Abstract
Background & Aims
CD4+ T cells are regulated by activating and inhibitory cues, and dysregulation of these proper regulatory inputs predisposes these cells to aberrant inflammation and exacerbation of disease. We investigated the role of the inhibitory receptor paired immunoglobulin-like receptor B (PIR-B) in the regulation of the CD4+ T-cell inflammatory response and exacerbation of the colitic phenotype.
Methods
We used Il10-/- spontaneous and CD4+CD45RBhi T-cell transfer models of colitis with PIR-B-deficient (Pirb-/-) mice. Flow cytometry, Western blot, and RNA sequencing analysis was performed on wild-type and Pirb-/- CD4+ T cells. In silico analyses were performed on RNA sequencing data set of ileal biopsy samples from pediatric CD and non–inflammatory bowel disease patients and sorted human memory CD4+ T cells.
Results
We identified PIR-B expression on memory CD4+ interleukin (IL)17a+ cells. We show that PIR-B regulates CD4+ T-helper 17 cell (Th17)-dependent chronic intestinal inflammatory responses and the development of colitis. Mechanistically, we show that the PIR-B– Src-homology region 2 domain-containing phosphatase-1/2 axis tempers mammalian target of rapamycin complex 1 signaling and mammalian target of rapamycin complex 1–dependent caspase-3/7 apoptosis, resulting in CD4+ IL17a+ cell survival. In silico analyses showed enrichment of transcriptional signatures for Th17 cells (RORC, RORA, and IL17A) and tissue resident memory (HOBIT, IL7R, and BLIMP1) networks in PIR-B+ murine CD4+ T cells and human CD4+ T cells that express the human homologue leukocyte immunoglobulin-like receptor subfamily B member 3 (LILRB3). High levels of LILRB3 expression were associated strongly with mucosal injury and a proinflammatory Th17 signature, and this signature was restricted to a treatment-naïve, severe pediatric CD population.
Conclusions
Our findings show an intrinsic role for PIR-B/LILRB3 in the regulation of CD4+ IL17a+ T-cell pathogenic memory responses.
Keywords: Paired Immunoglobulin Receptor, CD4+ T Cells, Interleukin 17, Inflammatory Bowel Disease
Abbreviations used in this paper: cCD, colonic-only involvement Crohn’s disease; CD, Crohn’s disease; DEG, differentially expressed gene; DU, deep ulcer; ERK, extracellular signal-regulated kinase; GAP, GTPase-activating protein; GFP, green fluorescent protein; GTP, guanosine triphosphate; IBD, inflammatory bowel disease; iCD, ileal involvement Crohn’s disease; IFN, interferon; IL, interleukin; ILC, innate lymphoid cell; ITIM, immunoreceptor tyrosine-based inhibitory motif; LILRB3, leukocyte immunoglobulin-like receptor subfamily B member 3; LP, lamina propria; mLN, mesenteric lymph node; mRNA, messenger RNA; mTORC1, mammalian target of rapamycin complex 1; PCR, polymerase chain reaction; p-ERK, phosphorylated extracellular signal-regulated kinase; PIR-B, paired immunoglobulin-like receptor B; Q, quartile; RPKM, reads per kb of transcript, per million mapped reads; SHP, Src-homology region 2 domain-containing phosphatase; Th, T-helper cell; TNF, tumor necrosis factor; TRM, tissue resident memory; TSC, tuberous sclerosis; WT, wild-type
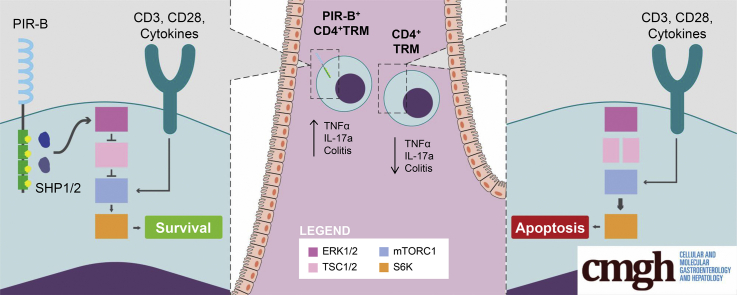
Graphical abstract
Summary.
The inhibitory receptor paired immunoglobulin-like receptor B regulates CD4+ T-helper 17 cell–dependent chronic intestinal inflammatory responses by tempering mammalian target of rapamycin complex 1 signaling and enhancing CD4+ interleukin 17a+ T-cell survival and regulates the outgrowth and maintenance of tissue resident memory CD4+ interleukin 17a+ T cells.
Inflammatory bowel diseases (IBDs), encompassing Crohn’s disease (CD) and ulcerative colitis (UC), are progressive chronic relapsing-remitting diseases, which result from an exaggerated inflammatory response to intestinal microbes in a genetically susceptible individual.1 Activation of innate immune receptors, such as Toll-like receptors and Nod-like receptors by pathogenic bacteria and viruses (dysbiosis), injury, or xenobiotic elements,2,3 leads to activation of intestinal macrophages and dendritic cells, and in turn drives proinflammatory cytokine (interleukin [IL]6, IL12, and IL23) production. These cytokines stimulate the development of effector CD4+-T-helper 1 cell (Th1), CD4+-Th2, and CD4+-Th17,4, 5, 6 activation of innate lymphoid cells (ILC1, ILC2, Natural cytotoxicity receptor (NCR)+ILC3, and NCR-ILC3 cells), which leads to an IL17A/IFNγ/TNFα-dominant proinflammatory response and the histopathologic manifestations of disease.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18
Paired immunoglobulin-like receptor B (PIR-B) is a immunoreceptor tyrosine-based inhibitory motif (ITIM)19,20 containing type I transmembrane glycoprotein expressed predominantly on myeloid cells, B cells, and granulocytes.21, 22, 23 Activation of PIR-B via major histocompatibility class I molecules in cis and trans fashion,24 and cell wall components of certain gram-negative and gram-positive bacteria,22,25,26 induces PIR-B ITIM domain engagement and activation of the intracellular phosphatases Src-homology region 2 domain-containing phosphatase (SHP)-1 and SHP-2. Subsequently, SHP-1 dephosphorylates p65 and extracellular signal-regulated kinase (ERK)1/2, resulting in the inhibition of downstream nuclear factor-κB– and mitogen-activated protein kinase–signaling pathways27 and inhibition of B-cell receptor, Toll-like receptor, and chemokine-receptor signaling.28 We previously reported that PIR-B restrains innate-immune–induced proinflammatory cytokine (IL1β, IL6, and TNFα) production by macrophages and limits acute intestinal inflammation and epithelial cell injury.29
Using PIR-B-deficient (Pirb-/-) mice and using the Il10-/- spontaneous, αCD3-mediated, and CD4+CD45RBhi T-cell transfer model of colitis, we show that loss of PIR-B expression protected mice from the development of CD4+ T-cell–dependent colitis. Notably, disease protection was associated with significantly reduced frequency of tissue resident memory (TRM) CD4+IL17a+ T cells. Adoptive transfer experiments showed that PIR-B expression on CD4+ T cells conferred a competitive advantage for T-cell survival and TRM CD4+ T-cell development. In vitro studies have shown that Pirb-/- naïve CD4+ T cells have decreased capacity to differentiate into Th17 cells, impaired cell-cycle entry into G1 and S phases, and enhanced cell death. Mechanistic analysis has shown that PIR-B acts as a rheostat, controlling mammalian target of rapamycin complex 1 (mTORC1) signaling in CD4+ T cells and limiting CD4+ IL17A+ T-cell outgrowth. Leukocyte immunoglobulin-like receptor subfamily B member 3 (LILRB3) expression was associated strongly with mucosal injury and a proinflammatory Th17 signature in a treatment-naïve endoscopically severe pediatric CD population. Flow cytometry and RNA sequencing analysis showed enhanced PIR-B and LILRB3 expression on a subset of memory CD4+ Th17 cells in mice and human beings, respectively. Collectively, these data suggest an intrinsic role for PIR-B in the regulation of the outgrowth and maintenance of TRM CD4+ IL17a+ T cells and the development of T-cell–dependent colitis.
Results
Decreased Susceptibility of Pirb-/- Mice to Il10-/- Spontaneous Colitis
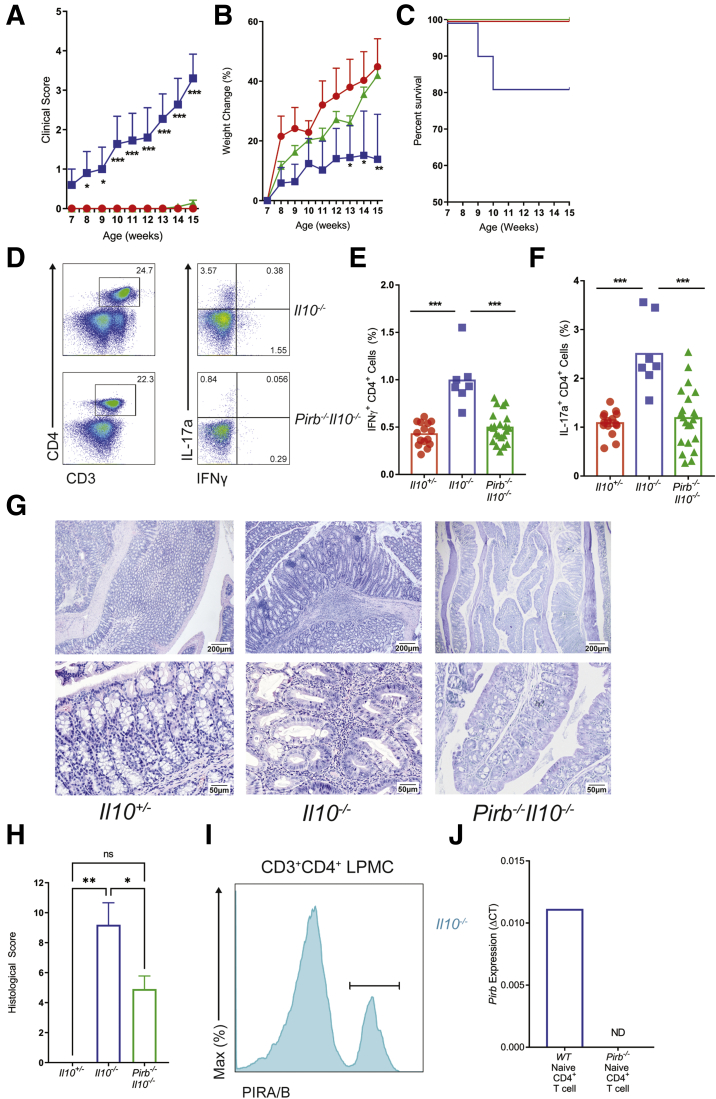
To define the role of PIR-B in T-cell–mediated colitis we backcrossed Pirb-/- mice (C57BL6) with Il10-/- mice (C57BL6), and monitored for the development of the spontaneous colitis phenotype. Pirbwild-type (WT)Il10-/- (Il10-/-) mice showed colitic symptoms including rectal prolapse, anal bleeding, diarrhea, and dehydration by 8 weeks of age, and disease progressed until 15 weeks of age (Figure 1A). These symptoms were associated with failure to thrive and an approximately 20% mortality rate (Figure 1B and C). Flow cytometry analysis showed increased frequency of CD4+ IFNγ+ T cells and CD4+ IL17a+ T cells in the mesenteric lymph nodes (mLN) of the colitic Il10-/- mice (Figure 1D–F). Histologic analysis of the colon from Il10-/- mice showed significant evidence of epithelial erosions, crypt abscesses, and transmural inflammation (Figure 1G and H). By contrast, Pirb-/-Il10-/- mice had significantly reduced symptoms (Figure 1A), with increased weight gain and survival rates comparable with healthy control Il10+/- mice (Figure 1B and C). Notably, the frequency of CD4+ IFNγ+ T cell and CD4+ IL17a+ T cell populations in the mLN of Pirb-/-Il10-/- mice was reduced significantly compared with colitic Il10-/- mice (Figure 1D–F). Congruent with the reduced disease phenotype, the colons of Pirb-/-Il10-/- mice showed reduced histopathologic disease phenotype possessing normal colonic epithelial architecture with reduced cellular infiltrate (Figure 1G and H). Using the PIR 6C1 antibody, which recognizes both PIR-A and PIR-B,19 we identified PIR-B expression on colonic lamina propria (LP) CD4+ T cells from PirbWTIl10-/- mice (Figure 1I). Additional quantitative reverse-transcription polymerase chain reaction (PCR) analyses showed PIR-B messenger RNA (mRNA) expression in naïve splenic CD4+ T cells (Figure 1J). Collectively, these data show that PIR-B is expressed on CD4+ T cells, and mice deficient in PIR-B are protected from Il10-/- spontaneous colitis.
Figure 1.
Loss of Pirb suppresses the development of spontaneous colitis in Il10-/-mice. (A) Clinical score, (B) percentage body weight change (relative to weight at start of the experiment), and (C) percentage survival rate of Il10+/- (red), Il10-/- (blue), Pirb-/-Il10-/- (green) mice corresponding to age (Il10+/-, n = 18; Il10-/-, n = 11; Pirb-/-Il10-/-, n = 22). (D) Flow cytometry analysis of CD4+IFNγ+ and CD4+IL17a+ T cells in the mLN of mice at 15 weeks of age. Percentage of (E) CD4+IFNγ+ or (F) CD4+IL17a+ T cells in the mLN of mice at 15 weeks of age (Il10+/-, n = 15; Il10-/-, n = 7; Pirb-/-Il10-/-, n = 22). (G) Representative image of colon histology (H&E staining) from Il10-/- and Pirb-/-Il10-/- mice; top row: 4× magnification. Bottom row: 20× magnification. (H) Colon histologic scoring from Il10+/-, Il10-/-, and Pirb-/-Il10-/- mice at 15 weeks of age (Il10+/-, n = 4; Il10-/-, n = 10; Pirb-/-Il10-/-, n = 22). (I) Representative flow cytometry histogram of the expression of PIRA/B on CD3+CD4+ T cells isolated from colonic lamina propria of Il10-/- mice. (J) Pirb mRNA expression in splenic naïve CD4+ T cells from WT and Pirb-/- mice (WT, n = 3; Pirb-/-, n = 3). Data are presented as means ± SEM. Statistical analysis was performed using 2-way analysis of variance (P < .05) followed by the (A and B) Dunnett multiple comparison test or the (E, F, and H) unpaired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (A–C, E, F, and H) Data shown encompass 3 independent experiments. LPMC, lamina propria mononuclear cell.
PIR-B Is Required for CD4+ T-Cell–Dependent Enteropathy
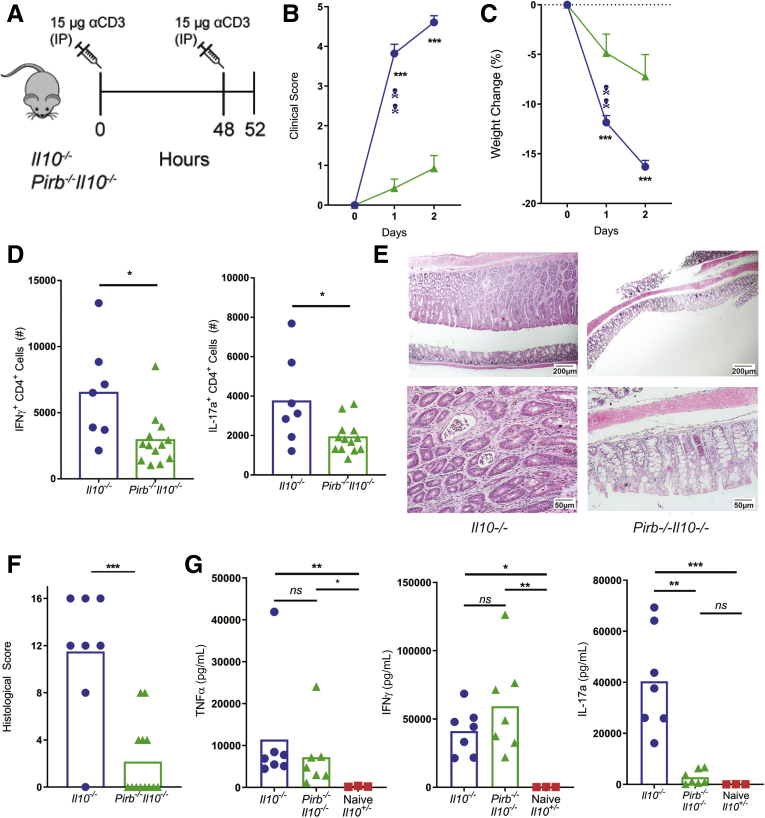
To directly assess the consequence of PIR-B deficiency on CD4+ T-cell activation and T-cell–dependent intestinal injury; we used the αCD3-mediated model of intestinal enteropathy (Figure 2A).30,31 Transient activation of T cells in Il10-/- mice by peritoneal injection of αCD3 resulted in the development of clinical signs of diarrhea, piloerection, decreased mobility, and exaggerated weight loss (Figure 2B and C) that was associated with a potent mLN CD4+ IFNγ+ T cell response and CD4+ IL17a+ T cell response (Figure 2D). Examination of colonic sections showed substantial epithelial injury including epithelial apoptosis, villus atrophy, and an inflammatory infiltrate (Figure 2E and F). In contrast, Pirb-/-Il10-/- mice showed limited evidence of αCD3-mediated disease, showing reduced clinical scores and weight loss, and this was associated with reduced numbers of mLN CD4+ IFNγ+ T cells and CD4+ IL17a+ T cells, and colonic injury (Figure 2B–F). Systemic levels of TNFα and IFNγ were comparable between Il10-/- and Pirb-/-Il10-/- mice; however, IL17a was reduced significantly in Pirb-/-Il10-/- mice compared with Il10-/- mice (Figure 2G). These studies suggest that PIR-B negatively regulates exacerbation of T-cell–dependent enteropathy and may have direct regulatory effects on the CD4+ Th17 compartment.
Figure 2.
Pirb-deficient CD4+T cells fail to induce acute intestinal enteropathy. (A) Schematic representation of αCD3 treatment protocol. (B) Clinical score and (C) percentage body weight change (relative to weight before initial αCD3 injection) of Il10-/- and Pirb-/-Il10-/- mice after αCD3 injection (Il10-/-, n = 14; Pirb-/-Il10-/-, n = 14). (D) Cell counts of CD4+IFNγ+ and CD4+IL17a+ T cells in the mLN of mice (Il10-/-, n = 7; Pirb-/-Il10-/-, n = 13). (E) Representative image of colon histology (H&E staining) from Il10-/- and Pirb-/-Il10-/- mice. Top row: 4× magnification; bottom row: 20× magnification. (F) Colon histologic scoring from Il10-/- and Pirb-/-Il10-/- mice (Il10-/-, n = 8; Pirb-/-Il10-/-, n = 13). (G) Systemic levels of TNFα, IFNγ, and IL17a in serum of Il10-/- and Pirb-/-Il10-/- mice at 52 hours after a CD3 injection. Cytokine levels were detected by in vivo cytokine capture assay (IVCCA) (Il10-/-, n = 7; Pirb-/-Il10-/-, n = 7; Il10+/-, n = 3). Data are presented as means ± SEM. Statistical analysis was performed using 2-way analysis of variance (P < .05) followed by the (B and C) Sidak multiple comparison test or (D, F, and G) the unpaired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. Data shown encompass 3 independent experiments. IP, intraperitoneally.
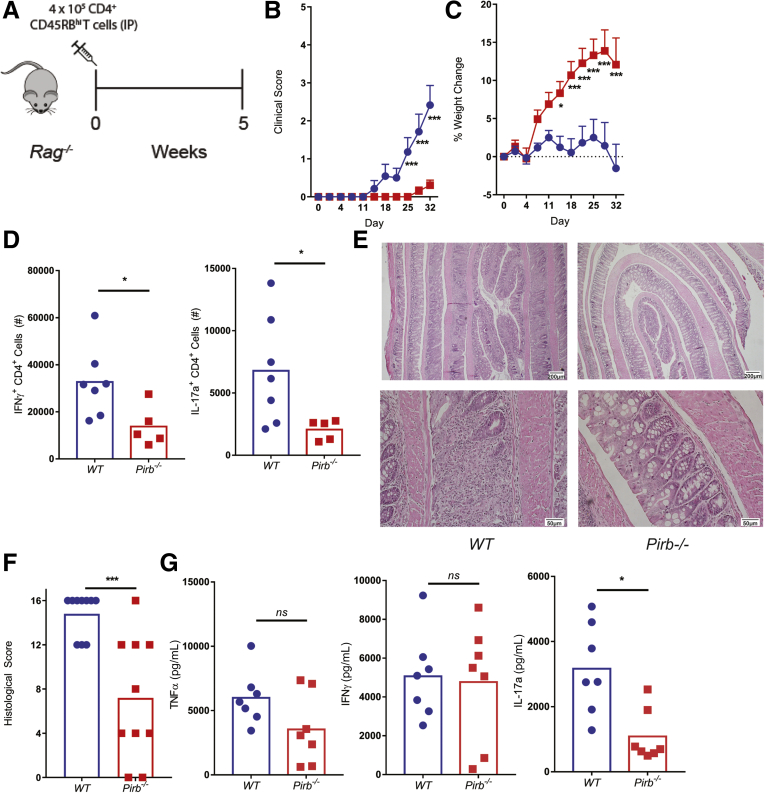
To test whether PIR-B intrinsically regulates CD4+ T cells, we used the CD4+CD45RBhi T-cell transfer model of colitis.32,33 Rag-/- mice that received naïve WT CD4+ T cells (400,000 cells) developed symptoms of colitis 2 weeks after T-cell transfer and these mice showed substantial weight loss by day 30 (Figure 3A–C). Development of colitis was associated with the presence of CD4+ IFNγ+ T cells and CD4+ IL17a+ T cells in the mLN (Figure 3D), colonic inflammation with severe crypt destruction (Figure 3E and F), and systemic TNFα, IFNγ, and IL17a cytokines (Figure 3G). By contrast, Rag-/- mice that received naive Pirb-/- CD4+ T cells developed a reduced colitic phenotype as evident by clinical score, weight loss, and colon histopathology (Figure 3B, C, E, and F). Consistent with this, the frequency of CD4+ IFNγ+ T cells and CD4+ IL17a+ T cells was reduced significantly in Rag-/- mice, which received Pirb-/- CD4+ T cells (Figure 3D). Importantly, Pirb deficiency in the CD4+ T-cell compartment resulted in the down-regulation of systemic levels of IL17a, but not TNFα and IFNγ (Figure 3G). Together, these data confirm that PIR-B intrinsically regulates IL17a+ inflammatory responses in vivo and indicate that PIR-B is required for archetypal CD4+ IL17a+ T-cell response.
Figure 3.
Pirb-deficient CD4+T cells fail to induce chronic colitis. (A) Schematic representation of T-cell–transfer protocol. Rag-/- mice received either 400,000 CD4+ CD45RBhi WT (blue) or Pirb-/- (red) T cells. (B) Clinical score and (C) percentage body weight change (relative to weight at start of the experiment) of Rag-/- mice after T-cell injection (WT, n = 13; Pirb-/-, n = 14). (D) Cell counts of CD4+IFNγ+ and CD4+IL17a+ T cells in the mLN of mice (WT, n = 7; Pirb-/-, n = 5). (E) Representative image of colon histology (H&E staining) from Rag-/- mice. Top row: 4× magnification; bottom row: 20× magnification. (F) Colon histologic scoring from Rag-/- mice (WT, n = 10; Pirb-/-, n = 10). (G) Systemic levels of TNFα, IFNγ, and IL17a in serum of Rag-/- mice at 5 weeks after T-cell transfer (WT, n = 7; Pirb-/-, n = 7). Cytokine levels were detected by IVCCA. Data are presented as means ± SEM. Statistical analysis was performed using 2-way analysis of variance (P < .05) followed by the (B and C) Sidak multiple comparison test or (D, F, and G) unpaired t test. ∗P < .05 and ∗∗∗P < .001. Data shown encompass 3 independent experiments. IP, intraperitoneally.
PIR-B Intrinsically Regulates Th17 Cell Survival In Vitro
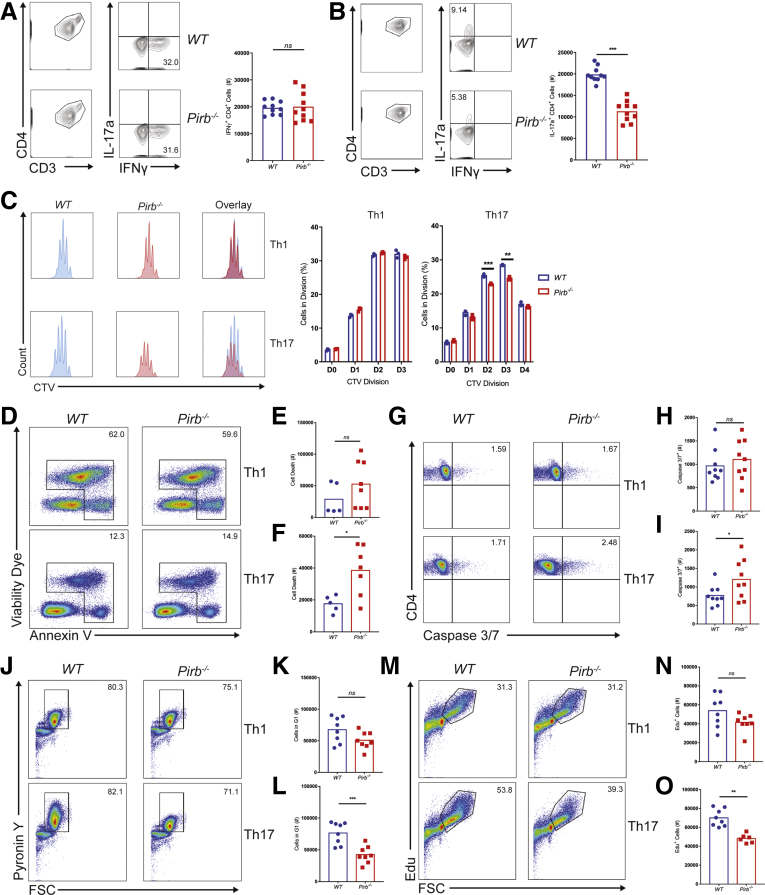
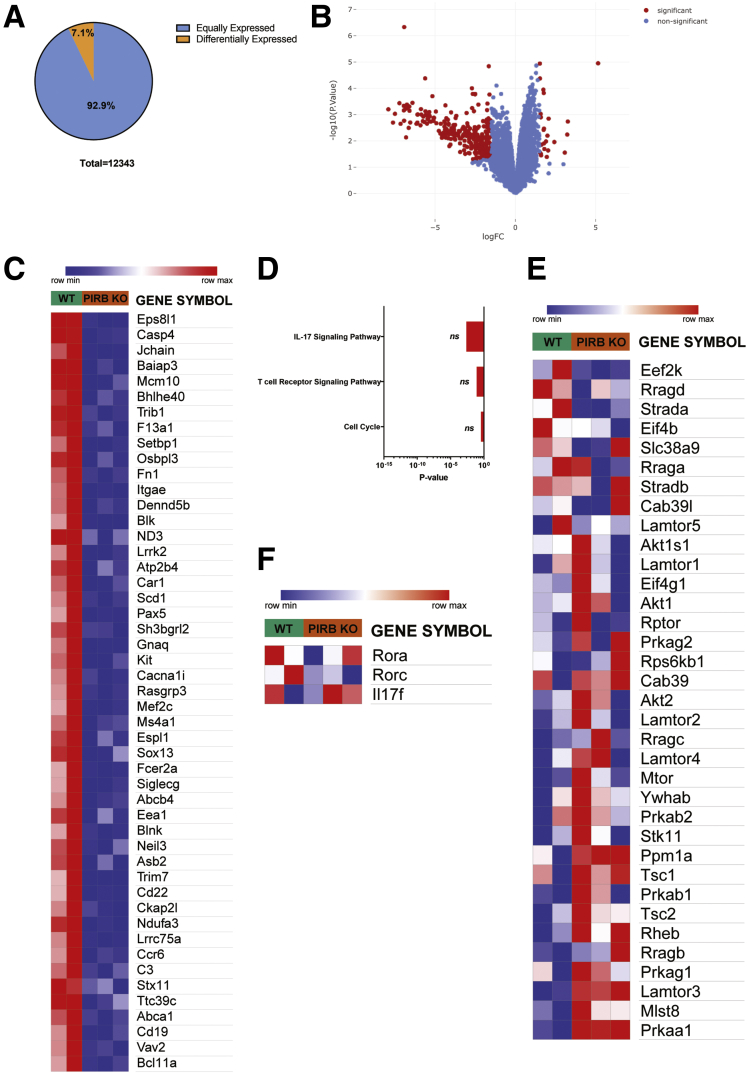
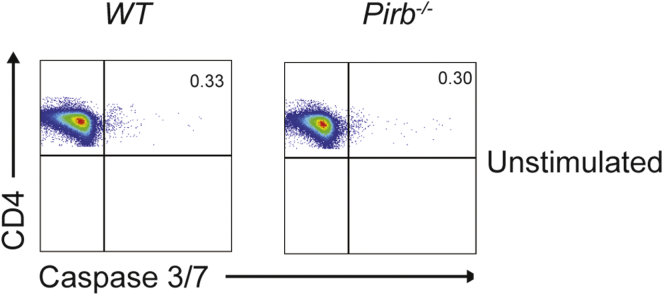
To gain mechanistic insight into PIR-B regulation of CD4+ T-cell function, we assessed the proliferation and differentiation capacity of WT and Pirb-/- naïve CD4+ T cells. Polyclonal activation of WT and Pirb-/- CD4+ T cells under Th1 polarizing conditions induced equivalent frequencies of CD4+ IFNγ+ T cells (Figure 4A). Consistent with this, we observed comparable number and cellular proliferative capacity in WT and Pirb-/- CD4+ Th1 cells (Figure 4C). Under Th17 polarizing conditions we observed a significant reduction in the frequency and number of CD4+ IL17a+ T cells generated from Pirb-/- CD4+ T cells compared with WT CD4+ T cells (Figure 4B). The reduced frequency in Pirb-/- CD4+ IL17a+ T cells was not a consequence of reduced proliferative capacity because we observed a similar number of cellular divisions (Figure 4C). However, quantification of the number of cells at each division showed a reduced frequency of Pirb-/- CD4+ T cells at each cell division compared with WT CD4+ T cells (Figure 4C), suggesting that PIR-B regulates CD4+ IL17a+ T-cell survival. Consistent with this, we observed increased cell death occurring via apoptosis, as evidenced by significantly increased caspase 3/7 activation in Pirb-/- CD4+ T cells compared with WT CD4+ T cells (Figure 4D–I). Importantly, this was specific to Th17 polarizing conditions because no significant differences in cell death or caspase 3/7 activation was observed in Pirb-/- CD4+ T cells compared with WT CD4+ T cells under Th1 polarizing conditions (Figure 4D, E, G, and H). To determine whether the effects of PIR-B were in part associated with CD4+ T-cell development, we performed RNA sequencing on unstimulated WT and Pirb-/- CD4+ naïve T cells. We identified a total of 12,343 genes expressed (reads per kb of transcript, per million mapped reads [RPKM], >5) by WT and Pirb-/- CD4+ naïve T cells (Supplementary Table 1, Figure 5A). A total of 11,463 genes (92.9% of totally expressed genes) were expressed equally (RPKM, >5; fold change, >1.0 and <1.5) between WT and Pirb-/- CD4+ naïve T cells and 880 differentially expressed genes (DEGs) (19 genes were up-regulated, 861 genes were down-regulated) (Supplementary Table 1, Figure 5). Gene network and pathway analyses showed no significant enrichment of T cell, cytokine and apoptosis, T-cell receptor, apoptosis and cell-cycle pathways, or differences in core Th17 or mTOR signaling genes (Supplementary Table 1, Figure 5A–F). Consistent with this, we observed no differences in live dead staining, Annexin V expression, or caspase 3/7 activation between WT and Pirb-/- naïve CD4+ T cells before stimulation, indicating that the observed induction of cell death and apoptosis was associated with CD4+ T-cell activation (Figure 6). We next investigated the impact of PIR-B on early CD4+ T-cell activation and cell-cycle entry under Th1 and Th17 polarizing conditions (Figure 4J–O). We observed significantly fewer Pirb-/- CD4+ T cells entering the G1 and S phase of the cell cycle by comparison with WT CD4+ T cells under Th17 polarizing conditions (Figure 4J, L, M, and O); importantly the cell-cycle G1 and S phase impairment of PIR-B–deficient cells was restricted to Th17 and not Th1 differentiation (Figure 4J–O). Cumulatively, these results indicate that PIR-B acts as a T-cell intrinsic factor regulating the outgrowth of Th17 cells by modulating cell survival and cell cycle.
Figure 4.
Pirb-/-CD4+T cells have impaired survival and differentiation under Th17 polarizing conditions. Gating strategy and cell counts of WT and Pirb-/- CD4+ T cells cultured under (A) Th1 polarizing and (B) Th17 polarizing conditions for 72 hours (WT, n = 10; Pirb-/-, n = 10). (C) Representative plots of Cell Trace Violet staining on Th1 polarized (top) and Th17 polarized (bottom) CD4+ T cells (left). Percentages of cells within each division (right) (WT, n = 3; Pirb-/-, n = 3). (D) Representative plots of the Annexin V expression and viability in CD4+ T cells activated under Th1 (top) and Th17 (bottom) conditions. Counts of dead (Annexin+ or Viability Dye+) CD4+ T cells cultured under (E) Th1 conditions or (F) Th17 conditions (WT, n = 5; Pirb-/-, n = 8). (G) Representative plots of the expression of active caspase 3/7 in CD4+ T cells activated under Th1 (top) and Th17 (bottom) conditions for 4 hours. Frequency of activated caspase 3/7+ CD4+ T cells cultured under (H) Th1 polarizing conditions or (I) Th17 polarizing conditions for 4 hours (WT, n = 9; Pirb-/-, n = 9). (J) Representative plots showing G1 cell-cycle entry via the expression of Pyronin Y in CD4+ T cells under Th1 (top) and Th17 (bottom) polarizing conditions for 12 hours. Frequency of CD4+ T cells that have entered G1 (Pyronin Y+) under (K) Th1 conditions or (L) Th17 conditions for 12 hours (WT, n = 8; Pirb-/-, n = 8). (M) Representative plots showing the incorporation of Edu in CD4+ T cells under Th1 (top) and Th17 (bottom) conditions for 28 hours. Frequency of CD4+ T cells that have entered S phase (Edu+) under (N) Th1 conditions or (O) Th17 conditions for 28 hours (WT, n = 8; Pirb-/-, n = 6). Data are presented as means ± SEM. Statistical analysis was performed using an unpaired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (A, B, E, F, H, I, K, L, N, and O) Data shown encompass 3 independent experiments. (C) Data shown are from 1 experiment, representative of 3 independent experiments. CTV, cell trace violet; FSC, forward scatter.
Figure 5.
RNA sequencing analysis of unstimulated WT and Pirb-/-naïve CD4+T cells. (A) Venn diagram indicating equal (blue: RPKM, ≥5; fold change, ≥1 and ≤1.5; n = 11,463) and differential (orange: RPKM, ≥5; Padjusted ≤ .05; absolute fold change, ≥1.5; n = 880) gene expression between WT naïve CD4+ T cells and Pirb-/- CD4+ naive T cells. (B) Volcano plot of RNA sequencing data analysis. Red indicates significantly enriched genes (|logFC| > 1; P < .05). (C) Heat map of differentially expressed genes comparing WT naïve CD4+ T cells and Pirb-/- CD4+ naive T cells. (D) Bar graphs of pathway analysis of differentially expressed genes in WT naïve CD4+ T cells relative to Pirb-/- CD4+ naive T cells; assessed via KEGG 2019, ranked by P value. Heat map showing relative expression of (E) mTORC1 core signaling and (F) Th17 transcriptome signature genes between WT naïve CD4+ T cells and Pirb-/- CD4+ naive T cells. Raw reads from WT (n = 2) and Pirb-/- CD4+ T cells (n = 3) were aligned to the reference mm9 mouse genome (GRCm38) using Hisat-build pipeline. Relative gene expression was quantified using the featureCounts function from the subread-2.0.0 package. The gene list was filtered for pseudogenes, RIKEN complementary DNA sequences, and immunoglobulin variable genes and downstream analysis of expressed genes was performed using IDEP 9.1. Raw data and the description of commonly expressed genes and DEGs are described in Supplementary Table 1.
Figure 6.
WT and Pirb-/-naïve CD4+T cells have limited caspase 3/7 activation before stimulation. Representative flow cytometry plots of the expression of active caspase 3/7 in unstimulated WT (left) and Pirb-/- (right) naive CD4+ T cells. Representative data of 3 independent experiments.
PIR-B Suppresses mTORC1 Signaling in CD4+ T Cells and Modulates Th17 Differentiation
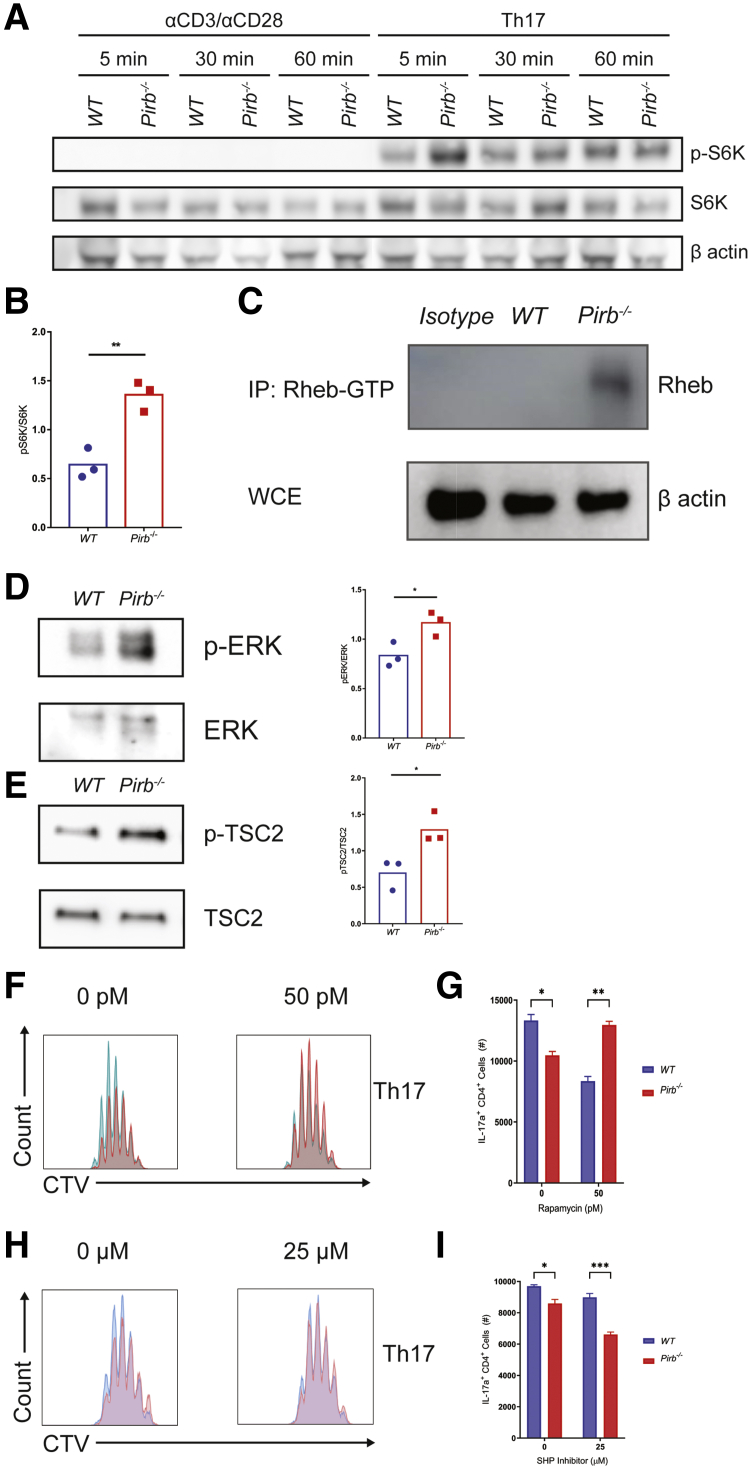
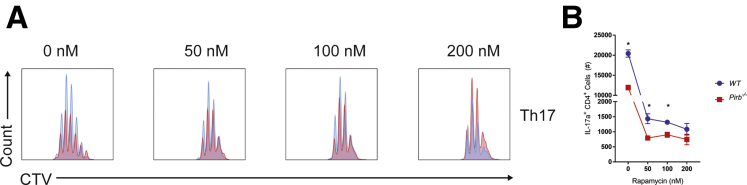
mTORC1 signaling regulates CD4+ T-cell fate decisions,34 and, in particular, CD4+ Th17 cell differentiation.35 Indeed, stimulation of WT and Pirb-/- naïve CD4+ T cells under Th17 conditions leads to rapid activation and phosphorylation of the downstream protein of mTORC1 signaling, S6 kinase (Figure 7A). Notably, we observed hyperphosphorylation of S6 kinase in Pirb-/- CD4+ T cells compared with WT CD4+ T cells (Figure 7A and B). Consistent with this, we observed increased levels of the mTOR activator, the guanosine triphosphate (GTP)-bound form of the small GTPase Ras homologue enriched in brain (Rheb) in Pirb-/- CD4+ T cells compared with WT CD4+ T cells (Figure 7C). The tuberous sclerosis (TSC1–TSC2) heterodimeric protein complex36 stimulates TSC2 GTPase-activating protein (GAP) activity toward Rheb, converting Rheb-GTP (active form) to Rheb-guanosine diphosphate (GDP_36,37 (inactive form), which in turn limits mTORC1 activity.38 TSC2-GAP activity on Rheb is regulated by extracellular signals through the phosphorylation of TSC1 and TSC2 by protein kinase B (AKT), AMP-activated protein kinase (AMPK), glycogen synthase kinase 3 (GSK3), ERK, serum and glucocorticoid (SGK), or ribosomal s6 kinase (RSK).36,38, 39, 40, 41 Notably, stimulation of Pirb-/- naïve CD4+ T cells under Th17 polarizing conditions lead to significantly increased phosphorylation of the mitogen-activated protein kinase ERK, compared with WT naïve CD4+ T cells (Figure 7D). Consistent with this, we observed significantly increased phosphorylated tuberous sclerosis 2 (p-TSC2) in Pirb-/- naïve CD4+ T cells compared with WT naïve CD4+ T cells (Figure 7E). To determine whether hyperactivated mTORC1 signaling may contribute to the heightened apoptosis and cell death in Pirb-/- naïve CD4+ T cells, we assessed Pirb-/- naïve CD4+ T-cell proliferation under Th17 polarizing conditions in the presence of the mTORC1 inhibitor rapamycin (Figure 7F and G). Consistent with previous observations,42,43 Th17 differentiation of naïve WT CD4+ T cells in the presence of increasing high concentrations of rapamycin that ablates mTOR activity led to a concentration-dependent reduction in the outgrowth of WT IL17a+ CD4+ T cells (Figure 8). Given the observation that abolition of mTORC1 activity with high concentration of rapamycin inhibited both WT and Pirb-/- IL17a+ CD4+ T cell outgrowth under Th17 polarizing conditions, we next examined whether partial inhibition of mTORC1 signaling could diminish mTOR activity and enhance Pirb-/- CD4+ T IL17+ cell frequency and reconstitute the WT phenotype. To do this we performed Th17 differentiation of naïve WT and Pirb-/- CD4+ T cells in the presence of 50 pmol/L rapamycin, which promotes partial inhibition of mTOR activity. Indeed, Th17 differentiation of naïve Pirb-/- CD4+ T cells in the presence of 50 pmol/L rapamycin significantly increased the frequency of Pirb-/- IL17a+ CD4+ T cells compared with that observed in the absence of rapamycin (Figure 7F and G). Notably, the frequency of Pirb-/- IL17a+ CD4+ T cells in the presence of 50 pmol/L rapamycin was comparable with that observed in WT CD4+ T cells in the presence of vehicle (Figure 7G). These studies suggest that tempering of mTORC1 activity in Pirb-/- naïve CD4+ T cells can enhance CD4+ Th17 differentiation and outgrowth and reconstitute the WT phenotype.
Figure 7.
Pirb regulates CD4+IL17a+cells via negative regulation of mTORC1 signaling. (A) Western blot analyses of phosphorylation of S6 kinase in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes. (B) Densitometry analyses of phosphorylation of S6 kinase in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes (WT, n = 3; Pirb-/-, n = 3). (C) WT and Pirb-/- CD4+ T-cell lysates were immunoprecipitated with Rheb-GTP antibody. Western blot was performed for Rheb. (D) Western blot analyses of phosphorylation of ERK in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes (left). Densitometry analyses of phosphorylation of ERK in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes (right) (WT, n = 3; Pirb-/-, n = 3). (E) Western blot analyses of phosphorylation of TSC2 in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes (left). Densitometry analyses of phosphorylation of TSC2 in purified CD4+ T cells cultured under Th17 polarizing conditions for 5 minutes (right) (WT, n = 3; Pirb-/-, n = 3). WT and Pirb-/- CD4+ T cells were cultured in the presence of rapamycin (50 pmol/L). (F) Representative flow cytometry plots of Cell Trace Violet staining on Th17 polarized cells and quantification of (G) IL17a+CD4+ T cells (WT, n = 3; Pirb-/-, n = 3). WT and Pirb-/- CD4+ T cells were cultured in the presence of SHP-1/2 inhibitor (25 μmol/L). (H) Representative flow cytometry plots of Cell Trace Violet staining on Th17 polarized cells and quantification of (I) IL17a+CD4+ T cells (WT, n = 3; Pirb-/-, n = 3). Data are presented as means ± SEM. Statistical analysis was performed using 2-way analysis of variance (P < .05) followed by the (G and I) Sidak multiple comparison test or (B, D, and E) unpaired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (B, D, and E) Data shown encompass 3 independent experiments. (A, C, and F–I) Data shown are from 1 experiment, representative of 3 independent experiments. CTV, cell trace violet; IP, immunoprecipitation; WCE, whole cell extract.
Figure 8.
Th17 polarization is impaired at high doses of rapamycin in WT and Pirb-/-naïve CD4+T cells. WT and Pirb-/- CD4+ T cells were cultured in the presence of different concentrations of rapamycin (0, 50, 100, and 200 nmol/L). (A) Representative flow cytometry plots of Cell Trace Violet staining on Th17 polarized cells and quantification of (B) IL17a+CD4+ T cells (n = 3). Data are presented as means ± SEM. Statistical analysis was performed using an unpaired t test. ∗P < .05. (A and B) Data shown are from 1 experiment, representative of 3 independent experiments. CTV, cell trace violet.
The regulatory function of PIR-B is mediated predominantly via SHP-1/2 inhibition of kinase activity.27,29 Given our demonstration of heightened kinase activity, ERK, and p-TSC2 in Pirb-/- naïve CD4+ T cells under Th17 polarizing conditions, we examined the requirement of SHP-1/2 function in the WT and Pirb-/- naïve CD4+ T-cell proliferation under Th17 polarizing conditions (Figure 7H and I). Th17 differentiation in the presence of SHP-1/2 inhibitor led to a reduction in the number of WT and Pirb-/- IL17a+CD4+ T cells (Figure 7H and I). Notably, at 25 μmol/L SHP-1/2 inhibitor the number of WT CD4+ IL17a+ cells were comparable with the number of Pirb-/- CD4+ IL17a+ cells generated in the absence of the inhibitor (Figure 7I). Collectively, these studies suggest a role for SHP-1/2 and mTORC1 signaling in PIR-B–mediated regulation of CD4+ IL17a+ cell maintenance.
PIR-B Expression Is Up-regulated on Memory CD4+ Th17 Cells
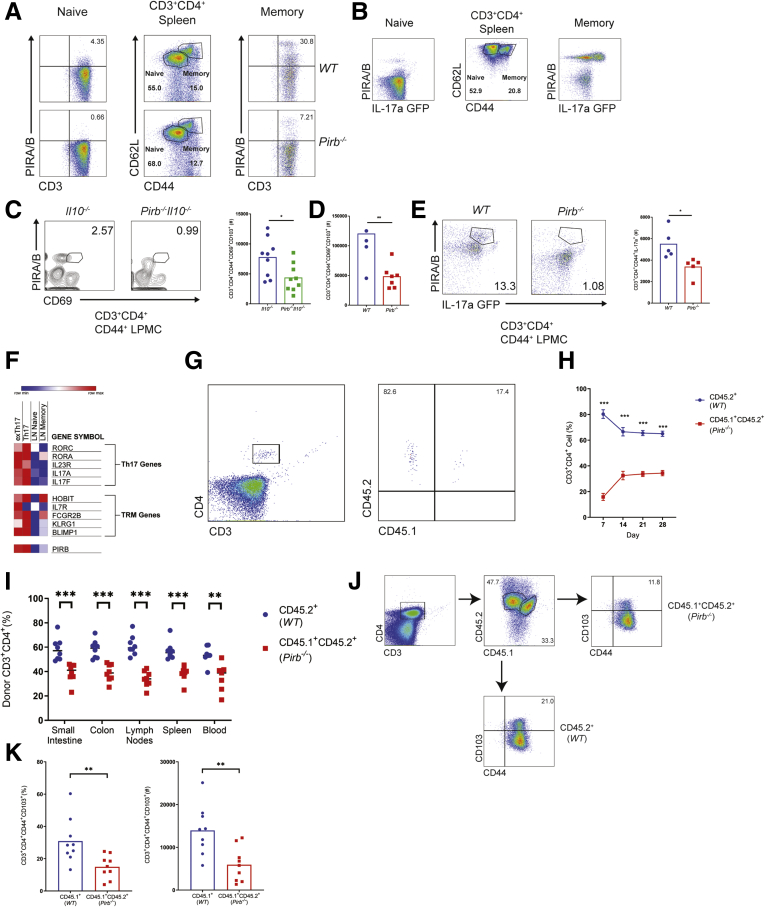
Flow cytometry analyses identified significant expression of PIR-B on the surface of CD3+ CD4+ CD44high CD62L+ memory splenic CD4+ T cells and limited expression on CD3+ CD4+ CD44low CD62L- naïve T cells from WT C57BL6 mice (Figure 9A). Notably, PIR-B+ splenic memory CD3+ CD4+ CD44high CD62L+ CD4+ cells were predominantly IL17a+ (Figure 9B). Given the recent demonstration that TRM CD4+ T cells are drivers of chronic inflammation in models of colitis18; we next examined PIR-B expression on colonic TRM CD4+ T cells from Il10-/- and Pirb-/-Il10-/- colitic mice. Indeed, PIR-B was expressed on a subset of CD3+CD4+CD44+CD69+ LP T cells in the colons of Il10-/- mice (Figure 9C). Furthermore, frequencies of TRM CD4+ T cells (CD3+CD4+CD44+ CD103+CD69+) were enriched in Il10-/- mice and Rag-/- mice, which received WT CD4+ T cells compared with Pirb-/-Il10-/- mice and Rag-/- mice, which received Pirb-/- CD4+ T cells, respectively (Figure 9C and D). Analyses with WT and Pirb-/- IL17a green fluorescent protein (GFP) mice showed PIR-B was expressed on a subset of CD3+CD4+CD44+IL17a+ lamina propria mononuclear cells and that the frequency of CD3+CD4+CD44+IL17a+ lamina propria mononuclear cells in Pirb-/- IL17a GFP mice was reduced compared with WT IL17a GFP mice (Figure 9E). These observations were validated in silico (GSE130446)44 by the demonstration that TRM CD4+ T IL17a+ cells identified by expression of HOBIT, FCGR2B, BLIMP1, RORC, RORCA, IL23R, and IL17A expressed high levels of PIR-B relative to naïve or memory CD4+ T cells from the draining lymph nodes of mice (Figure 9F). To determine if PIR-B expression intrinsically impacted CD4+ T-cell survival and TRM formation, we performed competitive adoptive transfer experiments in which equal numbers of congenically labeled WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells were co-transferred into the same Rag-/- recipient. Tracking the donor T-cell populations in the peripheral blood showed greater numbers of CD45.2+ WT CD4+ T cells than CD45.1+CD45.2+ Pirb-/- CD4+ T cells (Figure 9G and H). Furthermore, we observed significantly more CD45.2+ WT CD4+ T cells than CD45.1+CD45.2+ Pirb-/- CD4+ T cells in the secondary lymphoid organs and intestinal tissue in the same recipient mice (Figure 9I). Intriguingly, we observed a significant increase in the number of colonic TRM CD4+ T cells (CD3+ CD4+ CD44+ CD103+) derived from the CD45.2+ WT CD4+ T-cell donor population (Figure 9J and K). Collectively, our data show that PIR-B is expressed on a subset of TRM CD4+ Th17 cells and confers a competitive advantage for T-cell survival and TRM formation.
Figure 9.
Pirb is expressed on a subset of memory CD4+IL17a+cells. Flow cytometry strategy for gating on CD3+CD4+ naive and memory T cells. Splenocytes from (A) WT (top row), Pirb-/- (bottom row), and (B) IL17a GFP reporter mice were analyzed for PIRA/B expression on T-cell subsets. Plots were pregated on CD3+CD4+ T cells. (C) Flow cytometry of the expression of PIRA/B and CD69 on CD3+CD4+CD44+ T cells isolated from colonic lamina propria of Il10-/- and Pirb-/-Il10-/- at 10 weeks of age (left). Quantification of colonic lamina propria CD3+CD4+CD44+CD69+CD103+ T cells (right) (Il10-/-, n = 9; Pirb-/-Il10-/-, n = 9). (D) Quantification of colonic lamina propria CD3+CD4+CD44+CD69+CD103+ T cells from Rag-/- mice that received either 400,000 CD4+ CD45RBhi WT or Pirb-/- T cells (WT, n = 4; Pirb-/-, n = 7). (E) Flow cytometry of the expression of PIRA/B and IL17a on CD3+CD4+CD44+ T cells isolated from colonic lamina propria of WT IL17a GFP and Pirb-/- IL17a GFP reporter mice at 10 weeks of age (left). Quantification of CD3+CD4+CD44+IL17a+ T cells (right) (WT, n = 5; Pirb-/-, n = 5). (F) Heat map of differentially expressed genes based on RNA sequencing data (GSE130446) of a selected subset of genes known to be involved in resident memory T cells and Th17 cells comparing between former Th17 (exTh17) resident memory T cells, Th17 resident memory T cells, lymph node naïve CD4+ T cells, and lymph node memory CD4+ T cells. (G) Representative plots of donor WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells circulating in the peripheral blood of a Rag-/- recipient mouse. (H) Percentage of donor WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells circulating in the peripheral blood in Rag-/- recipient mice at different time points (WT, n = 10; Pirb-/-, n = 10). (I) Percentage of donor WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells that accumulated in different tissues in Rag-/- recipient mice (WT, n = 9; Pirb-/-, n = 9). (J) Representative plots of colonic CD3+CD4+CD44+ CD103+ T cells that have been derived from donor WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells injected in a Rag-/- recipient mouse. (K) Percentage (left) and cell counts (right) of colonic CD3+CD4+CD44+ CD103+ T cells that have been derived from donor WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) CD4+ T cells injected in a Rag-/- recipient mouse (WT, n = 9; Pirb-/-, n = 9). Data are presented as means ± SEM. Statistical analysis was performed using 2-way analysis of variance (P < .05) followed by the (H, I, and K) Sidak multiple comparison test or (C–E) unpaired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. Data shown encompass 3 independent experiments. LPMC, lamina propria mononuclear cell.
LILRB3 Expression Is Associated With Pathogenic IBD and Memory Th17 Responses
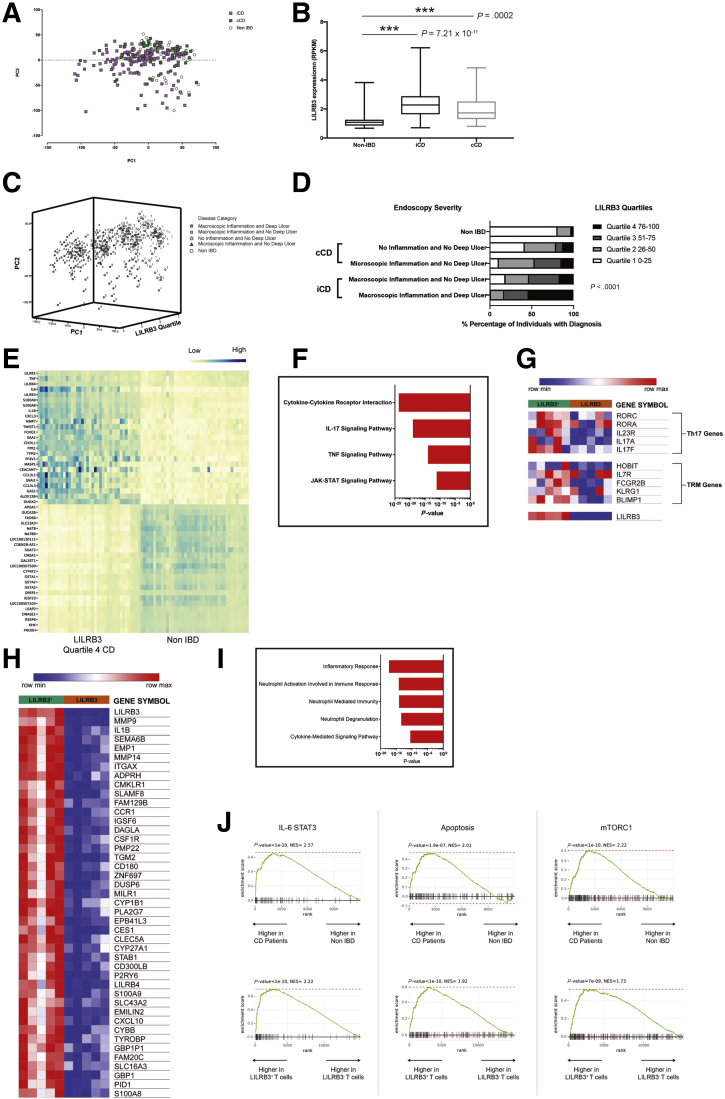
To identify if there was expression of the human homologue of PIR-B (LILRB3) in human CD4+ IL17A+ cells, and a relationship between LILRB3+ CD4+ IL17A+ function and the IBD phenotype, we next examined a RNA sequencing data set of ileal biopsy samples from a cohort of 259 pediatric individuals consisting of treatment naive CD and non-IBD patients (GSE57945).45 Principle component analysis of gene expression data between non-IBD (n = 42), pediatric ileal involvement CD (iCD) (n = 162), and colonic-only involvement CD (cCD) (n = 55) patients showed a distinct iCD transcriptome signature (Figure 10A). Pathway enrichment analyses of DEGs showed groups of related genes within the iCD gene signature associated with chemical carcinogenesis, IL17 signaling pathway, cytokine–cytokine receptor interaction (Table 1). LILRB3 mRNA expression was up-regulated significantly in cCD and iCD compared with non-IBD controls (Figure 10B). Stratification of the CD cohort based on endoscopic severity, control (Ctl, non-IBD), cCD no microscopic/macroscopic inflammation and no deep ulcer (DU), cCD with macroscopic inflammation and no DU, undetermined cCD, iCD with macroscopic inflammation with no DU and iCD macroscopic inflammation with DU,45,46 and principle component analysis of gene expression between non-IBD and the CD clinical subgroups with LILRB3 expression (RPKM values: quartile [Q]1, 0.59–1.44; Q2, 1.48–2.72; Q3, 2.73–5.47; and Q4, 5.56–73.39) showed a distinct segregation of the LILRB3 signature within the iCD-DU group compared with other CD disease subgroups (Figure 10C, Table 2). Forty of 65 patients in the Q4 LILRB3hi group were iCD-DU (Figure 10D). Evaluation of the differentially expressed genes (ranked by P value, n = 25 most up-regulated and down-regulated genes) between Q4 LILRB3hi iCD-DU patients and LILRB3low non-IBD patients identified significant enrichment of genes involved in cytokine–cytokine receptor interaction, IL17a signaling, and TNF signaling pathway (Figure 10E and F, Table 3) suggesting an interaction between LILRB3 and Th17 responses in CD. Consistent with this, LILRB3 mRNA expression within the iCD-DU group correlated positively with IL1B, IL17A, IL21, TNF, IL6, and S100A9 in the iCD-DU group; indicating that LILRB3 was related to mucosal inflammation and correlated with disease severity47,48 (Table 4). To determine whether there was a direct association between LILRB3 and memory CD4+Th17 cells in human beings, we examined a RNA sequencing data set on sorted human memory CD4+ T cells (GSE140244).44 Strikingly, we showed a subset of memory CD4+ T cells with high expression of LILRB3, and these LILRB3+ memory CD4+ T cells expressed heightened levels of genes that encode Th17 (Rorc, Rora) and TRM (Hobit, Blimp1, KLRG1) transcription factors (Figure 10G). Assessment of biological function by analyzing the differentially expressed genes (ranked by P value, n = 200) between LILRB3+ memory CD4+ T cells and LILRB3- memory CD4+ T cells showed that LILRB3+ memory CD4+ T cells were involved in Th17 inflammatory responses (Figure 10H and I). To determine which signaling pathways were impacted in IBD development and the memory CD4+ T-cell response, we performed gene set enrichment analysis on the most dysregulated genes from the GSE57945 and GSE140244 data sets. Analyses showed positive enrichment in hallmark gene sets for IL6 signal transducer and activator of transcription 3 signaling, apoptosis, and mTORC1 signaling in LILRB3hi CD patients and LILRB3+ memory CD4+ T cells relative to LILRB3low non-IBD patients and LILRB3- memory CD4+ T cells, respectively (Figure 10J). Cumulatively, our in silico results show that LILRB3 is highly expressed on a subset of memory CD4+Th17 cells in human beings and links LILRB3+ memory CD4+ T cells with pathogenic Th17 inflammatory responses in IBD.
Figure 10.
LILRB3 expression is up-regulated in IBD patients and memory CD4+T cells characterized by a Th17 signature. (A) Principle component analysis (PCA) of differentially expressed genes between non-IBD (NL, n = 42), pediatric iCD (n = 162), and cCD (n = 55) patients (GSE57945). (B) Box plots showing LILRB3 expression (RPKM) from CD or non-IBD tissue biopsy specimens. (C) Three-dimensional PCA plot of CD cohort stratified by endoscopic severity. (D) Stratification of the CD cohort based on endoscopic severity and LILRB3 expression (quartiles, RPKM). (E) Heat map of differentially expressed genes based on RNA sequencing data (GSE57945) comparing Crohn’s disease patients and healthy control individuals. (F) Pathway analysis of differentially expressed genes in Crohn’s disease patients relative to healthy individuals, assessed via KEGG 2019, ranked by P value. (G) Heat map of differentially expressed genes based on RNA sequencing data (GSE140244) of a selected subset of genes known to be involved in resident memory T cells and Th17 cells comparing between LILRB3+ CD4+ memory T cells and LILRB3-CD4+ memory T cells. (H) Heat map of differentially expressed genes based on RNA sequencing data (GSE140244) comparing LILRB3+ CD4+ memory T cells and LILRB3-CD4+ memory T cells collected from 10 donors. (I) Bar graphs of pathway analysis of differentially expressed genes in LILRB3+ CD4+ memory T cells relative to LILRB3-CD4+ memory T cells; assessed via Gene Ontology Biological Pathways, ranked by P value. (J) Gene set enrichment analysis (GSEA) for the indicated Hallmark genes comparing a ranked list of differentially expressed genes between Crohn’s disease patients and healthy control individuals (GSE57945) (top row) and comparing between LILRB3+ CD4+ memory T cells and LILRB3-CD4+ memory T cells (GSE140244) (bottom row). Statistical analysis was performed using the Student t test. ∗∗∗P < .001. JAK-STAT, janus kinases-signal transducer and activator of transcription protein; max, maximum; min, minimum; NES, normalized enrichment score.
Table 1.
Pathway Enrichment Analyses of iCD Gene Signature
| Direction | Adjusted P value | Pathways |
|---|---|---|
| Down-regulated | 2.00E-04 | Drug metabolism |
| Down-regulated | 2.00E-04 | Chemical carcinogenesis |
| Down-regulated | 2.00E-04 | Metabolism of xenobiotics by cytochrome P450 |
| Down-regulated | 2.00E-04 | Retinol metabolism |
| Down-regulated | 2.00E-04 | Fat digestion and absorption |
| Down-regulated | 2.00E-04 | Steroid hormone biosynthesis |
| Down-regulated | 2.00E-04 | Pentose and glucuronate interconversions |
| Down-regulated | 2.00E-04 | Mineral absorption |
| Down-regulated | 2.00E-04 | Drug metabolism |
| Up-regulated | 2.00E-04 | IL17 signaling pathway |
| Up-regulated | 2.00E-04 | Rheumatoid arthritis |
| Up-regulated | 2.00E-04 | Cytokine–cytokine receptor interaction |
| Up-regulated | 2.00E-04 | Leishmaniasis |
| Up-regulated | 2.00E-04 | Pertussis |
| Up-regulated | 2.00E-04 | TNF signaling pathway |
| Up-regulated | 2.00E-04 | Staphylococcus aureus infection |
| Up-regulated | 2.00E-04 | Osteoclast differentiation |
| Up-regulated | 2.00E-04 | Toll-like receptor signaling pathway |
| Up-regulated | 2.00E-04 | NOD-like receptor signaling pathway |
| Up-regulated | 2.00E-04 | Tuberculosis |
| Up-regulated | 2.00E-04 | Malaria |
| Up-regulated | 2.00E-04 | Legionellosis |
| Up-regulated | 2.00E-04 | Influenza A |
| Up-regulated | 2.00E-04 | Autoimmune thyroid disease |
| Up-regulated | 2.00E-04 | Phagosome |
| Up-regulated | 2.00E-04 | Salmonella infection |
| Up-regulated | 2.00E-04 | AGE-RAGE signaling pathway in diabetic complications |
| Up-regulated | 2.00E-04 | Chemokine signaling pathway |
| Up-regulated | 2.00E-04 | Amoebiasis |
| Up-regulated | 2.00E-04 | Measles |
NOTE. KEGG pathway analysis was used to identify important pathways altered by differentially regulated genes.
AGE-RAGE, advanced glycation end products-receptor for advanced glycation end products; NOD, Nucleotide-binding, oligomerization domain.
Table 2.
cCD and iCD Stratified by Endoscopic Severity and LILRB3 Expression
| Disease category | LILRB3_Q1 | LILRB3_Q2 | LILRB3_Q2 | LILRB3_Q4 | P value |
|---|---|---|---|---|---|
| Macroscopic inflammation and deep ulcer (iCD) | 1 (1.3%) | 12 (16%) | 22 (29.3%) | 40 (53.3%) | 2.09 × 10-22 |
| Macroscopic inflammation and no deep ulcer (iCD) | 16 (18.8%) | 24 (28.2%) | 31 (36.5%) | 14 (16.4%) | |
| No inflammation and no deep ulcer (cCD) | 10 (41.7%) | 9 (37.5%) | 2(8.3%) | 3 (12.5%) | |
| Undetermined and no deep ulcer (cCD) | 0 (0%) | 0 | 1 (33.3%) | 2 (66.7%) | |
| Microscopic inflammation and deep ulcer (iCD) | 0 | 1 (100%) | 0 | 0 | |
| Microscopic inflammation and no deep ulcer (cCD) | 3 (10.3%) | 12 (41.4%) | 9 (31.0%) | 5 (17.2%) | |
| Non-IBD | 34 (81%) | 7 (16.7%) | 0 | 1 (2.4%) |
NOTE. LILRB3 quartiles were established based on RPKM values: quartiles Q1 (0%–25%), 0.59–1.44; Q2 (26%–50%), 1.48–2.72; Q3 (51%–75%), 2.73–5.47; and Q4 (76%–100%), 5.56–73.39. Statistical analysis was performed using SPSS 17.0. The frequencies procedure was used for the LILRB3 quartile analysis. The χ2 test was used to assess the association of CD categories with 4 LILRB3 quartiles. Value (%) represents the number of patients (% of total of disease category).
Table 3.
Pathway Enrichment Analysis in Q4 LILRB3hi iCD Patients With Macroscopic Inflammation With Deep Ulcer Relative to LILRB3low Non-IBD Patients
| Direction | Adjusted P value | Pathways |
|---|---|---|
| Down-regulated | 5.00E-21 | Metabolic pathways |
| Down-regulated | 4.73E-16 | Drug metabolism |
| Down-regulated | 1.02E-15 | Chemical carcinogenesis |
| Down-regulated | 1.45E-14 | Metabolism of xenobiotics by cytochrome P450 |
| Down-regulated | 1.92E-14 | Retinol metabolism |
| Down-regulated | 4.32E-11 | Fat digestion and absorption |
| Down-regulated | 1.37E-09 | Steroid hormone biosynthesis |
| Down-regulated | 1.37E-09 | Mineral absorption |
| Down-regulated | 2.64E-09 | PPAR signaling pathway |
| Down-regulated | 5.98E-09 | Protein digestion and absorption |
| Down-regulated | 1.27E-07 | Vitamin digestion and absorption |
| Down-regulated | 1.08E-06 | Bile secretion |
| Down-regulated | 1.43E-06 | Arachidonic acid metabolism |
| Down-regulated | 3.14E-06 | Pentose and glucuronate interconversions |
| Down-regulated | 1.67E-05 | Drug metabolism |
| Up-regulated | 2.88E-24 | Cytokine–cytokine receptor interaction |
| Up-regulated | 1.51E-19 | IL17 signaling pathway |
| Up-regulated | 8.99E-18 | Rheumatoid arthritis |
| Up-regulated | 8.28E-17 | Staphylococcus aureus infection |
| Up-regulated | 6.72E-15 | Complement and coagulation cascades |
| Up-regulated | 6.72E-15 | Amoebiasis |
| Up-regulated | 1.39E-14 | TNF signaling pathway |
| Up-regulated | 3.45E-14 | Osteoclast differentiation |
| Up-regulated | 3.45E-14 | Pertussis |
| Up-regulated | 1.32E-13 | Leishmaniasis |
| Up-regulated | 1.37E-13 | Malaria |
| Up-regulated | 3.54E-13 | Tuberculosis |
| Up-regulated | 8.96E-13 | AGE-RAGE signaling pathway in diabetic complications |
| Up-regulated | 1.05E-11 | Chemokine signaling pathway |
| Up-regulated | 1.05E-11 | JAK-STAT signaling pathway |
NOTE. LILRB3 quartiles were established based on RPKM values: quartiles Q1 (0%–25%), 0.59–1.44; Q2 (26%–50%), 1.48–2.72; Q3 (51%–75%), 2.73–5.47; and Q4 (76%–100%), 5.56–73.39. KEGG pathway analysis was used to identify important pathways altered by DEG signature between Q4 LILRB3hi iCD-DU patients and LILRB3low non-IBD patients.
AGE-RAGE, advanced glycation end products-receptor for advanced glycation end products; JAK-STAT, janus kinases-signal transducer and activator of transcription protein; PPAR, peroxisome proliferator-activated receptor.
Table 4.
Correlation Analyses Between LILRB3 and LILRB5 mRNA Expression and Proinflammatory and Mucosal Injury Markers in iCD Patients Stratified by Endoscopic Severity and LILRB3 Expression
| LILRB3 | S100A9 | IL1B | IL17A | IL17B | IL21 | TNF | IL6 | LILRB5 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| LILRB3 | Pearson correlation | 1 | .929∗∗ | .863∗∗ | .730∗∗ | 0.112 | .454∗∗ | .843∗∗ | .842∗∗ | -0.195 |
| Signficance (2-tailed) | P < .005 | P < .005 | P < .005 | 0.339 | P < .005 | P < .005 | P < .005 | 0.093 | ||
| N | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | |
| LILRB5 | Pearson correlation | -0.195 | -0.177 | -0.163 | -0.191 | -0.181 | -.234∗ | -0.157 | -0.11 | 1 |
| Sig. (2-tailed) | 0.093 | 0.129 | 0.163 | 0.101 | 0.12 | 0.044 | 0.179 | 0.346 | ||
| N | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | |
NOTE. LILRB3 quartiles were established based on RPKM values: quartiles Q1 (0%–25%), 0.59–1.44; Q2 (26%–50%), 1.48–2.72; Q3 (51%–75%), 2.73–5.47; and Q4 (76%–100%), 5.56–73.39). Statistical analysis was performed using SPSS 17.0. The frequencies procedure was used for the LILRB3 quartile analysis. The χ2 test was used to assess the association of CD categories with 4 LILRB3 quartiles. The association of LILRB3 with Th17 pathway associated genes IL1B, IL17A, IL21, TNF, IL6, and S100A9 was evaluated by the Pearson correlation as quantitative variables. ∗∗P < .01.
Discussion
Herein, we have shown that PIR-B is a negative regulator of CD4+ T cells and loss of function inhibits the differentiation and outgrowth of TRM CD4+ Th17 cells, leading to the protection from CD4+ T-cell–dependent colitis. Mechanistically, we show that PIR-B is expressed by naïve and TRM CD4+ T cells and that PIR-B intrinsically regulates CD4+ IL17a+ T-cell survival. PIR-B modulates ERK and TSC1/2 heterodimeric protein complex activity restraining mTORC1 signaling and mTORC1-mediated CD4+ T-cell apoptosis. Finally, we showed that PIR-B expression is up-regulated in TRM CD4+ IL17a+ cells and that the human ortholog, LILRB3, is associated with a severe pediatric CD phenotype (iCD-DU) and memory CD4+ IL17A+ T-cell responses in human beings. Collectively, these studies show an intrinsic role for LILRB receptors in the regulation of the adaptive CD4+ T-cell inflammatory response and exacerbation of the TRM CD4+ IL17a+ T-cell driven CD phenotype.
By using multiple models of T-cell–dependent colitis we show that PIR-B deficiency leads to a loss of CD4+ IL17a+ cells, reduced systemic IL17a, and protects mice from colitis. Corroborative evidence from clinical and experimental studies support an important role for CD4+ IL17a+ cells in the induction and exacerbation of IBD,12,13,49,50 IL17a can be produced by multiple T-cell populations including CD4+ αβT cells, γδT cells, natural killer T cells, and non–T-cell populations including innate lymphoid cells.51, 52, 53, 54 Studies using the CD4+ CD45RBhigh T-cell transfer model of colitis supports a dominant role for IL17-producing CD4+ αβT cells in the augmentation of the colitic response.55 Analyses of draining LN and colonic LP from Il10-/- mice with spontaneous colitis showed that the IL17a signal is derived predominantly from CD3+ CD4+ T-cell receptor (TCR) αβ T cells. Furthermore, our demonstration that transfer of WT and not Pirb-/- CD4+ T cells to immunodeficient mice led to the generation of CD4+ IL17a+ cells, and colitis confirms that PIR-B expression in CD4+ αβT cells is important for the CD4+ T-cell–driven colitic response. Previous studies have reported that germ-free Il10-/- mice do not spontaneously develop colitis,56 indicating that the Il10-/- spontaneous model of colitis is dependent on the intestinal microbiota. We cannot rule out a possible role for the microbiome in the protected colitic phenotype observed in Pirb-/-Il10-/- mice. However, experiments using the WT and Pirb-/- CD4+ T-cell transfer model of colitis and competitive transfer experiments in co-housed recipient Rag-/- mice showed that loss of PIR-B expression specifically in the CD4+ T compartment resulted in significant protection from colitis development and suppressed the survival of the donor naive CD4+ T cells. Collectively, these experiments indicated that the phenotypic differences in the colitic phenotype likely are owing to the intrinsic regulation of PIR-B in the CD4+ T-cell compartment and not attributed to the microbiome.
We have shown high PIR-B expression on CD3+ CD4+ CD44high CD62L+ memory CD4+ IL17a+ cells. Genetic deletion of Pirb led to a marked reduction in the frequency of colonic LP CD3+CD4+CD44+IL17a+ TRM T cells and protection from CD4+ IL17a+ T-cell–driven colitis. Consistent with this, examination of an independent RNA sequencing data set showed PIR-B expression in sorted TRM CD4+ IL17a+ cells.44 Collectively, these studies suggest that PIR-B regulates TRM CD4+ IL17a+ T cells. We and others have identified an increased frequency of CD4+ TRM T cells with a proinflammatory Th17 phenotype in biopsy samples from active CD and ulcerative colitis patients compared with healthy control patients.17,18,57,58 Furthermore, increased levels of CD69+CD103+ CD4+ TRM T cells have been associated with clinical flares.18 Previous reports identified a link between CD4+ TRM IL17A+ T cells and CD by using biopsy specimens from CD patients undergoing surgery for severe, chronically active, or complicated disease.17
The contribution of IL17a to the IBD phenotype is complex.59 IL17a signaling has been shown to have beneficial effects on the integrity of the intestinal epithelial barrier and protect against the IBD phenotype.60 Furthermore, biologics targeting the IL17a cytokine led to worsening of intestinal inflammation in a subset of CD patients.61,62 In contrast, targeting of the cytokines IL6 and IL23 or their receptors, which are essential for the differentiation of pathogenic Th17 cells, led to positive outcomes in treating IBD patients.63, 64, 65 An emerging explanation to reconcile these divergent findings is that IL17a is produced by multiple cell populations including CD4+ Th17 cells and ILC3s. ILC3-derived IL17a in combination with IL22 is thought to promote intestinal epithelial barrier function and protect against colitis, whereas CD4+ T-cell–derived IL17a with cytokines such as TNFα and IFNγ are proinflammatory and drive intestinal inflammation and colitis.66 Our analyses of RNA sequencing data sets from pediatric CD patients at diagnosis showed the highest expression of LILRB3 to be restricted predominantly to the endoscopic severe subgroup (iCD-DU) and that LILRB3 mRNA expression in iCD-DU individuals correlated positively with mucosal inflammation and disease severity. Analyses of high LILRB3 expression individuals identified significant enrichment of genes involved in the IL17A signaling pathway in clinical distinct group iCD-DU. These studies support the notion that PIRB+ TRM CD4+ T cells may play a critical role in Th17 inflammatory response and the onset of severe mucosal injury observed in iCD-DU phenotype. Cumulatively, these studies suggest that specific targeting of CD4+ Th17 cells would have a greater beneficial clinical outcome for IBD patients than broadly targeting the IL17a signaling pathway. We provide multiple lines of evidence that PIR-B is expressed on CD4+ Th17 cells. Mining of the the Immunological Genome Project (ImmGen) data set (GSE109125) showed no expression of PIR-B on ILC3 cells, suggesting that targeting the PIR-B/LILRB3 pathway may permit selective targeting of CD4+ Th17 vs ILC3 cells.67
In vitro studies have shown a link between PIR-B deficiency, increased CD4+ T-cell apoptosis, and hyperactivation of mTORC1 signaling. Our demonstration that rapamycin led to reconstitution of the WT phenotype in Pirb-/- CD4+ IL17a+ T cells suggests that PIR-B negatively regulates CD4+ IL17a+ T-cell apoptosis via suppression of mTORC1 signaling. mTORC1 regulates CD4+ T-cell exit from quiescence and determination of CD4+ T-cell fate,68 particularly in CD4+ Th17 differentiation.43 mTORC1 functions as a CD4+ T-cell-intrinsic rheostat, with continual mTORC1 activity required to sustain CD4+ Th17 differentiation, however, oscillation of mTORC1 activity to a hypoactivation or hyperactivation status perturbs that differentiation process and triggers cell apoptosis and death. Consistent with this, diminished mTORC1 activation and signaling, such as that observed by genetic deletion of mTOR or S6 kinase, results in a failure of naïve CD4+ T cells to differentiate into Th17 cells.69, 70, 71 Conversely, exaggerated mTORC1 signaling as observed by deletion of TSC1 and loss of the TSC1–TSC2 heterodimeric protein inhibitory complex also results in impaired CD4+ T-cell survival.72
The formation and stabilization of the TSC1–TSC2 heterodimeric protein inhibitory complex stimulates TSC2 GAP activity and conversion of Rheb-GTP (active form) to Rheb-GDP37 (inactive form), switching mTOR into a catalytically inactive state and diminishing mTORC1 activity. The establishment and maintenance of the TSC1–TSC2 protein complex is tightly regulated by serine/threonine kinase activity. Phosphorylation of serine and threonine residues in TSC2 by kinases such as Akt and Erk destabilizes the TSC1–TSC2 protein complex, leading to TSC1–TSC2 dissociation and loss of TSC2-dependent activity. As a consequence, mTOR switches into a catalytically active state and induction of mTORC1 activity.36,38, 39, 40, 41 We showed heightened phosphorylated extracellular signal-regulated kinase (p-ERK) and p-TSC2 and increased mTORC1 signaling in Pirb-/- naïve CD4+ T cells under Th17 polarizing conditions. Furthermore, restraining mTORC1 activity using rapamycin lead to an increase in Pirb-/- CD4+ Th17 cell survival and the WT phenotype.
The PIR-B/LILRB ITIM domains are known to bind and activate intracellular phosphatases, including SHP-1 and SHP-2, which inhibit activating-type, receptor-mediated signaling.29,73 We speculate that activation of PIR-B promotes the recruitment and binding of SHP-1/2 phosphatases to the PIR-B ITIM domain. Activation of SHP-1/2 leads to dephosphorylation of p-ERK and loss of ERK-dependent regulation of the mTORC1 inhibitory complex (TSC1-TSC2), triggering hyperactivation of mTORC1 signaling in CD4+ T cells. Herein, we show significantly increased p-ERK and p-TSC2 in Pirb-/- naïve CD4+ T cells under Th17 polarizing conditions. Previous studies have reported that SHP-2 suppresses CD4+ T-cell activation by mediating inhibitory receptor signals, including those from cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1).74,75 Overexpression of catalytically inactive SHP-2 in T cells promoted T-cell differentiation,76 and inhibition of SHP-2 signaling in mice and human CD4+ T cells decreased IL17a/IL17f production.77 We predict that PIRB may act in a similar fashion to other T-cell–inhibitory receptors such as CTLA-4 and PD-1 to temper CD4+ T-cell activation and Th17 differentiation.
In conclusion, we show that PIR-B is expressed on TRM CD4+ Th17 cells and controls CD4+ Th17 inflammatory responses, and that loss of PIR-B activity leads to inhibition of CD4+ Th17 regulated inflammation and colitis. The demonstration of PIR-B expression on memory CD4+ Th17 cells and a requirement for this pathway in CD4+ Th17-mediated colitic development has promising implications for targeted therapy in IBD. Biologics focused on inhibitory receptors have been shown to be efficacious for the treatment of T-cell–mediated diseases including cancer and autoimmunity. Our data support the notion that targeting PIR-B could regulate pathogenic CD4+ Th17 cells and provide a new therapeutic approach for the treatment of TRM CD4+ Th17-driven diseases such as IBD.
Methods
Mice
Male and female, 5- to 10-week-old Il10-/- mice (C57BL/6) and IL17a GFP mice (C57BL/6) were obtained from Jackson Laboratories (Bar Harbor, ME). Male and female, 5- to 10-week-old Pirb-/- mice (C57BL/6) were kindly provided by Dr Hiromi Kubagawa.78 Male and female, 5- to 10-week-old Rag-/- mice (C57BL/6) were kindly provided by Dr Grace Chen. To generate the Pirb-/-Il10-/- mice (C57BL/6), Pirb-/- mice (C57BL/6) were intercrossed with Il10-/- mice (C57BL/6). C57BL/6 strain purity was confirmed (>99%) by DartMouse congenic analyses. To generate Pirb-/- IL17a GFP mice (C57BL/6), Pirb-/- mice (C57BL/6) were intercrossed with IL17a GFP mice (C57BL/6). All mice strains were co-housed in the same room.
Experimental Colitis Models
For the Il10-/- spontaneous model of colitis, co-housed Il10-/-, Il10+/-, and Pirb-/-Il10-/- mice were monitored for colitis development from 7 weeks of age. Euthanasia and postmortem analyses were performed at 15 weeks of age. For the αCD3-mediated model of intestinal enteropathy, 7- to 10-week-old co-housed mice were treated twice intraperitoneally with 15 μg αCD3 (145-2C11; Thermo Fisher, Waltham, MA) 48 hours apart. Euthanasia and postmortem analyses were performed 4 hours after the second injection. For the CD4+CD45RBhi T-cell transfer model of colitis, splenic naïve CD4+ T cells from WT and Pirb-/- donors were enriched by red blood cell lysis and magnetic bead depletion (BioLegend). A total of 400,000 naïve CD4+ T cells were transferred intraperitoneally into co-housed Rag-/- mice. Euthanasia and postmortem analyses were performed 32 days after injection of cells. The colitic mice were weighed twice every week to measure body weight, and the lymph nodes, spleen, small intestine, and colonic tissue were collected for analysis at the end of the experiment. Weight changes were calculated as a percentage of weight at the start of the experiment (day 0). Clinical disease was scored based on prior descriptions.79 In brief, mice were graded on a scale of 0 to 5 by assessment of symptoms including bump on nose, pilar erecti, rectal prolapse, anal bleeding, decreased activity, diarrhea, hunched back, excreted perianal mucus, shrunken eyes, and dehydration. For all animal experiments, mice that lost 20% or more of their body weight (relative to their weight at the start of the experiment) were killed in accordance with Institutional Animal Care and Use Committee protocols.
Competitive CD4+ T-Cell Transfer
We crossed Pirb-/- (CD45.2+) with WT mice that express the congenic CD45.1 allele to generate congenically labeled Pirb-/- (CD45.1+CD45.2+) mice. Splenic naïve CD4+ T cells from WT (CD45.2+) and Pirb-/- (CD45.1+CD45.2+) donors were enriched by red blood cell lysis and magnetic bead depletion (BioLegend). A total of 800,000 naïve CD4+ T cells (400,000 WT (CD45.2+) and 400,000 Pirb-/- (CD45.1+CD45.2+) were transferred intraperitoneally into co-housed Rag-/- mice. Blood was collected every 7 days after injection of cells and flow cytometry and intracellular cytokine staining was performed as described later. Euthanasia and postmortem analyses were performed 32 days after injection of cells and flow cytometry and intracellular cytokine staining was performed as described later.
Histology
Harvested tissues were washed with phosphate-buffered saline, Swiss-rolled, and fixed overnight in 4% paraformaldehyde. Paraffin-embedded tissues were stained with H&E and analyzed by bright field microscopy. Histologic scoring was performed on ascending, transverse, and descending colon and rectum segments as previously described.80
In Vivo Cytokine Capture Assay
Systemic TNFα, IFNγ, and IL17a levels were quantified in the serum of mice.81 Briefly, 10 μg of biotinylated detection antibodies against TNFα (clone TN3), IFNγ (clone R4-6A2), and IL17a (eBio17B7) were injected intravenously into mice and 24 hours later serum was collected. Luminescent enzyme-linked immunosorbent assays were performed using plates coated with a secondary capture antibody as previously described.
In Vitro CD4+ T-Cell Culture
Naïve CD4+ T cells were isolated from spleens using the naïve CD4+ T-cell isolation kit (BioLegend) and were stimulated with plate-bound αCD3 (1 μg/mL) and soluble αCD28 (2 μg/mL) alone or in the presence of polarizing cytokines and antibodies. For Th1 polarizing conditions, cells were cultured in the presence of IL12 (10 ng/mL) cytokines and neutralizing antibody αIL4 (1 μg/mL). For Th17 polarizing conditions, cells were cultured in the presence of IL6 (20 ng/mL), IL23 (20 ng/mL), and transforming growth factor β1 (2 ng/mL) cytokines and neutralizing antibodies αIFNγ (10 μg/mL) and αIL4 (1 μg/mL). Cells were cultured in supplemented RPMI media containing 10% fetal bovine serum, 2% penicillin/streptomycin, and 50 μmol/L 2-mercaptoethanol. To assess proliferation, naïve CD4+ T cells were labeled with 5 μmol/L cell trace violet (CTV) in 0.2% fetal bovine serum for 20 minutes. For chemical inhibitor experiments, either rapamycin (Sigma Aldrich, St. Louis, MO) (50 pmol/L) or SHP-1/2 inhibitor (NSC-87877; Millipore Sigma, Burlington, MA) (25 μmol/L) were added at the beginning of the experiment.
Flow Cytometry and Intracellular Cytokine Staining
Single-cell suspensions of spleen, mLNs, and colon lamina propria cells were surface stained ex vivo with fluorescent antibodies to T-cell markers (CD3, CD4, CD62L, CD44, CD69, CD103, CD45.1, and CD45.2) and for PIRA/B (6C1). For cytokine staining, cells were ex vivo stimulated at 37°C for 4 hours with phorbol 12-myristate 13-acetate (PMA) (50 ng/mL), ionomycin (1 mg/mL), and Brefeldin A. Cells then were processed and stained using an intracellular cytokine staining kit (BD Biosciences, Franklin Lakes, NJ) according to the manufacturer’s instructions with IFNγ and IL17a. For apoptosis staining, the dead cell apoptosis kit and live/dead viability assay (Thermo Fisher) was followed according to the manufacturer’s instructions. For activated caspase 3/7 staining, the caspase 3/7 kit (Thermo Fisher) was followed according to the manufacturer’s instructions. To assess cell cycling and entry into G1, Pyronin Y (Sigma Aldrich) staining was performed as previously described.82 To assess cell cycling and entry into S phase, the Edu flow cytometry kit (Sigma) was followed according to the manufacturer’s instructions. All flow cytometry samples were acquired on an Novocyte (ACEA Biosciences) and data were analyzed using FlowJo (Tree Star, San Carlos, CA) and Prism (GraphPad Software). Absolute cell numbers were calculated using Precision Count Beads (BioLegend) according to the manufacturer’s instructions.
LP Mononuclear Cell Isolation
Colons were cut longitudinally and incubated in Hank’s balanced salt solution with 5 mmol/L EDTA at 37°C for 30 minutes before vortexing to remove epithelial cells. The remaining tissues were minced and digested with 2.4 mg/mL collagenase A (Roche, Basel, Switzerland), and 0.2 mg/mL DNase I (Roche) at 37°C for 45 minutes. After removal of tissue debris, cells suspended in 44% Percoll were loaded above 67% Percoll before centrifugation. Colonic lamina propria cells were collected from the interface between 44% and 67% Percoll.
Western Blot and Immunoprecipitation
CD4+ T cells were lysed in protein lysis buffer (10% glycerol, 20 mmol/L Tris HCl [pH 7], 137 mmol/L NaCl, 2 mmol/L EDTA, and 1% NP-40) supplemented with proteinase inhibitor cocktail (Thermo Fisher) and PhoSTOP phosphatase inhibitors (Roche). Protein lysates were cleared of insoluble material through centrifugation, and the resulting protein lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Proteins were wet transferred to 0.2-mm nitrocellulose membranes (Thermo Fisher), which were blocked using 3% bovine serum albumin in Tris-buffered saline, 1% Tween 20 (TBST) buffer for 1 hour at room temperature. Membranes were incubated overnight using the following primary antibodies: α-β-actin, α-S6 kinase, α-pS6 kinase, α-TSC2, α–p-TSC2, α-ERK1/2, and α-pERK1/2. Primary antibodies were used at a 1:1000 dilution in blocking buffer. Membranes were washed in TBST and incubated with the secondary antibody, goat α-rabbit–horseradish peroxidase, at a 1:2000 dilution in blocking buffer. Rheb-GTP was immunoprecipitated using the Rheb activation assay kit (NewEast Biosciences, King of Prussia, PA). Immunoprecipitation was performed according to the manufacturer’s description. Protein bands were visualized after exposure of the membranes to enhanced chemiluminescence (ECL) substrate solution (Thermo Fisher) and quantified by densitometry analysis using Image Studio (Licor) software.
Quantitative Real-Time PCR
RNA was isolated from sorted naïve CD4+ T cells using the Quick RNA microprep kit (Zymo Research, Irvine, CA). Complementary DNA was generating by reverse-transcription using SuperScript II (Invitrogen) ad Oligo-dT (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was performed for Pirb and murine Gapdh using specific primers designed in Snapgene (Insightful Science) software. Samples were normalized to housekeeping expression of Gapdh using the delta cycle threshhold (2-ΔCt)method.
RNA Sequencing Analysis
RNA was isolated from sorted naïve CD4+ T cells using the Quick RNA microprep kit (Zymo Research). RNA was submitted to the University of Michigan Advanced Genomics Core for library preparation and sequencing (Illumina, San Diego, CA). Raw reads were aligned to the reference mm9 mouse genome (GRCm38) using the Hisat-build pipeline. Relative gene expression was quantified using featureCounts function from the subread-2.0.0 package. Pseudogenes, RIKEN complementary DNA sequences, and immunoglobulin variable genes were removed from downstream analysis of expressed genes. Downstream analysis was performed in R (R Core Team, Vienna, Austria) where the read counts were analyzed in IDEP 9.1 ( Brookings, SD) and DESeq (Heidelberg, Germany) was used to identify the DEGs.83 DEGs were identified with an adjusted P value of .05 or less and at least ±1.5-fold or more RPKM, and the heat map was generated using Python (Python Software Foundation, Wilmington, DE) on the normalized scale. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to identify important pathways altered by differentially regulated genes. Statistical analysis was performed using SPSS 17.0 (Chicago, IL). The frequencies procedure was used for the LILRB3 quartile analysis. The χ2 test was used to assess the association of CD categories with 4 LILRB3 quartiles. The association of LILRB3 with Th17 pathway–associated genes IL1B, IL17A, IL21, TNF, IL6, and S100A9 was evaluated by the Pearson correlation as quantitative variables.
Additional RNA sequencing data analyses were obtained from the NCBI GEO database, with accession numbers GSE130446, GSE57945, and GSE140244. With the Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn’s Disease (RISK) study (GSE57945), we examined a RNA sequencing data set of ileal biopsy samples from a cohort of 259 pediatric individuals consisting of treatment-naive CD and non-IBD patients. Principle component analysis of differentially expressed genes between non-IBD (n = 42), pediatric iCD (n = 162), and cCD (n = 55) patients showed a distinct CD transcriptome signature (results not shown). We next stratified the CD cohort based on endoscopic severity, cCD no microscopic/macroscopic inflammation and no DU, cCD with macroscopic inflammation and no DU, iCD with macroscopic inflammation with no DU, and iCD macroscopic inflammation with DU.45,46 LILRB3 quartiles were established based on RPKM values; quartiles Q1 (0%–25%), 0.59–1.44; Q2 (26%–50%), 1.48–2.72; Q3 (51%–75%), 2.73–5.47; and Q4 (76%–100%), 5.56–73.39. Differential expression was defined with a significant change in expression by limma.84 Heatmaps of gene expression were generated using Morpheus https://software.broadinstitute.org/morpheus (Cambridge, MA) and Phantasus (St. Louis, MO).85 Gene ontology analysis was performed using Enrichr (New York, NY) and Gene Set Enrichment Analysis.
Statistics
Statistical parameters are defined in the figure legends. Data are presented as means ± SEM. Data were considered significant at P < .05. Comparisons between 2 groups were made using a t test. Comparisons between more than 2 group were made using 2-way analysis of variance and where appropriate were followed with a Dunnett multiple comparison test or the Sidak multiple comparison test. Statistical analysis was performed in Prism (GraphPad Software).
Study Approval
All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Michigan (Ann Arbor, MI), and performed in accordance with University guidelines.
Acknowledgments
The authors would like to acknowledge members of the Divisions of Allergy and Immunology and Immunobiology at Cincinnati Children’s Hospital Medical Center, Mary H. Weiser Food Allergy Center, and Division of Experimental Pathology at the University of Michigan Medicine for thoughtful discussions.
CRediT Authorship Contributions
Jazib Uddin (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Sunil Tomar (Data curation: Supporting)
Ankit Sharma (Data curation: Supporting; Visualization: Supporting)
Lisa Waggoner (Data curation: Supporting)
Varsha Ganesan (Data curation: Supporting)
Sahiti Marella (Data curation: Supporting)
Yanfen Yang (Data curation: Supporting)
Taeko Noah (Data curation: Supporting)
Simone Vanoni (Writing – review & editing: Supporting)
Andrew Patterson (Methodology: Supporting)
Chang Zeng (Methodology: Supporting)
Paul S Foster (Writing – review & editing: Supporting)
Rodney Newberry (Writing – review & editing: Supporting)
Shrinivas Bishu (Writing – review & editing: Supporting)
John Y Kao (Writing – review & editing: Supporting)
Michael J Rosen (Writing – review & editing: Supporting)
Lee Denson (Writing – review & editing: Supporting)
Philip King (Methodology: Supporting; Writing – review & editing: Supporting)
Kasper Hoebe (Methodology: Supporting; Writing – review & editing: Supporting)
Senad Divanovic (Conceptualization: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Ariel Munitz (Methodology: Supporting; Writing – review & editing: Supporting)
Simon P Hogan, PhD (Funding acquisition: Lead; Writing – original draft: Supporting; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Institutes of Health grants DK073553, DK090119, DK125007, DK099222, AI138177, AI112626, and AI007413; Food Allergy Research and Education; Department of Defense grant W81XWH-15-1-051730; M-FARA; and the Mary H. Weiser Food Allergy Center (S.P.H.).
Supplementary Material
References
- 1.Kaser A., Zeissig S., Blumberg R.S. Inflammatory bowel disease. Ann Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahida Y.R., Patel S., Gionchetti P., Vaux D., Jewell D.P. Macrophage subpopulations in lamina propria of normal and inflamed colon and terminal ileum. Gut. 1989;30:826–834. doi: 10.1136/gut.30.6.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks D.J., Segal A.W. Innate immunity in inflammatory bowel disease: a disease hypothesis. J Pathol. 2008;214:260–266. doi: 10.1002/path.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neurath M.F., Weigmann B., Finotto S., Glickman J., Nieuwenhuis E., Iijima H., Mizoguchi A., Mizoguchi E., Mudter J., Galle P.R., Bhan A., Autschbach F., Sullivan B.M., Szabo S.J., Glimcher L.H., Blumberg R.S. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heller F., Fuss I.J., Nieuwenhuis E.E., Blumberg R.S., Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002;17:629–638. doi: 10.1016/s1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]
- 6.Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters C.P., Mjösberg J.M., Bernink J.H., Spits H. Innate lymphoid cells in inflammatory bowel diseases. Immunol Lett. 2016;172:124–131. doi: 10.1016/j.imlet.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Geremia A., Arancibia-Cárcamo C.V., Fleming M.P., Rust N., Singh B., Mortensen N.J., Travis S.P., Powrie F. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangan P.R., Harrington L.E., O'Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., Weaver C.T. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 10.McGeachy M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O'Shea J.J., Cua D.J. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettelli E., Korn T., Kuchroo V.K. Th17: the third member of the effector T cell trilogy. Curr Opin Immunol. 2007;19:652–657. doi: 10.1016/j.coi.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiekowski M.T., Leach M.W., Evans E.W., Sullivan L., Chen S.C., Vassileva G., Bazan J.F., Gorman D.M., Kastelein R.A., Narula S., Lira S.A. Ubiquitous transgenic expression of the IL-23 subunit p19 induces multiorgan inflammation, runting, infertility, and premature death. J Immunol. 2001;166:7563–7570. doi: 10.4049/jimmunol.166.12.7563. [DOI] [PubMed] [Google Scholar]
- 13.Yen D., Cheung J., Scheerens H., Poulet F., McClanahan T., McKenzie B., Kleinschek M.A., Owyang A., Mattson J., Blumenschein W., Murphy E., Sathe M., Cua D.J., Kastelein R.A., Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kullberg M.C., Jankovic D., Feng C.G., Hue S., Gorelick P.L., McKenzie B.S., Cua D.J., Powrie F., Cheever A.W., Maloy K.J., Sher A. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hue S., Ahern P., Buonocore S., Kullberg M.C., Cua D.J., McKenzie B.S., Powrie F., Maloy K.J. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elson C.O., Cong Y., Weaver C.T., Schoeb T.R., McClanahan T.K., Fick R.B., Kastelein R.A. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–2370. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 17.Hegazy A.N., West N.R., Stubbington M.J.T., Wendt E., Suijker K.I.M., Datsi A., This S., Danne C., Campion S., Duncan S.H., Owens B.M.J., Uhlig H.H., McMichael A., Oxford I.B.D.C.I., Bergthaler A., Teichmann S.A., Keshav S., Powrie F. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153:1320–1337 e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zundler S., Becker E., Spocinska M., Slawik M., Parga-Vidal L., Stark R., Wiendl M., Atreya R., Rath T., Leppkes M., Hildner K., López-Posadas R., Lukassen S., Ekici A.B., Neufert C., Atreya I., van Gisbergen K., Neurath M.F. Hobit- and Blimp-1-driven CD4(+) tissue-resident memory T cells control chronic intestinal inflammation. Nat Immunol. 2019;20:288–300. doi: 10.1038/s41590-018-0298-5. [DOI] [PubMed] [Google Scholar]
- 19.Kubagawa H., Chen C.C., Ho L.H., Shimada T.S., Gartland L., Mashburn C., Uehara T., Ravetch J.V., Cooper M.D. Biochemical nature and cellular distribution of the paired immunoglobulin-like receptors, PIR-A and PIR-B. J Exp Med. 1999;189:309–318. doi: 10.1084/jem.189.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long E.O. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 21.Masuda A., Nakamura A., Maeda T., Sakamoto Y., Takai T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J Exp Med. 2007;204:907–920. doi: 10.1084/jem.20060631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama M., Underhill D.M., Petersen T.W., Li B., Kitamura T., Takai T., Aderem A. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178:4250–4259. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- 23.Arita K., Endo S., Kaifu T., Kitaguchi K., Nakamura A., Ohmori H., Kohu K., Satake M., Takai T. Transcriptional activation of the Pirb gene in B cells by PU.1 and Runx3. J Immunol. 2011;186:7050–7059. doi: 10.4049/jimmunol.1001302. [DOI] [PubMed] [Google Scholar]
- 24.Takai T. A novel recognition system for MHC class I molecules constituted by PIR. Adv Immunol. 2005;88:161–192. doi: 10.1016/S0065-2776(05)88005-8. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama M., Kurokawa K., Nakamura K., Lee B.L., Sekimizu K., Kubagawa H., Hiramatsu K., Yagita H., Okumura K., Takai T., Underhill D.M., Aderem A., Ogasawara K. Inhibitory receptor paired Ig-like receptor B is exploited by Staphylococcus aureus for virulence. J Immunol. 2012;189:5903–5911. doi: 10.4049/jimmunol.1201940. [DOI] [PubMed] [Google Scholar]
- 26.Torii I., Oka S., Hotomi M., Benjamin W.H., Jr., Takai T., Kearney J.F., Briles D.E., Kubagawa H. PIR-B-deficient mice are susceptible to Salmonella infection. J Immunol. 2008;181:4229–4239. doi: 10.4049/jimmunol.181.6.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravetch J.V., Lanier L.L. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 28.Hayami K., Fukuta D., Nishikawa Y., Yamashita Y., Inui M., Ohyama Y., Hikida M., Ohmori H., Takai T. Molecular cloning of a novel murine cell-surface glycoprotein homologous to killer cell inhibitory receptors. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 29.Munitz A., Cole E.T., Beichler A., Groschwitz K., Ahrens R., Steinbrecher K., Willson T., Han X., Denson L., Rothenberg M.E., Hogan S.P. Paired immunoglobulin-like receptor B (PIR-B) negatively regulates macrophage activation in experimental colitis. Gastroenterology. 2010;139:530–541. doi: 10.1053/j.gastro.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musch M.W., Clarke L.L., Mamah D., Gawenis L.R., Zhang Z., Ellsworth W., Shalowitz D., Mittal N., Efthimiou P., Alnadjim Z., Hurst S.D., Chang E.B., Barrett T.A. T cell activation causes diarrhea by increasing intestinal permeability and inhibiting epithelial Na+/K+-ATPase. J Clin Invest. 2002;110:1739–1747. doi: 10.1172/JCI15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura N., Yamamoto M., Fukutake M., Ohtake N., Iizuka S., Ishige A., Sasaki H., Fukuda K., Yamamoto T., Hayakawa S. Anti-CD3 induces bi-phasic apoptosis in murine intestinal epithelial cells: possible involvement of the Fas/Fas ligand system in different T cell compartments. Int Immunol. 2005;17:513–522. doi: 10.1093/intimm/dxh231. [DOI] [PubMed] [Google Scholar]
- 32.Powrie F., Leach M.W., Mauze S., Menon S., Caddle L.B., Coffman R.L. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 33.Mottet C., Uhlig H.H., Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 34.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurebayashi Y., Nagai S., Ikejiri A., Ohtani M., Ichiyama K., Baba Y., Yamada T., Egami S., Hoshii T., Hirao A., Matsuda S., Koyasu S. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Gao X., Saucedo L.J., Ru B., Edgar B.A., Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 38.Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 39.Castel P., Ellis H., Bago R., Toska E., Razavi P., Carmona F.J., Kannan S., Verma C.S., Dickler M., Chandarlapaty S., Brogi E., Alessi D.R., Baselga J., Scaltriti M. PDK1-SGK1 signaling sustains AKT-independent mTORC1 activation and confers resistance to PI3Kalpha inhibition. Cancer Cell. 2016;30:229–242. doi: 10.1016/j.ccell.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C.Y., He X., MacDougald O.A., You M., Williams B.O., Guan K.L. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 41.Dibble C.C., Cantley L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopf H., de la Rosa G.M., Howard O.M., Chen X. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. Int Immunopharmacol. 2007;7:1819–1824. doi: 10.1016/j.intimp.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai S., Kurebayashi Y., Koyasu S. Role of PI3K/Akt and mTOR complexes in Th17 cell differentiation. Ann N Y Acad Sci. 2013;1280:30–34. doi: 10.1111/nyas.12059. [DOI] [PubMed] [Google Scholar]
- 44.Amezcua Vesely M.C., Pallis P., Bielecki P., Low J.S., Zhao J., Harman C.C.D., Kroehling L., Jackson R., Bailis W., Licona-Limon P., Xu H., Iijima N., Pillai P.S., Kaplan D.H., Weaver C.T., Kluger Y., Kowalczyk M.S., Iwasaki A., Pereira J.P., Esplugues E., Gagliani N., Flavell R.A. Effector TH17 cells give rise to long-lived TRM cells that are essential for an immediate response against bacterial infection. Cell. 2019;178:1176–1188 e15. doi: 10.1016/j.cell.2019.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haberman Y., Tickle T.L., Dexheimer P.J., Kim M.O., Tang D., Karns R., Baldassano R.N., Noe J.D., Rosh J., Markowitz J., Heyman M.B., Griffiths A.M., Crandall W.V., Mack D.R., Baker S.S., Huttenhower C., Keljo D.J., Hyams J.S., Kugathasan S., Walters T.D., Aronow B., Xavier R.J., Gevers D., Denson L.A. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allez M., Lemann M., Bonnet J., Cattan P., Jian R., Modigliani R. Long term outcome of patients with active Crohn's disease exhibiting extensive and deep ulcerations at colonoscopy. Am J Gastroenterol. 2002;97:947–953. doi: 10.1111/j.1572-0241.2002.05614.x. [DOI] [PubMed] [Google Scholar]
- 47.Ligumsky M., Simon P.L., Karmeli F., Rachmilewitz D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut. 1990;31:686–689. doi: 10.1136/gut.31.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunn S.K., Bisset W.M., Main M.J., Gray E.S., Olson S., Golden B.E. Fecal calprotectin: validation as a noninvasive measure of bowel inflammation in childhood inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2001;33:14–22. doi: 10.1097/00005176-200107000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Fujino S., Andoh A., Bamba S., Ogawa A., Hata K., Araki Y., Bamba T., Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng T., Qin H., Wang L., Benveniste E.N., Elson C.O., Cong Y. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J Immunol. 2011;186:6313–6318. doi: 10.4049/jimmunol.1001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rachitskaya A.V., Hansen A.M., Horai R., Li Z., Villasmil R., Luger D., Nussenblatt R.B., Caspi R.R. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol. 2008;180:5167–5171. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villanova F., Flutter B., Tosi I., Grys K., Sreeneebus H., Perera G.K., Chapman A., Smith C.H., Di Meglio P., Nestle F.O. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lockhart E., Green A.M., Flynn J.L. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 54.Park H., Li Z., Yang X.O., Chang S.H., Nurieva R., Wang Y.H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leppkes M., Becker C., Ivanov, Hirth S., Wirtz S., Neufert C., Pouly S., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Becher B., Littman D.R., Neurath M.F. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 56.Sellon R.K., Tonkonogy S., Schultz M., Dieleman L.A., Grenther W., Balish E., Rennick D.M., Sartor R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bishu S., Hou G., El Zaatari M., Bishu S.R., Popke D., Zhang M., Grasberger H., Zou W., Stidham R.W., Higgins P.D.R., Spence J.R., Kamada N., Kao J.Y. Citrobacter rodentium induces tissue-resident memory CD4(+) T cells. Infect Immun. 2019;87:7. doi: 10.1128/IAI.00295-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bishu S., El Zaatari M., Hayashi A., Hou G., Bowers N., Kinnucan J., Manoogian B., Muza-Moons M., Zhang M., Grasberger H., Bourque C., Zou W., Higgins P.D.R., Spence J.R., Stidham R.W., Kamada N., Kao J.Y. CD4+ Tissue-resident memory T cells expand and are a major source of mucosal tumour necrosis factor alpha in active Crohn's disease. J Crohns Colitis. 2019;13:905–915. doi: 10.1093/ecco-jcc/jjz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whibley N., Gaffen S.L. Gut-busters: IL-17 ain't afraid of no IL-23. Immunity. 2015;43:620–622. doi: 10.1016/j.immuni.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Connor W., Jr., Kamanaka M., Booth C.J., Town T., Nakae S., Iwakura Y., Kolls J.K., Flavell R.A. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hueber W., Sands B.E., Lewitzky S., Vandemeulebroecke M., Reinisch W., Higgins P.D., Wehkamp J., Feagan B.G., Yao M.D., Karczewski M., Karczewski J., Pezous N., Bek S., Bruin G., Mellgard B., Berger C., Londei M., Bertolino A.P., Tougas G., Travis S.P. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Targan S.R., Feagan B., Vermeire S., Panaccione R., Melmed G.Y., Landers C., Li D., Russell C., Newmark R., Zhang N., Chon Y., Hsu Y.H., Lin S.L., Klekotka P. A randomized, double-blind, placebo-controlled phase 2 study of brodalumab in patients with moderate-to-severe Crohn's disease. Am J Gastroenterol. 2016;111:1599–1607. doi: 10.1038/ajg.2016.298. [DOI] [PubMed] [Google Scholar]
- 63.Ito H., Takazoe M., Fukuda Y., Hibi T., Kusugami K., Andoh A., Matsumoto T., Yamamura T., Azuma J., Nishimoto N., Yoshizaki K., Shimoyama T., Kishimoto T. A pilot randomized trial of a human anti-interleukin-6 receptor monoclonal antibody in active Crohn's disease. Gastroenterology. 2004;126:989–996. doi: 10.1053/j.gastro.2004.01.012. discussion 47. [DOI] [PubMed] [Google Scholar]
- 64.Danese S., Vermeire S., Hellstern P., Panaccione R., Rogler G., Fraser G., Kohn A., Desreumaux P., Leong R.W., Comer G.M., Cataldi F., Banerjee A., Maguire M.K., Li C., Rath N., Beebe J., Schreiber S. Randomised trial and open-label extension study of an anti-interleukin-6 antibody in Crohn’s disease (ANDANTE I and II) Gut. 2019;68:40–48. doi: 10.1136/gutjnl-2017-314562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandborn W.J., Ferrante M., Bhandari B.R., Berliba E., Feagan B.G., Hibi T., Tuttle J.L., Klekotka P., Friedrich S., Durante M., Morgan-Cox M., Laskowski J., Schmitz J., D’Haens G.R. Efficacy and safety of mirikizumab in a randomized phase 2 study of patients with ulcerative colitis. Gastroenterology. 2020;158:537–549.e10. doi: 10.1053/j.gastro.2019.08.043. [DOI] [PubMed] [Google Scholar]
- 66.Withers D.R., Hepworth M.R., Wang X., Mackley E.C., Halford E.E., Dutton E.E., Marriott C.L., Brucklacher-Waldert V., Veldhoen M., Kelsen J., Baldassano R.N., Sonnenberg G.F. Transient inhibition of ROR-γt therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22:319–323. doi: 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida H., Lareau C.A., Ramirez R.N., Rose S.A., Maier B., Wroblewska A., Desland F., Chudnovskiy A., Mortha A., Dominguez C., Tellier J., Kim E., Dwyer D., Shinton S., Nabekura T., Qi Y., Yu B., Robinette M., Kim K.-W., Wagers A., Rhoads A., Nutt S.L., Brown B.D., Mostafavi S., Buenrostro J.D., Benoist C., Immunological Genome Project The cis-regulatory atlas of the mouse immune system. Cell. 2019;176:897–912.e20. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chapman N.M., Chi H. Hallmarks of T-cell exit from quiescence. Cancer Immunol Res. 2018;6:502–508. doi: 10.1158/2326-6066.CIR-17-0605. [DOI] [PubMed] [Google Scholar]
- 69.Delgoffe G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., Powell J.D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sasaki C.Y., Chen G., Munk R., Eitan E., Martindale J., Longo D.L., Ghosh P. p70S6K1 in the TORC1 pathway is essential for the differentiation of Th17 Cells, but not Th1, Th2, or Treg cells in mice. Eur J Immunol. 2016;46:212–222. doi: 10.1002/eji.201445422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang K., Neale G., Green D.R., He W., Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blery M., Kubagawa H., Chen C.C., Vely F., Cooper M.D., Vivier E. The paired Ig-like receptor PIR-B is an inhibitory receptor that recruits the protein-tyrosine phosphatase SHP-1. Proc Natl Acad Sci U S A. 1998;95:2446–2451. doi: 10.1073/pnas.95.5.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee K.M., Chuang E., Griffin M., Khattri R., Hong D.K., Zhang W., Straus D., Samelson L.E., Thompson C.B., Bluestone J.A. Molecular basis of T cell inactivation by CTLA-4. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 75.Chemnitz J.M., Parry R.V., Nichols K.E., June C.H., Riley J.L. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 76.Salmond R.J., Huyer G., Kotsoni A., Clements L., Alexander D.R. The src homology 2 domain-containing tyrosine phosphatase 2 regulates primary T-dependent immune responses and Th cell differentiation. J Immunol. 2005;175:6498–6508. doi: 10.4049/jimmunol.175.10.6498. [DOI] [PubMed] [Google Scholar]
- 77.Wang J., Mizui M., Zeng L.F., Bronson R., Finnell M., Terhorst C., Kyttaris V.C., Tsokos G.C., Zhang Z.Y., Kontaridis M.I. Inhibition of SHP2 ameliorates the pathogenesis of systemic lupus erythematosus. J Clin Invest. 2016;126:2077–2092. doi: 10.1172/JCI87037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ujike A., Takeda K., Nakamura A., Ebihara S., Akiyama K., Takai T. Impaired dendritic cell maturation and increased T(H)2 responses in PIR-B(-/-) mice. Nat Immunol. 2002;3:542–548. doi: 10.1038/ni801. [DOI] [PubMed] [Google Scholar]
- 79.Scheinin T., Butler D.M., Salway F., Scallon B., Feldmann M. Validation of the interleukin-10 knockout mouse model of colitis: antitumour necrosis factor-antibodies suppress the progression of colitis. Clin Exp Immunol. 2003;133:38–43. doi: 10.1046/j.1365-2249.2003.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berg D.J., Davidson N., Kühn R., Müller W., Menon S., Holland G., Thompson-Snipes L., Leach M.W., Rennick D. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finkelman F., Morris S., Orekhova T., Sehy D. The in vivo cytokine capture assay for measurement of cytokine production in the mouse. Curr Protoc Immunol. 2003 doi: 10.1002/0471142735.im0628s54. Chapter 6:Unit 6.28. [DOI] [PubMed] [Google Scholar]
- 82.Kim K.H., Sederstrom J.M. Assaying cell cycle status using flow cytometry. Curr Protoc Mol Biol. 2015;111:28. doi: 10.1002/0471142727.mb2806s111. 6.1–28.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zenkova D, Kamenev V, Sablina R, Artyomov M, Sergushichev A. Phantasus: visual and interactive gene expression analysis. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.