Abstract
Mucopolysaccharidosis type IIIA (MPS IIIA) is characterised by a progressive neurological decline leading to early death. It is caused by bi-allelic loss-of-function mutations in SGSH encoding sulphamidase, a lysosomal enzyme required for heparan sulphate glycosaminoglycan (HS GAG) degradation, that results in the progressive build-up of HS GAGs in multiple tissues most notably the central nervous system (CNS). Skin fibroblasts from two MPS IIIA patients who presented with an intermediate and a severe clinical phenotype, respectively, were reprogrammed into induced pluripotent stem cells (iPSCs). The intermediate MPS IIIA iPSCs were then differentiated into neural progenitor cells (NPCs) and subsequently neurons. The patient derived fibroblasts, iPSCs, NPCs and neurons all displayed hallmark biochemical characteristics of MPS IIIA including reduced sulphamidase activity and increased accumulation of an MPS IIIA HS GAG biomarker. Proliferation of MPS IIIA iPSC-derived NPCs was reduced compared to control, but could be partially rescued by reintroducing functional sulphamidase enzyme, or by doubling the concentration of the mitogen fibroblast growth factor 2 (FGF2). Whilst both control heparin, and MPS IIIA HS GAGs had a similar binding affinity for FGF2, only the latter inhibited FGF signalling, suggesting accumulated MPS IIIA HS GAGs disrupt the FGF2:FGF2 receptor:HS signalling complex. Neuronal differentiation of MPS IIIA iPSC-derived NPCs was associated with a reduction in the expression of neuronal cell marker genes βIII-TUBULIN, NF-H and NSE, revealing reduced neurogenesis compared to control. A similar result was achieved by adding MPS IIIA HS GAGs to the culture medium during neuronal differentiation of control iPSC-derived NPCs. This study demonstrates the generation of MPS IIIA iPSCs, and NPCs, the latter of which display reduced proliferation and neurogenic capacity. Reduced NPC proliferation can be explained by a model in which soluble MPS IIIA HS GAGs compete with cell surface HS for FGF2 binding. The mechanism driving reduced neurogenesis remains to be determined but appears downstream of MPS IIIA HS GAG accumulation.
Keywords: Induced pluripotent stem cells, MPS IIIA, Neurogenesis, Heparan sulphate glycosaminoglycan, Fibroblast growth factor 2
Abbreviations: GAG, glycosaminoglycan; HS, heparan sulphate; MPS IIIA, mucopolysaccharidosis type IIIA; FGF, Fibroblast growth factor; iPSC, induced pluripotent stem cell; NPC, neural progenitor cell; MEF, mouse embryonic fibroblast; CNS, central nervous system
Highlights
-
•
Induced pluripotent stem cells (iPSCs) were generated from MPS IIIA skin fibroblasts.
-
•
MPS IIIA iPSCs were successfully differentiated into neural progenitor cells (NPCs).
-
•
MPS IIIA NPC proliferation was reduced due to interference with FGF2 signalling.
-
•
Neurogenesis from MPS IIIA iPSC-derived NPCs is reduced via an unknown mechanism.
1. Introduction
MPS IIIA (Sanfilippo syndrome type A, OMIM# 252900) is an inherited metabolic disorder arising from bi-allelic mutations in SGSH resulting in a partial or total loss of sulphamidase activity (N-sulphoglucosamine sulphohydrolase, EC 3.10.1.1). Sulphamidase is one of a group of lysosomal enzymes that act in concert to degrade heparan sulphate (HS) glycosaminoglycan (GAG) [1]. The cumulative build-up of undegraded and/or partially degraded HS GAG within cells over time adversely affects tissue and organ function. Like the other MPS III subtypes, MPS IIIA is characterised by a severe neurological phenotype combined with a milder peripheral phenotype. Significant HS storage is observed in MPS IIIA patient brain [2] and can be detected in neurons as early as the second trimester [3]. An early clinical presentation of delayed development and hyperactivity is followed by increased aggression and a loss of motor and cognitive skills [4]. Behavioural issues become less prominent as neurodegeneration progresses and patients with the severe form of MPS IIIA die in their second or third decade from inanition [1], [4]. Patients with higher residual enzyme activity have an attenuated phenotype with a slower trajectory of neurodegenerative symptoms and a longer life span.
The carbohydrate structure of HS that builds up in MPS IIIA tissues and fluids is distinct to normal HS. In particular, a higher degree of sulphation is observed in both MPS IIIA patients and in MPS IIIA mice [5], [6]. Both a higher degree of sulphation [5] and higher levels of stored HS GAG [5], [7], [8] correlate to disease severity. Although the correlation between HS accumulation and CNS dysfunction is clear, the influence of increased lysosomal accumulation of HS and the increased extracellular level of partially degraded HS on disease pathogenesis is unknown.
Treatment for MPS IIIA is challenging as systemic approaches are unsuccessful, with sulphamidase unable to cross the mature blood brain barrier [9]. Bone marrow transplants (BMT) or hematopoietic stem cell transplants (HSCT) are also contraindicated for MPS IIIA although genetically modified HSCTs overexpressing sulphamidase have shown promise [10] and are in clinical trial (ClinicalTrials.gov Identifier: NCT04201405). Systemically administered gene therapy using an AAV9 vector capable of crossing the blood brain barrier is also under investigation and has been reported to improve a number of clinical biomarkers and functional outcome in behavior in preclinical trials in the MPS IIIA mouse [11], [12], [13]. The positive improvements observed in preclinical studies have enabled Phase I/II clinical trials in MPS III patients (NCT02716246). An important issue that may limit this type of gene therapy is pre-exposure to the vector in a number of patients which can lead to an acquired adverse immune response [14]. The ability to effectively treat the neurological symptoms experienced by MPS IIIA children would benefit from an improved understanding of how enzyme loss and the subsequent build-up of HS GAG affects CNS development and function.
Induced pluripotent stem cells (iPSCs) derived from patient somatic cells have the capacity to generate cells of the brain and are thus useful for modeling neurodegenerative diseases including the MPS disorders [15]. Skin fibroblasts are the most common somatic cell type used to generate iPSCs and are available from many MPS patients, as skin fibroblasts are commonly required for disease diagnosis. iPSCs have been successfully generated from somatic cells of patients with MPS I [16], MPS II [17], [18], [19], [20], [21], MPS IIIA [22], MPS IIIB [23], [24], [25], MPS IIIC [26], MPS IVA [27] and MPS VII [28], [29], an unaffected MPS II carrier [30] and a murine model of MPS VII [31]. iPSCs derived from human MPS I [16], MPS II [17], [21], MPS IIIB [23], [25], MPS IIIC [26] and MPS VII [28] skin fibroblasts maintained key molecular hallmarks of the disease phenotype, exhibiting a significant reduction in activity of the relevant enzyme and an increase in stored GAG. MPS IIIB, MPS IIIC and MPS VII iPSCs also displayed altered neuronal function upon differentiation [26], [28] with two cell lines unable to differentiate in NPCs and mature neurons in MPS IIIB [25]. To date this is the most comprehensive analysis of biochemical characteristics and differentiation capacity using MPS IIIA iPSCs. This study seeks to generate iPSCs from two different MPS IIIA skin fibroblast cell lines and to examine in detail the ability of one of the MPS IIIA iPSC lines to differentiate down the neurological lineage.
2. Methods
2.1. Generation of induced pluripotent stem cells from MPS IIIA skin fibroblasts
All procedures involving the acquisition of skin fibroblasts, their reprogramming into iPSCs and subsequent characterisation were approved by the Women's and Children's Hospital Network human ethics committee (Adelaide, SA, Australia). MPS IIIA fibroblasts from two patients were sourced from the National Referral Laboratory at the Women's and Children's Hospital, SA, Australia. The genotype of patient 1 involved compound heterozygous SGSH missense mutations c.892 T > C; c.1298G > A (p.S298P; p.R433Q), and was associated with an MPS IIIA intermediate phenotype [32], [33], [34]. The genotype of patient 2 involved a homozygous nonsense mutation c.672C > A (p.Y224*), and was associated with a severe phenotype as defined by residual enzyme activity and patient records (Genetics and Molecular Pathology, Women's and Children's Hospital patient records). Control fibroblasts derived from healthy individuals were purchased from the Coriell Institute, USA. Fibroblasts were cultured in DMEM (high glucose, Thermo-Fisher) supplemented with 10% (v/v) foetal calf serum (FCS, Thermo-Fisher), 50 U/mL penicillin and 50 μg/mL streptomycin and maintained at 37 °C in 5% CO2 and 90% humidity. Skin fibroblasts were seeded into 6-well plates and reprogrammed to iPSCs by transduction with Sendai virus vectors expressing the Yamanaka factors, Oct3/4, Sox2, Klf4 and c-Myc [35] using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Life Technologies). Three weeks post-transduction, single colonies with embryonic stem cell (ESC)-like morphology were manually picked and transferred to irradiated mouse embryonic fibroblast (MEF, Thermo Fisher) coated 6-well plates, with one colony per well. iPSCs were cultured in DMEM/F-12 (+ GlutaMAX, Life Technologies) supplemented with 20% (v/v) KnockOut ™ Serum Replacement (Life Technologies), 100 μM non-essential amino acids (Sigma), 55 μM β-mercaptoethanol (Sigma), 20 ng/mL FGF-2 (Life Technologies), 50 U/mL penicillin and 50 μg/mL streptomycin. Cells were maintained at 37 °C in 5% CO2 and 90% humidity, with media changed daily. Cells were passaged manually approximately every five days onto fresh MEF plates. After a minimum of 13 passages multiple clones were obtained from each genotype. Of these four control, two MPS IIIA-intermediate and three MPS IIIA-severe iPSC clones were sent for karyotype analysis. G-band chromosome analysis of iPSCs was performed by SA Health (Cytogenetics, Women's and Children's Hospital, Adelaide). One karyotypically normal clone of each cell line was chosen for further analysis.
2.2. Feeder-free iPSC culture
iPSC cultures were transferred to feeder-free culture conditions prior to characterisation and neural differentiation. 6-well plates were coated with 10 μg/mL vitronectin in CellAdhere Dilution Buffer (StemCell Technologies) for one hour at room temperature. Wells were washed once with CellAdhere Dilution Buffer, following which 2 mL TeSR-E8 media (StemCell Technologies) was added to each well. iPSCs were manually passaged from MEF plates to prepared vitronectin-coated plates and incubated overnight at 37 °C. Feeder-free iPSCs were maintained at 37 °C in 5% CO2 and 90% humidity, with TeSR-E8 media changed daily. Feeder-free iPSCs were passaged approximately every five days with Gentle Cell Dissociation Reagent (StemCell Technologies) onto fresh vitronectin-coated plates as per the manufacturer's instructions.
2.3. iPSC differentiation to NPCs
iPSCs maintained in feeder-free conditions were differentiated to NPCs as per the cortical neural differentiation protocol established by Shi et al. 2012 [36] and optimised by Homan et al. 2018 [37]. Following single cell dissociation of NPCs with Accutase (StemCell Technologies) [36], cells were plated on ECM-coated NUNC 6-well plates or flasks in neural expansion media consisting of a 1:1 ratio of Neurobasal and DMEM/F-12 (+ GlutaMAX) supplemented with 20 ng/mL FGF2 (Life Technologies), 1 x B27 supplement (Life Technologies), 1 x N2 supplement (Life Technologies), 5 μg/mL insulin, 100 μM non-essential amino acids, 100 μM β-mercaptoethanol, 1 mM l-glutamine, 50 U/mL penicillin and 50 μg/mL streptomycin. To prepare ECM-coated 6-well plates and flasks, ECM was diluted 1:100 in cold DMEM/F-12 (+ GlutaMAX), added to 6-well plates or flasks and incubated for two hours to overnight at 37 °C. Wells and flasks were washed once with PBS immediately prior to use. Cells were plated at 2.1 × 105 cells per cm2 and incubated at 37 °C. Cells were maintained at 37 °C in 5% CO2 and 90% humidity, with media changed every two days. At 90–100% confluency, iPSC-derived NPCs were passaged by washing once with PBS followed by incubation with 70 μL per cm2 pre-warmed Accutase per well/flask for up to ten minutes or until cells were forming single cells and detaching from the surface. An equal volume of neural expansion media was added to neutralise the Accutase. Cells were centrifuged at 120 xg for three minutes and resuspended in neural expansion media. Cells were plated at 2.1 × 105 cells per cm2 on ECM-coated plates and flasks for further expansion.
2.4. iPSC-derived NPC neurogenic differentiation
NUNC 6-well plates were coated with 100 μg/mL poly-L-ornithine (Sigma) for four hours at 37 °C. Wells were washed twice with PBS and coated in 20 μg/mL laminin (Life Technologies) overnight at 37 °C. Wells were washed once with Neurobasal media immediately prior to use. iPSC-derived NPCs were harvested at passage six with Accutase and plated at 5.0 × 104 cells per cm2 onto poly-L-ornithine/laminin-coated plates in neural expansion media. Cells were setup in triplicate at each time point and incubated at 37 °C overnight before transfer into neural maintenance media consisting of a 1:1 ratio of Neurobasal and DMEM/F-12 (+ GlutaMAX) supplemented with 1× B27 supplement, 1× N2 supplement, 5 μg/mL insulin, 100 μM non-essential amino acids, 100 μM β-mercaptoethanol, 1 mM l-glutamine, 50 U/mL penicillin and 50 μg/mL streptomycin. Cells were fed with neural maintenance media 3, 5, 7 and 10 days post-seeding. From 13 days post-seeding, cells were fed every three days by removing 1 mL old neural maintenance media and gently feeding with 1 mL fresh neural maintenance media. For gene expression analysis, RNA was isolated 14, 21 and 28 days post-induction with TRIzol (Life Technologies) followed by RNA clean-up with the RNeasy Mini Kit (Qiagen) as per the manufacturer's instructions and expression of neural marker genes was determined by real-time PCR. In addition HS isolated from MPS IIIA urine as previously described [38] was added to the culture medium of control iPSC-derived NPCs undergoing neural differentiation at a concentration of 3 μg/mL for the entire culture period. On day 28 only RNA was isolated as described above for gene expression analysis.
2.5. Sulphamidase activity assay and measurement of primary and secondary storage product
Sulphamidase activity was determined on cell extracts of human fibroblasts, iPSCs, iPSC-derived NPCs and differentiated neurons. Harvested cells were sonicated in PBS for ~10 s and frozen at −80 °C prior to analysis. Protein content was determined using the Bicinchoninic Acid (BCA) Assay Kit (Sigma Aldrich) or micro Lowry assay [39]. Sulphamidase activity was determined on samples containing 5–10 μg protein using the fluorogenic substrate 4-methylumbelliferyl-alpha-D-N-sulphoglucosaminide as described [40], with the first incubation decreased from 17 h to 4.5 h. Activity was expressed as pmol substrate converted/h per mg protein. Samples corresponding to 0.1 mg protein were derivatised with 1-phenyl-3-methyl-5-pyrazolone (PMP) and the concentration of the monosulphated disaccharide (HN-UA (1S)), a specific biomarker of MPS IIIA HS [41] was determined by mass spectrometry as previously described [42]. Results were expressed as pmol HN-UA (1S) per mg protein. Gangliosides were quantified by mass spectrometry as previously described [43] and expressed as pmol total GM2 or GM3 ganglioside/mg protein.
2.6. iPSC-NPC proliferation and rescue assay
MPS IIIA-intermediate iPSC-derived NPCs (MPS IIIA-intermed iPSC-derived NPCs) were transduced with lentivirus encoding sulphamidase at an MOI of 1. After two days cells were harvested with Accutase and set up in the proliferation assay along with control and untransduced MPS IIIA-intermed iPSC-derived NPCs. All cells were plated at a density of 1.50 × 105 cells per 24-well coated with ECM in 0.5 mL neural expansion media. Cells were maintained at 37 °C in 5% CO2 and 90% humidity, with media changed every two days. Additionally, triplicate wells of MPS IIIA-intermed NPCs were incubated in the presence of a 2-fold concentration of FGF2 to give a final concentration of 40 ng/mL. On day seven cells were harvested with Accutase and cell number counted using trypan blue and the automated cell counter Countess™.
2.7. Fibroblast growth factor (FGF2) binding to and signalling in the presence of MPS IIIA GAGs
The binding of MPS IIIA GAGs to FGF2 was analysed in the Blitz ® system (ForteBio) using protein G sensors and operated with PBS, pH 7.4 containing 1% (v/v) Tween-20. Sensors were equilibrated in buffer for 30 s and then exposed to anti-FGF2 antibody (1:1000, rabbit polyclonal anti-FGF2, R&D Systems) for 120 s followed by rinsing in buffer for 30 s. Sensors were exposed to 50 μg/mL FGF2 for 120 s followed by rinsing in buffer for 30 s and then exposed to MPS IIIA GAGs or heparin (1–20 μg/mL; Sigma H3393) for 120 s and rinsed with buffer for 30 s. The affinity, KD, between FGF2 and either MPS IIIA GAGs or heparin was determined assuming 1:1 binding. Signalling via the fibroblast growth factor receptor (FGFR) was determined using the BaF32 cell proliferation assay which relies on the formation of an active ternary complex between GAG, FGF2 and FGFR1c [44]. The assay was performed as described previously [45] in the presence of 0.03 nM FGF2 and either unfractionated heparin (0–0.5 μg/mL; Sigma H3393) or MPS IIIA GAG (0–2 μg/mL). The relative number of cells present was determined using the MTS reagent (Promega, Madison, Wisconsin, USA) by addition to the cell cultures for 6 h prior to measurement of absorbance at 490 nm.
2.8. Pluripotency gene expression
Fibroblasts were plated at 2.0 × 104 cells per cm2 in triplicate in 6-well plates in DMEM (high glucose) supplemented with 10% (v/v) foetal calf serum (FCS), 50 U/mL penicillin and 50 μg/mL streptomycin. iPSC colonies were plated in triplicate on vitronectin-coated 6-well plates in TeSR-E8 media. iPSC-derived NPCs were plated at 2.1 × 105 cells per cm2 in triplicate in ECM-coated 6-well plates in neural expansion media. RNA was isolated as described previously and concentration determined using a nanodrop spectrophotometer (Thermo Scientific, USA). Between 100 ng and 500 ng of RNA was reverse transcribed to cDNA using the QuantiTect Reverse Transcriptase Kit (Qiagen). Exon-exon boundary gene specific primers were designed for all neural genes. Primers for OCT-4, NANOG and SOX-2 were as previously described [37] and βIII-TUBULIN as per [46]. All other primer sequences are shown in Table 1, with cyclophilin A included as the housekeeping gene. Each 25 μL real-time PCR reaction included 1 μL cDNA, 1× SYBR™ Green PCR Mastermix (Life Technologies), 0.45 μM forward primer and 0.45 μM reverse primer. Real-time reactions were carried out on an ABI 7300 thermocycler (Applied Biosystems) with initial steps of one cycle for two minutes at 50 °C and one cycle for ten minutes at 95 °C. cDNA amplification consisted of 40 cycles of 15 s at 95 °C and one minute at 60 °C. A dissociation step of one cycle for 15 s at 95 °C, 30 s at 60 °C and 15 s at 95 °C was added after the amplification step. The 2-ΔΔCt method was used to determine the fold change in gene expression [47]. The relative expression method was used to determine mean normalised gene expression for iPSC-derived NPC gene expression [48].
Table 1.
Primer sequences.
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| PAX6 | CCAGGGCAACCTACGCAA | CTGAATCTTCTCCGTTGGAACT |
| NESTIN | CGCACCTCAAGATGTCCCTC | CAGCTTGGGGTCCTGAAAGC |
| NF-H | GGACCTGCTCAATGTCAAGATG | GCCAAAGCCAATCCGACAC |
| NSE | GGGCACTCTACCAGGACTTTG | CCCTACATTGGCTGTGAACT |
| CYCLOPHILIN A | TCCTAAAGCATACGGGTCCT | CTTGCCATCCAACCACTCA |
2.9. Immunofluorescence
iPSCs were plated in triplicate on vitronectin-coated 35 mm dishes (Nunc) in TeSR-E8 media and iPSC-NPCs were plated in triplicate at 5.0 × 104 cells per cm2 on ECM-coated coverslips in 12-well plates. Cells were fixed in 4% (w/v) paraformaldehyde for 15 min at room temperature and washed three times in PBS. Immunofluorescent detection of OCT-4 and SSEA4 on 4% (w/v) paraformaldehyde-fixed iPSCs was undertaken using the PSC 4-Marker Immunocytochemistry Kit (Life Technologies), while immunofluorescent detection of PAX6 and NESTIN was as previously described [37].
2.10. Statistics
The statistical significance of differences between means was determined using a Student's t-test, a one-way analysis of variance (ANOVA) or a two-way ANOVA followed by Tukey's HSD post-hoc test as appropriate in GraphPad Prism Version 7.03 for Windows (GraphPad Software., USA).
3. Results
3.1. Generation and characterisation of iPSCs from MPS IIIA skin fibroblasts
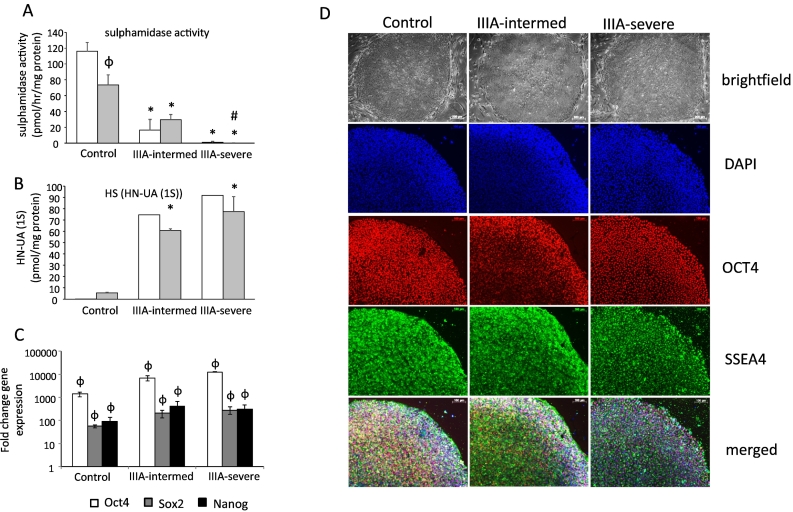
iPSCs were successfully formed from control and MPS IIIA skin fibroblasts. Sulphamidase activity was significantly reduced in MPS IIIA fibroblasts compared to control fibroblasts, mirroring the enzyme deficiency of MPS IIIA. Control sulphamidase activity was 116.1 ± 11.31 pmol/h/mg protein, MPS IIIA intermediate sulphamidase activity was 16.4 ± 13.14 pmol/h/mg protein and MPS IIIA severe sulphamidase activity was 0.85 ± 1.13 pmol/h/mg protein (Fig. 1A). Conversely, the level of HN-UA (1S), a marker of HS storage, increased from undetectable in control fibroblasts to 75 pmol/mg protein in MPS IIIA intermediate fibroblasts and 92 pmol/mg in MPS IIIA severe fibroblasts (Fig. 1B). This pattern of decreased sulphamidase activity and increased HS storage was maintained when fibroblasts were reprogrammed to iPSCs. Control iPSC sulphamidase activity and HS storage was 73.46 ± 12.74 pmol/h/mg protein and 5.5 pmol/mg protein, respectively. MPS IIIA-intermediate sulphamidase activity and HS storage was 29.43 ± 6.62 pmol/h/mg protein and 61 pmol/mg protein respectively. MPS IIIA-severe sulphamidase activity and HS storage was 0.07 ± 0.12 pmol/h/mg protein and 77.5 pmol/mg protein, respectively (Fig. 1A-B). All resulting iPSCs displayed typical colony morphology (Fig. 1D). Significant increases in the expression of pluripotency marker genes OCT-4, SOX2 and NANOG compared to the original fibroblasts from which the iPSCs were derived provided evidence of pluripotency (Fig. 1C) and this was supported by positive OCT-4 and SSEA4 immunostaining of colonies (Fig. 1D). Collectively, these data reveal no difference in iPSC morphologies or pluripotency marker gene profiles between control and MPS IIIA iPSCs.
Fig. 1.
Generation and characterisation of iPSCs. Biochemical markers of sulphamidase activity (A) and the monsulphated dissacharide HS storage biomarker HN-UA(1S) (B) were measured in cell extracts of skin fibroblasts (clear boxes) and iPSCs (grey boxes) and expressed relative to cell protein. Quantitative data is expressed as the mean ± std. dev of n = 3 replicates (A and C). HN-UA(1S) biomarker (B) was expressed as a single value for skin fibroblasts or the average of 2 replicates for iPSCs where the individual values are:- Control skin fibroblasts 0.2 and 0.4 pmol/mg protein; MPS IIIA-intermed fibroblasts 75 pmol/mg protein; MPS IIIA-severe fibroblasts 92 pmol/mg protein. Control iPSCs 5 and 6 pmol/mg protein; MPS IIIA-intermed iPSCs 62 and 60 pmol/mg protein; MPS IIIA-severe iPSCs 87 and 68 pmol/mg protein.
iPSC gene expression of pluripotency markers Oct4, Sox2 and Nanog were all significantly elevated compared in iPSCs compared to skin fibroblasts (C) and morphology was shown by brightfield microscopy while pluripotency markers typical of iPSCs were assessed by immunofluorescence (D). Scale bars are 100 μm as indicated.
φ denotes significant difference between iPSCs and skin fibroblasts, P < 0.05.
* denotes significant difference between MPS IIIA and control, P < 0.05.
# denotes significant difference between MPS IIIA-intermed and MPS IIIA-severe, P < 0.05.
3.2. Generation and characterisation of iPSC-derived neural progenitor cells
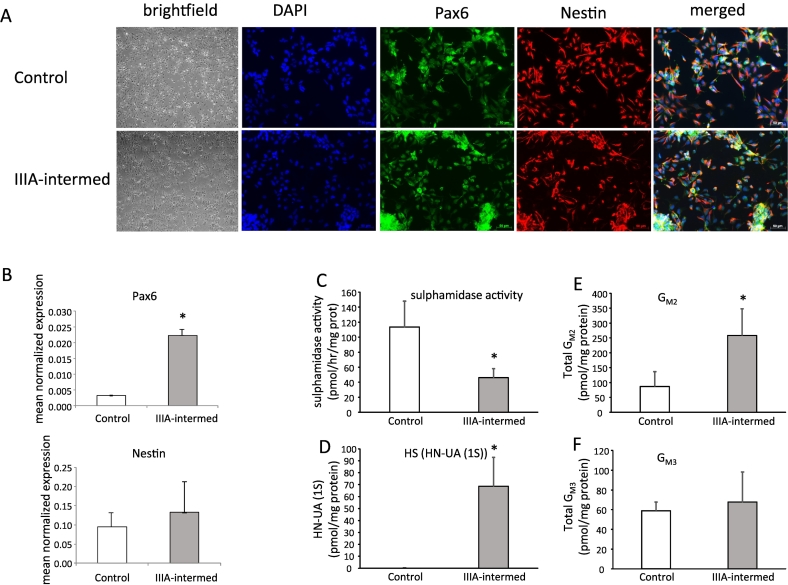
iPSCs from patient 1 with an intermediate severity phenotype: (MPS IIIA-intermed iPSCs) were chosen for further analysis and along with control iPSCs were differentiated along the neural lineage to form neural progenitor cells (NPCs). Both control and MPS IIIA-intermed iPSCs were successfully differentiated into NPCs as initially evidenced by the formation of hallmark ‘neural rosette’ colonies. Unfortunately, and for currently unknown reasons, across multiple attempts, the MPS IIIA-severe iPSC clone from patient 2 was unable to be differentiated into NPCs (data not shown). At this stage of differentiation, no overt morphological difference was observed between control and MPS IIIA-intermed iPSC-derived NPCs. The NPCs were then dissociated from the neural rosette structures and cultured in the presence of FGF2. After two passages, morphology typical of neural progenitors was evident in both control and MPS IIIA-intermed cells (Fig. 2A). The formation of NPCs was confirmed through immunofluorescent detection of NPC marker proteins PAX6 and NESTIN. Positive staining was detected in both control and MPS IIIA-intermed cells after six passages (Fig. 2A). These cells were thus designated iPSC-derived NPCs. PAX6 and NESTIN expression in iPSC-derived NPCs was confirmed by real-time PCR. Both control and MPS IIIA-intermed iPSC-derived NPCs exhibited significant expression of PAX6; with PAX6 expression higher in MPS IIIA-intermed iPSC-derived NPCs than in control (Fig. 2B). In line with positive immunofluorescent staining, both control and MPS IIIA-intermed iPSC-derived NPCs expressed NESTIN; however, expression levels were not significantly different. Sulphamidase activity was reduced in MPS IIIA-intermed iPSC-derived NPCs compared to control (46.1 ± 11.8 pmol/h/mg protein versus 113.5 ± 34.5 pmol/h/mg protein) (Fig. 2C), while the HS biomarker increased from undetectable in control cells to 68.6 pmol/mg protein in MPS IIIA-intermed iPSC-derived NPCs (Fig. 2D). A key feature of the MPS IIIA phenotype is the elevation of secondary storage products; the gangliosides GM2 and GM3 in the brain. A significant increase of 197% in total GM2 storage was observed in MPS IIIA-intermed iPSC-derived NPCs (Fig. 2E), however total GM3 levels were the same in both control and MPS IIIA-intermed iPSC-derived NPCs (Fig. 2F).
Fig. 2.
Characterisation of iPSC-derived NPCs. iPSC-derived NPC morphology was assessed by brightfield microscopy (A). PAX6 and NESTIN both markers characteristic of NPCs were assessed by immunofluorescence (A) and gene expression (B). Sulphamidase activity (C), the monosulphated dissacharide HS storage biomarker (D), ganglioside GM2 (E) and ganglioside GM3 (F) were measured in cell extracts and normalised to cell protein. Quantitative data is expressed as the mean ± std. dev of n = 3 (sulphamidase activity) or n = 6 (GM2 and GM3) replicates. HN-UA(1S) is expressed as the average of 2 replicates where the individual values are:- Control 0.2 and 0.25 pmol/mg protein; MPS IIIA-intermed 86.5 and 50.5 pmol/mg protein.
* denotes significant difference between MPS IIIA-intermed and control, P < 0.05.
Scale bars are 50 μm as indicated.
3.3. Reduced proliferation of MPS IIIA-intermed iPSC-derived NPCs is partially rescued by FGF2
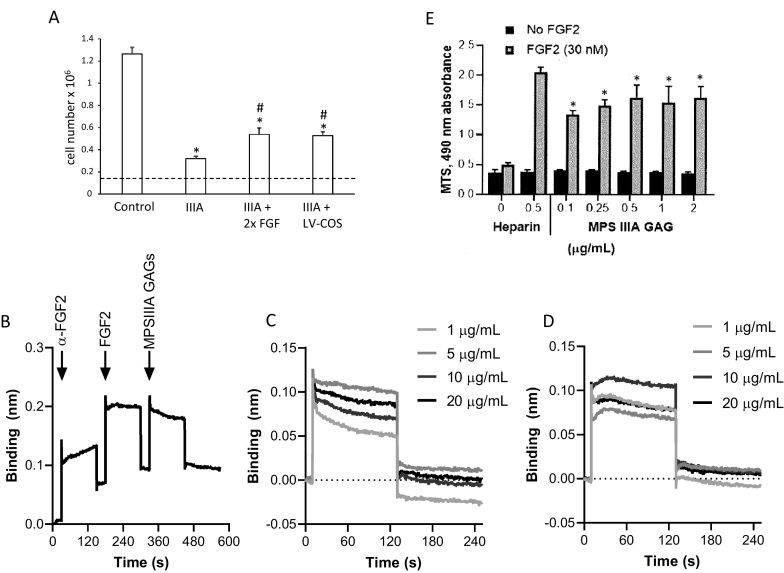
MPS IIIA-intermed iPSC-derived NPCs grew more slowly than control iPSC-derived NPCs (Fig. 3A). Control iPSC-derived NPC numbers increased by 8.3-fold over seven days while MPS IIIA–intermed iPSC-derived cell numbers only doubled in the same time. MPS IIIA-intermed iPSC-derived NPC cell division was stimulated by adding additional FGF2 to the culture medium (increasing the concentration from 20 ng/mL to 40 ng/mL) or by transduction of cells with lentivirus encoding sulphamidase, which corrected the enzyme defect. Although this represented a significant 2-fold increase in cell number over untreated MPS IIIA cells in both cases, proliferation was not normalised during the seven day experimental period.
Fig. 3.
Rescue of iPSC-derived NPC proliferation and role of MPS IIIA GAG on FGF2 binding and signalling. (A) Control iPSC-derived NPCs, MPS IIIA-intermed iPSC-derived NPCs and MPS IIIA-intermed iPSC-derived NPCs transduced with a lentivirus encoding sulphamidase (LV-COS) were seeded at 150,000 cells per 24-well and maintained in neural expansion medium. Additional cultures of MPS IIIA-intermed iPSC-derived NPCs received a 2-fold increase in FGF2 in the culture medium (i.e., 40 ng/mL instead of 20 ng/mL). Cell number was determined 7 days after seeding and results expressed as the mean ± std. dev of n = 3 replicates. *denotes significant difference to control and # denotes significant difference between MPS IIIA-intermed iPSC-derived NPCs treated with additional FGF2 or transduced with a lentivirus encoding sulphamidase and MPS IIIA-intermed iPSC-derived NPCs (P < 0.05). (B) MPS IIIA GAG binding to FGF2 was determined using FGF2 immobilised to a protein G sensor via an anti-FGF2 antibody followed by the addition of FGF2 and then MPSIIIA GAGs as shown or heparin (not shown). (C) Binding curves for MPSIIIA GAG at concentrations between 1 and 20 μg/mL. (D) Binding curves for heparin at concentrations between 1 and 20 μg/mL. (E) Proliferation of BaF32 cells expressing FGFR1c in the presence of 0.03 nM FGF2 and either heparin or MPSIIIA GAG. Results are expressed as the mean ± std. dev of n = 3 replicates. * denotes significant difference to cells grown in the absence of FGF2 and ** denotes significant difference to cells grown in the presence of FGF2 and all concentrations of MPSIIIA GAGs (P < 0.05).
3.4. MPS IIIA GAGs display similar FGF2 binding but reduced FGF2 signalling compared to heparin
Because FGF2 increased MPS IIIA-intermed iPSC-derived NPC proliferation and binding of FGF2 to HS GAG is a known prerequisite for FGF2 signalling [49], the relationship between FGF2, MPS IIIA HS and NPC proliferation was further investigated by assessing the binding affinity of MPS IIIA GAG for FGF2 and the effect of MPS IIIA GAG on FGF2 signalling. The binding of MPS IIIA GAGs to FGF2 was analysed in real-time using a dip-and-read biosensor, where FGF2 was immobilised via a polyclonal anti-FGF2 antibody to a protein G functionalised sensor. In the last step of the experimental workflow, either MPS IIIA GAGs (Fig. 3B) or heparin (data not shown) were exposed to the FGF2 functionalised sensors. The binding of MPS IIIA GAGs to FGF2 was similar in both association and dissociation profiles across the concentration range of 1 to 20 μg/mL (Fig. 3C) with similar responses also observed for heparin (Fig. 3D). Both MPS IIIA GAGs and heparin exhibited transient binding with little permanent binding once the sensor was returned to the buffer. These binding curves were modelled as Langmuir binding, which assumes 1:1 interactions, and established similar equilibrium dissociation constants (KD) in the nanomolar range of 11.65 nM for MPS IIIA GAG and 14.05 nM for heparin indicating a similar affinity for FGF2. In addition, the Х2 values form the model for both MPS IIIA GAGs and heparin were both greater than two indicating that the interactions were multivalent.
The BaF32 cell assay was used to analyse the ability of the MPS IIIA GAGs to form biologically active ternary complexes with FGF2 and FGFR1c. Proliferation of the cells was indicative of the formation of active ternary complexes that was measured by the MTS assay (Fig. 3E). MPS IIIA GAGs supported the proliferation of the BaF32 cells above the negative controls for each of the MPS IIIA GAG concentrations used in the absence of FGF2, indicating that biologically active ternary complexes were formed. However, no dose-dependent level of activity was observed over the MPS IIIA concentration range of 0–2 μg/mL. The positive control for the assay was heparin in the presence of FGF2 which was shown to support significantly higher cell proliferation that the MPS IIIA samples suggesting that the complexes formed between heparin, FGF2 and FGFR1c were more active than the complexes formed between the MPS IIIA GAGs, FGF2 and FGFR1c.
3.5. MPS IIIA iPSC-derived neural progenitors display impaired neurogenesis
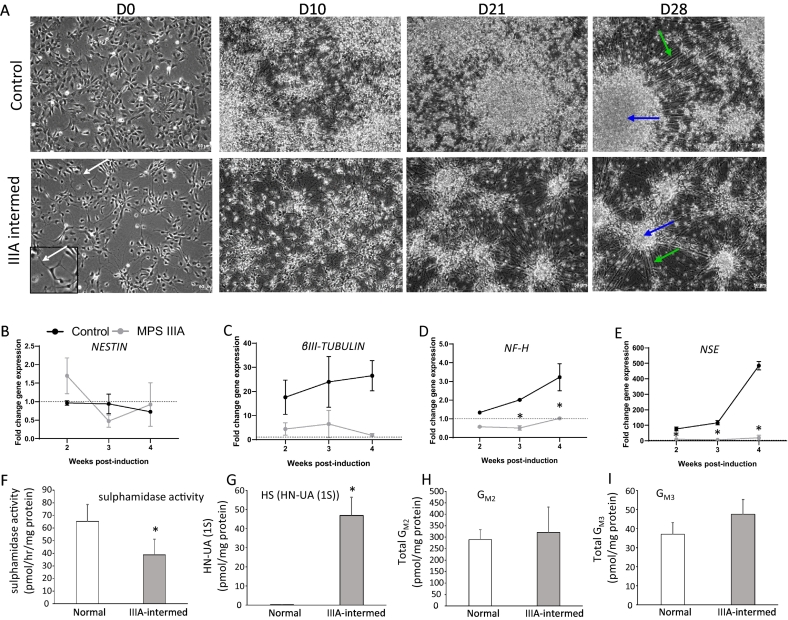
Neurogenesis of control and MPS IIIA iPSC-derived NPCs was induced by the withdrawal of FGF2 for a total of four weeks. On D0, prior to the removal of FGF2, both control and MPS IIIA-intermed iPSC-derived NPCs displayed a neural progenitor-like morphology (Fig. 4A). A pronounced change in morphology developed in both control and MPS IIIA-intermed cultures over the 28 days of culture resulting in aggregation of cell bodies and large bundles of cell extensions emanating across the cultures (Fig. 4A). It was noted that the cellular aggregates were less frequent, smaller and with less elaborate extensions in MPS IIIA-intermed cultures compared to control, a morphological phenomenon that has been noted in other stem cells models of neurological disorders [50], [51].
Fig. 4.
Neural differentiation of iPSC-derived NPCs. (A) Brightfield images of the neuronal differentiation of control and MPS IIIA-intermed iPSC-derived NPCs over 28 days following FGF2 withdrawl. Green arrows represent cellular extensions and blue arrows represent cell aggregates. Scale bars are 50 μm as indicated. (B-E) Gene expression by cells undergoing neuronal differentiation presented as the fold change in mRNA levels compared to undifferentiated NPCs, i.e. prior to FGF2 removal. Results are expressed as the mean ± std. dev of n = 3 replicates. (F-I) Sulphamidase activity, the monosulphated dissacharide HS storage biomarker and gangliosides GM2 and GM3 were measured in cell lysates. Results are expressed as the mean ± std. dev of n = 3 (sulphamidase) or 4 replicates (ganglioside). HN-UA(1S) is expressed as the average of 2 replicates where the individual values are:- Control 0.2 and 0.2 pmol/mg protein; MPS IIIA-intermed 40 and 54 pmol/mg protein.
*denotes a significant difference between MPS IIIA-intermed and control.
The expression of neural marker genes was then examined to quantify any differences between the ability of control and MPS IIIA-intermed iPSC-derived NPCs to differentiate into neurons. The expression was determined relative to undifferentiated control and MPS IIIA-intermed iPSC-derived NPCs prior to FGF2 removal (i.e., undifferentiated NPCs). Both control and MPS IIIA-intermed cells expressed NESTIN a marker of neural progenitors, throughout the course of neural induction (Fig. 4B); however, little change in expression was seen compared to undifferentiated NPC controls, indicating that a NPC population was maintained over the time course in both cultures. βIII-TUBULIN expression increased in control cultures undergoing neural differentiation from 14 days post-induction onwards, with 17.58 ± 7.10, 23.94 ± 10.52 and 26.51 ± 6.31 fold increases in expression 14, 21 and 28 days post-induction compared to undifferentiated NPC controls, respectively, indicating the formation of immature neurons (Fig. 4C). βIII-TUBULIN expression was also upregulated in MPS IIIA-intermed cultures compared to undifferentiated NPC controls following the initiation of neural induction; however, expression was consistently lower than in control iPSC-derived NPCs, with 4.43 ± 2.59, 6.48 ± 5.65 and 1.76 ± 0.78 fold increases in βIII-TUBULIN expression compared to undifferentiated controls 14, 21 and 28 days post-induction, respectively (Fig. 4C). NF-H expression increased steadily over the course of neural induction in control cultures, with expression peaking with a significant 3.23 ± 0.27 fold increase compared to undifferentiated NPC controls 28 days post-induction (Fig. 4D). In contrast, NF-H expression was never significantly elevated above undifferentiated controls in MPS IIIA-intermed cultures; with NF-H expression significantly lower than both undifferentiated MPS IIIA NPC controls and control cultures 21 days post-induction, indicating a dysfunction in neuron formation or survival. This was supported by the decreased expression of NSE in MPS IIIA-intermed cultures compared to control. NSE expression was significantly increased in control cells compared to undifferentiated NPC controls at all time points, with expression peaking at 28 days post-induction with a 485.1 ± 26.87 fold increase in expression. NSE expression did increase in MPS-intermed cultures compared to undifferentiated controls, with the highest increase of 20.34 ± 15.68 fold seen 28 days post-induction, indicating the presence of neurons; however, expression was consistently lower than what was seen in control cultures indicating a comparative disruption in neuron formation or survival (Fig. 4E).
At the end of the differentiation protocol (day 28), sulphamidase activity was significantly reduced in MPS IIIA-intermed cultures compared to control cultures (36.64 ± 12.52 pmol/h/mg protein versus 62.27 ± 13.42 pmol/h/mg protein) (Fig. 4F), while the MPS IIIA HS marker HN-UA (1S) increased from undetectable in control cells to 46.9 pmol/mg protein in MPS IIIA-intermed cultures (Fig. 4G). Ganglioside storage also increased in MPS IIIA-intermed cultures, although the increase was not significant. GM2 storage was 321 ± 10.31 pmol/mg protein in MPS IIIA-intermed neurons compared to 289.5 ± 43.62 pmol/mg protein in control (Fig. 4H), while GM3 storage was 47.5 ± 7.78 pmol/mg protein in MPS IIIA-intermed neurons compared to 37 ± 6.16 pmol/mg protein in control (Fig. 4I).
Partially degraded and undegraded HS GAG progressively builds up in MPS IIIA patients. To investigate the effect of elevated MPS IIIA GAG on neural differentiation, it was added to the culture medium of control iPSC-derived NPCs during the neuronal differentiation period. On day 28 the expression of NESTIN, βIII-TUBULIN and NSE in control iPSC-derived NPCs cultured in the presence of MPS IIIA GAG was not significantly different to expression in the undifferentiated controls, with a 1.45 ± 0.18, 1.59 ± 0.08 and 1.1 ± 0.07 fold change in expression respectively (data not shown). In the presence of MPS IIIA GAG, the expression of NF-H was significantly higher than undifferentiated with a 2.23 ± 0.63 fold change in expression but was still lower than in control cells undergoing neuronal differentiation in the absence of MPS IIIA GAG (Fig. 4D).
4. Discussion
Despite the CNS being the main site of pathology in MPS IIIA the consequence of impaired HS turnover on the development and function of neural cells has not been studied in detail; due no doubt in part to the challenge inherent in isolating neurons/neural stem cells from MPS IIIA patients or animal models. Although there is one previous report of iPSC generated MPS IIIA cell lines [22], which showed ability to form the three germ cell layers including ectoderm, the precursor for neural cells, the study presented here undertakes characterisation at a biochemical level, as well as a comparison of neural differentiation kinetics using well characterised and accepted methodology for generating neural cells against controls. Reprogramming MPS IIIA patient somatic cells to iPSCs generates a renewable cell resource that can be used to investigate neurogenesis and neuronal cell function. In this study iPSCs were successfully generated from skin fibroblasts isolated from a control individual and individuals presenting with both intermediate and severe MPS IIIA clinical phenotypes. The pluripotent state of the iPSCs was evidenced by typical iPSC morphology and the expression of key pluripotency marker genes OCT4, SOX2, NANOG and SSEA4. MPS IIIA iPSCs maintained their hallmark biochemical characteristics of reduced sulphamidase activity and increased storage of the HS GAG marker HN-UA (1S) compared to control iPSCs. Unlike iPSCs derived from MPS IIIB fibroblasts [23], visually no obvious difference was observed in the proliferation of MPS IIIA iPSCs and cultures were not supplemented with sulphamidase. No impairment in the proliferation of iPSCs derived from MPS I, II, IIIC, IVA or VII patient fibroblasts has been reported in the literature [16], [17], [18], [19], [20], [26], [27], [28], [29]. The N-acetylglucosamine moiety that accumulates in MPS IIIB may play a role in maintaining iPSCs. However, Vallejo-Diez [24] did not report proliferation problems in two other MPS IIIB iPSC cell lines thus different mutations and differing methodology may have played a role in the discrepancy between reports. A very recent report by Huang et al., 2021 to generate additional patient iPSC MPS IIIB stem cell lines does not report any abnormalities in proliferation [25].
NPCs were then generated from the control and MPS IIIA-intermed iPSC cell lines. Unfortunately, across several attempts, the MPS IIIA-severe iPSC cell clone was unable to be successfully differentiated in iPSCs derived from the severely affected individual into NPCs (data not shown), implying that the process may be particularly sensitive to SGSH function in vitro. Similarly, studies in MPS IIIB iPSCs only two of their four cell lines were able to be into successfully differentiated into NPCs and mature neurons suggesting that heparan sulphate may have an important role in neurodifferentiation potential [25]. Both lines expressed the NPC markers PAX6 and NESTIN. MPS IIIA iPSC-derived NPCs displayed reduced sulphamidase activity and increased accumulation of the HS GAG biomarker HN-UA (1S), consistent with MPS IIIA. An increase in GM2 accumulation was also observed consistent with phenotype although GM3 accumulation appeared unaffected. Unlike MPS IIIA iPSCs, MPS IIIA-intermed iPSC-derived NPCs were visually observed to proliferate slower than control and this was shown to be a significant decrease on quantification. Proliferation was partially rescued by overexpression of sulphamidase in the affected iPSC-derived NPCs within the seven-day experimental time frame. Correcting the MPS IIIA enzyme defect was expected to improve proliferation and we anticipate that a longer timeframe would result in complete restoration of proliferative capacity.
Likewise, partial rescue was achieved by doubling the concentration of FGF2 in the culture medium and again a longer experimental timeframe may have completely restored proliferation. FGF2 is known to play an important role in neurogenesis and its temporospatial expression corresponds to areas of neurogenesis in the developing CNS [52], [53]. During development, and in vitro, FGF2 is a major NPC mitogen that stimulates neural progenitor cell proliferation and regulates neural cell differentiation [54], [55], [56]. This is underscored by the observation that FGF2 deficient mice display impaired neuronal proliferation and differentiation [57], [58]. FGF2 is also implicated in the regulation of adult neurogenesis [58], [59]. FGF2 forms a complex with its receptor FGFR and co-receptor HS GAG, located on cell surface proteoglycans, to stimulate cell proliferation [49]. HS containing proteoglycans are themselves regulated during NPC proliferation and differentiation [60] and play important roles in axon guidance, synaptogenesis and plasticity [61], [62], [63], [64]. The ability of HS GAG chains to bind to and regulate the biological activity of a range of growth factors, including FGF2 [65], places HS containing proteoglycans in a central role in neurogenesis, CNS development and homeostasis. The HS GAG that accumulates in MPS IIIA reduced cell proliferation in the BaF32 cell assay compared to a heparin control, but its affinity for FGF2 was similar to the heparin positive control. Because MPS IIIA HS GAG has unaltered affinity for FGF2, we suggest that the vastly increased amount of partially degraded/undegraded HS GAG that accumulates in MPS IIIA alters the distribution of FGF2. That is, FGF2 is just as likely to bind to MPS IIIA HS GAG as it is to cell surface proteoglycan HS or to the FGFR, but as there is a large excess of MPS IIIA HS GAG compared to the other two molecules FGF2 is more likely to bind to MPS IIIA HS GAG. As MPS IIIA HS GAG is freely diffusible the FGF2 bound to it may no longer be localized where it can bind to the FGFR. This finding is also supported by de Risi (2021) who showed using the same MTS assay in Baf32 cells that MPS IIIA HS extracts failed to induce proliferation mediated by FGF2 in comparison to wildtype HS [66]. Thus the abnormal turnover of HS in MPS IIIA and the accumulation of high levels of abnormally structured HS in the CNS clearly interrupts the delicate balance between FGF2 growth factor/growth factor receptor/growth factor co-receptor in NPC proliferation. This is supported by the observation that MPS I bone marrow stromal progenitor cell proliferation is decreased in association with aberrant FGF2 signalling [67]. Common to MPS I and MPS IIIA is the accumulation of HS GAG providing further evidence of the role of this GAG in supporting progenitor cell proliferation. A very recent study generated a SH-SY5Y neuroblastoma cell line based MPS IIIA model using CRISPR-Cas9 disruption of SGSH. These neural progenitor-like cells displayed increased proliferation which was reduced when wildtype heparan sulphate was introduced into the cell media [66], suggesting a role for HS in proliferative ability and neurodifferention of SH-SY5Y cells. Further evidence from de Pasquale et al. (2021) using a NAGLU silenced neuroblastoma model of MPS IIIB highlights a key role for HSPGs in neurite outgrowth using the specific growth factor NK1 which is proposed to bind with high affinity the extracellular accumulated HS [68]. This same group has also characterised NK1 in skin fibroblast models [69], suggesting that targeting excess HS reduces lysosomal burden and is able to restore specific downstream FGF signalling.
The initial ‘neural induction’ of MPS IIIA-intermed iPSCs appeared unaffected, with the overt formation of an integral neuroepithelial sheet, and subsequently formation of hallmark ‘neural rosettes’ colony structures unchanged between control and MPS IIIA-intermed cultures. Both control and MPS IIIA cultures displayed NPC morphology and expressed markers of dorsal cortical NPCs, PAX6 and NESTIN, indicating the successful formation of equivalent NPCs from MPS IIIA patient-derived iPSCs and the control. Whilst expression of the dorsal cortical progenitor marker PAX6 varied between control and MPS IIIA-intermed iPSC-derived NPCs, expression of the pan NPC marker, NESTIN, was comparable between control and MPS IIIA iPSC-derived NPCs, indicating a similar global neural progenitor population and formation capacity. However, whilst NPCs were successfully formed from MPS IIIA-intermed iPSCs, their subsequent ability to undergo neurogenesis did appear to be impacted. Upon differentiation, several lines of evidence suggested that MPS IIIA-intermed iPSC-derived NPC cultures had reduced numbers of neurons compared to control cultures. MPS IIIA-intermed iPSC-derived NPC cultures did not display the same degree of overt neuronal morphology, with delayed formation and reduced presence of neuronal body aggregates (ganglion-like structures) and less prominent neurite bundles. The expression of several post-mitotic neuron marker genes, βIII-TUBULIN, NF-H and NSE were decreased in MPS IIIA-intermed iPSC-derived NPCs compared to control, with the difference becoming more prominent as neurogenesis progressed. In contrast, no significant change in NESTIN expression was seen between control and MPS IIIA-intermed cultures throughout neural induction, indicating that neural progenitor formation was unaffected in MPS IIIA-intermed iPSC-derived NPCs. This data does not distinguish between reduced neurogenic capacity and reduced neuronal survival. While loss of Purkinje cells is a feature in MPS IIIA mice [70] and dogs [71], it is observed in older animals and to our knowledge neuronal cell death is not a feature of MPS IIIA neurodevelopment. If MPS IIIA cell death should be proven in our system, it may be indicative of a degenerative rather than developmental process. While the expression of all post-mitotic genes tested were reduced in differentiating MPS IIIA cultures compared to control, the expression of NF-H and NSE were beginning to increase by day 28 of culture, compared to control cultures in which gene expression increased after day 21. Thus neuronal differentiation of MPS IIIA-intermed iPSC-derived NPCs may be temporally delayed in culture, which could lead to a population of cells at different neurogenic stages rather than a flagrant loss of neurodifferentiation capacity.
Sulphamidase activity was decreased in MPS IIIA cultures on day 28 of neural differentiation and accumulation of the HS GAG biomarker HN-UA (1S) increased consistent with the MPS IIIA phenotype, however, levels of GM2 and GM3 ganglioside were not different to control. This latter was unexpected as increased ganglioside storage is consistently observed in MPS IIIA patient brain and in the IIIA mouse model [6], [72], [73], [74], [75]. This may reflect the reduction in neurogenesis observed from MPS IIIA-intermed iPSC-derived NPCs or that the experimental time frame while permitting the accumulation of the primary storage product HS GAG was not sufficient for the production and subsequent accumulation of the secondary storage product ganglioside.
The addition of MPS IIIA GAG to the culture medium of control iPSC-derived NPCs impaired subsequent neurogenesis. HS is a highly bioactive GAG with the ability to bind to a range of growth factors implicated in brain development and function such as Wnts, BMP and members of the hedgehog family in addition to FGF2 [65], [76], [77], [78], [79]. While we have shown that MPS IIIA GAG structures impede MPS IIIA iPSC-derived NPC proliferation, it is unclear if the GAG is acting with FGF2 to impair neurogenesis [54], [56] or through a different growth factor pathway or combination of pathways.
This study focussed on neurogenesis from MPS IIIA iPSC-derived NPCs but the cells are also useful tools for studying mature neuronal cell function in this disorder. iPSC-derived neuronal networks generated from MPS IIIC and MPS VII iPSCs have been shown to display abnormal activity and connectivity [26], [28] and an extension of this study could address similar questions in MPS IIIA. iPSCs can also be used to examine pathogenesis of disease, to screen therapeutics and develop/test treatment strategies [80], [81], [82]. To this end the availability of MPS IIIA skin fibroblasts in diagnostic laboratories are a useful resource for producing iPSCs and gene editing techniques can be employed to generate additional SGSH mutations from control iPSC lines. Whilst these results are preliminary, validation of multiple clones and mutations, including isogenic CRISPR repaired cell lines would allow a further understanding of the effect of MPS IIIA disease pathology on neurodifferentiation in iPSC-derived stem cell models and correlation of genotype/phenotype. Overall, this study provides an initial insight into the mechanism of CNS dysfunction in MPS IIIA. The reduction in HS GAG turnover and associated disruption to lysosomal function and accumulation of unique HS GAG structures in MPS IIIA do not greatly affect iPSC production from MPS IIIA skin fibroblasts or their differentiation to NPCs. MPS IIIA iPSC-derived NPC proliferation is however affected by GAG accumulation that interferes with FGF2 signalling. Additionally the formation of neurons from MPS IIIA iPSC-derived NPCs and/or their survival was reduced via an as yet unknown mechanism.
Acknowledgements
This study was supported by the Million Dollar Bike Ride Grant, Orphan Diseases Centre, University of Pennsylvania to ADR, Sanfilippo Children's Foundation Incubator Grant to LAJ and ADR and Robinson Research Institute Seed Grant to LAJ and ADR. LAJ is supported by Australian Research Council ARC DE160100620 grant. The Molecular Surface Interaction Network Laboratory, UNSW funded in part by the Research Infrastructure Program, UNSW supported ML and HNK. RL is the recipient of an Australian Postgraduate Award (APA)/ Research Training Program (RTP) scholarship.
Contributor Information
Lachlan A. Jolly, Email: lachlan.jolly@adelaide.edu.au.
Ainslie L.K. Derrick-Roberts, Email: ainslie.derrickroberts@adelaide.edu.au.
References
- 1.Neufeld E., Muenzer J. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C., Beaudet A., Sly W., Valle D., editors. McGraw Hill; New York: 2001. The Mucopolysaccharidoses; pp. 3421–3452. [Google Scholar]
- 2.Constantopoulos G., McComb R.D., Dekaban A.S. Neurochemistry of the mucopolysaccharidoses: brain glycosaminoglycans in normals and four types of mucopolysaccharidoses. J. Neurochem. 1976;26:901–908. doi: 10.1111/j.1471-4159.1976.tb06471.x. [DOI] [PubMed] [Google Scholar]
- 3.Ceuterick C., Martin J.J., Libert J., Farriaux J.P. Sanfilippo a disease in the fetus–comparison with pre- and postnatal cases. Neuropadiatrie. 1980;11:176–185. doi: 10.1055/s-2008-1071387. [DOI] [PubMed] [Google Scholar]
- 4.Barone R., Pellico A., Pittala A., Gasperini S. Neurobehavioral phenotypes of neuronopathic mucopolysaccharidoses. Ital. J. Pediatr. 2018;44:121. doi: 10.1186/s13052-018-0561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochuli M., Wuthrich K., Steinmann B. Two-dimensional NMR spectroscopy of urinary glycosaminoglycans from patients with different mucopolysaccharidoses. NMR Biomed. 2003;16:224–236. doi: 10.1002/nbm.833. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson F.L., Holley R.J., Langford-Smith K.J., Badrinath S., Liao A., Langford-Smith A., Cooper J.D., Jones S.A., Wraith J.E., Wynn R.F., Merry C.L., Bigger B.W. Neuropathology in mouse models of mucopolysaccharidosis type I IIIA and IIIB. 2012;7 doi: 10.1371/journal.pone.0035787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppa G.V., Gabrielli O., Zampini L., Maccari F., Mantovani V., Galeazzi T., Santoro L., Padella L., Marchesiello R.L., Galeotti F., Volpi N. Mental retardation in mucopolysaccharidoses correlates with high molecular weight urinary heparan sulphate derived glucosamine. Metab. Brain Dis. 2015;30:1343–1348. doi: 10.1007/s11011-015-9684-y. [DOI] [PubMed] [Google Scholar]
- 8.de Ruijter J., Ijlst L., Kulik W., van Lenthe H., Wagemans T., van Vlies N., Wijburg F.A. Heparan sulfate derived disaccharides in plasma and total urinary excretion of glycosaminoglycans correlate with disease severity in sanfilippo disease. J. Inherit. Metab. Dis. 2013;36:271–279. doi: 10.1007/s10545-012-9535-5. [DOI] [PubMed] [Google Scholar]
- 9.Gliddon B.L., Hopwood J.J. Enzyme-replacement therapy from birth delays the development of behavior and learning problems in mucopolysaccharidosis type IIIA mice. Pediatr. Res. 2004;56:65–72. doi: 10.1203/01.PDR.0000129661.40499.12. [DOI] [PubMed] [Google Scholar]
- 10.Langford-Smith A., Wilkinson F.L., Langford-Smith K.J., Holley R.J., Sergijenko A., Howe S.J., Bennett W.R., Jones S.A., Wraith J., Merry C.L., Wynn R.F., Bigger B.W. Hematopoietic stem cell and gene therapy corrects primary neuropathology and behavior in mucopolysaccharidosis IIIA mice. Mol. Ther. 2012;20:1610–1621. doi: 10.1038/mt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu H., Cataldi M.P., Ware T.A., Zaraspe K., Meadows A.S., Murrey D.A., McCarty D.M. Functional correction of neurological and somatic disorders at later stages of disease in MPS IIIA mice by systemic scAAV9-hSGSH gene delivery. Mol. Ther. Methods Clin. Dev. 2016;3:16036. doi: 10.1038/mtm.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruzo A., Marco S., Garcia M., Villacampa P., Ribera A., Ayuso E., Maggioni L., Mingozzi F., Haurigot V., Bosch F. Correction of pathological accumulation of glycosaminoglycans in central nervous system and peripheral tissues of MPSIIIA mice through systemic AAV9 gene transfer. Hum. Gene Ther. 2012;23:1237–1246. doi: 10.1089/hum.2012.029. [DOI] [PubMed] [Google Scholar]
- 13.Saville J., Derrick-Roberts A., McIntyre C., Fuller M. Systemic scAAV9.U1a.hSGSH delivery corrects brain biochemistry in mucopolysaccharidosis type IIIA at early and later stages of disease. Hum Gene Ther. 2021 Apr;32(7-8):420–430. doi: 10.1089/hum.2020.253. [DOI] [PubMed] [Google Scholar]
- 14.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X., Bhattacharyya A. Human models are needed for studying human neurodevelopmental disorders. Am. J. Hum. Genet. 2018;103:829–857. doi: 10.1016/j.ajhg.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaroop M., Brooks M.J., Gieser L., Swaroop A., Zheng W. Patient iPSC-derived neural stem cells exhibit phenotypes in concordance with the clinical severity of mucopolysaccharidosis I. Hum. Mol. Genet. 2018;27:3612–3626. doi: 10.1093/hmg/ddy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybova J., Ledvinova J., Sikora J., Kuchar L., Dobrovolny R. Neural cells generated from human induced pluripotent stem cells as a model of CNS involvement in mucopolysaccharidosis type II. J. Inherit. Metab. Dis. 2018;41:221–229. doi: 10.1007/s10545-017-0108-5. [DOI] [PubMed] [Google Scholar]
- 18.Varga E., Nemes C., Bock I., Varga N., Feher A., Dinnyes A., Kobolak J. Generation of mucopolysaccharidosis type II (MPS II) human induced pluripotent stem cell (iPSC) line from a 1-year-old male with pathogenic IDS mutation. Stem Cell Res. 2016;17:482–484. doi: 10.1016/j.scr.2016.09.033. [DOI] [PubMed] [Google Scholar]
- 19.Varga E., Nemes C., Bock I., Varga N., Feher A., Kobolak J., Dinnyes A. Generation of mucopolysaccharidosis type II (MPS II) human induced pluripotent stem cell (iPSC) line from a 3-year-old male with pathogenic IDS mutation. Stem Cell Res. 2016;17:479–481. doi: 10.1016/j.scr.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Varga E., Nemes C., Bock I., Varga N., Feher A., Kobolak J., Dinnyes A. Generation of mucopolysaccharidosis type II (MPS II) human induced pluripotent stem cell (iPSC) line from a 7-year-old male with pathogenic IDS mutation. Stem Cell Res. 2016;17:463–465. doi: 10.1016/j.scr.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Kobolak J., Molnar K., Varga E., Bock I., Jezso B., Teglasi A., Zhou S., Lo Giudice M., Hoogeveen-Westerveld M., Pijnappel W.P., Phanthong P., Varga N., Kitiyanant N., Freude K., Nakanishi H., Laszlo L., Hyttel P., Dinnyes A. Modelling the neuropathology of lysosomal storage disorders through disease-specific human induced pluripotent stem cells. Exp. Cell Res. 2019;380:216–233. doi: 10.1016/j.yexcr.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Vallejo S., Fleischer A., Martin J.M., Sanchez A., Palomino E., Bachiller D. Generation of two induced pluripotent stem cells lines from mucopolysaccharydosis IIIA patient: IMEDEAi004-a and IMEDEAi004-B. Stem Cell Res. 2018;32:110–114. doi: 10.1016/j.scr.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Lemonnier T., Blanchard S., Toli D., Roy E., Bigou S., Froissart R., Rouvet I., Vitry S., Heard J.M., Bohl D. Modeling neuronal defects associated with a lysosomal disorder using patient-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:3653–3666. doi: 10.1093/hmg/ddr285. [DOI] [PubMed] [Google Scholar]
- 24.Vallejo-Diez S., Fleischer A., Martin-Fernandez J.M., Sanchez-Gilabert A., Bachiller D. Generation of two induced pluripotent stem cells lines from a mucopolysaccharydosis IIIB (MPSIIIB) patient. Stem Cell Res. 2018;33:180–184. doi: 10.1016/j.scr.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Huang W., Cheng Y.S., Yang S., Swaroop M., Xu M., Huang W., Zheng W. Disease modeling for mucopolysaccharidosis type IIIB using patient derived induced pluripotent stem cells. Exp. Cell Res. 2021;407 doi: 10.1016/j.yexcr.2021.112785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canals I., Soriano J., Orlandi J.G., Torrent R., Richaud-Patin Y., Jimenez-Delgado S., Merlin S., Follenzi A., Consiglio A., Vilageliu L., Grinberg D., Raya A. Activity andhigh-order effective connectivity alterations in Sanfilippo C patient-specific neuronal networks. Stem Cell Rep. 2015;5:546–557. doi: 10.1016/j.stemcr.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong B.A., Zou J., Liu C., Almeciga-Diaz C., Zheng W. Generation of an induced pluripotent stem cell line (TRNDi005-A) from a mucopolysaccharidosis type IVA patient carrying compound heterozygous p.R61W and p.WT405del mutations in the GALNS gene. Stem Cell Res. 2019;36:101408. doi: 10.1016/j.scr.2019.101408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayo-Puxan N., Terrasso A.P., Creyssels S., Simao D., Begon-Pescia C., Lavigne M., Salinas S., Bernex F., Bosch A., Kalatzis V., Levade T., Cuervo A.M., Lory P., Consiglio A., Brito C., Kremer E.J. Lysosomal and network alterations in human mucopolysaccharidosis type VII iPSC-derived neurons. Sci. Rep. 2018;8:16644. doi: 10.1038/s41598-018-34523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin T.A., Anderson H.C., Wolfe J.H. Ex vivo gene therapy using patient iPSC-derived NSCs reverses pathology in the brain of a homologous mouse model. Stem Cell Rep. 2015;4:835–846. doi: 10.1016/j.stemcr.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga E., Nemes C., Kovacs E., Bock I., Varga N., Feher A., Dinnyes A., Kobolak J. Generation of human induced pluripotent stem cell (iPSC) line from an unaffected female carrier of mucopolysaccharidosis type II (MPS II) disorder. Stem Cell Res. 2016;17:514–516. doi: 10.1016/j.scr.2016.09.035. [DOI] [PubMed] [Google Scholar]
- 31.Meng X.L., Shen J.S., Kawagoe S., Ohashi T., Brady R.O., Eto Y. Induced pluripotent stem cells derived from mouse models of lysosomal storage disorders. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7886–7891. doi: 10.1073/pnas.1002758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Natale P., Villani G.R., Di Domenico C., Daniele A., Dionisi Vici C., Bartuli A. Analysis of SanfilippoAgene mutations in a large pedigree. Clin. Genet. 2003;63:314–318. doi: 10.1034/j.1399-0004.2003.00053.x. [DOI] [PubMed] [Google Scholar]
- 33.Meyer A., Kossow K., Gal A., Steglich C., Muhlhausen C., Ullrich K., Braulke T., Muschol N. The mutation p.Ser298Pro in the sulphamidase gene (SGSH) is associated with a slowly progressive clinical phenotype in mucopolysaccharidosis type IIIA (Sanfilippo A syndrome) Hum. Mutat. 2008;29:770. doi: 10.1002/humu.20738. [DOI] [PubMed] [Google Scholar]
- 34.Valstar M.J., Neijs S., Bruggenwirth H.T., Olmer R., Ruijter G.J., Wevers R.A., van Diggelen O.P., Poorthuis B.J., Halley D.J., Wijburg F.A. Mucopolysaccharidosis type IIIA: clinical spectrum and genotype-phenotype correlations. Ann. Neurol. 2010;68:876–887. doi: 10.1002/ana.22092. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 37.Homan C.C., Pederson S., To T.H., Tan C., Piltz S., Corbett M.A., Wolvetang E., Thomas P.Q., Jolly L.A., Gecz J. PCDH19 regulation of neural progenitor cell differentiation suggests asynchrony of neurogenesis as a mechanism contributing to PCDH19 girls clustering epilepsy. Neurobiol. Dis. 2018;116:106–119. doi: 10.1016/j.nbd.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Byers S., Rozaklis T., Brumfield L.K., Ranieri E., Hopwood J.J. Glycosaminoglycan accumulation and excretion in the mucopolysaccharidoses: characterization and basis of a diagnostic test for MPS. Mol. Genet. Metab. 1998;65:282–290. doi: 10.1006/mgme.1998.2761. [DOI] [PubMed] [Google Scholar]
- 39.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 40.Karpova E.A., Voznyi Ya V., Keulemans J.L., Hoogeveen A.T., Winchester B., Tsvetkova I.V., van Diggelen O.P. A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease type A(MPS IIIA) J. Inherit. Metab. Dis. 1996;19:278–285. doi: 10.1007/BF01799255. [DOI] [PubMed] [Google Scholar]
- 41.Saville J.T., Flanigan K.M., Truxal K.V., McBride K.L., Fuller M. Evaluation of biomarkers for sanfilippo syndrome. Mol. Genet. Metab. 2019;128:68–74. doi: 10.1016/j.ymgme.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Saville J.T., McDermott B.K., Fletcher J.M., Fuller M. Disease and subtype specific signatures enable precise diagnosis of the mucopolysaccharidoses. Genet Med. 2019;21:753–757. doi: 10.1038/s41436-018-0136-z. [DOI] [PubMed] [Google Scholar]
- 43.Saville J.T., Fuller M. Sphingolipid dyshomeostasis in the brain of the mouse model of mucopolysaccharidosis type IIIA. Mol. Genet. Metab. 2020;129:111–116. doi: 10.1016/j.ymgme.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Ornitz D.M., Xu J., Colvin J.S., McEwen D.G., MacArthur C.A., Coulier F., Gao G., Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 45.Lord M.S., Chuang C.Y., Melrose J., Davies M.J., Iozzo R.V., Whitelock J.M. The role of vascular-derived perlecan in modulating cell adhesion, proliferation and growth factor signaling. Matrix Biol. 2014;35:112–122. doi: 10.1016/j.matbio.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson M., Derrick Roberts A., Martin E., Rout-Pitt N., Gronthos S., Byers S. Mucopolysaccharidosis enzyme production by bone marrow and dental pulp derived human mesenchymal stem cells. Mol. Genet. Metab. 2015;114:584–593. doi: 10.1016/j.ymgme.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chua C.C., Rahimi N., Forsten-Williams K., Nugent M.A. Heparan sulfate proteoglycans function as receptors for fibroblast growth factor-2 activation of extracellular signal-regulated kinases 1 and 2. Circ. Res. 2004;94:316–323. doi: 10.1161/01.RES.0000112965.70691.AC. [DOI] [PubMed] [Google Scholar]
- 50.Jolly L.A., Taylor V., Wood S.A. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol. Biol. Cell. 2009;20:2015–2029. doi: 10.1091/mbc.E08-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mincheva-Tasheva S., Guil A.F.Nieto, Homan C.C., Gecz J., Thomas P.Q. Disrupted excitatory synaptic contacts and altered neuronal network activity underpins the neurological phenotype in PCDH19-clustering epilepsy (PCDH19-CE) Mol. Neurobiol. 2021;58:2005–2018. doi: 10.1007/s12035-020-02242-4. [DOI] [PubMed] [Google Scholar]
- 52.Murphy M., Reid K., Ford M., Furness J.B., Bartlett P.F. FGF2 regulates proliferation of neural crest cells, with subsequent neuronal differentiation regulated by LIF or related factors. Development. 1994;120:3519–3528. doi: 10.1242/dev.120.12.3519. [DOI] [PubMed] [Google Scholar]
- 53.Powell P.P., Finklestein S.P., Dionne C.A., Jaye M., Klagsbrun M. Temporal, differential and regional expression of mRNA for basic fibroblast growth factor in the developing and adult rat brain. Brain Res. Mol. Brain Res. 1991;11:71–77. doi: 10.1016/0169-328x(91)90023-q. [DOI] [PubMed] [Google Scholar]
- 54.Chen C.W., Liu C.S., Chiu I.M., Shen S.C., Pan H.C., Lee K.H., Lin S.Z., Su H.L. The signals of FGFs on the neurogenesis of embryonic stem cells. J. Biomed. Sci. 2010;17:33. doi: 10.1186/1423-0127-17-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Israsena N., Hu M., Fu W., Kan L., Kessler J.A. The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev. Biol. 2004;268:220–231. doi: 10.1016/j.ydbio.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Qian X., Davis A.A., Goderie S.K., Temple S. FGF2 concentration regulates the generation of neurons and glia from multipotent cortical stem cells. Neuron. 1997;18:81–93. doi: 10.1016/s0896-6273(01)80048-9. [DOI] [PubMed] [Google Scholar]
- 57.Raballo R., Rhee J., Lyn-Cook R., Leckman J.F., Schwartz M.L., Vaccarino F.M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Werner S., Unsicker K., von Bohlen O., Halbach Fibroblast growth factor-2 deficiency causes defects in adult hippocampal neurogenesis, which are not rescued by exogenous fibroblast growth factor-2. J. Neurosci. Res. 2011;89:1605–1617. doi: 10.1002/jnr.22680. [DOI] [PubMed] [Google Scholar]
- 59.Rai K.S., Hattiangady B., Shetty A.K. Enhanced production and dendritic growth of new dentate granule cells in the middle-aged hippocampus following intracerebroventricular FGF-2 infusions. Eur. J. Neurosci. 2007;26:1765–1779. doi: 10.1111/j.1460-9568.2007.05820.x. [DOI] [PubMed] [Google Scholar]
- 60.Oikari L.E., Okolicsanyi R.K., Qin A., Yu C., Griffiths L.R., Haupt L.M. Cell surface heparan sulfate proteoglycans as novel markers of human neural stem cell fate determination. Stem Cell Res. 2016;16:92–104. doi: 10.1016/j.scr.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 61.Bandtlow C.E., Zimmermann D.R. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol. Rev. 2000;80:1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 62.Schwartz N.B., Domowicz M.S. Proteoglycans in brain development and pathogenesis. FEBS Lett. 2018;592:3791–3805. doi: 10.1002/1873-3468.13026. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi Y. Heparan sulfate proteoglycans in the nervous system: their diverse roles in neurogenesis, axon guidance, and synaptogenesis. Semin. Cell Dev. Biol. 2001;12:99–106. doi: 10.1006/scdb.2000.0238. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi Y., Inatani M., Matsumoto Y., Ogawa J., Irie F. Roles of heparan sulfate in mammalian brain development current views based on the findings from Ext1 conditional knockout studies. Prog. Mol. Biol. Transl. Sci. 2010;93:133–152. doi: 10.1016/S1877-1173(10)93007-X. [DOI] [PubMed] [Google Scholar]
- 65.Zhang F., Zheng L., Cheng S., Peng Y., Fu L., Zhang X., Linhardt R.J. Comparison of the interactions of different growth factors and glycosaminoglycans. Molecules. 2019;24 doi: 10.3390/molecules24183360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Risi M., Tufano M., Alvino F.G., Ferraro M.G., Torromino G., Gigante Y., Monfregola J., Marrocco E., Pulcrano S., Tunisi L., Lubrano C., Papy-Garcia D., Tuchman Y., Salleo A., Santoro F., Bellenchi G.C., Cristino L., Ballabio A., Fraldi A., De Leonibus E. Altered heparan sulfate metabolism during development triggers dopamine-dependent autistic-behaviours in models of lysosomal storage disorders. Nat. Commun. 2021;12:3495. doi: 10.1038/s41467-021-23903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan C., Nelson M.S., Reyes M., Koodie L., Brazil J.J., Stephenson E.J., Zhao R.C., Peters C., Selleck S.B., Stringer S.E., Gupta P. Functional abnormalities of heparan sulfate in mucopolysaccharidosis-I are associated with defective biologic activity of FGF-2 on human multipotent progenitor cells. Blood. 2005;106:1956–1964. doi: 10.1182/blood-2005-02-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Pasquale V., Scerra G., Scarcella M., D'Agostino M., Pavone L.M. Competitive binding of extracellular accumulated heparan sulfate reduces lysosomal storage defects and triggers neuronal differentiation in a model of mucopolysaccharidosis IIIB. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2021.119113. [DOI] [PubMed] [Google Scholar]
- 69.De Pasquale V., Sarogni P., Pistorio V., Cerulo G., Paladino S., Pavone L.M. Targeting heparan sulfate proteoglycans as a novel therapeutic strategy for mucopolysaccharidoses. Mol. Ther. Methods Clin. Dev. 2018;10:8–16. doi: 10.1016/j.omtm.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruzo A., Garcia M., Ribera A., Villacampa P., Haurigot V., Marco S., Ayuso E., Anguela X.M., Roca C., Agudo J., Ramos D., Ruberte J., Bosch F. Liver production of sulfamidase reverses peripheral and ameliorates CNS pathology in mucopolysaccharidosis IIIA mice. Mol. Ther. 2012;20:254–266. doi: 10.1038/mt.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hassiotis S., Jolly R.D., Hemsley K.M. Development of cerebellar pathology in the canine model of mucopolysaccharidosis type IIIA (MPS IIIA) Mol. Genet. Metab. 2014;113:283–293. doi: 10.1016/j.ymgme.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 72.Dawson G., Fuller M., Helmsley K.M., Hopwood J.J. Abnormal gangliosides are localized in lipid rafts in sanfilippo (MPS3a) mouse brain. Neurochem. Res. 2012;37:1372–1380. doi: 10.1007/s11064-012-0761-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viana G.M., Priestman D.A., Platt F.M., Khan S., Tomatsu S., Pshezhetsky A.V. Brain pathology in mucopolysaccharidoses (MPS) patients with neurological forms. J. Clin. Med. 2020;9 doi: 10.3390/jcm9020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walkley S.U. Secondary accumulation of gangliosides in lysosomal storage disorders. Semin. Cell Dev. Biol. 2004;15:433–444. doi: 10.1016/j.semcdb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 75.McGlynn R., Dobrenis K., Walkley S.U. Differential subcellular localization of cholesterol, gangliosides, and glycosaminoglycans in murine models of mucopolysaccharide storage disorders. J. Comp. Neurol. 2004;480:415–426. doi: 10.1002/cne.20355. [DOI] [PubMed] [Google Scholar]
- 76.Guo Z., Wang Z. The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development. 2009;136:3627–3635. doi: 10.1242/dev.036939. [DOI] [PubMed] [Google Scholar]
- 77.Kim M.S., Saunders A.M., Hamaoka B.Y., Beachy P.A., Leahy D.J. Structure of the protein core of the glypican dally-like and localization of a region important for hedgehog signaling. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13112–13117. doi: 10.1073/pnas.1109877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song H.H., Shi W., Xiang Y.Y., Filmus J. The loss of glypican-3 induces alterations in wnt signaling. J. Biol. Chem. 2005;280:2116–2125. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
- 79.Tkachenko E., Rhodes J.M., Simons M. Syndecans: new kids on the signaling block. Circ. Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 80.Elitt M.S., Barbar L., Tesar P.J. Drug screening for human genetic diseases using iPSC models. Hum. Mol. Genet. 2018;27:R89–R98. doi: 10.1093/hmg/ddy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farkhondeh A., Li R., Gorshkov K., Chen K.G., Might M., Rodems S., Lo D.C., Zheng W. Induced pluripotent stem cells for neural drug discovery. Drug Discov. Today. 2019;24:992–999. doi: 10.1016/j.drudis.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moradi S., Mahdizadeh H., Saric T., Kim J., Harati J., Shahsavarani H., Greber B., J.B.t. Moore 2019;10:341. doi: 10.1186/s13287-019-1455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]