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Casanova J.L., Puel A. Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol. 2018;11:581–589. doi: 10.1038/mi.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Estes J.D., Schlievert P.M., Duan L., Brosnahan A.J., Southern P.J., Reilly C.S., Peterson M.L., Schultz-Darken N., Brunner K.G. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberatore R.A., Bieniasz P.D. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc. Natl. Acad. Sci. U S A. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau C., Requena M., Mavigner M., Cazabat M., Carrere N., Suc B., Barange K., Alric L., Marchou B., Massip P. CCR6(-) regulatory T cells blunt the restoration of gut Th17 cells along the CCR6-CCL20 axis in treated HIV-1-infected individuals. Mucosal Immunol. 2016;9:1137–1150. doi: 10.1038/mi.2016.7. [DOI] [PubMed] [Google Scholar]
- Margolis D.M., Archin N.M., Cohen M.S., Eron J.J., Ferrari G., Garcia J.V., Gay C.L., Goonetilleke N., Joseph S.B., Swanstrom R. Curing HIV: seeking to target and clear persistent infection. Cell. 2020;181:189–206. doi: 10.1016/j.cell.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis L., Shattock R. Selective transmission of CCR5-utilizing HIV-1: the 'gatekeeper' problem resolved? Nat. Rev. Microbiol. 2006;4:312–317. doi: 10.1038/nrmicro1387. [DOI] [PubMed] [Google Scholar]
- Masson L., Salkinder A.L., Olivier A.J., McKinnon L.R., Gamieldien H., Mlisana K., Scriba T.J., Lewis D.A., Little F., Jaspan H.B. Relationship between female genital tract infections, mucosal interleukin-17 production and local T helper type 17 cells. Immunology. 2015;146:557–567. doi: 10.1111/imm.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavigner M., Cazabat M., Dubois M., L'Faqihi F.E., Requena M., Pasquier C., Klopp P., Amar J., Alric L., Barange K. Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. J. Clin. Invest. 2012;122:62–69. doi: 10.1172/JCI59011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary C.S., Alvarez X., Harrington S., Cervasi B., Ryan E.S., Iriele R.I., Paganini S., Harper J.L., Easley K., Silvestri G. The loss of CCR6+ and CD161+ CD4+ T-cell homeostasis contributes to disease progression in SIV-infected rhesus macaques. Mucosal Immunol. 2017;10:1082–1096. doi: 10.1038/mi.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S., Poles M.A., Tenner-Racz K., Horowitz A., Hurley A., Hogan C., Boden D., Racz P., Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G., Wei X., Wu X., Sellers M.T., Decker J.M., Moldoveanu Z., Orenstein J.M., Graham M.F., Kappes J.C., Mestecky J. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat. Med. 2002;8:150–156. doi: 10.1038/nm0202-150. [DOI] [PubMed] [Google Scholar]
- Micci L., Ryan E.S., Fromentin R., Bosinger S.E., Harper J.L., He T., Paganini S., Easley K.A., Chahroudi A., Benne C. Interleukin-21 combined with ART reduces inflammation and viral reservoir in SIV-infected macaques. J. Clin. Invest. 2015;125:4497–4513. doi: 10.1172/JCI81400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Karasuyama H. Defects in Jak-STAT-mediated cytokine signals cause hyper-IgE syndrome: lessons from a primary immunodeficiency. Int. Immunol. 2009;21:105–112. doi: 10.1093/intimm/dxn134. [DOI] [PubMed] [Google Scholar]
- Monteiro P., Gosselin A., Wacleche V.S., El-Far M., Said E.A., Kared H., Grandvaux N., Boulassel M.R., Routy J.P., Ancuta P. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J. Immunol. 2011;186:4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- Mudd J.C., Brenchley J.M. Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J. Infect. Dis. 2016;214:S58–S66. doi: 10.1093/infdis/jiw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P.M. Neutrophil receptors for interleukin-8 and related CXC chemokines. Semin. Hematol. 1997;34:311–318. [PubMed] [Google Scholar]
- Nasr N., Alshehri A.A., Wright T.K., Shahid M., Heiner B.M., Harman A.N., Botting R.A., Helbig K.J., Beard M.R., Suzuki K. Mechanism of interferon-stimulated gene induction in HIV-1-Infected macrophages. J. Virol. 2017;91 doi: 10.1128/JVI.00744-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr N., Maddocks S., Turville S.G., Harman A.N., Woolger N., Helbig K.J., Wilkinson J., Bye C.R., Wright T.K., Rambukwelle D. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120:778–788. doi: 10.1182/blood-2012-01-407395. [DOI] [PubMed] [Google Scholar]
- Nazli A., Chan O., Dobson-Belaire W.N., Ouellet M., Tremblay M.J., Gray-Owen S.D., Arsenault A.L., Kaushic C. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. Plos Pathog. 2010;6:e1000852. doi: 10.1371/journal.ppat.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J., Sandrin V., Sundquist W.I., Bieniasz P.D. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Niwa M., Fujisawa T., Mori K., Yamanaka K., Yasui H., Suzuki Y., Karayama M., Hozumi H., Furuhashi K., Enomoto N. IL-17A attenuates IFN-lambda expression by inducing suppressor of cytokine signaling expression in airway epithelium. J. Immunol. 2018;201:2392–2402. doi: 10.4049/jimmunol.1800147. [DOI] [PubMed] [Google Scholar]
- Ochsenbauer C., Edmonds T.G., Ding H., Keele B.F., Decker J., Salazar M.G., Salazar-Gonzalez J.F., Shattock R., Haynes B.F., Shaw G.M. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M.B., Brenner D.A., de Grandpre L.Y., Roebuck K.A., Richman D.D., Kagnoff M.F. HIV-1 infection and expression in human colonic cells: infection and expression in CD4+ and CD4- cell lines. AIDS (London, England) 1991;5:275–281. doi: 10.1097/00002030-199103000-00005. [DOI] [PubMed] [Google Scholar]
- Pallikkuth S., Micci L., Ende Z.S., Iriele R.I., Cervasi B., Lawson B., McGary C.S., Rogers K.A., Else J.G., Silvestri G. Maintenance of intestinal Th17 cells and reduced microbial translocation in SIV-infected rhesus macaques treated with interleukin (IL)-21. Plos Pathog. 2013;9:e1003471. doi: 10.1371/journal.ppat.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish N.F., Wilen C.B., Banks L.B., Iyer S.S., Pfaff J.M., Salazar-Gonzalez J.F., Salazar M.G., Decker J.M., Parrish E.H., Berg A. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. Plos Pathog. 2012;8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau M., Pantaleo G., Kremer E.J. Activation of a dendritic cell-T cell axis by Ad5 immune complexes creates an improved environment for replication of HIV in T cells. J. Exp. Med. 2008;205:2717–2725. doi: 10.1084/jem.20081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Routy J.P., Ancuta P. New Th17-specific therapeutic strategies for HIV remission. Curr. Opin. HIV AIDS. 2019;14:85–92. doi: 10.1097/COH.0000000000000522. [DOI] [PubMed] [Google Scholar]
- Planas D., Zhang Y., Monteiro P., Goulet J.P., Gosselin A., Grandvaux N., Hope T.J., Fassati A., Routy J.P., Ancuta P. HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms. JCI Insight. 2017;2 doi: 10.1172/jci.insight.93230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Le Duff Y., Wang Y., Pan Q., Ding S., Zheng Y.M., Liu S.L., Liang C. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology. 2015;474:10–18. doi: 10.1016/j.virol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M., Santos R.L., Verhoeven D.E., George M.D., Wilson R.P., Winter S.E., Godinez I., Sankaran S., Paixao T.A., Gordon M.A. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real F., Sennepin A., Ganor Y., Schmitt A., Bomsel M. Live imaging of HIV-1 transfer across T cell virological synapse to epithelial cells that promotes stromal macrophage infection. Cell Rep. 2018;23:1794–1805. doi: 10.1016/j.celrep.2018.04.028. [DOI] [PubMed] [Google Scholar]
- Ribeiro Dos Santos P., Rancez M., Pretet J.L., Michel-Salzat A., Messent V., Bogdanova A., Couedel-Courteille A., Souil E., Cheynier R., Butor C. Rapid dissemination of SIV follows multisite entry after rectal inoculation. PLoS ONE. 2011;6:e19493. doi: 10.1371/journal.pone.0019493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J., Prevost J., Baxter A.E., von Bredow B., Ding S., Medjahed H., Delgado G.G., Brassard N., Sturzel C.M., Kirchhoff F. Uninfected bystander cells impact the measurement of HIV-specific antibody-dependent cellular cytotoxicity responses. mBio. 2018;9 doi: 10.1128/mBio.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M. Compensation in flow cytometry. Curr. Protoc. Cytom. 2002;Chapter 1:Unit 1 14. doi: 10.1002/0471142956.cy0114s22. [DOI] [PubMed] [Google Scholar]
- Ryzhakov G., Lai C.C., Blazek K., To K.W., Hussell T., Udalova I. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J. Immunol. 2011;187:5357–5362. doi: 10.4049/jimmunol.1100917. [DOI] [PubMed] [Google Scholar]
- Sandler N.G., Bosinger S.E., Estes J.D., Zhu R.T., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., del Prete G.Q. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankapal S., Gupta P., Ratner D., Ding M., Shen C., Sanyal A., Stolz D., Cu-Uvin S., Ramratnam B., Chen Y. HIV exposure to the epithelia in ectocervical and colon tissues induces inflammatory cytokines without tight junction disruption. AIDS Res. Hum. retroviruses. 2016;32:1054–1066. doi: 10.1089/AID.2015.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T., Huang W., Hall J.A., Yang Y., Chen A., Gavzy S.J., Lee J.Y., Ziel J.W., Miraldi E.R., Domingos A.I. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Takeuchi J.S., Misawa N., Izumi T., Kobayashi T., Kimura Y., Iwami S., Takaori-Kondo A., Hu W.S., Aihara K. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. Plos Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A., Deleage C., Sereti I., Rerknimitr R., Phanuphak N., Phuang-Ngern Y., Estes J.D., Sandler N.G., Sukhumvittaya S., Marovich M. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. Plos Pathog. 2014;10:e1004543. doi: 10.1371/journal.ppat.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Siliciano R.F. Targeting the latent reservoir for HIV-1. Immunity. 2018;48:872–895. doi: 10.1016/j.immuni.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L., Duan L., Perkey K.E., Wietgrefe S., Zupancic M., Smith A.J., Southern P.J., Johnson R.P., Haase A.T. Epithelium-innate immune cell axis in mucosal responses to SIV. Mucosal Immunol. 2017;10:508–519. doi: 10.1038/mi.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugasundaram U., Critchfield J.W., Pannell J., Perry J., Giudice L.C., Smith-McCune K., Greenblatt R.M., Shacklett B.L. Phenotype and functionality of CD4+ and CD8+ T cells in the upper reproductive tract of healthy premenopausal women. Am. J. Reprod. Immunol. 2014;71:95–108. doi: 10.1111/aji.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsouk M., Estes J.D., Deleage C., Dunham R.M., Albright R., Inadomi J.M., Martin J.N., Deeks S.G., McCune J.M., Hunt P.W. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS (London, England) 2015;29:43–51. doi: 10.1097/QAD.0000000000000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K.E., Wagh K., Korber B., Barouch D.H. Vaccines and broadly neutralizing antibodies for HIV-1 prevention. Annu. Rev. Immunol. 2020;38:673–703. doi: 10.1146/annurev-immunol-080219-023629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieh D.J., Matias E., Xu H., Fought A.J., Blanchard J.L., Marx P.A., Veazey R.S., Hope T.J. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe. 2016;19:529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tincati C., Douek D.C., Marchetti G. Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res. Ther. 2016;13:19. doi: 10.1186/s12981-016-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotard M., Tsopoulidis N., Tibroni N., Willemsen J., Binder M., Ruggieri A., Fackler O.T. Sensing of HIV-1 infection in tzm-bl cells with reconstituted expression of STING. J. Virol. 2016;90:2064–2076. doi: 10.1128/JVI.02966-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N., Goff D., Katsura C., Jorgenson R.L., Mitchell R., Johnson M.C., Stephens E.B., Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey R.S. Intestinal CD4 depletion in HIV/SIV infection. Curr. Immunol. Rev. 2019;15:76–91. doi: 10.2174/1573395514666180605083448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacleche V.S., Goulet J.P., Gosselin A., Monteiro P., Soudeyns H., Fromentin R., Jenabian M.A., Vartanian S., Deeks S.G., Chomont N. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology. 2016;13:59. doi: 10.1186/s12977-016-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacleche V.S., Landay A., Routy J.P., Ancuta P. The Th17 lineage: from barrier surfaces homeostasis to autoimmunity, cancer, and HIV-1 pathogenesis. Viruses. 2017;9 doi: 10.3390/v9100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Kang S.G., Lee J., Sun Z., Kim C.H. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2009;2:173–183. doi: 10.1038/mi.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche Salinas T.R., Zheng B., Routy J.P., Ancuta P. Targeting the interleukin-17 pathway to prevent acute respiratory distress syndrome associated with SARS-CoV-2 infection. Respirology. 2020 doi: 10.1111/resp.13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Yang X.O., Chung Y., Fukunaga A., Nurieva R., Pappu B., Martin-Orozco N., Kang H.S., Ma L., Panopoulos A.D. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Pappu B.P., Nurieva R., Akimzhanov A., Kang H.S., Chung Y., Ma L., Shah B., Panopoulos A.D., Schluns K.S. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasen A., Herrera R., Rosbe K., Lien K., Tugizov S.M. Release of HIV-1 sequestered in the vesicles of oral and genital mucosal epithelial cells by epithelial-lymphocyte interaction. Plos Pathog. 2017;13:e1006247. doi: 10.1371/journal.ppat.1006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Xu L., Sennepin A., Federici C., Ganor Y., Tudor D., Damotte D., Barry Delongchamps N., Zerbib M., Bomsel M. The HIV-1 viral synapse signals human foreskin keratinocytes to secrete thymic stromal lymphopoietin facilitating HIV-1 foreskin entry. Mucosal Immunol. 2018;11:158–171. doi: 10.1038/mi.2017.23. [DOI] [PubMed] [Google Scholar]
- Zhyvoloup A., Melamed A., Anderson I., Planas D., Lee C.H., Kriston-Vizi J., Ketteler R., Merritt A., Routy J.P., Ancuta P. Digoxin reveals a functional connection between HIV-1 integration preference and T-cell activation. Plos Pathog. 2017;13:e1006460. doi: 10.1371/journal.ppat.1006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-Sequencing data sets have been deposited in the Gene Expression Omnibus (GEO) database: GSE147045 and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
-
•
All original code has been included in Data S5 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.