Abstract
By PCR screening, we found an extremely high prevalence of TT virus (TTV) in the general populations from different geographic regions. This suggests that TTV may be a common DNA virus with no clear disease association in humans. TTV genotyping by phylogenetic analysis was also performed.
In 1997, the genome of a novel DNA virus, termed the TT virus (TTV), isolated from the sera of patients with posttransfusion non-A-G hepatitis, was sequenced by representational difference analysis (6, 7). Very recently, the complete nucleotide sequence of TTV was reported by two different groups (2, 3). TTV is an unenveloped circular single-stranded DNA virus and comprises 3,852 nucleotides, with an isopycnic density of 1.31 to 1.34 g/ml in CsCl (2, 3). The TTV genome has three possible open reading frames, capable of encoding 770, 202, and 105 amino acids, respectively (2). Due to the genome structure and its banding in buoyant density gradient centrifugation, TTV is related among the known animal virus families to the Circoviridae family (2, 3, 7, 9). Despite TTV being a DNA virus, its sequence has a wide range of sequence divergence, allowing classification into several genotypes (7, 11). TTV sequences were detected in sera and liver tissues from liver disease patients, suggesting that TTV could be responsible for some acute and chronic liver disease of unknown etiology (1, 7). On the other hand, it has been reported elsewhere that TTV infection does not induce significant liver damage (5). However, the epidemiology, clinical significance, and transmission patterns of TTV remain unclear. To clarify the characterization of seroepidemiology of TTV, we carried out PCR screening for TTV in individuals, including healthy populations from different geographic regions.
We collected serum samples from individuals in Japan (233 individuals without liver disease), Myanmar (51 healthy individuals and 92 liver disease patients), Nepal (177 blood donors), Egypt (95 blood donors), Bolivia (95 blood donors), Vietnam (62 high-risk individuals consisting of medical staff), Korea (73 hemodialysis patients), Cambodia (8 human immunodeficiency virus [HIV]-infected patients), Ghana (95 HIV-infected patients), and the United States (68 HIV-infected patients). Informed consent was obtained from participants in this study. The serum samples were stored at −20°C or below until assayed.
DNA was extracted from 100 μl of serum samples with a nucleic acid extraction kit (SepaGene RV-R; Sanko Junyaku Co., Ltd., Tokyo, Japan). The resulting pellet was resuspended in RNase- and DNase-free water and then subjected to PCR as described by Takahashi et al. (10). In brief, the thermocycler was programmed first to preheat at 95°C for 10 min to activate AmpliTaq Gold DNA polymerase (Perkin-Elmer, Norwalk, Conn.), and then samples were subjected to 55 cycles consisting of 94°C for 20 s, 60°C for 20 s, and 72°C for 30 s with a Perkin-Elmer 9600 or 9700 thermal cycler. The sequences of the TTV-specific primers were 5′-GCTACGTCACTAACCACGTG-3′ (T801, sense primer, nucleotides 6 to 25) and 5′-CTBCGGTGTGTAAACTCACC-3′ (T935, antisense primer, nucleotides 185 to 204; B = G, C, or T) as designed by Takahashi et al. (10) in the 5′-end region of the TA278 isolate. The PCR products were detected by electrophoresis on 2% agarose gels, stained with ethidium bromide, and photographed under UV light.
To determine the genotype, TTV DNA was amplified by nested PCR with primers NG059 and RD038 for the outer primer pairs (377 bases) and NG061 and NG063 for the inner primer pairs (271 bases) as designed by Okamoto et al. (7) in the ORF1 region of the TA278 isolate. Amplified PCR products were subjected to direct sequencing, and then phylogenetic analysis was performed as reported previously (4). Twenty previously reported TTV sequences were obtained from the GenBank database and used for comparison with the sequence of the isolate in this study.
Statistical analyses were performed by the chi-square test or Fisher’s exact test. A difference with a P value of <0.05 was considered significant.
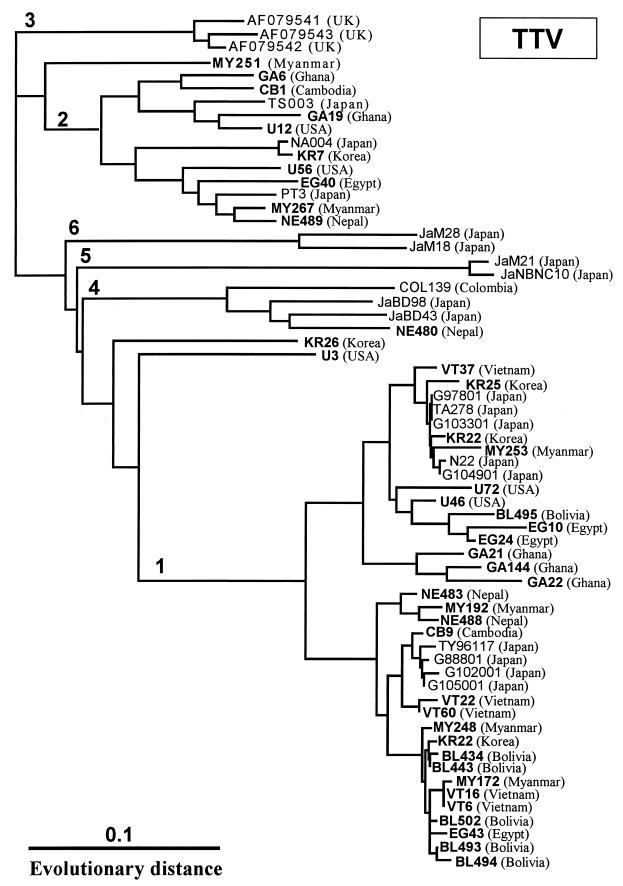
As shown in Table 1, a very high prevalence of TTV infection was found in tested individuals, including healthy populations, from 10 different countries. In the Japanese study, TTV DNA was detected significantly more often in the groups of people over 10 years of age (P < 0.05) (Table 2). On the other hand, the prevalence of TTV in individuals from other countries had already reached nearly 80% or more in the age groups over 10 years old (differences between the age groups were not statistically significant). Furthermore, the TTV genome could be classified into at least six different genotypes by phylogenetic analysis, and the major genotypes are type 1 and type 2 (Fig. 1). However, there is no correlation between major genotypes and geographic origin.
TABLE 1.
Prevalence of TTV infection in different countries
| Country | Category | n | No. with TTV DNA (%) |
|---|---|---|---|
| Japan | Without liver disease | 195a | 137 (70) |
| Myanmar | Healthy individual | 51 | 49 (96)b |
| With liver disease | 92 | 90 (98)b | |
| Nepal | Blood donor | 177 | 145 (82) |
| Egypt | Blood donor | 95 | 81 (85) |
| Bolivia | Blood donor | 95 | 78 (82) |
| Vietnam | High-risk group | 62 | 53 (86) |
| Korea | Hemodialysis | 73 | 71 (97) |
| Cambodia | HIV infection | 8 | 8 (100) |
| Ghana | HIV infection | 95 | 84 (88) |
| USAc | HIV infection | 68 | 58 (85) |
Not including individuals under 10 years old.
Statistically not significant.
USA, United States of America.
TABLE 2.
Age prevalence of TTV in different geographic regionsa
| Country | n | No. with TTV/total no. (%) in age group (yr):
|
|||
|---|---|---|---|---|---|
| ≤9 | 10–29 | 30–49 | ≥50 | ||
| Japan | 233 | 3/38 (8) | 14/28 (50) | 83/113 (74) | 40/54 (74) |
| Myanmar | 143 | None | 29/31 (94) | 60/61 (98) | 50/51 (98) |
| Nepal | 177 | None | 78/93 (84) | 45/58 (78) | 22/26 (85) |
| Vietnam | 62 | None | 29/31 (94) | 23/30 (77) | 1/1 (100) |
| Korea | 73 | None | 8/8 (100) | 34/34 (100) | 29/31 (94) |
| Cambodia | 8 | None | 3/3 (100) | 4/4 (100) | 1/1 (100) |
| Egypt | 95 | None | 11/14 (79) | 50/55 (91) | 20/26 (77) |
| Bolivia | 95 | None | 44/50 (80) | 31/42 (74) | 3/3 (100) |
| USA | 68 | None | 12/16 (75) | 42/47 (89) | 4/5 (80) |
Data for Ghana are not shown due to lack of detailed information on age. USA, United States of America.
FIG. 1.
Phylogram generated by neighbor-joining analysis of genetic distances in the ORF1 region of TTV isolates. Isolates determined in this study are presented in boldface. The database-derived isolates and their accession numbers were as follows: G97801 (AB011486), TA278 (AB008394), G103301 (AB011487), N22 (AB017767), G10491 (AB011489), TY96117 (AB011494), G88801 (AB011491), G102001 (AB011488), G105001 (AB011490), TS003 (AB017770), NA004 (AB017771), PT3 (AB017768), AF079541 (AF079541), AF079542 (AF079542), AF079543 (AF079543), COL139 (AB01635), JaM21 (AB017887), JaNBNC10 (AB018961), JaM28 (AB017888), and JaM18 (AB017886). UK, United Kingdom; USA, United States of America.
Although the TTV PCR system has been established already by Nishizawa et al. (6) and Okamoto et al. (7), there have been problems associated with low sensitivity and unstable reaction. To resolve this problem, Takahashi et al. recently reported that TTV PCR with a new primer combination is more sensitive and stable than the previous system (10). Using the new PCR system, they reported that TTV DNA was identified in 92% of 100 healthy individuals in Japan (10). In the present study, we found that TTV viremia is widespread in the general populations in many countries. Such an extremely high prevalence of TTV infection in the general population suggests that TTV may be transmissible not only in the blood but also by a nonparenteral route. Indeed, Okamoto et al. reported that TTV was excreted into the feces, thereby suggesting that TTV may be transmitted not only parenterally but also nonparenterally by a fecal-oral route (8). Molecular analysis of TTV genomes showed that genotypes 1 and 2 of TTV were widespread worldwide.
In conclusion, TTV viremia is widespread, with a very high incidence in general populations worldwide. This suggests that TTV is a common virus and may be a nonpathogenic DNA virus in humans, although the pathogenic role of TTV still remains to be investigated.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB023311 through AB023352.
Acknowledgments
We thank Takeshi Kurata for his continuous encouragement during this study and Nami Konomi, Hideo Naito, Mari Yamaguchi, and Mitsugu Usui for their kind cooperation.
This study was supported in part by grants-in-aid for science research from the Ministry of Education, Science and Culture of Japan and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 2.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Dexai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naito H, Win K M, Abe K. Identification of a novel genotype of hepatitis G virus in southeast Asia. J Clin Microbiol. 1999;37:1217–1220. doi: 10.1128/jcm.37.4.1217-1220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naoumov N V, Petrova E P, Thomas M G, Williams R. Presence of a new described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352:195–197. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 6.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 8.Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J Med Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- 9.Takahashi K, Ohta Y, Mishiro S. Partial ∼2.4kb sequence of TT virus genome from eight Japanese isolates: diagnostic and phylogenetic implications. Hepatol Res. 1998;12:111–120. [Google Scholar]
- 10.Takahashi K, Hoshino H, Ohta Y, Yoshida N, Mishiro S. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol Res. 1998;12:233–239. [Google Scholar]
- 11.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Kato H, Mukaide M, Park Y-M, Kim B-S, Ueda R. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201–206. doi: 10.1016/s0014-5793(98)01231-9. [DOI] [PubMed] [Google Scholar]