Abstract
Our current health applications do not adequately take into account contextual and personalized knowledge about patients. In order to design “Personalized Coach for Healthcare” applications to manage chronic diseases, there is a need to create a Personalized Healthcare Knowledge Graph (PHKG) that takes into consideration a patient’s health condition (personalized knowledge) and enriches that with contextualized knowledge from environmental sensors and Web of Data (e.g., symptoms and treatments for diseases). To develop PHKG, aggregating knowledge from various heterogeneous sources such as the Internet of Things (IoT) devices, clinical notes, and Electronic Medical Records (EMRs) is necessary. In this paper, we explain the challenges of collecting, managing, analyzing, and integrating patients’ health data from various sources in order to synthesize and deduce meaningful information embodying the vision of the Data, Information, Knowledge, and Wisdom (DIKW) pyramid. Furthermore, we sketch a solution that combines: 1) IoT data analytics, and 2) explicit knowledge and illustrate it using three chronic disease use cases – asthma, obesity, and Parkinson’s.
Keywords: Healthcare, Knowledge Graph (KG), Personalized Knowledge Graph, Data Management, Reasoning and Integration, Contextualization, Ontology, Linked Open Data (LOD)
1. Introduction
World Health Organization (WHO)3 estimates the number of people suffering from asthma, obesity and Parkinson’s disease as 235 million, 650 million, and 4–6 million respectively [1]. Chronic diseases such as obesity and asthma are multifactorial diseases and have become epidemics of the current century according to the WHO. Recent studies have shown the need for a variety of effective solutions to maintain a healthy lifestyle (e.g., Hapifork4 is an electronic fork that helps monitor and track eating) and are the basis for emerging applications such as “Personalized Preventive Coach” and “Digital Health Advisor” that track and interpret health and well-being data [2]. Other related areas contributing to these applications include Wireless Body Area Networks (WBANs) [3], medical Internet of Things (mIoT), m-health, e-health, and Ambient Assisted Living5 (AAL). Specifically, these applications utilize: 1) inexpensive sensors to collect raw data generated by the IoT devices (e.g., weight scale device), 2) a domain model (e.g., asthma or obesity ontology6) to structure and abstract the data, and 3) a reasoning mechanism to deduce insights and recommendations (e.g., using Body Mass Index (BMI) and environmental data with a rule-based inference engine to provide relevant information). However, the current-state-of-the-art lacks appropriate exploitation of: 1) contextual data, 2) personalized data, and 3) their integration with background medical knowledge bases for monitoring user health. In the following, we discuss these limitations.
Context-Awareness refers to the use of external data that can impact the user’s situation. For instance, IoT devices can be used to monitor the surrounding environment. Interpretation of IoT data using a background model for abstraction can provide contextual awareness to physicians. Clinical protocols should take these into account to determine a patient’s condition. For example, each patient can react differently when exposed to different environmental factors (e.g., air pollution or pollen level).
Personalization adjusts the treatment to each patient’s condition. Patient’s data is obtained by harnessing multi-modal data7 from clinical documents (demographic information, clinician’s observations, lab tests, data collected during clinical visits), patient generated health data including sensor and social data (see Figure 1 for an example involving more than 25 types of data for each patient). It permits one to tailor judgments and treatments based on patient’s vulnerabilities, triggers, and symptoms [4]. Personalization, in conjunction with predictive analytics, enables actionable insights. IoT devices can be utilized for personalization. For instance, the dosage of long-term allergy medication prescribed for an asthmatic patient to control the symptoms is tailored to a person’s asthma severity, potential for environmental triggers, and the past history.
Fig. 1.
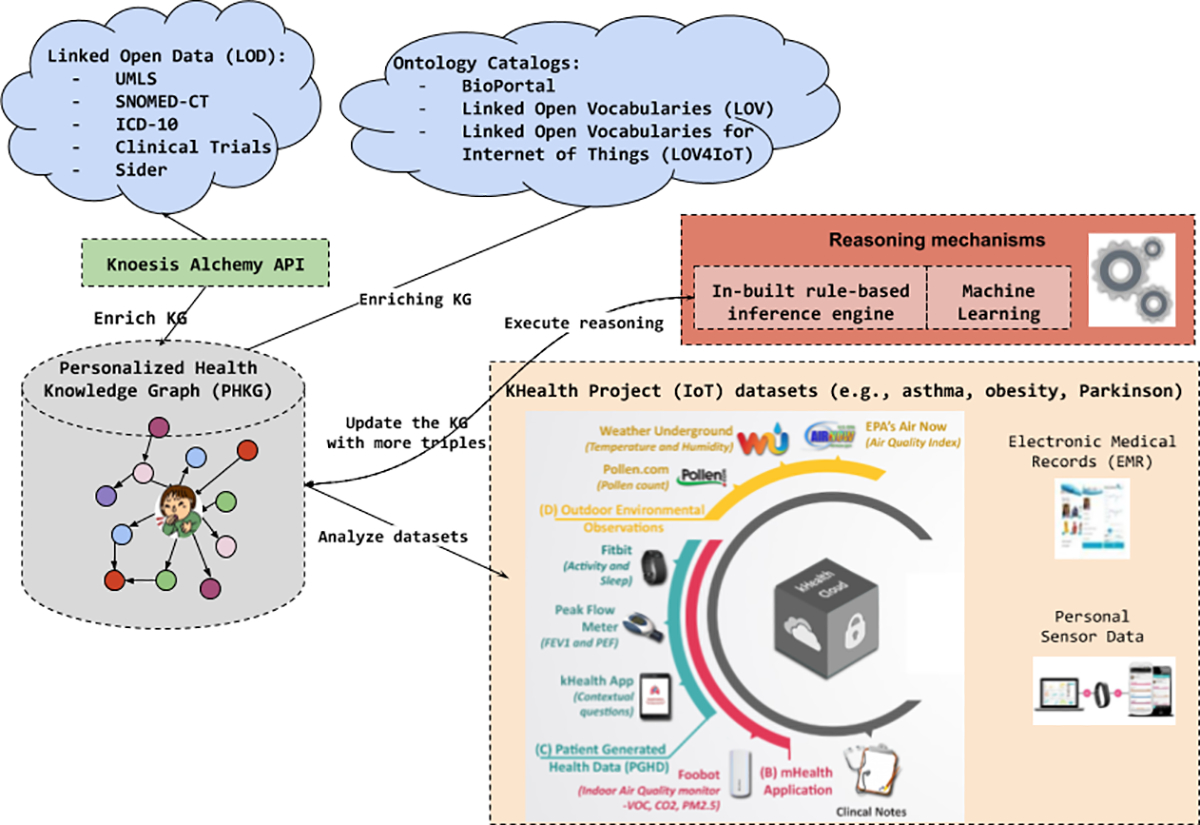
PHKG Architecture
Personalized Healthcare Knowledge Graph (PHKG)8 is a representation of all relevant medical knowledge and personal data for a patient. PHKG can support development of innovative applications such as digitalized personalized coach applications that can keep patients informed and help manage their chronic condition, and empower the physicians to make effective decisions on health-related issues or receive timely alerts as needed through continuous monitoring. Typically, PHKG formalizes medical information in terms of relevant relationships between entities. For instance, a knowledge graph (KG) for asthma can describe causes, symptoms and treatments for asthma, and PHKG can be the subgraph containing just those causes, symptoms, and treatments that are applicable to a given patient.
State-of-the-art of Health Knowledge Graphs.
Google Healthcare Knowledge Graph is a manually curated health knowledge graph that integrates ICD-9 and UMLS9 along with probabilistic machine learning and physician support to provide relevant information upon a user search [5]. In [6], a KG is applied to the pneumonia use case [6], by performing a contextual pruning algorithm on knowledge graphs. DepressionKG is a disease-specific KG [7] that can benefit representation and reasoning about Major Depressive Disorder (MDD) requiring overcoming challenges in: 1) heterogeneity of datasets, 2) highly contextual text processing, 3) incompleteness and inconsistency in datasets, and 4) expression, representation, and reasoning of medical knowledge.
PHKG is one solution to achieve the vision of the Data - Information Knowledge - Wisdom (DIKW) pyramid. DIKW describes a hierarchical relationship between Data, Information, Knowledge, and Wisdom, an example of which has been applied to the healthcare domain in the context of managing blood pressure [8]. At each layer of the DIKW pyramid, the contextualization becomes finer and becomes finest at the Wisdom stage. In our study, we incorporate relevant domain-specific medical knowledge bases for contextualizing the information on health diseases. We aim to design the methodology to achieve this DIKW vision and provide a set of easy-to-use tools [9].
In this paper, we explain the following Research Challenges (RC) to achieve this DIKW vision and to design the PHKG: (RC1) How to model a knowledge graph for healthcare and chronic disease management? (RC2) How to model personalization and context-awareness to understand patient’s symptoms and derive actionable insights? (RC3) How to analyze datasets generated by IoT devices to deduce meaningful information? (RC4) How to promote reproduceable experiments from previous projects (e.g., datasets, data models, and reasoning mechanisms)? (RC5) How to customize and instantiate relevant knowledge from existing publicly available health knowledge bases to obtain insights from health-related social media text? In the next section, we provide our vision to address these challenges through the PHKG. Then, we conclude the paper and provide directions for future work.
2. Designing PHKG
We explain the methodology to build the PHKG in terms of: 1) its architecture, 2) the use cases considered, 3) the medical datasets obtained from the LOD cloud, 4) the reasoning mechanism to deduce high-level information from IoT datasets, and 5) an online ontology catalog tool to reuse and share the domain knowledge. The architecture designed to build our PHKG is introduced in Figure 1. PHKG uses heterogeneous sources of knowledge: 1) IoT data provided by sensors, 2) medical datasets from Alchemy API that provides access to SNOMED-CT10, UMLS, and ICD-1011, 3) ontology catalogs to reuse models (e.g., asthma ontology), and 4) a set of unified rules to interpret data.
kHealth project12 developed at Kno.e.sis Research Center, is a framework for continuous monitoring of the patient’s personal data and for generating notifications as needed to assist the clinicians [10]. kHealth integrates data from three different sources: 1) Electronic Medical Records, 2) Environment using IoT devices (e.g., Foobot) and querying Web Services (for weather data), and 3) Personal health signals using IoT devices (e.g., Fitbit) to provide data on sleep, activity, and heart rate, etc. The knoesis Asthma Ontology (kAO)13 integrates: 1) W3C SOSA ontology to semantically annotate sensor observations (e.g., peak flow meter is a subclass of the sosa:Sensor class), 2) the Asthma Ontology (AO) from BioPortal to reuse relevant concepts, 3) FOAF ontology to describe people, and 4) weather ontology to deduce meaningful information from weather datasets. The asthma dataset consists of data generated by IoT devices such as peak flow meter, Foobot, Fitbit, AirNow, and from Web Services obtaining air quality parameters14, pollen index and type15, outside humidity, and temperature. The obesity dataset consists of data generated by IoT devices such as weighing scale, pill and water bottle, and Fitbit to obtain parameters such as weight, medication consumption, heart rate and sleep activity. The Parkinson dataset16 from Kaggle consists of mobile sensor data from accelerometer, compass, and microphone etc. on smartphone to synthesize patient symptom information such as unsteady walk, lacks balance, has a fall, and has slurred speech. This information can be used to both diagnose and monitor progression of Parkinson’s disease.
Kno.e.sis Alchemy API17 addresses RC2 and RC5, it identifies healthcare-related entities, entity types, and relations from social media text (e.g., Reddit) to define the context. Figure 2 demonstrates the utilization of medical datasets such as SNOMED-CT, ICD-10, and Clinical Trials to achieve entity extraction (e.g., cough concept is a taxonomy itself within SNOMED-CT). Furthermore, SIDER (a drug and side-effect knowledge base) is utilized to identify treatment, disorder, side-effects, drugs, drug-dosage form, drug-dosage level, and adverse drug reactions using the entities and their type defined in the context.
Fig. 2.
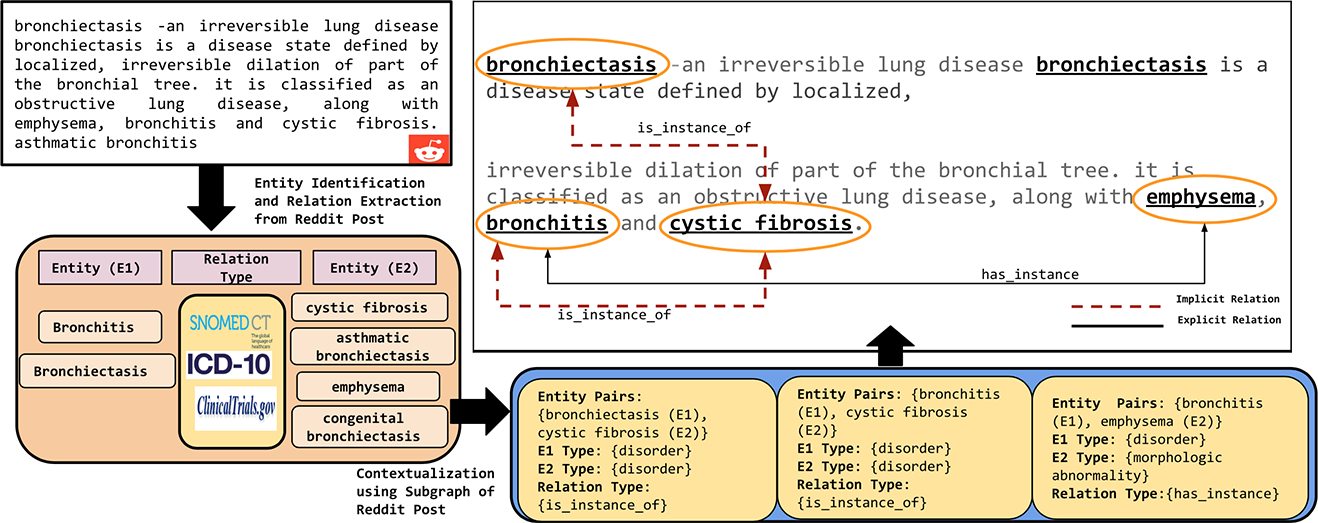
Sample annotation of entities and relations identified from Reddit Post on r/Health subreddit. Implicit relations are discovered after 1 hop and Explicit relations are present in 1 hop.
kHealth reasoner is a rule-based reasoning engine (an extension of the reasoner explained in [9]18) which deduces meaningful insights from heterogeneous data provided by clinicians and patient questionnaire responses, and obtained from IoT devices (as depicted in Figure 3). The reasoner addresses RC2 and RC3.
Fig. 3.
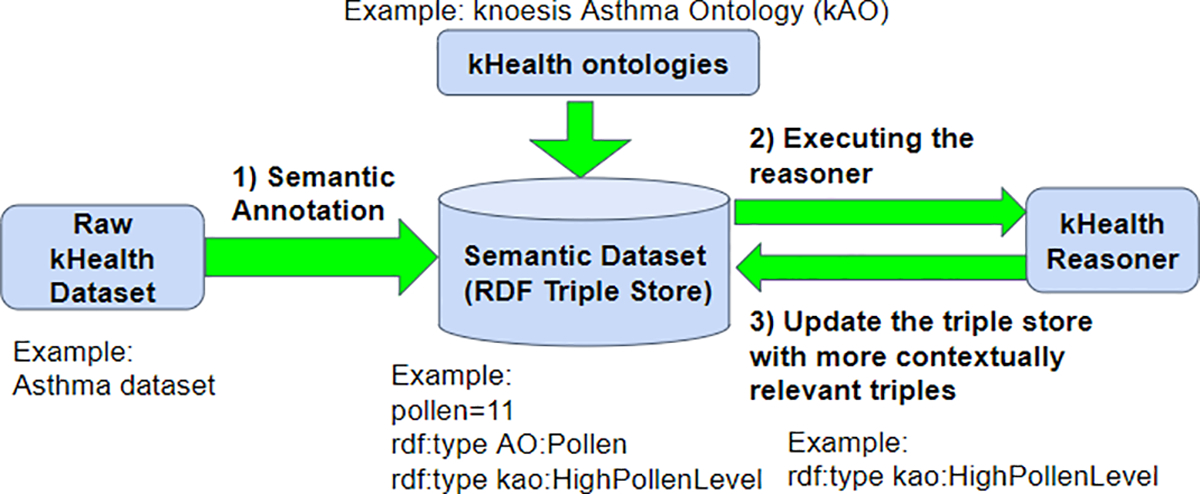
kHealth Reasoner Framework
A kHealth IoT dataset is semantically annotated using an appropriate ontology (e.g., the asthma dataset is annotated according to the kAO ontology) to make its meaning explicit and later deduce abstractions. The rules to support reasoning reflect domain knowledge and are mainly extracted from scientific publications, or from web services explicitly describing the domain expertise, or manually curated as required to interpret the data. The formalism is inspired by the Jena inference grammar that we enrich to be compliant with a dictionary of IoT devices (e.g., thermometer) and IoT observations types (e.g., outside temperature) classified within the kAO ontology. The execution of the rule provides meaningful abstractions from IoT observations (e.g., high temperature) and links the IoT data to specific domain ontologies (e.g., weather) from ontology catalogs or datasets from the LOD cloud.
Ontology Catalogs (e.g., LOV4IoT-Health19 [9], BioPortal20, Linked Open Vocabularies (LOV)21) address RC1, RC2 and RC4 because catalogs provide domain-specific knowledge already structured to enrich the PHKG. LOV4IoT covers IoT domains (e.g., healthcare and weather) to deduce abstractions from sensor data.
However, reusing existing ontologies is challenging. For instance, the AO ontology22: 1) is a taxonomy rather than an ontology, 2) has concept URIs that are opaque for humans to decipher (e.g., AO:MOCHA-Asthma_000073), and 3) has common pitfalls detected and explained by the OOPS ontology validation tool (e.g., merging different concepts in the same class).
Conclusion and Future Work.
Designing the PHKG is critical to achieve Digitalized Personalized Health coaches (e.g., chatbots) to assist doctors and patients, especially given that generic knowledge should be tailored for each patient. Designing PHKG is challenging because it requires semantic integration of heterogeneous data from: healthcare providers, IoT devices, and the Web, taking into account context and personal history for reasoning and deducing high-level abstractions and effective actionable insights. PHKG can be serve as a foundation for assisting physicians in understanding the symptoms, hypothesizing and explaining disease progression, and then inferring a potential management and treatment plan.
Acknowledgments
Thanks to the kHealth team for fruitful discussions and feedback. This work is partially funded by KHealth NIH 1 R01 HD087132-01 and Hazards SEES NSF Award EAR 1520870. The opinions expressed are those of the authors and do not reflect those of the sponsors.
Footnotes
Position Paper, Copyright ©2018 for this paper by its authors. Copying permitted for private and academic purposes.
References
- 1.Anantharam P, Thirunarayan K, Taslimi V, Sheth AP: Predicting parkinson’s disease progression with smartphone data. (2013) [Google Scholar]
- 2.Dimitrov DV: Medical internet of things and big data in healthcare. Healthcare informatics research; (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negra R, Jemili I, Belghith A: Wireless body area networks: Applications and technologies. Procedia Computer Science; (2016) [Google Scholar]
- 4.Sheth A, Jaimini U, Thirunarayan K, Banerjee T: Augmented personalized health: How smart data with iots and ai is about to change healthcare. In: IEEE RTSI. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotmensch M, Halpern Y, Tlimat A, Horng S, Sontag D: Learning a health knowledge graph from electronic medical records. Scientific reports; (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi L, Li S, Yang X, Qi J, Pan G, Zhou B: Semantic health knowledge graph: Semantic integration of heterogeneous medical knowledge and services. BioMed Research International; (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Z, Yang J, van Harmelen F, Hu Q: Constructing knowledge graphs of depression. In: ICHIS. (2017) [Google Scholar]
- 8.Sheth A, Anantharam P, Henson C: Physical-cyber-social computing: An early 21st century approach. IEEE Intelligent Systems; (2013) [Google Scholar]
- 9.Gyrard A: Designing Cross-Domain Semantic Web of Things Applications. PhD thesis, Telecom ParisTech, Eurecom (2015) [Google Scholar]
- 10.Sheth A, Anantharam P, Thirunarayan K: khealth: Proactive personalized actionable information for better healthcare. In: PDA@ IOT at VLDB. (2014) [Google Scholar]


