Abstract
Cotton is a globally important cash crop. Verticillium wilt (VW) is commonly known as “cancer” of cotton and causes serious loss of yield and fiber quality in cotton production around the world. Here, we performed a BSA-seq analysis using an F2:3 segregation population to identify the candidate loci involved in VW resistance. Two QTLs (qvw-D05-1 and qvw-D05-2) related to VW resistance in cotton were identified using two resistant/susceptible bulks from the F2 segregation population constructed by crossing the resistant cultivar ZZM2 with the susceptible cultivar J11. A total of 30stop-lost SNPs and 42 stop-gained SNPs, which included 17 genes, were screened in the qvw-D05-2 region by SnpEff analysis. Further analysis of the transcriptome data and qRT-PCR revealed that the expression level of Ghir_D05G037630 (designated as GhDRP) varied significantly at certain time points after infection with V. dahliae. The virus-induced gene silencing of GhDRP resulted in higher susceptibility of the plants to V. dahliae than the control, suggesting that GhDRP is involved in the resistance to V. dahlia infection. This study provides a method for rapid mining of quantitative trait loci and screening of candidate genes, as well as enriches the genomic information and gene resources for the molecular breeding of disease resistance in cotton.
Keywords: upland cotton, BSA-seq, verticillium wilt, QTL, resistance gene
Introduction
As a globally important cash crop, cotton provides approximately 35% of total fiber used worldwide (Zhang et al., 2014a) and is also an important source of oilseed (Hulse-Kemp et al., 2015). Verticillium wilt (VW) is commonly known as a “cancer” of cotton, and it is one of the most destructive diseases caused by the soil-borne fungus Verticillium dahliae Kleb (Burpee and Bloom, 1978; Fradin et al., 2011). VW not only reduces the yield of cotton but also causes significant degradation of the fiber quality (Zhang et al., 2014a). Development of new cotton varieties resistant to VW has been considered as the most effective and feasible way to control VW (Zhao et al., 2017).
It has been widely recognized that VW is a quantitative trait controlled by multiple genes (Devey and Roose, 1987; Wang et al., 2004, 2010). The quantitative trait loci (QTL) related to VW in cotton have been located using F2 or recombinant inbred line (RIL) populations. For example, Fang et al. (2001) found a RAPD marker with OPB-191300 that is related to wilt resistance in upland cotton by bulked segregant analysis (BSA). Zhen et al. (2006) screened 768 pairs of SSR primers with BSA by using the upland cotton cultivar CCRI8 and the island cotton cultivar Pima90-53 as parents, and identified the genetic distance between the locus of VW resistance and BNL3255-208 marker as 13.7cM. Shi et al. (2016) used the BC1F1, BC1S1, and BC2F1 populations generated from the cross between Hai1 (an island cotton cultivar with high VW resistance) and CCRI36 (a susceptible upland cotton cultivar) to locate the QTLs related to VW resistance of cotton. Further, they constructed a linkage map containing 2,292 SSR marker sites with a total length of 5115.16cM, and detected 48 QTLs related to VW resistance. Localization of these QTLs lays a solid research basis for the fine mapping and cloning of disease resistant genes. To date, many genes related to VW resistance have been cloned in cotton, such as GbRVd (Yang et al., 2016), GbaNA1 (Li et al., 2018), GbSTK (Zhang et al., 2013), GbRLK (Jun et al., 2015), and Gbvdr3 (Chen et al., 2016), which were derived from island cotton, and GhBAK1 (Gao et al., 2013), GhPAO (Mo et al., 2015), GhSKIP35 (Zhang et al., 2017), and GhPGIP1 (Liu et al., 2017), which were derived from upland cotton. Most of these genes encode enzymes and receptor proteins with certain roles in disease resistance. Most research on VW resistance genes has been performed using reverse genetics; however, only a few genes have been cloned through forward genetics.
BSA, a method of forward genetics, is a practical technique of gene/marker mapping to identify the genomic regions that contain the loci affecting a certain trait. As early as 1991, Michelmore et al. (1991) used BSA to successfully screen three markers closely associated with the downy mildew resistance gene DM5/8 in an isolated lettuce population. Combined with deep sequencing, BSA-seq analysis can realize efficient and accurate fine mapping of QTLs or key genes related to some important traits of crops. Currently, BSA-seq analysis is widely used in the research on various agronomic traits of different crops such as maize (Klein et al., 2018), apple (Jia et al., 2018), chickpea (Singh et al., 2016), pepper (Lee et al., 2020), and cotton (Zhu et al., 2017), and the candidate gene for pepper fruit color (CaPRR2) and the bud yellow gene (GhCHL1) of cotton have been successfully identified.
In the research on plant disease resistance, QTLs for related traits have been identified and even the candidate genes have been screened by BSA-seq analysis, such as peanut rust and late spot (Pandey et al., 2017; Clevenger et al., 2018), tomato yellow rot (Zhao et al., 2016), corn gray leaf spot (Cui et al., 2018), and crustal blight of chickpea (Deokar et al., 2019). However, BSA-seq analysis has not been used in the detection of candidate genes for VW resistance in cotton, possibly due to the complexity of the cotton genome.
With the availability of the draft cotton genome (Wang et al., 2018), map-based gene cloning technology has been greatly developed in cotton. On this basis, we combined BSA-seq analysis and virus-induced gene silencing (VIGS) for the rapid mapping and identification of candidate genes associated with VW resistance in cotton. This work provides a paradigm for rapid map-based gene cloning in plants with large and polyploid genomes.
Materials and Methods
Plant Materials and Trait Evaluation
The cotton F2 segregation population was developed using the VW resistant cultivar Zhongzhimian2 (ZZM2) and susceptible cultivar Jimian11 (J11) as cross parents in 2014. To evaluate the VW resistance, 232 F2:3 cotton lines together with their parents were planted in a field in Xinjiang (41.29°N, 80.24°E, a serious verticillium disease plot), where the phenotypes were observed from July to September at the adult-plant stage in 2017, and in a greenhouse in Henan Anyang (36.1°N, 114.35°E), where the phenotypes were observed at the seedling stage in 2018. In the greenhouse experiments, phenotypes were observed at 15–35days post inoculation (dpi) with the V. dahliae isolate Vd080, a defoliating strain with moderate pathogenicity to cotton in the two-leaf stage. The greenhouse had a controlled 12h photoperiod and temperature variation of 23–30°C. The experiments in both environments were of a randomized block design with three replications. VW resistance was evaluated with the disease index (DI), which could present a comprehensive and objective measurement of plant health and was classified into five grades according to the symptoms on the cotyledons and true leaves (Jian et al., 2001; Zhu et al., 2010; Zhang et al., 2012).
The susceptible cultivar J11 was used as a susceptible control to estimate the severity of disease and determine the optimal time for investigation. The DI was further adjusted into the relative disease index (RDI) according to previous descriptions (Zhao et al., 2014). A higher DI or RDI value indicates a more advanced infection by V. dahliae. Descriptive statistics, analysis of variance (ANOVA), and correlation analysis were performed to evaluate the traits performance in the greenhouse and field using the SAS system (v8.02). Origin8.0 software was used to present the phenotypic differences between the two extreme pools.
BSA-Seq Library Construction and Analysis
To detect variations in the genome related to VW resistance, a BSA-seq analysis was implemented using two extreme pools. Eighteen resistant F2 individuals (R-bulk) and 18 susceptible F2 individuals (S-bulk) in both environments were selected for pooling and sequencing. DNA from the two pools was extracted with the SDS method as previously described (Kuang et al., 2010), quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher, United States), detected by 1% agarose gel electrophoresis, and then mixed at equal concentrations, respectively. The DNA of the two extreme pools and their parents was used to prepare four DNA libraries for Illumina sequencing on the Illumina HiSeq2500 platform at BGI (Beijing Genomics Institute, Beijing, China). The quality of the raw data obtained from sequencing was evaluated and clean reads were obtained through filtration. Clean data were compared to the reference genome (Wang et al., 2018) using the BWA software (Md et al., 2019). Based on the results, SNPs and InDels were detected and annotated using the GATK4.0 software.1 The QTL interval was obtained according to the △SNP-index and G-value, which were calculated using the QTLseqr software (Mansfeld and Grumet, 2018).
SnpEff Analysis
Stop-gained and stop-lost SNPs, respectively, with generation of new stop codons and loss of stop codons may have significant effects on gene function. Therefore, in this study, stop-gained and stop-lost SNPs in the QTL interval were screened using the SnpEff software (Klein et al., 2018; https://pcingola.github.io/SnpEff/).
Structural Analysis of Candidate Genes
In order to analyze the structure of the candidate gene Ghir_D05G037630 and its difference between ZZM2 and J11, the DNA was divided into three segments for amplification because of the long length of the gene. Primer Premier 5.0 software was used for primer design and the primer sequences are shown in Supplementary Table 1. The DNA was amplified using high fidelity enzyme KOD-Plus-Neo (KOD-401, Toyobo, Japan) and its PCR products were purified by 1% concentration agarose gel electrophoresis and recycled using a Vazyme’s FastPure Gel DNA Extraction Mini Kit (DC301, Vazyme, Nanjing, China). The target fragment was connected and recombined with the carrier using a 5-min TA/Blunt-Zero Cloning Kit (C601, Vazyme). The recombinant product was transformed into competent cells DH5a, and monoclones grown from expanded culture were sent to Sangon (Sangon Biotech, Zhengzhou Branch, China) for sequencing. The SeqMan software (v7.1.0) was used to splice and align the sequences.
Preparation of Samples Infected by V. dahliae
The cultivars ZZM2 and J11 were planted in an incubator under a 16h/light/25°C and 8h/dark/22°C regimen and infected with Vd080 at the three-leaf stage as previously described (Zhu et al., 2010). Root tissue samples were taken at 0, 6, 12, 24, 36, 48, 72, and 96h after infection and then stored at −80°C. Total RNA was extracted using an RNA prep Pure Plant Kit (DP441, Tiangen, Beijing, China) and used to generate cDNA with a PrimeScript RT reagent kit (RR037A, Takara, Japan) following the manufacturer’s instructions.
Quantitative Real-Time PCR
The Gossypium hirsutum histone H3.3 gene (Ghir_D03G004040) was used as an internal control for cotton. Diluted cDNA was used for qRT-PCR with ChamQ Universal SYBR qPCR Master Mix (Q711, Vazyme) on a QuantStudio 6 Flex fluorescence quantitative PCR instrument (Applied Biosystems, USA). A two-step method was used with the following PCR conditions: 95°C for 30s, 40cycles of 95°C for 10s, and 60°C for 30s. The dissociation curves of each reaction were checked, and the cycle threshold (CT) 2−△△CT method (Livak and Schmittgen, 2001) was used to calculate the expression level of each target gene. Each reaction was performed with at least three biological replicates.
VIGS Vector Construction and Pathogen Inoculation
The VIGS experiments were carried out with the method described by Pang et al. (2013). The primer pair Gh_V037630-2F/R was designed at 1931–2280bp of GhDRP (Supplementary Table 1) and used to amplify the specific fragment by polymerase chain reaction (PCR) from ZZM2. The target fragment was linked to the Tobacco Rattle Virus (TRV)-based vector (Bachan and Dinesh-Kumar, 2012; Zeng et al., 2019) at the BamHI and SacI restriction sites by homologous recombination using a ClonExpress II One Step Cloning Kit (C112, Vazyme). All vectors used in this study were transformed into Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method (Gong et al., 2018). The cotyledons of 11-day-old seedlings of ZZM2 were injected with equal amounts of TRV vectors. After 24h of incubation in darkness, the cotton seedlings were transferred to the greenhouse (Gong et al., 2017). Approximately 10days after VIGS injection, when the true leaves in TRV-PDS (positive control) began to fade and turn white, leaves from the TRV-target and TRV-empty seedlings were collected for each of the three replicates. Total RNA was extracted and reversed into cDNA for qRT-PCR to detect the silencing efficiency using the primer pair Ghir_RT037630F/R in Supplementary Table 1. After the silencing of the target gene was confirmed, the cotton seedlings were inoculated with V. dahliae.
Quantification of V. dahliae Colonization and Recovery Assay
qRT-PCR approach was performed to detect and quantify V. dahliae colonization. Leaves from each of three independent plants of TRV::00 and TRV::GhDRP were sampled at 20 and 25 dpi, rapidly frozen in liquid nitrogen, and stored at −80°C. The primers VdActin-F/R (Supplementary Table 1) and the qRT-PCR program were used as described by Atallah et al. (2007). To visualize the degree of V. dahliae infection, a recovery assay was performed using fragments from the first node of the stem (Zhang et al., 2011). The stem segment surface was sterilized with 70% ethanol for 20–30s, sodium hypochlorite for 1min, and sterile water for 4–5 times, and then placed on potato dextrose agar (PDA) culture medium, which were incubated at 25°C under dark conditions for 2–3 d. The sensitivity of cotton to V. dahliae was evaluated according to the degree of fungal growth in stem segments.
Cell Death Assay
Cell death in cotton leaves at 25 dpi was visualized by trypan blue staining. Leaves were soaked in trypan blue dye (10ml phenol, 10ml glycerol, 10ml lactic acid, 10ml water, and 0.02g of trypan blue; then dilution with 96% ethanol in a ratio of 1:2), boiled for 1min, cooled to room temperature, and soaked overnight. The next day, the samples were decolorized with a chloral hydrate solution (2.5g/ml) for phenotype observation and photo taking.
Safranin-Fixed Green Dyeing Assay
Safranin-fixed green dyeing could highlight the tissues that have undergone lignification or suberification, along with the cellulose cell walls of vascular plants. An upper stem segment of approximately 4cm was taken at 25 dpi and the cross section was stained with safranin-fixed green dye to observe cell staining and determine the sensitivity of cotton to V. dahliae. The experiment was performed by Servicebio (Wuhan, China).
Results
Phenotyping of Verticillium Wilt Resistance in F2:3 Population
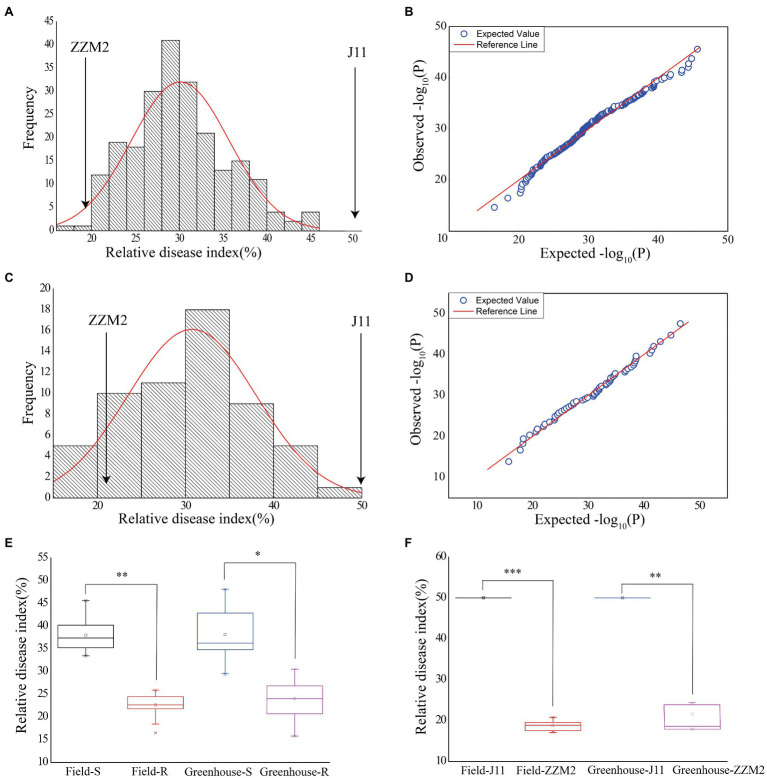
Assessment of VW severity showed that the RDI of the parent ZZM2 was 18.71 in the field and 21.25 in the greenhouse, respectively; and that of J11 was adjusted to 50 in both environments, which was used as a susceptible control (Figure 1F). There were considerable variations in RDI in the F2:3 population, ranging from 16.31 to 45.31 (30.81 in average) in the field, and from 15.79 to 46.56 (31.17 in average) in the greenhouse (Table 1). The ANOVA results revealed that different genotypes had significantly different RDI whether in the field (F value=3.24, p<0.0001) or greenhouse (F value=11.35, p<0.0001; Table 1). The values of RDI in the two environments followed a normal distribution (Figures 1A,C). A quantile-quantile (QQ) plot showed that the observed p values were consistent with the expected p values in the field (Figure 1B) and greenhouse (Figure 1D). Correlation analysis showed that the RDI value in the greenhouse was strongly correlated with that in the field (r=0.628, p<0.001), which revealed that RDI investigated in seedling and adult stages could reflect the resistance of cotton to VW.
Figure 1.
Phenotypic variations of RDI in the field and greenhouse. (A) Normal distribution diagram of the RDI in the field. (B) Quantile-quantile plot for RDI in the field. (C) Normal distribution diagram of the RDI in the greenhouse. (D) Quantile-quantile plot for RDI in the greenhouse. (E) Box plot for the RDI of selected individuals in two pools in the two environments. Greenhouse-R stands for the RDI of 18 resistant individuals in the greenhouse. Field-R stands for the RDI of 18 resistant individuals in the field. Greenhouse-S stands for the RDI of 18 susceptible individuals in the greenhouse. Field-S stands for the RDI of 18 susceptible individuals in the field. (F) Box plot for the RDI of the two parents in the two environments. Asterisk indicates statistically significant differences determined by Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001).
Table 1.
Descriptive statistics and ANOVA results of VW resistance (indicated by RDI) in the field and greenhouse environments.
| Environment | Mean | SD | Variance | Min | Max | CV (%) | Skewness | Kurtosis | DF | Anova SS | Mean Square | F Value | Pr>F |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| field | 30.81 | 5.58 | 31.15 | 16.31 | 45.31 | 18.54 | 0.40 | −0.05 | 223 | 11374.34 | 51.00 | 3.24 | <0.0001 |
| greenhouse | 31.17 | 7.30 | 53.34 | 15.79 | 46.56 | 23.75 | −0.01 | −0.63 | 58 | 7005.58 | 120.79 | 11.35 | <0.0001 |
Eighteen F2 plants with the lowest and highest RDI, respectively, were selected to construct two extreme pools, in which the individuals had stable performance regardless of the two assessment method in the field and greenhouse, and the RDI of ZZM2 and the R-bulk was significantly lower than that of J11 and the S-bulk (Figure 1E; Supplementary Table 2). These results indicated that the experimental materials met the requirements of BSA-seq analysis.
Selection of Candidate QTLs
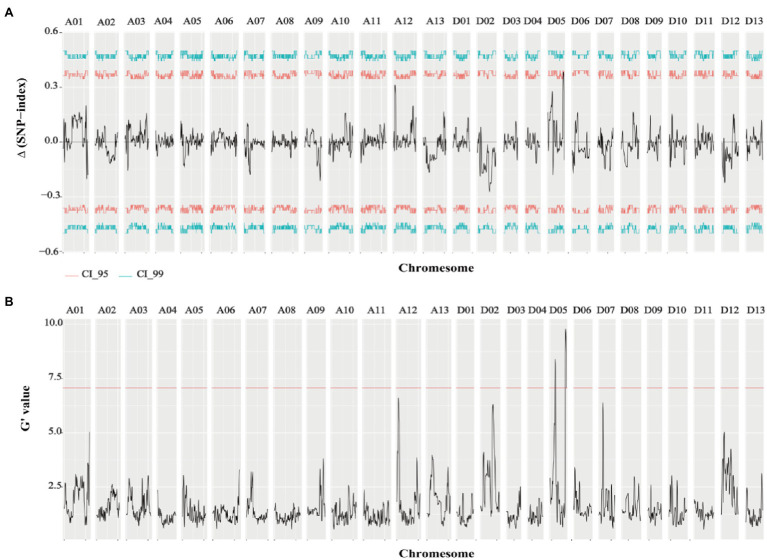
Genome-wide sequencing was carried out in the two extreme pools with a 20x depth, as well as for the two parents. Clean data (294.19G) were obtained from the raw data (306.41G) by filtering, including 80.62G reads for ZZM2, 69.54G reads for J11, 72.36G reads for R-bulk, and 71.67G reads for S-bulk (Supplementary Table 3). After the sequencing, 99.42–99.81% of sequences could be successfully mapped to the reference genome (Wang et al., 2018; Supplementary Table 4). Therefore, this protocol could be used for subsequent variation detection and gene mapping of traits. QTLseqr is an R package used to identify QTLs by combining QTL-seq and G’ approach. Candidate QTL segments were determined by using a q-value ≥0.01 in G’ or an △SNP-index ≥99% in the QTL-seq. As a result, two QTLs (18.67–20.74Mb, 2.06Mb in length; 58.19–62.88Mb, 4.69Mb in length) located on chromosome D05 related to VW resistance were obtained (Figure 2; Table 2).
Figure 2.
Results of △SNP-index and G-value of VW resistance extreme pools. (A) Results of △SNP-index in VW resistant extreme pools. (B) Results of G-value in VW resistant extreme pools (q=0.01).
Table 2.
Information of the candidate QTLs of verticillium wilt resistance.
| CHROM | qtl | start | end | length | nSNPs | avgSNPs_Mb | meanPval | meanQval |
|---|---|---|---|---|---|---|---|---|
| Ghir_D05 | qvw-D05-1 | 18,676,998 | 20,741,557 | 2,064,559 | 1,001 | 485 | 2.74E-05 | 0.002605994 |
| Ghir_D05 | qvw-D05-2 | 58,195,994 | 62,887,284 | 4,691,290 | 7,777 | 1,658 | 1.30E-05 | 0.001653478 |
SnpEff Analysis
A total of 123 stop-gained SNPs and 99 stop-lost SNPs were found through the annotation of 69,642 SNPs on chromosome D05 using the SnpEff software. There were 42 stop-gained SNPs and 30stop-lost SNPs in the region of qvw-D05-2, while none located within the qvw-D05-1 interval. These 72 mutation sites, which involve 17 genes in total, were distributed between 60,016,518bp and 61,342,887bp (Table 3).
Table 3.
Seventeen genes involving 72 mutation sites in the qvw-D05-2 interval.
| Gene ID | Stop-lost SNPs | Stop-gained SNPs |
|---|---|---|
| Ghir_D05G037600 | 60,016,518 | |
| Ghir_D05G037610 | 60,028,687,60,029,472,60,029,916, 60,029,917,60,029,993,60,030,399, 60,030,407,60,031,346,60,031,431, 60,031,591,60,031,644,60,031,655, 60,031,718,60,031,855 |
60,029,070,60,029,308,60,029,311, 60,029,352,60,029,362,60,029,845, 60,029,983,60,030,137,60,030,170, 60,030,389,60,030,484,60,030,599, 60,031,032,60,031,295,60,031,762, 60,031,928, |
| Ghir_D05G037620 | 60,038,613 | 60,038,430 |
| Ghir_D05G037630 | 60,063,746,60,069,431 | |
| Ghir_D05G037640 | 60,098,573,60,098,639 | |
| Ghir_D05G037650 | 60,116,797 | 60,118,071 |
| Ghir_D05G037670 | 60,277,261,60,280,293,60,280,721, 60,280,827 |
60,283,114,60,283,152 |
| Ghir_D05G037690 | 60,304,322 | 60,308,519,60,308,605 |
| Ghir_D05G037740 | 60,392,358 | 60,392,305,60,392,611 |
| Ghir_D05G037820 | 60,471,685 | |
| Ghir_D05G037850 | 60,574,375,60,575,359,60,575,360, 60,575,362,60,575,429 |
60,572,956,60,574,925,60,575,169, 60,575,298,60,576,319,60,576,329 |
| Ghir_D05G037880 | 60,686,560 | |
| Ghir_D05G038110 | 61,099,420,61,099,920,61,100,203 | |
| Ghir_D05G038190 | 61,173,648 | |
| Ghir_D05G038210 | 61,196,009 | 61,196,394 |
| Ghir_D05G038220 | 61,200,375 | |
| Ghir_D05G038360 | 61,342,887 |
Screening of Candidate Genes
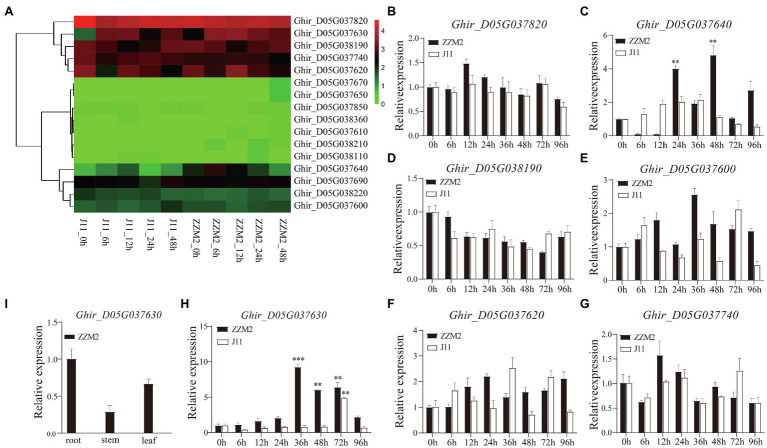
Combined with the transcriptome data from our previous study (Zhao et al., 2021), seven genes were found to be significantly differentially expressed, including Ghir_D05G037820, Ghir_D05G037630, Ghir_D05G038190, Ghir_D05G037740, Ghir_D05G037620, Ghir_D05G037640, and Ghir_D05G037600 (Figure 3A). To determine whether these seven genes were related to VW resistance, we performed a qRT-PCR analysis to detect candidate genes expression at 0, 6, 12, 24, 36, 48, 72, and 96h after infection with V. dahliae in root of ZZM2 and J11. The qRT-PCR results showed that two genes (Ghir_D05G037630 and Ghir_D05G037640) were up-regulated significantly after V. dahliae inoculation in ZZM2, of which Ghir_D05G037630 showed the most significant differential expression between the two parents at different stages (Figures 3B–H). Tissue pattern analysis showed that the expression level of Ghir_D05G037630 in root was higher than that in leaf and stem in ZZM2 (Figure 3I). Ghir_D05G037630 is homologous to AT4G27220 in Arabidopsis, which is annotated as a disease-resistant protein. Therefore, we mainly focused on the candidate gene Ghir_D05G037630 and designated it as GhDRP.
Figure 3.
Differential expression of candidate genes. (A) Heatmap of the 17 candidate genes in transcriptome analysis. (B–H) Expression patterns of the seven candidate genes at 0, 6, 12, 24, 36, 48, 72, and 96h after infection with V. dahliae in root of ZZM2 and J11. (I) Expression patterns of Ghir_D05G037630 in root, stem, and leaf in ZZM2. The error bar represents the standard deviation of three biological replicates. Asterisk indicates statistically significant differences determined by Student’s t-test (**p<0.01; ***p<0.001).
The DNA of GhDRP had a total length of 7,558bp including six exons and five introns, while the CDS sequence had a total length of 3,876bp, and the protein sequence contained 1,291 amino acids, including one NB-ARC and seven LRR domains. GhDRP was cloned from ZZM2 and J11, respectively, and the results showed that there was a mutation of one base, namely base “C” in ZZM2 and base “A” in J11 at 301 bp of the gene, which caused a change in amino acids to produce proline in ZZM2 and glutamine in J11 (Supplementary Figure 1). The mutated amino acid was located on the Pfam domain PF00931 (NB-ARC domain), which is related to plant disease resistance. These results indicated that GhDRP may play an important role in VW resistance of cotton.
Effect of GhDRP Silencing on VW Resistance of Cotton
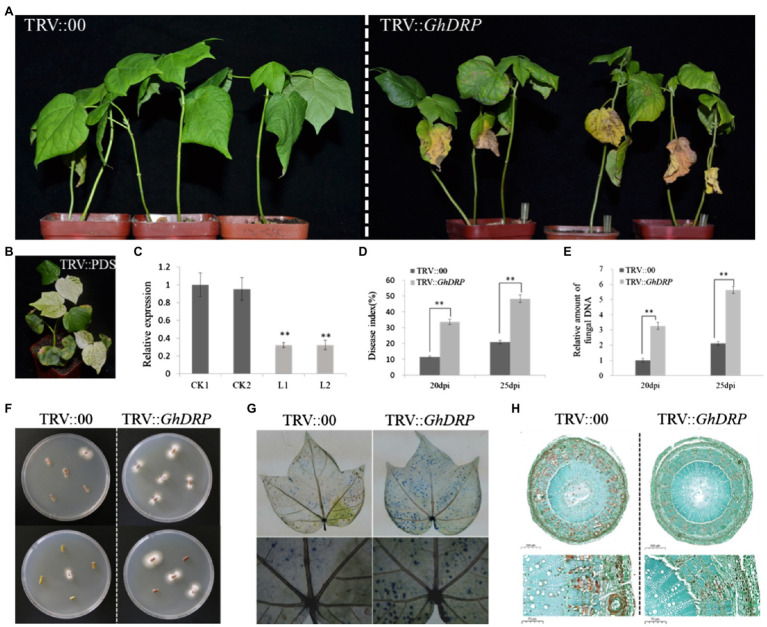
In order to further investigate the function of GhDRP in the VW resistance of cotton, the VIGS system was used to generate GhDRP-knockdown plants. Approximately 10days after the injection of the VIGS vector, the newly grown true leaves of the PDS control plants faded and gradually turned white (Figure 4B), indicating successful injection of the VIGS vector. Leaves of TRV::GhDRP plants and TRV::00 plants were randomly selected to measure the expression level of GhDRP by qRT-PCR. The results showed that the expression level of GhDRP in TRV::GhDRP plants was significantly lower than that in the control (Figure 4C), indicating the successful silencing of GhDRP. After inoculation with V. dahliae for 25days, yellowing and then peeling of leaves were observed in TRV::GhDRP plants, while no apparent symptoms of disease were observed in TRV::00 plants (Figure 4A).
Figure 4.
Effect of GhDRP silencing on V. dahliae resistance of cotton. (A) Phenotypes of TRV::GhDRP and TRV::00 plants induced by V. dahliae at 25 dpi. (B) Phenotype of positive control. (C) Transcript levels of GhDRP in the leaves of TRV::GhDRP and TRV::00 plants. CK1 and CK2 represent the control plants TRV::00; L1 and L2 represent the gene silencing plants TRV::GhDRP. Asterisk indicates statistically significant differences determined by Student’s t-test (**p<0.01), and the same below. (D) DI in TRV::GhDRP and TRV::00 plants at 20 dpi and 25 dpi. (E) Relative amount of fungal DNA in the leaves of TRV::GhDRP and TRV::00 plants at 20 dpi and 25 dpi. (F) The fungi in the stem segment were restored on PDA culture medium. (G) Trypan blue staining of the leaves of TRV::GhDRP and TRV::00 plants at 25 dpi. (H) Cell morphology of the stem cross section of TRV::GhDRP and TRV::00 plants at 25 dpi.
The DI value of TRV::GhDRP plants was significantly higher than that of the control at 20 and 25 dpi (Figure 4D). The qRT-PCR results showed that V. dahliae biomass in TRV::GhDRP plants was significantly higher than that in the control (Figure 4E), indicating that the plants with the silencing of GhDRP were more susceptible to V. dahliae. The recovery assay revealed that significantly more fungal colonies were present in TRV::GhDRP plants than in the control plants (Figure 4F). Trypan blue staining was then performed to compare the leaves of plants under different treatments at 25 dpi. The leaves from TRV::GhDRP plants exhibited large areas of staining and significant cell damage or death, while those from the control plants showed smaller areas of staining and less severe cell damage (Figure 4G). Examination of the cell morphology after pathogen inoculation showed that the cells of TRV::00 plants underwent more severe lignification than those of TRV::GhDRP plants (Figure 4H), suggesting that the plants might resist V. dahliae infection through lignification.
In summary, our results confirmed that knockdown of GhDRP could transform a resistant accession into a susceptible one. Therefore, GhDRP may play an important role in the resistance of cotton to V. dahliae.
Discussion
QTLs and Candidate Genes Can Be Rapidly Mined and Screened Using the F2:3 Segregation Population and BSA-Seq Analysis
Upland cotton is one of the most widely cultivated cotton species in the world. It has the advantages of high yield and adaptability, but with a relatively weaker resistance to VW. Cotton VW is a soil-borne fungal disease caused by V. dahliae and the most serious disease in cotton production to affect both the yield and fiber quality. Numerous studies have been performed to identify the QTLs for VW resistance in cotton using different populations such as F2 segregation population, RIL, and backcross population with the protocols of SSR, SNP, or GWAS (Jiang et al., 2009; Zhang et al., 2014b, 2015; Shi et al., 2016; Palanga et al., 2017; Abdelraheem et al., 2019). However, these QTLs have been seldom used in breeding. Michelmore et al. (1991) for the first time used the BSA method to screen three markers closely associated with the downy mildew resistance gene DM5/8 in an isolated lettuce population. With the development of high-throughput sequencing technology, BSA-seq analysis has been applied in wheat (Yin et al., 2018), soybean (Song et al., 2017), tomato (Zhao et al., 2016), rice (Ma et al., 2018; Liang et al., 2020), groundnut (Pandey et al., 2017), pepper (Lee et al., 2020), and cotton (Chen et al., 2015; Zhu et al., 2017; Zhao et al., 2018) for the rapid mining of QTLs and screening of candidate genes. However, the method has not been used in research related to VW resistance of cotton.
In the present study, an F2:3 segregation population was planted in a field in Xinjiang and a greenhouse in Anyang. There was a significant correlation between RDI at seedling stage in the greenhouse and adult stage in the field, which indicated that the RDI investigated at seedling stage could represent the resistance of cotton. Extremely resistant and susceptible materials with stable performance in both environments were selected, providing a material basis for BSA-seq analysis to mine the QTLs related to cotton VW resistance. Two QTLs (qvw-D05-1 and qvw-D05-2) related to VW resistance were obtained by BSA-seq analysis, which have large mapping intervals and many candidate genes. However, it is still necessary to develop molecular markers or bioinformatics methods to further fine-map the QTLs or mine the candidate genes.
We found that SnpEff could effectively annotate variations and predict the influence of genetic variations, which may facilitate the quick anchoring of a wide range of SNP mutation sites. Therefore, we performed SnpEff analysis and transcriptome analysis to further fine-mine the candidate genes. Cingolani et al. (2012) took the SNPs in the genome of Drosophila melanogaster strain w1118 as an example to elaborate and verify the principle and application of the SnpEff software. They identified 28 stop-gained SNPs and five stop-lost SNPs, which may have a great influence on gene expression. Some other researchers analyzed the genotype diversity of Brassica napus, potato and tomato using SnpEff as well (Kevei et al., 2015; Malmberg et al., 2018; Caruana et al., 2019). In the present study, SnpEff was used to narrow the QTL (qvw-D05-2) interval from 4.69Mb to 1.32Mb and decrease the number of candidate genes from 262 to 17. Hence, this study provides and verifies an analytical method for rapid mining of candidate genes.
GhDRP Can Be Used as a Candidate Gene for VW Resistance
In this study, GhDRP was identified to be associated with VW resistance. Its DNA length was 7,558bp, including six exons and five introns. The total length of the CDS sequence was 3,876bp and the protein sequence contained 1,291 amino acids, including one NB-ARC and seven LRR domains. These results suggest that GhDRP belongs to the NBS-LRR family and its homologue in Arabidopsis thaliana was annotated as a disease resistance protein. NBS-LRR genes are the most important resistance gene family in plants, with the largest number of members and the most abundant types of subfamilies. Generally, it has been considered that the LRR domain plays a specific role in pathogen recognition and an important role in downstream signal transduction (Anderson et al., 1997; Parker et al., 1997; Luck et al., 2000; Dangl and Jones, 2001). The NBS-LRR genes are ubiquitous in plants: there are approximately 149 NBS-LRR genes in Arabidopsis (Meyers et al., 2003), 500 in rice (Monosi et al., 2004), 245 in Solanum pimpinellifolium (Wei et al., 2020), 333 in Medicago truncatula (Ameline-Torregrosa et al., 2008), 99 in Raphanus sativus L. (Ma et al., 2021), 400 in Populus trichocarpa (Kohler et al., 2008), 459 in grapevine (Yang et al., 2008), 330 in poplar (Yang et al., 2008), and 536 in upland cotton.2 These R genes play very important roles in plant disease resistance. For example, GbRVd, GbaVd1, GbaVd2, GhPGIP1, and GbaNA1 have been cloned from G. barbadense or G. hirsutum and verified to be involved in the process of plant resistance to VW (Yang et al., 2016; Chen et al., 2017; Liu et al., 2017; Li et al., 2018).
In the present study, the qRT-PCR results indicated that after V. dahliae infection, there is a quick induction of GhDRP and high expression level during 36–72 h in ZZM2, while that in J11 roots was only significantly up-regulated at 72h (Figure 4B). There was the different induction pattern of GhDRP in resistant cultivar ZZM2 and susceptive cultivar J11. In some other studies, GhARPL18A-6, GhDSC1, and GhCAMTA3, which had been proved to be related to cotton disease resistance, were up-regulated especially during the early infection stages 6–24h after inoculation with V. dahliae in resistant cultivar cotton, but no induction was observed in susceptible cultivar (Li et al., 2019; Zhang et al., 2019a), which has the similar induction pattern with GhDRP. Therefore, we infer that activation of cotton immune response may depend on the duration higher expression of resistance gene instead of short-duration stress response or non-expression. Furthermore, GhDRP was cloned from ZZM2 and J11, and it was found that one base change in the DNA sequence led to the change of one amino acid, resulting in the production of proline in ZZM2 and glutamine in J11. Proline is a non-polar R-based amino acid with very low solubility in water and an isoelectric point of 6.30. Glutamine is an uncharged polar R-based amino acid easily soluble in water with an isoelectric point of 5.65. However, it remains unclear whether the alteration of this amino acid is related to the difference between resistant and susceptible varieties of V. dahlia infection, which requires further validation. Next, we will focus on developing molecular markers to validate the mutant sites in other populations, and expect to differentiate disease resistant/susceptible materials through genotyping.
The results of the VIGS experiment revealed that the plants with the silencing of GhDRP were more susceptible to V. dahliae infection than the control according to the phenotypes of plants, DI, fungal biomass, stem recovery assay, and trypan blue staining, indicating that GhDRP may play an important role in resistance to V. dahliae. However, the molecular mechanism underlying the effect requires further exploration. Furthermore, cells in TRV::00 plants showed a higher level of lignification than those in TRV::GhDRP plants after pathogen inoculation, suggesting that the resistance of the plants to V. dahliae infection may be mainly dependent on lignification, which can help to prevent fungal invasion by increasing the mechanical barrier of cell walls (Hückelhoven, 2007). It has been previously confirmed that the lignin content is positively correlated with cotton resistance to V. dahliae (Shi et al., 2012; Guo et al., 2016; Zhang et al., 2019b). However, the molecular mechanism and signal pathway GhDRP involved in remains to be elucidated. And we will conduct further functional verification of GhDRP by transferring the gene into cotton, and clarify its metabolic pathway and mechanism of action. In summary, GhDRP may be used as a candidate gene for VW resistance and has potential application in the breeding of VW resistant cotton varieties.
Data Availability Statement
CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb; https://db.cngb.org/) with accession number CNP0001983.
Author Contributions
YC, QG, HW, and QC conceived and designed the experiments. YC implemented the experiments and prepared the manuscript. PZ guided the molecular experiments. WC and XS collected the field data. QG and YC analyzed the results. QG, PZ, YZ, and HW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Natural Science Foundation of Henan Province (No. 202300410548) and Central Public-Interest Scientific Institution Basal Research Fund (No. 1610162021005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was supported by China National GeneBank (CNGB). We thank Zhu Heqin (Cotton Research Institute, CAAS) for providing verticillium wilt strains. We also thank all for supporting our work.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.703011/full#supplementary-material
Cloning and differential expression of GhDRP in ZZM2 and J11.
Primer sequences used in the study.
RDI of individuals in resistant and susceptible pool in the field and greenhouse environments.
Quality evaluation of raw data and clean data.
Alignment assessment.
References
- Abdelraheem A., Elassbli H., Zhu Y., Kuraparthy V., Hinze L., Stelly D., et al. (2019). A genome-wide association study uncovers consistent quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US upland cotton. Theor. Appl. Genet. 133, 563–577. doi: 10.1007/s00122-019-03487-x, PMID: [DOI] [PubMed] [Google Scholar]
- Ameline-Torregrosa C., Wang B. -B., O'Bleness M. S., Deshpande S., Zhu H., Roe B., et al. (2008). Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 146, 5–21. doi: 10.1104/pp.107.104588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. A., Lawrence G. J., Morrish B. C., Ayliffe M. A., Finnegan E. J., Ellis J. G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. doi: 10.1105/tpc.9.4.641, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah Z. K., Bae J., Jansky S. H., Rouse D. I., Stevenson W. R. (2007). Multiplex real-time quantitative PCR to detect and quantify Verticillium dahliae colonization in potato lines that differ in response to Verticillium wilt. Phytopathology 97, 865–872. doi: 10.1094/PHYTO-97-7-0865, PMID: [DOI] [PubMed] [Google Scholar]
- Bachan S., Dinesh-Kumar S. P. (2012). Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol. Biol. 894, 83–92. doi: 10.1007/978-1-61779-882-5_6 [DOI] [PubMed] [Google Scholar]
- Burpee L. L., Bloom J. R. (1978). The influence of pratylenchus penetrans on the incidence and severity of Verticillium wilt of potato. J. Nematol. 10, 95–99. PMID: [PMC free article] [PubMed] [Google Scholar]
- Caruana B. M., Pembleton L. W., Constable F., Rodoni B., Slater A. T., Cogan N. O. I. (2019). Validation of genotyping by sequencing using transcriptomics for diversity and application of genomic selection in tetraploid potato. Front. Plant Sci. 10:670. doi: 10.3389/fpls.2019.00670, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Kan J., Yang Y., Ling X., Chang Y., Zhang B. (2016). A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. 98, 101–111. doi: 10.1016/j.plaphy.2015.11.015, PMID: [DOI] [PubMed] [Google Scholar]
- Chen J., Li N., Ma X., Gupta V. K., Zhang D., Li T., et al. (2017). The ectopic overexpression of the cotton Ve1 and Ve2-homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 8:844. doi: 10.3389/fpls.2017.00844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yao J., Chu L., Yuan Z., Li Y., Zhang Y. (2015). Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor. Appl. Genet. 128, 539–547. doi: 10.1007/s00122-014-2452-2, PMID: [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 80–92. doi: 10.4161/fly.19695, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger J., Chu Y., Chavarro C., Botton S., Culbreath A., Isleib T. G., et al. (2018). Mapping late leaf spot resistance in peanut (Arachis hypogaea) using QTL-seq reveals markers for marker-assisted selection. Front. Plant Sci. 9:83. doi: 10.3389/fpls.2018.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Wang P., Liu X., Zou C. (2018). QTL mapping for gray leaf spot resistance in maize based on bulk sequencing. Curr. Biotechnol. 8, 317–323. doi: 10.19586/j.2095-2341.2018.0015 [DOI] [Google Scholar]
- Dangl J. L., Jones J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161, PMID: [DOI] [PubMed] [Google Scholar]
- Deokar A., Sagi M., Daba K., Tar’an B. (2019). QTL sequencing strategy to map genomic regions associated with resistance to ascochyta blight in chickpea. Plant Biotechnol. J. 17, 275–288. doi: 10.1111/pbi.12964, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devey M. E., Roose M. L. (1987). Genetic analysis of Verticillium wilt tolerance in cotton using pedigree data from three crosses. Theor. Appl. Genet. 74, 162–167. doi: 10.1007/BF00290099, PMID: [DOI] [PubMed] [Google Scholar]
- Fang W., Xu S., Sun Y., Tang Z., Wang J. (2001). The RAPD maker linked with Verticillium wilt resistance in cotton. Henan Agri. Sci. 9, 11–13. doi: 10.3969/j.issn.1004-3268.2001.09.005 [DOI] [Google Scholar]
- Fradin E. F., Abd-El-Haliem A., Masini L., van den Berg G. C., Joosten M. H., Thomma B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Li F., Li M., Kianinejad A. S., Dever J. K., Wheele T. A., et al. (2013). Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55, 586–596. doi: 10.1111/jipb.12064, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q., Yang Z., Chen E., Sun G., He S., Butt H. I., et al. (2018). A phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 59, 275–289. doi: 10.1093/pcp/pcx180, PMID: [DOI] [PubMed] [Google Scholar]
- Gong Q., Yang Z., Wang X., Butt H. I., Chen E., He S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 17:59. doi: 10.1186/s12870-017-1007-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Jin L., Miao Y., He X., Hu Q., Guo K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6, PMID: [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. (2007). Cell wall–associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325 [DOI] [PubMed] [Google Scholar]
- Hulse-Kemp A. M., Lemm J., Plieske J., Ashrafi H., Buyyarapu R., Fang D. D., et al. (2015). Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3 5, 1187–1209. doi: 10.1534/g3.115.018416, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Shen F., Wang Y., Wu T., Xu X., Zhang X., et al. (2018). Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII. Plant J. 95, 427–443. doi: 10.1111/tpj.13957, PMID: [DOI] [PubMed] [Google Scholar]
- Jian G., Sun W., Ma C. (2001). A new method for identifing the Verticillium wilt resistance: sprinkle the strain suspention on root in non-bottom plastic film bottle. Cotton Sci. 13, 67–69. doi: 10.3969/j.issn.1002-7807.2001.02.001 [DOI] [Google Scholar]
- Jiang F., Zhao J., Zhou L., Guo W., Zhang T. (2009). Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton. Sci. China Life Sci. 52, 872–884. doi: 10.1007/s11427-009-0110-8, PMID: [DOI] [PubMed] [Google Scholar]
- Jun Z., Zhang Z., Gao Y., Zhou L., Fang L., Chen X., et al. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5:15048. doi: 10.1038/srep15048, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z., King R. C., Mohareb F., Sergeant M. J., Awan S. Z., Thompson A. J. (2015). Resequencing at ≥40-fold depth of the parental genomes of a Solanum lycopersicum × S. pimpinellifolium recombinant inbred line population and characterization of frame-shift InDels that are highly likely to perturb protein function. G3 5, 971–981. doi: 10.1534/g3.114.016121, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., Xiao Y., Conklin P. A., Govindarajulu R., Kelly J. A., Scanlon M. J., et al. (2018). Bulked-segregant analysis coupled to whole genome sequencing (BSA-Seq) for rapid gene cloning in maize. G3 8, 3583–3592. doi: 10.1534/g3.118.200499, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Rinaldi C., Duplessis S., Baucher M., Geelen D., Duchaussoy F., et al. (2008). Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 66, 619–636. doi: 10.1007/s11103-008-9293-9, PMID: [DOI] [PubMed] [Google Scholar]
- Kuang M., Yang W., Xu H., Wang Y., Zhou D., Feng X. (2010). A rapid method of DNA extraction from single cotton seed. Mol. Plant Breed. 8, 827–831. doi: 10.3969/mpb.008.000827 [DOI] [Google Scholar]
- Lee S. B., Kim J. E., Kim H. T., Lee G. M., Kim B. S., Lee J. M. (2020). Genetic mapping of the c1 locus by GBS-based BSA-seq revealed pseudo-response regulator 2 as a candidate gene controlling pepper fruit color. Theor. Appl. Genet. 133, 1897–1910. doi: 10.1007/s00122-020-03565-5, PMID: [DOI] [PubMed] [Google Scholar]
- Li N., Ma X., Short D. P. G., Li T., Zhou L., Gui Y., et al. (2018). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. doi: 10.1111/mpp.12630, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wang B., Yin C., Zhang D., Wang D., Song J., et al. (2019). The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol. Plant Pathol. 20, 857–876. doi: 10.1111/mpp.12797, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Chi W., Huang L., Qu M., Zhang S., Chen Z., et al. (2020). Bulked segregant analysis coupled with whole-genome sequencing (BSA-Seq) mapping identifies a novel pi21 haplotype conferring basal resistance to rice blast disease. Int. J. Mol. Sci. 21:2162. doi: 10.3390/ijms21062162, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhang X., Sun Y., Wang P., Li X., Pei Y., et al. (2017). Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 7:39840. doi: 10.1038/srep39840, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262, PMID: [DOI] [PubMed] [Google Scholar]
- Luck J. E., Lawrence G. J., Dodds P. N., Shepherd K. W., Ellis J. G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12, 1367–1377. doi: 10.1105/tpc.12.8.1367, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Chhapekar S. S., Lu L., Oh S., Singh S., Kim C. S., et al. (2021). Genome-wide identification and characterization of NBS-encoding genes in Raphanus sativus L. and their roles related to Fusarium oxysporum resistance. BMC Plant Biol. 21:47. doi: 10.1186/s12870-020-02803-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zheng Z., Lin F., Ge T., Sun H. (2018). Genetic analysis and gene mapping of a low stigma exposed mutant gene by high-throughput sequencing. PLoS One 13:e0186942. doi: 10.1371/journal.pone.0186942, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg M. M., Shi F., Spangenberg G. C., Daetwyler H. D., Cogan N. O. I. (2018). Diversity and genome analysis of Australian and global oilseed Brassica napus L. germplasm using transcriptomics and whole genome re-sequencing. Front. Plant Sci. 9:508. doi: 10.3389/fpls.2018.00508, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld B. N., Grumet R. (2018). QTLseqr: An R package for bulk segregant analysis with nextygeneration sequencing. Plant Genome 11:180006. doi: 10.3835/plantgenome2018.01.0006 [DOI] [PubMed] [Google Scholar]
- Md V., Misra S., Li H., Aluru S. (2019). “Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems.” in 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS); May 20–24, 2019, 314–324.
- Meyers B. C., Kozik A., Griego A., Kuang H., Michelmore R. W. (2003). Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H., Wang X., Zhang Y., Zhang G., Zhang J., Ma Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941, PMID: [DOI] [PubMed] [Google Scholar]
- Monosi B., Wisser R. J., Pennill L., Hulbert S. H. (2004). Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 109, 1434–1447. doi: 10.1007/s00122-004-1758-x, PMID: [DOI] [PubMed] [Google Scholar]
- Palanga K. K., Jamshed M., Rashid M. H. O., Gong J., Li J., Iqbal M. S., et al. (2017). Quantitative trait locus mapping for Verticillium wilt resistance in an upland cotton recombinant inbred line using SNP-based high density genetic map. Front. Plant Sci. 8:382. doi: 10.3389/fpls.2017.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M. K., Khan A. W., Singh V. K., Vishwakarma M. K., Shasidhar Y., Kumar V., et al. (2017). QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol. J. 15, 927–941. doi: 10.1111/pbi.12686, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J., Zhu Y., Li Q., Liu J., Tian Y., Liu Y., et al. (2013). Development of agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One 8:e73211. doi: 10.1371/journal.pone.0073211, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. E., Coleman M. J., Szabò V., Frost L. N., Schmid R., Biezen E. A., et al. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. doi: 10.1105/tpc.9.6.879, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Liu Z., Zhu L., Zhang C., Chen Y., Zhou Y., et al. (2012). Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 44, 555–564. doi: 10.1093/abbs/gms035, PMID: [DOI] [PubMed] [Google Scholar]
- Shi Y., Zhang B., Liu A., Li W., Li J., Lu Q., et al. (2016). Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum×Gossypium barbadense. BMC Genomics 17:877. doi: 10.1186/s12864-016-3128-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. K., Khan A. W., Jaganathan D., Thudi M., Roorkiwal M., Takagi H., et al. (2016). QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 14, 2110–2119. doi: 10.1111/pbi.12567, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Li Z., Liu Z., Guo Y., Qiu L. (2017). Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 8:919. doi: 10.3389/fpls.2017.00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qi F., Zhang W., Jian G. (2010). Analysis on the inheritance of resistance to Verticillium wilt of Zhongzhimian KV-3. Cotton Sci. 22, 501–504. doi: 10.3969/j.issn.1002-7807.2010.05.019 [DOI] [Google Scholar]
- Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., et al. (2018). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x, PMID: [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang X., Li Y., Nie Y. (2004). Analysis on the inheritance of Verticillium dahliae resistance in G. hirsutum. Cotton Sci. 16, 84–88. doi: 10.3969/j.issn.1002-7807.2004.02.004 [DOI] [Google Scholar]
- Wei H., Liu J., Guo Q., Pan L., Chai S., Cheng Y., et al. (2020). Genomic organization and comparative phylogenic analysis of NBS-LRR resistance gene family in Solanum pimpinellifolium and Arabidopsis thaliana. Evol. Bioinforma. 16:1176934320911055. doi: 10.1177/1176934320911055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ma Q., Zhang Y., Wang X., Zhang G., Ma Z. (2016). Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694. doi: 10.1016/j.gene.2015.09.046 [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang X., Yue J., Tian D., Chen J. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Gen. Genomics. 280, 187–198. doi: 10.1007/s00438-008-0355-0, PMID: [DOI] [PubMed] [Google Scholar]
- Yin J., Fang Z., Sun C., Zhang P., Zhang X., Lu C., et al. (2018). Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 8:3086. doi: 10.1038/s41598-018-21489-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Xie Y., Liu G., Wei Y., Hu W., Shi H. (2019). Agrobacterium-mediated gene transient overexpression and tobacco rattle virus (TRV)-based gene silencing in cassava. Int. J. Mol. Sci. 20:3976. doi: 10.3390/ijms20163976, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fang H., Zhou H., Sanogo S., Ma Z. (2014a). Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 54, 1289–1303. doi: 10.2135/cropsci2013.08.0550 [DOI] [Google Scholar]
- Zhang Y., Jin Y., Gong Q., Li Z., Zhao L., Han X., et al. (2019a). Mechanismal analysis of resistance to Verticillium dahliae in upland cotton conferred by overexpression of RPL18A-6 (ribosomal protein L18A-6). Ind. Crop. Prod. 141:111742. doi: 10.1016/j.indcrop.2019.111742 [DOI] [Google Scholar]
- Zhang Y., Wang X., Li Y., Wu L., Zhou H., Zhang G., et al. (2013). Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 32, 1703–1713. doi: 10.1007/s00299-013-1481-7, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang P., Luo X., Yang C., Tang Y., Wang Z., et al. (2019b). Cotton plant defence against a fungal pathogen is enhanced by expanding BLADE-ON-PETIOLE1 expression beyond lateral-organ boundaries. Commun. Biol. 2:238. doi: 10.1038/s42003-019-0468-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Yang S., Chi J., Zhang G., Ma Z. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. doi: 10.1007/s00299-011-1115-x, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Yu J., Pei W., Li X., Said J., Song M., et al. (2015). Genetic analysis of Verticillium wilt resistance in a backcross inbred line population and a meta-analysis of quantitative trait loci for disease resistance in cotton. BMC Genomics 16:577. doi: 10.1186/s12864-015-1682-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y., Wei Z., Guo X., Guo Y., Zhang S., et al. (2014b). Molecular mapping and validation of a major QTL conferring resistance to a defoliating isolate of Verticillium wilt in cotton (Gossypium hirsutum L.). PLoS One 9:e96226. doi: 10.1371/journal.pone.0096226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang H., Liu K., Jian G., Qi F., Ning S. (2017). Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis. PLoS One 12:e0181609. doi: 10.1371/journal.pone.0181609, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang B., Yang Y., Chen T., Yu W., Liu T., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One 7:e51091. doi: 10.1371/journal.pone.0051091, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen W., Cui Y., Sang X., Lu J., Jing H., et al. (2021). Detection of candidate genes and development of KASP markers for Verticillium wilt resistance by combining genome-wide association study, QTL-seq and transcriptome sequencing in cotton. Theor. Appl. Genet. 134, 1063–1081. doi: 10.1007/s00122-020-03752-4, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao T., Jiang J., Liu G., He S., Zhang H., Chen X., et al. (2016). Mapping and candidate gene screening of tomato Cladosporium fulvum-resistant gene Cf-19, based on high-throughput sequencing technology. BMC Plant Biol. 16:51. doi: 10.1186/s12870-016-0737-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang H., Chen W., Li Y. (2014). Genetic structure, linkage disequilibrium and association mapping of Verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PLoS One 9:e86308. doi: 10.1371/journal.pone.0086308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang H., Chen W., Zhao P., Gong H., Sang X., et al. (2017). Regional association analysis-based fine mapping of three clustered QTL for Verticillium wilt resistance in cotton (G. hirsutum. L). BMC Genomics 18:661. doi: 10.1186/s12864-017-4074-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhao G., Geng Z., Wang Z., Wang K., Liu S., et al. (2018). Physical mapping and candidate gene prediction of fertility restorer gene of cytoplasmic male sterility in cotton. BMC Genomics 19:6. doi: 10.1186/s12864-017-4406-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen R., Wang X., Ma Z., Zhang G., Wang X. (2006). A SSR marker linked with the gene of Verticillium wilt resistance in Gossypium barbadense. Cotton Sci. 18, 269–272. doi: 10.3969/j.issn.1002-7807.2006.05.003 [DOI] [Google Scholar]
- Zhu J., Chen J., Gao F., Xu C., Wu H., Chen K., et al. (2017). Rapid mapping and cloning of the virescent-1 gene in cotton by bulked segregant analysis–next generation sequencing and virus-induced gene silencing strategies. J. Exp. Bot. 68, 4125–4135. doi: 10.1093/jxb/erx240, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Feng Z., Li Z., Zhao L., Shi Y. (2010). Identification of Verticillium wilt resistance of cotton variety (line) by quantitative dipping bacterial solution in vermiculite sand soil. China Cotton 37, 15–17. doi: 10.3969/j.issn.1000-632X.2010.12.006 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning and differential expression of GhDRP in ZZM2 and J11.
Primer sequences used in the study.
RDI of individuals in resistant and susceptible pool in the field and greenhouse environments.
Quality evaluation of raw data and clean data.
Alignment assessment.
Data Availability Statement
CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb; https://db.cngb.org/) with accession number CNP0001983.