Abstract
Marek's disease virus (MDV) causes malignant lymphomas in chickens (Marek's disease; MD). Although MD has caused significant economic losses to the poultry industry, currently, its occurrence in the field is effectively controlled by vaccination. However, the genetic characteristics of MDV strains have changed, and the poultry industry has experienced MD outbreaks in vaccinated chickens because of enhanced virulence. Meq, an oncoprotein of MDV, is a key transcription factor correlated with the tumorigenesis in MD. Animal experiments using recombinant MDV revealed that distinct polymorphisms in Meq affect the virulence of MDV strains. Meq containing an insertion or deletion is present in some MDV strains. In the 2010s, field strains that encode Meq containing the deletion (S-Meq) were reported. In this study, we characterized the genetic features of S-Meq detected in a Japanese MDV strain and analyzed its transactivation activity to investigate S-Meq's protein function. S-Meq lacked 41 amino acids, and the deletion was at the same position as those observed in other countries. In addition, S-Meq exhibited higher transactivation activity than Meq from other MDV strains circulating in Japan. These results suggest that the deletion in the transactivation domain may enhance the Meq protein's function. Further investigation is needed to clarify whether the deletion in the transactivation domain of Meq affects MDV's virulence.
Key words: deletion, Marek's disease virus, Meq, S-Meq, transactivation
INTRODUCTION
Marek's disease (MD) is a lymphoproliferative disease of chickens caused by Marek's disease virus (MDV or Gallid alphaherpesvirus 2 [GaHV-2]), a cell-associated alphaherpesvirus which belongs to the Herpesviridae family (subfamily: Alphaherpesvirinae, genus: Mardivirus) (https://talk.ictvonline.org). Although MD previously caused significant economic losses to the poultry industry, its occurrence is well-controlled by applying live nonpathogenic and attenuated vaccines (Osterrieder et al., 2006). However, the field MDV strains’ virulence tends to increase over time, possibly due to the relationship between the selection of highly virulent strains in the field and the introduction of vaccines (Trimpert et al., 2017). Although MD cases have dramatically reduced since the introduction of vaccines, several MD cases in vaccinated chickens are sporadically reported, suggesting that a highly virulent MDV strains could potentially induce future outbreaks (Osterrieder et al., 2006).
A distinct diversity has been observed in the meq genes of MDV strains, correlating with its virulence, and animal experiments using recombinant MDV (rMDV) revealed that the polymorphisms in meq could affect the virulence of MDV in vivo (Conradie et al., 2020). The meq gene product, Meq, can regulate the expression of several host and virus genes as a basic leucine zipper transcription factor and has 2 basic regions (BR) and a leucine zipper (ZIP) at the N-terminal and a transactivation domain at the C-terminal (Osterrieder et al., 2006). Unique polymorphisms in meq have been reported in MDV strains in various countries, and therefore, it may reflect on the evolution of the pathogenicity of MDV strains circulating in each country (Renz et al., 2012; Murata et al., 2013; Wajid et al., 2013; Zhuang et al., 2015; Mescolini et al., 2019; Deng et al., 2020).
Besides polymorphisms in meq, insertions and deletions in Meq's transactivation domain have been detected in some MDV strains (Renz et al., 2012; Wajid et al., 2013; Mescolini et al., 2019). For example, in addition to meq, at least 2 other meq variants namely the long-meq (L-meq) and short-meq (S-meq) genes containing a 180-bp-insertion and a 123-bp-deletion in the transactivation domain respectively, were detected in Rispens CVI988, an attenuated vaccine strain of MDV. Since the L-meq gene was first detected in CVI988 as L-Meq, the L-meq gene's product, this possibly contributed to CVI988’s low virulence. Subsequently, however, it was reported that the pathogenic MDV strains circulating in Australia encoded L-Meq (Renz et al., 2012). Moreover, when the pathogenicity of rMDV encoding CVI988 Meq or CVI988 L-Meq was compared, the L-Meq-encoding rMDV indicated a higher pathogenicity (Conradie et al., 2019), suggesting that the insertion in meq (or deletion in L-meq) contributes to the increase (or decrease) in virulence.
In the 2010s, S-Meq-encoding field strains were reported in some countries (Wajid et al., 2013; Mescolini et al., 2019). Therefore, the deletion in meq could be involved in the MDV strains’ evolution in the field. However, whether the deletion in meq affects protein functions and the virulence of the MDV strains is yet to be determined. In Japan, most of the MDV strains encode Meq (Murata et al., 2013). However, in one instance, an MDV strain encoding S-Meq was detected from diseased chickens on a poultry farm in Japan. In this study, we determined the nucleotide sequence of the MDV strain Kgw-c2. In addition, we analyzed the transactivation activity of Kgw-c2 and compared it to that of Meq, which is encoded for most of the MDV strains distributed in Japan to examine whether the deletion in Meq affects the protein functions.
MATERIALS AND METHODS
Background of the Specimen
Several chickens from a small-scale poultry farm in Japan showed signs of depression and neurological symptoms were diagnosed with MD at the livestock hygiene service center. No vaccine against MD was used at this poultry farm. During the diagnostic process at the livestock hygiene service center, a PCR for the meq gene was performed on kidney, liver, and spleen samples sourced from the diseased chickens. Short fragments of meq genes were detected in all of these samples, thus our laboratory was requested to characterize them.
DNA Sequencing
Total cellular DNA samples were extracted from the internal organs using SepaGene (Sankojunyaku, Tokyo, Japan) according to the manufacturer's protocol. The latter were used as templates for PCR reactions using meq-specific primer sets, and amplifications were performed as outlined by Murata et al. (2013). Nucleotide sequences of the amplified meq genes were determined using the GenomeLab GeXP Genetic Analysis System (Beckman Coulter Inc., Brea, CA). The accession numbers of the sequences used for the analysis are indicated in Table 1.
Table 1.
Comparison of the S-Meq protein amino acid sequences among field isolates.
| Basic region |
Leucine zipper |
Transactivation domaina |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDV strain | Country | Accession number | 71 | 77 | 80 | 110 | 114 | 115 | 119 | 150 | 168 | -/176 | 176/217b | 177/218 | 180/221 | 285/326 |
| Kgw-c2 | Japan | LC385874 | A | E | Y | C | C | A | C | H | S | - | A | P | A | T |
| CVI988(S-Meq) | Netherlands | AY243338 | A | E | D | C | R | V | C | H | S | - | P | P | A | I |
| 855/17 | Italy | MK139678 | A | E | Y | S | R | V | C | H | S | - | P | S | T | T |
| MDV/2/SA | Saudi Arabia | KJ949618 | A | E | Y | S | R | V | C | P | S | - | P | P | A | T |
| Iraq3A | Iraq | KC243262 | A | E | Y | C | R | V | R | H | P | - | A | P | A | T |
| Iraq10A | Iraq | KC243264 | S | E | D | C | R | V | C | H | P | - | A | P | A | T |
| Iraq6F | Iraq | KC243263 | S | E | D | C | R | V | C | H | S | - | A | P | A | T |
| Nr-c1c | Japan | LC385871 | A | E | Y | C | R | A | C | H | S | S | A | P | A | T |
S-Meq has 41 aa-deletions in the transactivation domain.
Interruptions at position 2 of the direct proline repeats.
Meq amino acid sequence from a Japanese strain is indicated and its sequence is the most frequent among the field strains in Japan (Murata et al., 2013).
Plasmids
The expression plasmid for S-Meq was constructed according to the instructions set by Murata et al. (2013). The open reading frame (ORF) of the S-meq gene was amplified and cloned into the pCI-neo vector (Promega, Madison, WI). In addition, we introduced a cysteine-to-arginine substitution at position 114 in S-Meq as previously exemplified by Murata et al. (2013). To compare the transactivation activity of S-Meq with that of the Meq protein, we constructed an expression plasmid for Meq from the MDV strain, Nr-c1, which encodes the Meq sequence that is most frequently detected in Japan (Murata et al., 2013). In addition, the c-Jun expression plasmid was constructed to observe the difference in transactivation activity (Murata et al., 2013). For the assay to measure the transactivation activity, a reporter plasmid was prepared by inserting the meq promoter region upstream of the firefly-luciferase-coding region in the pGL3-Basic vector (Promega) (Murata et al., 2013); pRL-TK Renilla luciferase plasmid (Promega) was used as the control plasmid.
Cell Lines and Transfection
Cells from DF-1—a chicken fibroblast cell line—were seeded in 24-well plates at 2.0 × 105 cells per well in 0.5 mL of Dulbecco's modified Eagle's medium, containing 10% fetal bovine serum, and incubated at 39°C under 5% CO2 for 24 h. The cells in each well were transfected with plasmids, S-Meq or Meq (300 ng), c-Jun (200 ng), reporter (500 ng), and control pRL-TK (10 ng) using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions.
Dual-Luciferase Reporter Assay
Dual-luciferase reporter assays were performed according to the instructions set by Murata et al. (2013). The lysates of transfected cells were prepared 24-h post-transfection with a 1 × Passive Lysis Buffer (Promega). Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) and a Luminescencer-JNR AB-2100 (Atto corp., Tokyo, Japan). The luminescence intensity of firefly luciferase was normalized to that of Renilla luciferase, and the results were indicated as relative to the value of the luciferase activity in cells transfected with the pCI-neo vector.
Statistics
All values are expressed as mean ± standard deviation, and statistical comparisons were performed using the Tukey–Kramer test using EZR (https://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html). Results with P < 0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Amino Acid Sequence Comparison of S-Meq
We determined the nucleotide sequence of Kgw-c2 S-Meq, and compared its amino acid sequence with those detected in other countries. S-Meq lacked 41 amino acids in Meq's transactivation domain, and its position was the same as observed in previous reports (Wajid et al., 2013; Mescolini et al., 2019). The cysteine residue at position 114 was the unique feature observed in Kgw-c2 S-Meq (Table 1). Furthermore, Kgw-c2 S-Meq had a PXPP sequence at position 176 (Table 1), which is a proline-to-X substitution in the direct proline repeats in the transactivation domain. The interruptions at position 2 in the direct repeats of 4 prolines are found in highly virulent MDV strains (Conradie et al., 2020). This proline-to-alanine substitution at position 176 was also detected in MDV strains in Iraq (Wajid et al., 2013).
An earlier report suggested that the reduced number of PPPP sequences—direct repeats of 4 prolines—in the transactivation domain in Meq correlated with the high virulence of MDV strains in Australia (Renz et al., 2012). Due to the deletion in the transactivation domain, S-Meq had fewer PPPP sequences than Meq, and L-Meq. Therefore, based on the hypothesis that the number of PPPP sequences correlates with virulence, S-Meq-encoding MDV strains’ pathogenicity is predicted to be high. However, the pathogenicity of MDV encoding CVI988 L-Meq was higher than that of CVI988 Meq (Conradie et al., 2019), although L-Meq has more PPPP sequences than Meq. Some amino acid substitutions in PPPP sequences were observed in L-Meq-encoding MDV strains circulating in Australia, resulting in fewer PPPP sequences than the other L-Meq-encoding MDV strains (Renz et al., 2012). Thus, MDV strains in Australia may have shown a high virulence because of insertions in the transactivation domain with PPPP-to-PXPP substitutions. The insertion (and perhaps deletion) in the transactivation domain possibly affects virulence. Although the reduced number of PPPP sequences may affect MDV's virulence, it cannot predict S-Meq-encoding MDV strains’ virulence or facilitate a comparison with those encoding Meq or L-Meq, respectively.
Transactivation Activity of Kgw-c2 S-Meq
To examine the transactivation activity of Kgw-c2 S-Meq, we compared it with Nr-c1 Meq which is observed in most of the MDV strains circulating in Japan (Murata et al., 2013). The transactivation activity of Kgw-c2 S-Meq was higher than that of Nr-c1 Meq (Figure 1). There was an amino acid difference at position 114 between Kgw-c2 S-Meq and Nr-c1 Meq, including the differences in the transactivation domain (Table 1). Therefore, we introduced a cysteine-to-arginine substitution at position 114 in Kgw-c2 S-Meq, and compared the transactivation of the mutant, Kgw-c2 S-Meq C114R, with those of Kgw-c2 S-Meq and Nr-c1 Meq. The transactivation activity of the mutant was lower than that of wild-type Kgw-c2 S-Meq, however it was still higher than that of Nr-c1 Meq (Figure 1). These data suggest that the deletion in Meq, in addition to the amino acid substitution at position 114, affects the transactivation activity of Meq. Thus, Kgw-c2 S-Meq seems to have a higher transactivation activity than the Meq predominantly found in MDV strains circulating in Japan. Based on the transactivation activity, Kgw-c2’s protein function seems higher than Nr-c1 and could be a factor affecting variation among their virulence.
Figure 1.
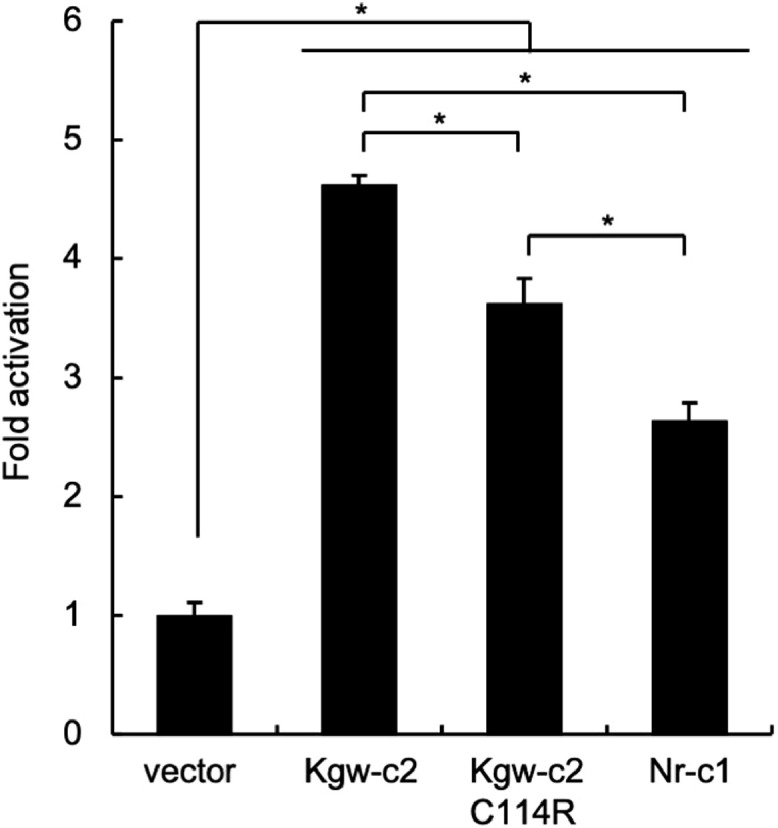
Transactivation activity of Kgw-c2 S-Meq. The transactivation activity of Kgw-c2 S-Meq on the Meq promoter-driven luciferase activities was compared with those of Nr-c1 Meq and Kgw-c2 C114R, which were introduced to a cysteine-to-arginine substitution at position 114. DF-1 cells were transfected with S-Meq or Meq-expression plasmids (300 ng), c-Jun-expression plasmid (200 ng), reporter plasmid (500 ng), and pRL-TK internal control plasmid (10 ng). Luciferase activities were analyzed 24 h post-transfection. Firefly luciferase activity is expressed relative to the mean basal activity in the presence of pCI-neo after normalization to Renilla luciferase activity. Error bars indicate standard deviations. *P < 0.05.
In the 2010s, some MDV strains whose Meq contains a 41 aa-deletion in the transactivation domain have emerged in the field (Wajid et al., 2013; Mescolini et al., 2019). The current study suggests that the position of the deletion in S-Meq of Kgw-c2, which caused the disease on a poultry farm in Japan, was similar to those reported in other countries. In addition, we clarified that the transactivation activity of Kgw-c2 S-Meq was higher than that of Meq predominantly found in MDV strains circulating in Japan. These data suggest that the deletion in the transactivation domain could be a factor contributing to the virulence of MDV strains and may support the possibility that the deletion in Meq is an aspect of MDV strains’ evolution. However, Kgw-c2 was detected in unvaccinated chickens; therefore, Kgw-c2’s potential to break the vaccination-induced immunity requires further investigation. In addition, the pathogenicity of S-Meq-encoding MDV strains in other countries is unclear. Thus, we have limited information regarding S-Meq-encoding MDV strains’ potential to cause MD outbreaks. Therefore, animal experiments using rMDV encoding Kgw-c2 S-Meq should be conducted to decide whether the deletion in meq affects MD pathogenicity.
Acknowledgments
ACKNOWLEDGMENTS
This research was supported in part by Grants-in-Aid for Scientific Research (B:18H02332 and B:20H03137) and Grant-in-Aid for Challenging Research (Exploratory) (20K21357) from the Japan Society for the Promotion of Science. We would also like to thank Editage (www.editage.jp) for their English language editing services.
DISCLOSURES
The authors have no conflicts of interest.
REFERENCES
- Conradie A.M., Bertzbach L.D., Bhandari N., Parcells M., Kaufer B.B. A common live-attenuated avian herpesvirus vaccine expresses a very potent oncogene. mSphere. 2019;4 doi: 10.1128/mSphere.00658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradie A.M., Bertzbach L.D., Trimpert J., Patria J.N., Murata S., Parcells M.S., Kaufer B.B. Distinct polymorphisms in a single herpesvirus gene are capable of enhancing virulence and mediating vaccinal resistance. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Q., Shi, M., Li, Q., Wang, P., Li, M., Wang, W., Gao, Y., Li, H., Lin, L., Huang, T. and Wei, P. 2020. Analysis of the evolution and transmission dynamics of the field MDV in China during the years 1995–2020, indicating the emergence of a unique cluster with the molecular characteristics of vv+ MDV that has become endemic in southern China [e-pub ahead of print]. Transbound. Emerges. Dis. doi: 10.1111/tbed.13965, accessed December 23, 2020. [DOI] [PubMed]
- Mescolini G., Lupini C., Felice V., Guerrini A., Silveira F., Cecchinato M., Catelli E. Molecular characterization of the meq gene of Marek's disease viruses detected in unvaccinated backyard chickens reveals the circulation of low- and high-virulence strains. Poult. Sci. 2019;98:3130–3137. doi: 10.3382/ps/pez095. [DOI] [PubMed] [Google Scholar]
- Murata S., Hashiguchi T., Hayashi Y., Yamamoto Y., Matsuyama-Kato A., Takasaki S., Isezaki M., Onuma M., Konnai S., Ohashi K. Characterization of Meq proteins from field isolates of Marek's disease virus in Japan. Infect. Genet. Evol. 2013;16:137–143. doi: 10.1016/j.meegid.2012.12.032. [DOI] [PubMed] [Google Scholar]
- Osterrieder N., Kamil J.P., Schumacher D., Tischer B.K., Trapp S. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- Renz K.G., Cooke J., Clarke N., Cheetham B.F., Hussain Z., Fakhrul Islam A.F.M., Tannock G.A., Walkden-Brown S.W. Pathotyping of Australian isolates of Marek's disease virus and association of pathogenicity with meq gene polymorphism. Avian Pathol. 2012;41:161–176. doi: 10.1080/03079457.2012.656077. [DOI] [PubMed] [Google Scholar]
- Trimpert J., Groenke N., Jenckel M., He S., Kunec D., Szpara M.L., Spatz S.J., Osterrieder N., McMahon D.P. A phylogenomic analysis of Marek's disease virus reveals independent paths to virulence in Eurasia and North America. Evol. Appl. 2017;10:1091–1101. doi: 10.1111/eva.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajid S.J., Katz M.E., Renz K.G., Walkden-Brown S.W. Prevalence of Marek's disease virus in different chicken populations in Iraq and indicative virulence based on sequence variation in the ecoRI-q (meq) gene. Avian Dis. 2013;57(2 Suppl):562–568. doi: 10.1637/10342-083112-Reg.1. [DOI] [PubMed] [Google Scholar]
- Zhuang X., Zou H., Shi H., Shao H., Ye J., Miao J., Wu G., Qin A. Outbreak of Marek's disease in a vaccinated broiler breeding flock during its peak egg-laying period in China. BMC Vet. Res. 2015;11:157. doi: 10.1186/s12917-015-0493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


