Abstract
Objective: To assess the characteristics of Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations and investigated whether all KRAS mutations predict poor prognosis in patients with unresectable colorectal liver metastasis (CRLM). Methods: Correlations between KRAS-mutation status and clinicopathological characteristics of 93 patients with unresectable CRLM at our institution between 2010 and 2018 were retrospectively analyzed. Kaplan–Meier and Cox proportional hazard models were used to evaluate the prognostic significance of KRAS mutations. Results: KRAS were primarily single-point mutations, identified in 41.9% of patients. There were no significant differences in clinicopathological characteristics between wild-type KRAS and mutant KRAS. Patients with mutant KRAS had significantly worse overall survival (OS) and progression-free survival (PFS) than those with wild-type KRAS. Moreover, patients with codon 12 mutations had worse OS and PFS than those with wild-type KRAS, whereas mutations in codon 13 were not associated with a worse prognosis. Among the 5 most common mutations in codons 12, G12V, and G12D were associated with worse OS, furthermore, G12C mutation seemed to associated with worse PFS than patients with wild-type KRAS. Conclusion: KRAS codon 12 mutations were predictive for a poor prognosis in patients with unresectable CRLM. G12D and G12V mutations were associated with worse OS, whereas G12C mutation seemed to be associated with decreased PFS.
Keywords: KRAS mutation, colorectal cancer, liver metastasis, prognosis
Introduction
The liver is the most common target organ for hematogenous metastasis of colorectal cancer (CRC), and hepatic metastases are found among 15%-25% of patients suffering from CRC at the time of primary diagnosis. 1 Several reports have shown that the long-term survival of patients with colorectal liver metastasis (CRLM) is significantly shorter than that in patients without liver metastasis,2,3 additionally, some investigators have reported on the prognostic value of Kirsten rat sarcoma viral oncogene homolog (KRAS) in patients with resectable CRLM.4-8 However, a considerable proportion of patients with CRLM cannot undergo resection of liver metastasis at the time of diagnosis. Thus, it is necessary to evaluate the prognostic value of KRAS mutations in patients who cannot undergo curative intent liver resection for CRLM.
The mutation status of KRAS in primary tumors in patients with CRLM, which is closely related to the biological characteristics of the tumor, has a significant impact on prognosis.9,10 Moreover, a recent study suggested that the mutation status of KRAS plays a more important role in the prognosis of patients than the surgical technique, which emphasized the importance of KRAS in evaluating the prognosis of patients with resectable colorectal liver metastases. 11 However, the prognostic role of KRAS in patients with unresectable liver metastasis remains unclear.
Few studies have examined the relationship between prognosis and KRAS status in patients with unresectable liver metastases. The KRAS gene, which has been shown to harbor several mutations in various types of cancer, is a key component of several signaling pathways. Specific KRAS mutations have been found to lead to different activation states of KRAS protein, thereby affecting different signaling pathway molecules. Moreover, specific KRAS mutations result in different postoperative prognosis in patients following curative intent liver resection of CRLM thereby providing insights in predicting prognosis in patients and establishing a basis for optimal treatment.4,12 Several studies have shown that patients who are unable to undergo resection of liver metastases exhibit prolonged survival after resection of the primary tumor.13-17 However, even patients with the same KRAS mutations who receive the same surgical treatment may experience different postoperative survival times. This indicates that the different codons of KRAS and even different site mutations, may have diverse effects on tumor biological behavior.
Accordingly, we investigated the characteristics of 7 common mutations in codons 12 and 13 of KRAS exon 2 in the primary tumors of patients with unresectable CRLM and identified the prognostic value of distinct codon-specific KRAS mutations and their association with clinicopathological characteristics for evidence of potential clinical application in unresectable CRLM in our study.
Materials and Methods
Study Population
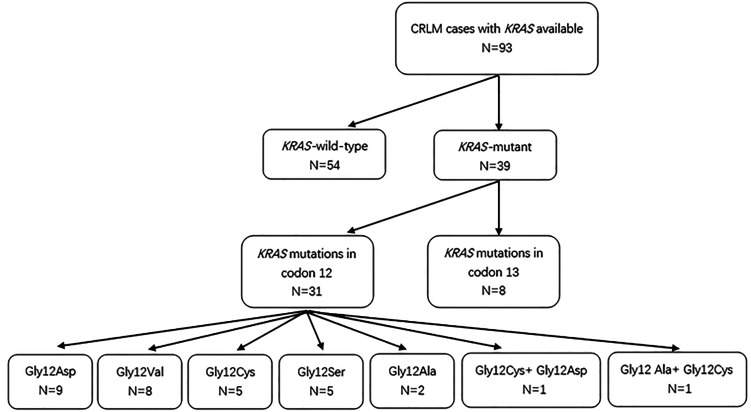
Based on the database of Fujian Provincial Hospital, a total of 93 patients with CRLM who underwent resection of primary tumors between June 2010 and January 2018 were identified (Figure 1). According to the American Joint Committee on Cancer (AJCC) (eighth edition) CRC staging, all 93 cases were of stage IV cancer. The inclusion criteria were as follows: (1) primary lesions were surgically resected and proved to be CRC by histopathology; (2) hepatic metastases of people meet the following conditions: (a) the accumulation or location of metastases near major vessels, including all main branches of retrohepatic inferior vena cava, bifurcation of the portal vein, and 3 hepatic veins; (b) the involvement of 1 branch of portal vein and its opposite branches of hepatic vein; (c) multiple metastases and distribution of 2 lobes of the liver; (d) multiple metastases or large hepatic metastases, which resulting in residual liver volume was <30% to 50%, and liver failure even occurred after surgical resection. The exclusion criteria were as follows: (1) patients reported to have tumors with BRAF mutations; (2) a history of other tumors in addition to CRLM; (3) severe heart or cerebrovascular disease; (4) prior administration of radiotherapy or chemotherapy for other diseases; (5) patients received neoadjuvant therapy or antiepidermal growth factor receptor agents in the perioperative period. Standard demographic and clinicopathologic data were collected on each patient, including gender, age, disease status, tumor characteristics, perioperative status, date of last follow-up, date of disease progression, and date of death. All patients underwent surgical resection of the primary lesion and received postoperative chemotherapy: the regimens included oral Xeloda and intravenous systemic chemotherapy (XELOX). The Fujian Provincial Hospital institutional review board approved the study. Patient informed consent specific to this study was not required given its retrospective nature.
Figure 1.
Flow chart.
KRAS Gene Detection
DNA was extracted from fresh CRC tissues using a Qiagen DNeasy Blood and Tissue Kit (Qiagen). The DNA concentration and purity were measured by using a UV–Vis spectrophotometer and an appropriate amount of TE buffer (pH 8.0) was added to adjust the DNA concentration to 0.4 to 1 ng/μL. As previously described, 18 specific primers required for amplification of codons 12 and 13 were selected, and polymerase chain reaction (PCR) was performed. PCR products were subjected to electrophoresis and the results were observed and analysed using a gel imaging analysis system. The PCR-positive DNA gel strip was selected as the target fragment and the PCR product was recovered using a QIA quick PCR Purification Kit (Qiagen). A human KRAS gene 7 mutation fluorescent PCR detection kit (Xiamen Aide Biomedical Technology Co., Ltd) was used to detect KRAS mutations.
Postoperative Follow-up
Follow-up was conducted by telephone and outpatient review through June 15, 2018. OS was defined as from the date of resection of the primary to the date of death or the end of the study period. PFS was defined as the time from the date of resection of the primary tumor to the date of progression of the primary tumor or liver metastasis.
Statistical Analysis
Data were analyzed using Chi-squared (χ2) tests or Fisher's exact tests to compare proportions. The Kaplan–Meier method was performed for survival analysis, and log rank tests were used to compare the survival distributions. Cox's proportional hazards regression model was used to identify the impact of factors on OS and PFS. Hazard ratios (HRs) were calculated with 95% confidence intervals (CIs). Differences with P values of less than .05 were considered statistically significant. All statistical analyses were performed using SPSS statistical software (version 22.0; IBM Corp.).
Results
Patient Clinicopathological Data
The clinicopathological data of the patients are summarized in Table 1. Complete clinical and pathological data for patients were obtained and all patients had simple simultaneous liver metastases. There were 61 men and 32 women. The 31 patients were over 60 years old and 62 patients were <60 years old. Overall, 29 patients had primary tumor lesions ≥5 cm, whereas 64 patients had primary tumors <5 cm. As for the primary tumor site, 11 patients were in the right colon and 40 patients were in left colon, whereas 41 patients were in rectum. There were 58 cases of multiple liver metastases, which were all multiple metastases and distribution of 2 lobes of the liver. Besides, although 35 cases were of single liver metastases, there were 8 cases of liver metastases involving 1 branch of the portal vein and the involvement of 1 branch of portal vein and its opposite branches of hepatic vein, 10 cases of liver metastasis locating near the main blood vessels, including all main branches of retrohepatic inferior vena cava, bifurcation of the portal vein among them and 3 hepatic veins and residual liver volume of another 17 cases was less than 30%. There were 73 cases of lymph node metastasis and 20 cases without lymph node metastasis (Table 1). According to the AJCC (eighth edition) CRC staging, all 93 cases were of stage IV cancer.
Table 1.
Characteristics of Patients With CRLM According to KRAS Mutation Status.
| KRAS status | KRAS mutant status | ||||||
|---|---|---|---|---|---|---|---|
| Total | Wild | Mutant | P | Codon 12 | Codon 13 | P | |
| Gender | .254 | 1 | |||||
| Male | 61 | 38 | 23 | 16 | 5 | ||
| Female | 32 | 16 | 16 | 13 | 3 | ||
| Age | .656 | .391 | |||||
| ≥60 | 62 | 35 | 27 | 19 | 7 | ||
| <60 | 31 | 19 | 12 | 10 | 1 | ||
| Tumor size (cm) | .404 | .683 | |||||
| ≥5 | 29 | 15 | 14 | 12 | 2 | ||
| <5 | 64 | 39 | 25 | 17 | 6 | ||
| Position | .823 | .471 | |||||
| Right colon | 11 | 7 | 4 | 4 | 0 | ||
| Left colon | 40 | 24 | 16 | 10 | 4 | ||
| Rectum | 42 | 23 | 19 | 15 | 4 | ||
| Tumor type | .231 | .655 | |||||
| Raised | 23 | 10 | 13 | 11 | 2 | ||
| Ulcerative | 66 | 41 | 25 | 17 | 6 | ||
| Infiltrative | 4 | 3 | 1 | 1 | 0 | ||
| Number of liver metastases | .111 | .091 | |||||
| Solitary | 35 | 24 | 11 | 7 | 3 | ||
| Multiple | 58 | 30 | 28 | 22 | 5 | ||
| Maximum diameter of liver metastasis (cm) | .052 | .530 | |||||
| ≤5 | 77 | 41 | 36 | 27 | 7 | ||
| >5 | 16 | 13 | 3 | 2 | 1 | ||
| T stage | .306 | .530 | |||||
| T1 or T2 | 4 | 1 | 3 | 2 | 1 | ||
| T3 orT4 | 89 | 53 | 36 | 27 | 7 | ||
| N stage | .843 | .332 | |||||
| N0 | 20 | 12 | 8 | 5 | 3 | ||
| N1 or N2 | 73 | 42 | 31 | 24 | 5 | ||
| CEA (ng/mL) | .233 | 1 | |||||
| <5 | 25 | 12 | 13 | 10 | 2 | ||
| ≥5 | 68 | 42 | 26 | 19 | 6 | ||
| CA199 (U/mL) | .496 | .423 | |||||
| <27 | 42 | 26 | 16 | 14 | 2 | ||
| ≥27 | 51 | 28 | 23 | 15 | 6 | ||
Abbreviations: CEA, carcinoembryonic antigen; CA 199, carbohydrate antigen 199; KRAS, Kirsten rat sarcoma viral oncogene homolog; CRLM, colorectal liver metastasis.
KRAS Mutation Status and Clinicopathological Data
Mutated KRAS was identified in 39 patients (41.9%) and were mainly found in codons 12 and 13 (31 in codon 12 only, 8 in codon 13 only). KRAS mutations were mainly single-point mutation (37 cases, 94.8%), and multiple-point mutations were found only in 2 cases only (5.2%) as G12C + G12D and G12A + G12C mutations. Among patients with KRAS codon 12 mutations, we identified 9 cases with G12D, 8 cases with G12V, 5 cases with G12S, 5 cases with G12C, and2 cases with G12A. The G13D mutation was only detected in patients with KRAS codon 13 mutations (8 cases). All codon 12 and 13 mutations were missense mutations, although the base changes for each mutation were different. Among the observed mutations, G12S, G12D, and G13D were transitions (G > A), accounting for 59.5% (22/37), and G12C, G12 V, and G12A were transversions (G > T for 13 cases, G > C for 2 cases), accounting for 40.5% (15/37; Table 2). There were no significant differences in any of the features examined between patients with mutant and wild-type KRAS or between patients with mutations in codons 12 and 13 (Table 1).
Table 2.
Characteristics of Specific Mutations in KRAS.
| KRAS | N/total | Mutant rate (%) |
|---|---|---|
| Single site mutation | ||
| KRAS codon 12 mutation | ||
| G12A(GGT>GCT) | 2/39 | 5.1 |
| G12D(GGT>GAT) | 9/39 | 23.1 |
| G12C(GGT>TGT) | 5/39 | 12.8 |
| G12S(GGT>AGT) | 5/39 | 12.8 |
| G12V(GGT>GTT) | 8/39 | 20.5 |
| KRAS codon 13 mutation | ||
| G13D (GGC>GAC) | 8/39 | 20.5 |
| Multiple site mutations | ||
| G12C + G12D | 1/39 | 2.6 |
| G12A + G12C | 1/39 | 2.6 |
Abbreviations: KRAS, Kirsten rat sarcoma viral oncogene homolog.
Overall Survival
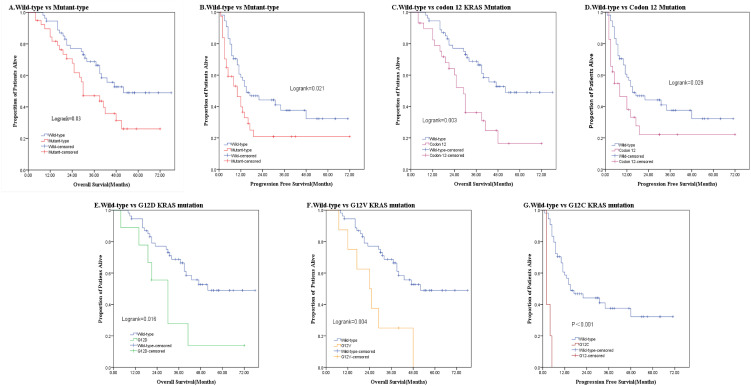
The median OS for patients with mutant KRAS was 30.0 months, and the 3- and 5-year OS rates for 39 patients with mutant KRAS were 47.1% and 26.1%, respectively. In contrast, in patients with wild-type KRAS, the median OS was 52.0 months, with 3- and 5-year OS rates of 68.8% and 49.0%, respectively. The OS of patients with mutant KRAS was worse than that in patients with wild-type KRAS (HR = 1.947; 95% CI, 1.079-3.513; P = .027; Figure 2, Table 3). Further analysis showed that the median OS and PFS for patients with KRAS 12 codon mutations were 29 and 8 months, whereas those in patients with wild-type KRAS were 52 and 15 months, respectively; the differences were statistically significant. In both Kaplan–Meier analysis (log-rank P = .003) and Cox regression analysis (univariate HR = 2.455; 95% CI, 1.339-4.501; P = .004; and multivariate HR = 2.527; 95% CI, 1.349-4.734; P = .004), OS was worse in patients with KRAS codon 12 mutations than in patients with wild-type KRAS. In contrast, KRAS codon 13 mutations were not associated with a worse OS than wild-type KRAS (univariate analysis: HR = .873; 95% CI, 0.262-2.911; P = .825; multivariate analysis: HR = .822; 95% CI, 0.243-2.776; P = .752; Figure 2, Table 4). In univariate analysis, multiple liver metastases (HR = 3.033; 95% CI, 1.495-6.155; P = .002), palliative surgery (HR = 2.715; 95% CI, 1.149-6.415; P = .023), and KRAS codon 12 mutations were risk factors for worse prognosis in patients with unresectable CRLM (Tables 3 and 4). Multivariate analysis using the Cox proportional hazard model revealed that codon 12 mutations were an independent negative prognostic factor in patients with unresectable CRLM. Futhermore, we analysed the 5 most common KRAS codon 12 mutations. G12D (HR = 2.568; 95% CI, 1.087-6.070; P = .032) and G12 V (HR = 3.400; 95% CI, 0.1.432-8.073; P = .006; Figure 2, Table 5) mutations were associated with a significantly higher risk of long-term death compared with wild-type KRAS. There were no significant differences in OS between patients with the remaining 4 KRAS mutations and wild-type KRAS (P > .05).
Figure 2.
Kaplan–Meier curves from patients with colorectal cancer with unresectable liver metastases according to KRAS mutation status. (A) OS according to wild-type or mutant KRAS. (B) PFS according to wild-type or mutant KRAS. (C) OS according to KRAS codon 12 or wild-type. (D) PFS according to KRAS codon 12 or wild-type. (E) OS according to KRAS c.35G>A (p.G12D) mutation status. (F) OS according to KRAS c.35G>T (p.G12V) mutation status. (G) PFS according to KRAS c.34G>T (p.G12C) mutation status.
Abbreviation: KRAS, Kirsten rat sarcoma viral oncogene homolog; OS, overall survival; PFS, progression-free survival.
Table 3.
Univariate and Multivariate Cox Proportional Hazard Analysis for Overall Survival and Progression Free Survival.
| Prognostic factor | Overall survival | Progression free survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate HR (95% CI) |
P | Multivariate HR (95% CI) |
P | Univariate HR (95% CI) |
P | Multivariate HR (95% CI) |
P | |
| KRAS status | .022 | .027 | .026 | .016 | ||||
| Wild type | 1(Referent) | 1(Referent) | 1(Referent) | 1(Referent) | ||||
| Mutant type | 1.974(1.105-3.529) | 1.947(1.079-3.513) | 1.810(1.073-3.053) | 1.906(1.127-3.223) | ||||
| Gender | .738 | .895 | ||||||
| Male | 1(Referent) | … | 1(Referent) | … | ||||
| Female | 0.903(0.495-1.646) | … | 0.964(0.606-1.773) | … | ||||
| Age(years) | .949 | .657 | ||||||
| <60 | 1(Referent) | … | 1(Referent) | … | ||||
| ≥60 | 1.021(0.544-1.914) | … | 0.884(0.514-1.522) | … | ||||
| Tumor size(cm) | .295 | .415 | ||||||
| <5 | 1(Referent) | … | 1(Referent) | … | ||||
| ≥5 | 1.384(0.753-2.542) | … | 0.790(0.448-1.392) | … | ||||
| Location of tumor | .199 | .029 | .764 | |||||
| Right colon | 1(Referent) | 1(Referent) | 1(Referent) | … | ||||
| Left colon | 0.779(0.345-1.761) | 0.817(0.358-1.867) | 1.347(0.552-3.285) | … | ||||
| Rectum | 0.497(0.214-1.153) | 0.382(0.162-0.904) | 1.163(0.480-2.819) | … | ||||
| Tumor type | .472 | .744 | ||||||
| Raised | 1(Referent) | … | 1(Referent) | … | ||||
| Ulcerative | 1.337(0.675-2.650) | … | 1.246(0.677-2.295) | … | ||||
| Infiltrative | 0.500(0.065-3.877) | … | 0.949(0.214-4.205) | … | ||||
| Number of liver metastases | .002 | .002 | .423 | |||||
| Solitary | 1(Referent) | 1(Referent) | 1(Referent) | … | ||||
| Multiple | 3.033(1.495-6.155) | 2.797(1.174-6.662) | 1.247(0.727-2.138) | … | ||||
| Maximum diameter of metastasis(cm) | .999 | .677 | ||||||
| <5 | 1(Referent) | … | 1(Referent) | |||||
| ≥5 | 1.001(0.466-2.148) | … | 0.859(0.420-1.756) | … | ||||
| AJCC eighth edition T stage | .353 | .817 | ||||||
| T1/T2 | … | … | 1(Referent) | … | ||||
| T3/T4 | … | … | 0.872(0.271-2.800) | … | ||||
| Regional lymph node metastasis | .292 | … | .042 | .029 | ||||
| Negative | 1(Referent) | … | 1(Referent) | 1(Referent) | ||||
| Positive | 0.693(0.351-1.370) | … | 2.170(1.027-4.586) | 2.302(1.087-4.878) | ||||
| CEA (ng/mL) | .271 | .755 | ||||||
| <5 | 1(Referent) | … | 1(Referent) | … | ||||
| ≥5 | 0.702(0.374-1.318) | … | 0.912(0.512-1.624) | … | ||||
| CA199 (U/mL) | .956 | .947 | ||||||
| <27 | 1(Referent) | … | 1(Referent) | … | ||||
| ≥27 | 1.017(0.570-1.814) | … | 0.947(0.566-1.587) | … | ||||
| Surgery | .023 | .020 | .444 | |||||
| Radical resection | 1(Referent) | 1(Referent) | 1(Referent) | … | ||||
| Palliative resection | 2.715(1.149-6.415) | 3.154(1.524-6.528) | 0.798(0.448-1.422) | … | ||||
Abbreviations: CEA, carcinoembryonic antigen; CA 19 to 9, carbohydrate antigen 19 to 9; HR, hazard ratio; 95% CI, 95% confidence interval; KRAS, Kirsten rat sarcoma viral oncogene homolog.
Table 4.
Univariate and Multivariate Cox Proportional Hazard Analysis for Overall Survival and Progression Free Survival According to Specific KRAS Codon Mutation.
| Overall survival | Progression free survival | |||
|---|---|---|---|---|
| Univariate HR (95% CI) |
Multivariate HR (95% CI) |
Univariate HR (95% CI) |
Multivariate HR (95% CI) |
|
| Wild type | 1(Referent) | 1(Referent) | 1(Referent) | 1(Referent) |
| All codon 12 mutations | 2.455(1.339-4.501) P = .004 |
2.527(1.349-4.734) P = .004 |
1.840(1.047-3.232) P = .034 |
1.830(1.041-3.216) P = .036 |
| All codon 13 mutations | 0.873(0.262-2.911) P = .825 |
0.822(0.243-2.776) P = .752 |
1.717(0.715-4.126) P = .227 |
2.238(0.907-5.522) P = .080 |
Abbreviations: HR, hazard ratio; 95% CI, 95% confidence interval; KRAS, Kirsten rat sarcoma viral oncogene homolog.
Table 5.
Univariate and Multivariate Cox Proportional Hazard Analysis for Overall Survival and Progression Free Survival According to Specific KRAS Mutation in the Whole Group.
| KRAS | Overall survival | Progression free survival | ||
|---|---|---|---|---|
| Univariate HR (95% CI) |
Multivariate HR (95% CI) |
Univariate HR (95% CI) |
Multivariate HR (95% CI) |
|
| Wild type | 1(Referent) | 1(Referent) | 1(Referent) | 1(Referent) |
| G12D | 2.702(1.152-6.337) P = .022 |
2.568(1.087-6.070) P = .032 |
1.104(0.429-2.844) P = .838 |
1.207(0.467-3.115) P = .698 |
| G12A | 2.346(0.313-17.606) P = .407 |
1.440(0.191-10.885) P = .724 |
1.190(0.161-8.804) P = .865 |
1.039(0.140-7.703) P = .970 |
| G12C | 1.663(0.497-5.565) P = .409 |
1.326(0.391-4.491) P = .651 |
9.318(3.522-24.653) P < .001 |
8.676(3.274-22.99) P < .001 |
| G12V | 3.292(1.401-7.738) P = .006 |
3.400(0.1.432-8.073) P = .006 |
1.913(0.795-4.607) P = .148 |
1.655(0.684-4.005) P = .264 |
| G12S | 1.698(0.400-7.209) P = .473 |
3.872(0.806-18.589) P = .091 |
2.258(0.678-7.524) P = .185 |
2.679(0.790-9.084) P = .114 |
| G13D | 0.873(0.262-2.910) P = .825 |
0.701(0.209-2.354) P = .566 |
1.801(0.747-4.344) P = .190 |
2.362(0.949-5.881) P = .065 |
Abbreviations: HR, hazard ratio; 95% CI, 95% confidence interval; KRAS, Kirsten rat sarcoma viral oncogene homolog.
Progression Free Survival
The median PFS for 39 patients with KRAS mutations was significantly shorter (10 months) than that in patients with wild-type KRAS (15 months; HR = 1.947; 95% CI, 1.079-3.513; P = .027; Table 3). Moreover, the PFS of patients with KRAS codon 12 mutations (8 months) was significantly shorter than that of patients with wild-type KRAS (log-rank = 0.029; multivariate analysis: HR = 1.830; 95% CI, 1.041-3.216; P = .036). In contrast, there were no significant differences in PFS between patients with KRAS codon 13 mutations and wild-type KRAS (HR = 2.238; 95% CI, 0.907-5.522; P = .080; Figure 2, Table 4). In multivariate analysis, regional lymph node metastasis (HR = 2.302; 95% CI, 1.087-4.878; P = .029) was also associated with a worse prognosis in patients with unresectable CRLM (Table 3). Further analysis revealed that G12C was associated with a statistically significant decrease in PFS compared with all other KRAS mutants or with wild-type KRAS (HR = 8.676; 95% CI, 3.274-22.99; P < .001; Figure 2, Table 5).
Discussion
The importance of KRAS in assessing the prognosis of patients with hepatic metastasis has been confirmed in previous studies. However, there are many mutation sites in KRAS and the biological effects caused by mutations at the different sites are still controversial. Moreover, few studies have examined the prognostic roles of specific point mutations in KRAS codons in patients with unresectable CRLM. Therefore, we evaluated specific mutations in KRAS and analyzed the relationships between the mutations and the clinicopathological characteristics and the prognosis of unresectable CRLM, with the goal of providing information for improvement of prognoses and treatments.
Similar to previous studies demonstrating that the incidence of KRAS mutations ranged from 30% to 50%,19-21 we found that 41.9% of tumor specimens from our cohort with unresectable CRLM harboring KRAS mutations. Moreover, the mutations in KRAS were point mutations and were mostly observed in exon 2, including codon 12 and codon 13,22-26 which is consistent with our findings. Among patients with mutations in codons 12 and 13, G12D, G12 V, and G13D were detected in more than 20% of specimens, whereas G12S, G12A, and G12C were less common. These findings were similar to the results of a previous study by Thierry and colleagues. 27
In present study, we found that CRLM patients with KRAS mutations had a significant increased risk of postoperative tumor recurrence and death compared with that of patients with a wild-type KRAS. However, the impact of KRAS-specific codons and even specific sites within the gene on the prognosis of CRLM patients is still not clear. Specifically, our current study indicated that OS and PFS in patients with codon 12 mutations, but not codon 13 mutation, were worse than those in patients with wild-type KRAS, resulting in ∼150% increased risk of death and 83% increased risk of tumor progression. According to the results from cell experiments by Guerrero et al 28 KRAS codon 12 mutations are associated with a more aggressive tumor phenotype than codon 13 mutations owing to alterations in the threshold for apoptosis induction. Additionally, Renaud et al 29 confirmed that codon 12 mutations are associated with increased upregulation of vascular endothelial growth factor (VEGF) and more stable bonds between RatSarcoma (RAS) and guanosine triphosphate (GTP). These findings indicate that the codon 12 and 13 mutations represent different tumor clones, although they are both KRAS mutations. Compared with the codon 13 mutation, the KRAS codon 12 mutation conferred a more aggressive tumor phenotype. Therefore, we may be able to determine the prognosis of CRLM patients by evaluating specific mutation subtypes in KRAS and formulate different treatment strategies according to particular genetic conditions.
Notably, we also confirmed that not all codon 12 mutations led to worse survival outcomes than wild-type KRAS. Among the 5 most common codon 12 mutations, only G12 V and G12D mutations increased the risk of postoperative death, with a worse OS than in patients with other KRAS mutations or with wild-type KRAS. Moreover, the G12C mutation increased the risk of tumor progression with a decreased PFS. The aggressive behaviors of tumors harboring the G12V and G12D mutations has been explained by Al-Mulla et al 30 who showed that both G12V and G12D mutations produce proteins with lower basal GTPase activity that are refractory to stimulation by GAP binding. Moreover, more stable complexes formed from combining with GTP and the lower affinity of the G12V mutant for GAP provided Ras a more constant positive signal. Consistent with the results of Margonis and Imamural,12,28 we confirmed that the G12 V and G12D mutation conferred cancer cells with more aggressive biologic behaviors after primary resection in CRLM. Additionally, we also found that G12C was associated with poor prognosis. These findings indicate that even if they are all located in codon 12, the mechanism that affect tumor biological behavior is different in distinct mutation sites. To the best of our knowledge, various type of KRAS point mutations may activate different downstream signaling events. As previously described by Ihle and colleagues, 31 both G12C and G12V mutations activate Ral signalling and decrease growth factor-dependent Akt activation, whereas the G12D mutation activates phosphatidylinositol 3-kinase and mitogen-activated protein kinase/extracellular signal-regulated kinase signaling. Additionally, Floyd and Dance-Barnes reported the G12C mutation preferentially induces anchorage-independent growth by activating Raf and Ral.32,33 These also prove that even different mutations in a single gene may affect unique biological behavior, which further emphasizes the importance of tumor heterogeneity in the diagnosis and treatment of cancer.
Furthermore, according to our study, not all patients with unresectable CRLM had a short survival period, which may encourage patients not to give up treatment, cooperate with the examination and treatment to prolong survival, and improve quality of life. Besides, the value of specific KRAS point mutations in accessing prognosis of CRLM patients will improve the accuracy of prognosis, which may be superior to tumor size and tumor infiltration depth in clinical practice. Moreover, the use of KRAS status as a prognostic indicator at the molecular biology level may be more objective and accurate than previous prognostic indicators.
Despite these positive findings, there were some limitations to the current study. Firstly, the follow-up period was relatively short, and the median OS was not reached in the partial subgroup. Secondly, due to being a retrospective study, the sample population was limited, which may have introduced some selection bias. Finally, the patient number with specific mutation was low (such as G12C mutation) and its prognostic value needs further study. Thus, we need additional studies using larger sample sizes with longer follow-up times to provide more reliable results in the future.
Conclusion
In conclusion, specific mutations in codon 12 of KRAS, specifically G12 V and G12D, were independent prognostic factors of worse OS after resection, whereas G12C seemed to be associated with worse PFS. The determination of specific mutations in KRAS could help clinicians develop therapeutic strategies and even provide important information for developing molecular targeted drugs to improve prognosis in patients with unresectable CRLM.
Acknowledgments
The authors thank all study participants of the Department of Pathology for their contributions to this project. The authors also thank Amoy Diagnostics Co., Ltd for providing KRAS Mutation Detection Kit and the corresponding primers (patent no. CN200910111501.6). The authors would also like to thank Editage (www.editage.cn) for English language editing.
Abbreviations
- CEA
carcinoembryonic antigen
- CI
confidence interval
- CRC
colorectal cancer
- CRLM
colorectal liver metastasis
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- OS
overall survival
- PFS
progression-free survival
- HR
hazard ratio
Footnotes
Authors’ Note: The name of the committee is Fujian Provincial Hospital Ethics Committee and the approval number is K2018-08-034.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship and/or publication of this article.
ORCID iD: Weihua Li https://orcid.org/0000-0003-4581-2240
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Weber SM, Jarnagin WR, DeMatteo RP, et al. Survival after resection of multiple hepatic colorectal metastases. Ann Surg Oncol. 2000;7:643-650. doi: 10.1007/s10434-000-0643-3. [DOI] [PubMed] [Google Scholar]
- 2.Swan PJ, Welsh FK, Chandrakumaran K, et al. Long-term survival following delayed presentation and resection of colorectal liver metastases. Br J Surg. 2011;98(9):1309-1317. doi: 10.1002/bjs.7527. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Liu YF, Cheng Y, et al. Prognosis of colorectal cancer with liver metastasis: value of a prognostic index. Brazilian J Med Biol Res=Revista Brasileira de Pesquisas Medicas E Biologicas. 2010;43(11):1116-1122. doi: 10.1590/S0100-879X2010007500103. [DOI] [PubMed] [Google Scholar]
- 4.Margonis GA, Kim Y, Spolverato G, et al. Association between specific mutations in KRAS Codon 12 and colorectal liver metastasis. JAMA Surg. 2015;150(8):722-729. doi: 10.1001/jamasurg.2015.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margonis GA, Kim Y, Sasaki K, et al. Codon 13 KRAS mutation predicts patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Cancer. 2016;122(17):2698-2707. doi: 10.1002/cncr.30085. [DOI] [PubMed] [Google Scholar]
- 6.Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS Mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2013;258(4):619-627; discussion 626-617. doi: 10.1097/SLA.0b013e3182a5025a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel TL, Vakiani E, Nathan H, et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer. 2017;123(4):568-575. doi: 10.1002/cncr.30351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki K, Margonis GA, Wilson A, et al. Prognostic implication of KRAS status after hepatectomy for colorectal liver metastases varies according to primary colorectal tumor location. Ann Surg Oncol. 2016;23:3736-3743. doi: 10.1245/s10434-016-5361-6. [DOI] [PubMed] [Google Scholar]
- 9.Coulson R. Molecular profiling in resectable colorectal liver metastases: the role of KRAS mutation status in assessing prognosis in the preoperative setting. J Adv Pract Oncol. 2015;6:470-474. doi: 10.6004/jadpro.2015.6.5.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margonis GA, Spolverato G, Kim Y, et al. Effect of KRAS mutation on long-term outcomes of patients undergoing hepatic resection for colorectal liver metastases. Ann Surg Oncol. 2015;22:4158-4165. doi: 10.1245/s10434-015-4587-z. [DOI] [PubMed] [Google Scholar]
- 11.Margonis GA, Sasaki K, Kim Y, et al. Tumor biology rather than surgical technique dictates prognosis in colorectal cancer liver metastases. J Gastrointestinal Surgery. 2016;20:1821-1829. doi: 10.1007/s11605-016-3198-8. [DOI] [PubMed] [Google Scholar]
- 12.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753-4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shida D, Hamaguchi T, Ochiai H, et al. Prognostic impact of palliative primary tumor resection for unresectable stage 4 colorectal cancer: using a propensity score analysis. Ann Surg Oncol. 2016;23:3602-3608. doi: 10.1245/s10434-016-5299-8. [DOI] [PubMed] [Google Scholar]
- 14.Winner M, Mooney SJ, Hershman DL, et al. Incidence and predictors of bowel obstruction in elderly patients with stage IV colon cancer: a population-based cohort study. JAMA Surg. 2013;148(8):715-722. doi: 10.1001/jamasurg.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishihara S, Nishikawa T, Tanaka T, et al. Benefit of primary tumor resection in stage IV colorectal cancer with unresectable metastasis: a multicenter retrospective study using a propensity score analysis. Int J Colorectal Dis. 2015;30:807-812. doi: 10.1007/s00384-015-2228-4. [DOI] [PubMed] [Google Scholar]
- 16.Anwar S, Peter MB, Dent J, et al. Palliative excisional surgery for primary colorectal cancer in patients with incurable metastatic disease. Is there a survival benefit? A systematic review. Colorectal Disease. 2012;14(8):920-930. doi: 10.1111/j.1463-1318.2011.02817.x. [DOI] [PubMed] [Google Scholar]
- 17.Eisenberger A, Whelan RL, Neugut AI. Survival and symptomatic benefit from palliative primary tumor resection in patients with metastatic colorectal cancer: a review. Int J Colorectal Dis. 2008;23:559-568. doi: 10.1007/s00384-008-0456-6. [DOI] [PubMed] [Google Scholar]
- 18.Jhawer M, Goel S, Wilson AJ, et al. PIK3CA Mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68(6):1953-1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Annals of Oncol. 2008;19(3):508-515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 20.Bell SM, Scott N, Cross D, et al. Prognostic value of p53 overexpression and c-Ki-ras gene mutations in colorectal cancer. Gastroenterology. 1993;104(1):57-64. doi: 10.1016/0016-5085(93)90835-Z. [DOI] [PubMed] [Google Scholar]
- 21.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 22.Khosravi-Far R, Der CJ. The Ras signal transduction pathway. Cancer Metastasis Rev. 1994;13:67-89. doi: 10.1007/BF00690419. [DOI] [PubMed] [Google Scholar]
- 23.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293-297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 24.Boughdady IS, Kinsella AR, Haboubi NY, et al. K-ras gene mutations in adenomas and carcinomas of the colon. Surg Oncol. 1992;1(4):275-282. doi: 10.1016/0960-7404(92)90088-3. [DOI] [PubMed] [Google Scholar]
- 25.Neumann J, Zeindl-Eberhart E, Kirchner T, et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205(12):858-862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Finkelstein SD, Sayegh R, Christensen S, et al. Genotypic classification of colorectal adenocarcinoma. Biologic behavior correlates with K-ras-2 mutation type. Cancer. 1993;71(12):3827-3838. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430-435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 28.Guerrero S, Casanova I, Farré L, et al. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750-6756. [PubMed] [Google Scholar]
- 29.Renaud S, Guerrera F, Seitlinger J, et al. KRAS Exon 2 codon 13 mutation is associated with a better prognosis than codon 12 mutation following lung metastasectomy in colorectal cancer. Oncotarget. 2017;8:2514-2524. doi: 10.18632/oncotarget.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Mulla F, Milner-White EJ, Going JJ, et al. Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J Pathol. 1999;187(4):433-438. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104(3):228-239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floyd HS, Farnsworth CL, Kock ND, et al. Conditional expression of the mutant Ki-rasG12C allele results in formation of benign lung adenomas: development of a novel mouse lung tumor model. Carcinogenesis. 2005;26(12):2196-2206. doi: 10.1093/carcin/bgi190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dance-Barnes ST, Kock ND, Floyd HS, et al. Effects of mutant human Ki-ras(G12C) gene dosage on murine lung tumorigenesis and signaling to its downstream effectors. Toxicol Appl Pharmacol. 2008;231(1):77-84. doi: 10.1016/j.taap.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]