Abstract
A seminested PCR typing assay has been extended to identify rotavirus strains with the P[14] genotype. The specificity of the method was confirmed by Southern hybridization and by restriction analysis with the enzyme AluI. One out of four human rotavirus (HRV) strains with unusual subgroup-electropherotype linkage but none out of 50 HRV strains with usual linkage was typed as P[14].
The two proteins on the outer capsid of group A rotaviruses, VP7 and VP4, are independently involved in virus neutralization. Knowledge about the antigenic specificity carried on VP7, determining the G serotype, and that carried on VP4, determining the P serotype, is crucial to vaccine development, because the response to both proteins is mostly homotypic (13). Whereas epidemiological studies on the distribution of G specificities among human rotavirus (HRV) strains have been easily carried out with serological reagents, the search for information on P specificities has been hampered by the lack of available typing antibodies, and only genetic studies on the VP4 gene have permitted the differentiation of HRV strains into different P genotypes (4). The circulation of strains with P[8] and P[4] types and a sporadic presence of strains with the P[6] type have been recognized all over the world (5, 14). Other P types have been more rarely investigated and detected (4).
The first identification of HRV isolates with the P[14] type was reported in 1992 by Gerna et al. (6, 7), who sequenced two rotavirus strains recovered from infants with diarrhea. One of these strains, PA169 (P[14]G6), was isolated in our laboratory in Palermo (Italy), and the other, HAL 1166 (P[14]G8), was isolated in Finland. HRV strains with the P[14] type were later recovered, by nucleic acid sequencing or hybridization analysis, from a child from Thailand (strain Mc35, P[14]G10), from six infants from South Africa, and from a child from Australia (strain MG6, P[14]G6) (10–12). Such findings indicate a global distribution of HRV strains with the P[14] type and potential epidemiological significance. Recently, Ciarlet et al. found that four lapine rotavirus strains from Japan and America possessed the P[14] genotype (3).
In previous studies, the characterization of rotavirus strains has been carried out by revealing the migration pattern of the 11 viral genomic RNA segments by polyacrylamide gel electrophoresis (electropherotype) and by determining the subgroup I and II specificities, located on the major inner capsid protein VP6 (1). Usually, HRV strains with a long electropherotype (faster-moving gene segment 11) have been found to be associated with subgroup II specificity, whereas HRV strains with a short electropherotype (slower-moving gene segment 11) exhibit subgroup I specificity. It is interesting that HRV strains with the P[14] type isolated in Italy, Finland, Thailand, and Australia all showed an unusual electropherotype-subgroup linkage, having a long electropherotype associated with subgroup I specificity (10–12).
Here, we describe a valid genotyping approach to identifying the P[14] genotype of rotavirus strains and the four major HRV P genotypes (P[8], P[4], P[6], and P[9]) by seminested PCR typing. Furthermore, we report the identification of a new HRV strain with P[14] type that also shows an unusual electropherotype-subgroup linkage.
The study was carried out with (i) 8 cell culture-adapted reference strains, including Wa (P[8]G1), DS1 (P[4]G2), YO (P[8]G3), VA70 (P[8]G4), WI61 (P[8]G9), AU-1 (P[9]G3), PA169 (P[14]G6), and PA151 (P[9]G6), and (ii) 54 HRV strains selected from 903 strains obtained from 1985 to 1996 from 2,500 fecal samples of children admitted with acute gastroenteritis to the G. Di Cristina Children’s Hospital, Palermo, Italy. Selection was performed on the basis of a previous characterization of clinical isolates by electropherotyping, subgrouping, and G serotyping (2): 50 strains were representative of 896 strains with usual subgroup-electropherotype linkage and the G1 to G4 serotypes, and 4 strains were representative of 7 strains with long electropherotypes associated with subgroup I specificity and the G3 or G6 serotype.
The PCR typing method first described by Wu et al. (14), with some modifications, was used for identification of P[14] genotype.
Viral RNA, extracted from rotavirus-positive clarified stool supernatants by the glass powder method, was used as the template for reverse transcription (RT) and a first PCR amplification, in the presence of a pair of common primers corresponding to nucleotide sequences of the VP4 gene which were found to be well conserved in numerous HRV strains. The DNA fragment of 1,084 bp obtained after the first amplification was then used as a template in a second PCR with a pool of primers flanking the P genotype-specific divergent regions of gene 4 and generating fragments with type-specific lengths. The primer mixture selected for the second amplification consisted of primers C, D, and E, described by Wu et al. (14), to determine the P[8], P[4], and P[6] genotypes, respectively; primer 4T-1, described by Gentsch et al. (5), to determine the P[9] genotype; and primer SE-1, selected in our laboratory, to determine the P[14] genotype, as corresponding to a hyperdivergent region of PA169 gene 4. The sequence of the oligonucleotide primer SE-1 (5′ to 3′) was CTCTGCTACTCTACCTATTTG (complementary to nucleotides [nt] 271 to 291).
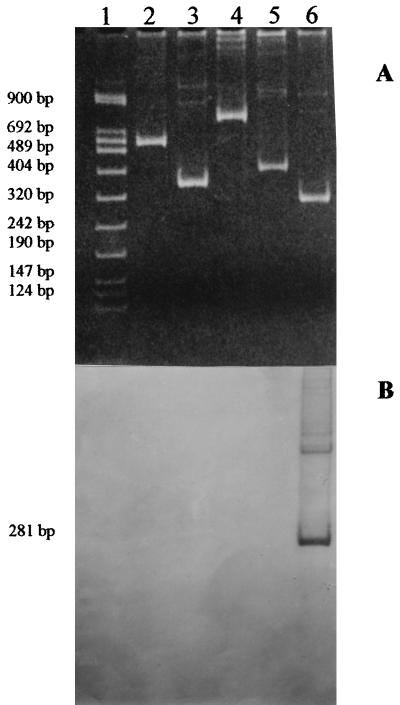
The cell culture-adapted strains were used to test the specificity of the PCR-based P typing assay. As shown in Fig. 1A, DNA fragments with the expected different sizes of 498 bp for the P[8] type, 338 bp for the P[4] type, 745 bp for the P[6] type, 391 bp for the P[9] type, and 281 bp for the P[14] type were detected.
FIG. 1.
Electrophoretic analysis of PCR products of HRV reference strains. Lane 1, DNA molecular weight marker VIII (a mixture of pUCBM21 DNA HpaII and DraI-plus-HindIII digest; Boehringer Mannheim); lane 2, strain Wa (G1, P[8]); lane 3, strain DS1 (G2, P[4]); lane 4, strain ST3 (G4, P[6]); lane 5, strain AU-1 (G3, P[9]); lane 6, strain PA169 (G6, P[14]). (A) Ethidium bromide-stained gel showing RT-PCR products after second amplification, visualized under UV light. (B) Results of Southern hybridization with biotinylated HRV P[14] genotype-specific PR-1 probe.
Southern hybridization and nonisotopic detection with an oligonucleotide probe, labeled at the 5′ end with biotin, were carried out according to the protocols described by R. Helmuth (8), with some modifications, to confirm the genotyping assay of P[14] strains. The probe, which we designed to have high homology with an internal nucleotide sequence of PA169 gene 4 and low homology with that of other genotypes, was called PR-1, and its sequence (5′ to 3′) was CACATTTAATTTACCAATTGAC (complementary to nt 225 to 246 of PA169 gene 4). Briefly, the 281-bp fragment obtained after RT-PCR was blotted onto a nylon membrane (Qiabrane, Qiagen Nylon plus) by electrophoretic transfer for 3 h at 80 V in 0.5× TBE (1× TBE is 90 mM Tris-borate–2 mM EDTA [pH 8.0]); prehybridization was performed at 52°C for 2 h in a buffer containing 0.5 M sodium phosphate (pH 7.2) plus 7% sodium dodecyl sulfate; and hybridization was carried out at 52°C overnight in plastic bags containing 200 ng of the PR-1 probe per ml. Primers and probe were synthesized by GIBCO-BRL Life Technologies. As shown in Fig. 1B, the PR-1 probe hybridized only to the 281-bp PCR product derived from PA169 gene 4.
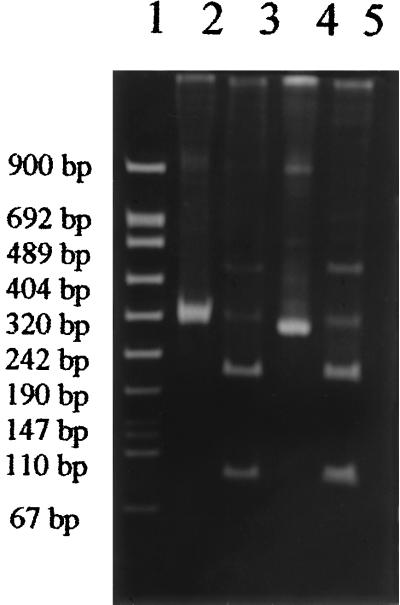
The specificity of P[14] genotyping was further confirmed by restriction analysis with the enzyme AluI, which had previously been demonstrated to be capable of recognizing a unique site at the level of nt 87 on the PA169 gene 4 sequence with the use of the DNA Strider 2.1 software program. After phenol-chloroform extraction of PCR products, 4.5 μg of DNA was incubated overnight at 37°C with 10 U of AluI enzyme (Boehringer Mannheim Biochemical GmbH, Mannheim, Germany). The digested fragments of 76 and 204 bp were separated on a 10% polyacrylamide gel in 0.5× TBE and visualized under UV light following ethidium bromide staining.
The use of the two-step amplification typing technique enabled us to identify P[14] type specificity in one strain, designated PA-5/89, out of the four strains with unusual subgroup-electropherotype linkage and G6 specificity but in none of the 50 strains with usual correlations (Table 1). In fact, after the second PCR amplification, only DNA from strain PA-5/89 yielded a product of the size (281 bp) expected for P[14] viruses. Confirmation of the P[14] type of strain PA-5/89 was obtained by Southern hybridization with the PR-1 probe. Furthermore, following digestion with AluI, amplified gene 4 cDNA of strain PA-5/89 yielded two fragments (76 and 204 bp in length) identical to those found in gene 4 cDNA of the PA169 strain (Fig. 2). PA-5/89 was isolated in January 1989 from a two-year-old child hospitalized in Palermo with acute gastroenteritis. Two out of the other three strains which exhibited unusual subgroup-electropherotype linkage and G3 specificity had a P[9] type, whereas one was not genotypeable. Our results confirmed that, as reported by Kaga et al. (9), viruses carrying the P[9] type always possess G3 specificity, as in the prototype strain AU-1. P[14] and P[9] types have never been detected in strains with usual electropherotype-subgroup linkage. For the 50 strains with usual linkage, the P[8] type was associated with G1, G3, and G4 specificities, the P[4] type was associated with G2 specificity, and the P[6] type was associated with G4 specificity (Table 1). Such combinations of G and P specificities are most commonly identified all over the world (4). From 1985 to 1996, only three other HRV strains, obtained from symptomatic children living in Palermo, Italy, showed the characteristics of the PA-5/89 strain, but they could not be tested in this study because they had been collected in amounts too small for analysis.
TABLE 1.
P genotypes in HRV strains with usual and unusual linkage among electropherotype, subgroup, and G serotype
| Genotype | No. of strainsa
|
|||||
|---|---|---|---|---|---|---|
| Sg II
|
Sg I
|
|||||
| G1 | G3 | G4 | G3 | G6 | G2 | |
| P[8] | 18 | 6 | 22 | 0 | 0 | 0 |
| P[6] | 0 | 0 | 2 | 0 | 0 | 0 |
| P[4] | 0 | 0 | 0 | 0 | 0 | 2 |
| P[9] | 0 | 0 | 0 | 2 | 0 | 0 |
| P[14] | 0 | 0 | 0 | 0 | 1 | 0 |
| NTb | 0 | 0 | 0 | 1 | 0 | 0 |
All strains were characterized as having a long electropherotype, except strains of serotype G2, which were characterized as having a short electropherotype. Sg, subgroup.
NT, not typeable.
FIG. 2.
Electrophoresis profiles of PCR-amplified gene 4 cDNA of strain PA169 (lanes 2 and 3) and of fecal isolate PA-5/89 (lanes 4 and 5), before and after, respectively, digestion with AluI, visualized by ethidium bromide staining of the gel. Lane 1, DNA molecular weight marker VIII (Boehringer Mannheim).
So far, reports on the identification of HRV strains with P[14] type have been limited. The adoption of the RT-PCR technique for HRV strains with unusual electropherotype-subgroup linkages could be useful for detecting the presence of a P[14] specificity in HRV strains circulating worldwide and for better defining the epidemiological significance of such a genotype, as well as for evaluating the occurrence of type P[14] rotavirus strains in different animal populations. Viruses with the P[14] type could have a potential role in generating by reassortment viral strains with unusual antigenic and genetic characteristics. Such a hypothesis requires further studies.
Acknowledgments
We thank Sheila McIntyre for revision of the English.
This work was supported by Ministero Pubblica Istruzione, Ricerca Scientifica 60%.
REFERENCES
- 1.Arista S, Giovannelli L, Passarani N, Titone L, Gerna G. Electropherotyping of human rotaviruses: an epidemiological survey of rotavirus infections in Sicily. Eur J Epidemiol. 1986;2:104–107. doi: 10.1007/BF00157019. [DOI] [PubMed] [Google Scholar]
- 2.Arista S, Giovannelli L, Pistoia D, Cascio A, Parea M, Gerna G. Electropherotypes, subgroups and serotypes of human rotavirus strains causing gastroenteritis in infants and young children in Palermo, Italy, from 1985 to 1989. Res Virol. 1990;141:435–446. doi: 10.1016/0923-2516(90)90044-j. [DOI] [PubMed] [Google Scholar]
- 3.Ciarlet M, Estes M K, Conner M E. Comparative amino acid sequence analysis of the outer capsid protein VP4 from four lapine rotavirus strains reveals identity with genotype P[14] human rotaviruses. Arch Virol. 1997;142:1059–1069. doi: 10.1007/s007050050142. [DOI] [PubMed] [Google Scholar]
- 4.Gentsch J R, Woods P A, Ramachandran M, Das B K, Leite J P, Alfieri A, Kumar R, Bhan M K, Glass R I. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174(Suppl. 1):S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 5.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brüssow H, Hoshino Y, Flores J. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol. 1992;30:9–16. doi: 10.1128/jcm.30.1.9-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerna G, Sears J, Hoshino Y, Steele A D, Nakagomi O, Sarasini A, Flores J. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- 8.Helmuth R. Nonisotopic detection of PCR products. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. New York, N.Y: Academic Press, Inc.; 1990. pp. 119–128. [Google Scholar]
- 9.Kaga E, Iuzuka M, Nakagomi T, Nakagomi O. The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol Immunol. 1994;38:317–320. doi: 10.1111/j.1348-0421.1994.tb01784.x. [DOI] [PubMed] [Google Scholar]
- 10.Mphahlele M J, Steele A D. Relative frequency of human rotavirus VP4 (P) genotypes recovered over a ten-year period from South African children with diarrhea. J Med Virol. 1995;47:1–5. doi: 10.1002/jmv.1890470102. [DOI] [PubMed] [Google Scholar]
- 11.Palombo E A, Bishop R F. Genetic and antigenic characterization of a serotype G6 human rotavirus isolated in Melbourne, Australia. J Med Virol. 1995;47:348–354. doi: 10.1002/jmv.1890470410. [DOI] [PubMed] [Google Scholar]
- 12.Urasawa T, Taniguchi K, Kobayashi N, Mise K, Hasegawa A, Yamazi Y, Urasawa S. Nucleotide sequence of VP4 and VP7 genes of a unique human rotavirus strain Mc35 with subgroup I and serotype 10 specificity. Virology. 1993;195:766–771. doi: 10.1006/viro.1993.1428. [DOI] [PubMed] [Google Scholar]
- 13.Ward R L. Mechanisms of protection against rotavirus in humans and mice. J Infect Dis. 1996;174(Suppl. 1):S51–S58. doi: 10.1093/infdis/174.supplement_1.s51. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Taniguchi K, Wakasugi F, Ukae S, Chiba S, Osheto M, Hasegawa A, Urasawa T, Urasawa S. Survey on the distribution of the gene 4 alleles of human rotavirus by polymerase chain reaction. Epidemiol Infect. 1994;112:615–622. doi: 10.1017/s0950268800051311. [DOI] [PMC free article] [PubMed] [Google Scholar]