Abstract
Background
Detection of SARS-CoV-2 infections relies on the use of sensitive, accurate and high throughput RT-PCR assays.
Objectives
We assessed the analytical performance of the Abbott RealTime SARS-CoV-2 (RT-SARS), Alinity m SARS-CoV-2 (AlinSARS) assays and compared the clinical performance of the RT-SARS, AlinSARS, and Alinity m Resp-4-Plex (Alin4Plex) assays to the Seegene Allplex assay (Allplex) and an inhouse test (Inhouse).
Results
We found 100 % positive percent agreement (PPA) and 100 % negative percent agreement (NPA) comparing RT-SARS and Allplex. RT-SARS, AlinSARS and Inhouse showed 100 % NPA and 100 % PPA across all assays, except for the RdRp target of Inhouse (PPA = 84 %). Similarly, Alin4Plex and Allplex showed high agreement with specimens containing either SARS-CoV-2, influenza A, influenza B, or RSV. Detection rates of 100 % for SARS-CoV-2 at 50 copies/mL, high precision, and no cross-reactivity with non-SARS-CoV-2 respiratory pathogens were observed for RT-SARS and AlinSARS. AlinSARS detected SARS-CoV-2 in spiked throat washes and in specimens infected with SARS-CoV-2 Alpha or Beta variants.
Conclusions
The newly developed RT-SARS, AlinSARS, and Alin4Plex assays proved to be useful for detecting SARS-CoV-2 RNA in clinical samples.
Keywords: SARS-CoV-2, Real-Time PCR, Alinity m, multiplex, Turnaround time
1. Introduction
Rapid and accurate diagnostic testing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is essential for the management of the COVID-19 pandemic (Watson et al., 2020). Reverse transcriptase-polymerase chain reaction (RT-PCR) test methods targeting SARS-CoV-2 RNA are the gold standard for diagnosing suspected cases of COVID-19, patient care, contact tracing and outbreak investigations and are expected to reliably detect also newly emerging variants of concern (VOC) (European Centre for Disease Prevention and Control, 2020a; European Centre for Disease Prevention and Control, 2021a). Testing strategies are increasingly complemented by active surveillance of individuals at high risk of infection, like healthcare workers or teachers and children at school (European Centre for Disease Prevention and Control, 2021b; European Centre for Disease Prevention and Control, 2020b). Recommendations include saliva or throat wash as alternative specimen types besides oro- and nasopharyngeal swabs, especially when testing children (European Centre for Disease Prevention and Control, 2020b). As case definitions for influenza and acute respiratory infections overlap with those of COVID-19, the use of multiplex RT-PCR assays, e.g. for SARS-CoV-2, influenza, and RSV, can be considered (European Centre for Disease Prevention and Control, 2020c).
Testing of specimens for the presence and discrimination of respiratory viruses relies on the use of accurate molecular diagnostic assays. High throughput molecular diagnostic analyzers, with high level automation returning the RT-PCR test results within 24 h, support coping with the high testing demand for SARS-CoV-2 (European Centre for Disease Prevention and Control, 2021c). We evaluated the analytical performance of the Abbott RealTime™ SARS-CoV-2 assay for use on the automated m2000™ batch analyzer system and the Abbott Alinity m SARS-CoV-2 assay for use on the Alinity m system, a fully automated continuous and random-access analyzer reporting approximately 1000 results in 24 h. We also compared the clinical performance of the RealTime SARS-CoV-2, the Alinity m SARS-CoV-2, and the Alinity m Resp-4-Plex assays to the Seegene Allplex assay and an inhouse test.
2. Methods
2.1. Molecular SARS-CoV-2 detection methods
The Abbott RealTime SARS-CoV-2 (“RT-SARS”) and the Alinity m SARS-CoV-2 (“AlinSARS”) assays (both Abbott Molecular Inc., Des Plaines, IL, USA) are intended for the qualitative detection of SARS-CoV-2 in nasopharyngeal and oropharyngeal swabs from patients suspected of COVID-19 infection (Abbott Molecular Inc., 2020a; Abbott Molecular Inc., 2020b). The Alinity m Resp-4-Plex assay (“Alin4Plex”; Abbott Molecular Inc., Des Plaines, IL, USA) is intended for the qualitative detection and identification of influenza A virus (FluA), influenza B virus (FluB), respiratory syncytial virus (RSV), and SARS-CoV-2 from nasopharyngeal swabs (Abbott Molecular Inc., 2020c). All three assays detect highly conserved and SARS-CoV-2-specific target regions in the RdRp and N genes of the SARS-CoV-2 genome. Additionally, Alin4Plex targets the Matrix genes of RSV and FluA and the Nonstructural 1 gene of FluB.
The comparator method Allplex™ SARS-CoV-2 (“Allplex”, Seegene Inc., Seoul, Korea) targets the E gene for the detection of the sarbecovirus group and additionally two SARS-CoV-2-specific sequences in the RdRp and N genes (Seegene Inc., 2020). Extraction and master-mix preparation was performed on the Seegene NIMBUS automated liquid handling workstation. Abbott and Seegene assays were performed according to the manufacturers' instructions. Furthermore, the TIB MolBiol LightMix® SarbecoV E-gene plus EAV control assay (TIB MolBiol Syntheselabor GmbH, 2020) (TIB Molbiol Syntheselabor GmbH, Berlin, Germany) was extended to an inhouse multiplex assay by adding primers and probe targeting the RdRp gene according to Corman et al (Corman et al., 2020) (“Inhouse”). It was run on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories GmbH, Feldkirchen, Germany) and was used as a second molecular comparator test. All samples positive for either one or two targets of Inhouse were retested with Allplex for confirmation.
The study was performed in accordance with the principles of Good Clinical Practice and conducted in adherence with the Declaration of Helsinki. Only residual samples from routine SARS-CoV-2 testing were used.
2.2. SARS-CoV-2 detection rates and precision of RT-SARS and AlinSARS
To assess the detection rates of RT-SARS, an RT-SARS dilution panel with target concentrations of 1000, 500, 250, 100, 50, 10, 5, and 2.5 cps/mL was prepared by diluting the Abbott RealTime SARS-CoV-2 positive control with 0.9 % NaCl solution. An AlinSARS dilution panel was prepared by diluting the Alinity m SARS-CoV-2 positive control, which is identical to the RealTime positive control, to target concentrations of 400, 300, 200, 100, 50, 25, 10, and 5 cps/mL to evaluate the detection rates of AlinSARS. Each panel member was tested in 9–20 replicates, respectively. Assay precision was assessed by calculating means, standard deviations (SD), and coefficients of variation (CV) of the cycle threshold (Ct) values for each test.
2.3. Cross-reactivity with other respiratory pathogens
A total of 40 swab samples from individuals infected with various non-SARS-CoV-2 respiratory pathogens was used to assess cross-reactivity of RT-SARS and AlinSARS assays: Influenza A (n = 25), Influenza B (n = 5), RSV A (n = 2), RSV B (n = 2), M. pneumoniae (n = 4), B. parapertussis (n = 1), H. influenzae (n = 1). Samples had been pretested with Seegene Allplex™ Respiratory Panels 1&4 (Seegene Inc., Seoul, Korea) and had been stored at −20 °C for up to 48 weeks prior to retesting.
2.4. Clinical performance of RT-SARS, AlinSARS and Alin4Plex
A total of 197 de-identified swab samples with sufficient residual volume were selected based on their previous test results to evaluate the clinical performance of RT-SARS, AlinSARS, and Alin4Plex assays: Twenty-nine SARS-CoV-2 negative and 29 SARS-CoV-2 positive specimens by Allplex (Ct values 17–34, 18–37, and 20–38 for the E, RdRp, and N genes, respectively) were retested with RT-SARS. A second set of 50 SARS-CoV-2 negative and 50 SARS-CoV-2 positive specimens by Inhouse were retested with RT-SARS and AlinSARS. Positive samples by Inhouse (Ct values 17–46 and 21–43 for the E and RdRp genes, respectively) had been confirmed to be positive by Allplex. Finally, a third set of 20 SARS-CoV-2 positive (Ct values 12–36) and 19 SARS-CoV-2 negative samples that were positive for FluA (n = 11), FluB (n = 5), or RSV (n = 3) by Allplex was retested with Alin4Plex. Specimens with sufficient sample volume and previously tested positive for SARS-CoV-2 by Allplex, were also retested with AlinSARS (n = 11). All samples were negative for the other three pathogens using Allplex. After routine testing, samples had been stored at −20 °C for up to 48 weeks before retesting.
2.5. Detectability of SARS-CoV-2 VOCs with AlinSARS
Since February 2021, increasing percentages of the SARS-CoV-2 VOCs Alpha (B.1.1.7, UK) and Beta (B.1.351, South Africa) were observed in our laboratory. Therefore, 23 samples with Alpha and one sample with Beta were tested with both AlinSARS and Allplex.
2.6. Testing throat washes with AlinSARS
Spiked throat wash specimens were prepared by spiking 10 μL of leftover media from SARS-CoV-2 positive swab samples into 1.1 mL of either different residual negative patient throat wash specimens (n = 10) or 0.9 % NaCl solution as control (n = 1), respectively. Samples with either high positive (about 6 log cps/mL) or low positive (around the detection limit) target concentrations, respectively, were tested with AlinSARS.
3. Results
3.1. SARS-CoV-2 RNA detection rates and precision of RT-SARS and AlinSARS
The detection rates of RT-SARS were assessed by testing multiple replicates of the RT-SARS dilution panel with target concentrations between 1000 and 2.5 cps/mL. RT-SARS exhibited 100 % detection rate at a concentration of 50 cps/mL (Table 1 ). Probit analysis provided a limit of detection (LOD) of 95 % at 37.73 cps/mL (95 % CI 15.61–91.19). The detection rates of AlinSARS were evaluated by testing multiple replicates of the AlinSARS dilution panel with target concentrations ranging from 400 to 5 cps/mL. AlinSARS showed 100 % detection rate at 50 cps/mL (Table 2 ). Probit analysis revealed an LOD of 95 % at 23.44 cps/mL (95 % CI 10.81–50.86). High precision was observed at concentrations around the LOD with CVs ≤2.8 % with RT-SARS and ≤1.9 % with AlinSARS. The mean Ct values of RT-SARS were lower than those of AlinSARS due to the first 10 cycles being unread.
Table 1.
Detection rates of RT-SARS at low SARS-CoV-2 RNA concentrations.
| Target concentration (cps/mL) | Replicates tested (n) | Replicates detected (n) | Percentage of replicates detected | Ct values |
|
|---|---|---|---|---|---|
| (mean ± SD) | (CV) | ||||
| 1000 | 10 | 10 | 100 % | 22.74 ± 0.40 | 1.8 % |
| 500 | 10 | 10 | 100 % | 23.71 ± 0.36 | 1.5 % |
| 250 | 10 | 10 | 100 % | 24.93 ± 0.24 | 1.0 % |
| 100 | 20 | 20 | 100 % | 26.22 ± 0.72 | 2.8 % |
| 50 | 10 | 10 | 100 % | 27.37 ± 0.74 | 2.7 % |
| 10 | 10 | 5 | 50 % | 29.11 ± 0.50 | 1.7 % |
| 5 | 10 | 3 | 30 % | 29.62 ± 0.28 | 0.9 % |
| 2.5 | 10 | 2 | 20% | 29.13 ± 0.79 | 2.7 % |
cps = copies; n = number of samples; Ct = cycle threshold; SD = standard deviation; CV = coefficient of variation.
Table 2.
Detection rates of AlinSARS at low SARS-CoV-2 RNA concentrations.
| Target concentration (cps/mL) | Replicates tested (n) | Replicates detected (n) | Percentage of replicates detected | Ct values |
|
|---|---|---|---|---|---|
| (mean ± SD) | (CV) | ||||
| 400 | 10 | 10 | 100 % | 37.32 ± 0.46 | 1.2 % |
| 300 | 10 | 10 | 100 % | 37.48 ± 0.52 | 1.4 % |
| 200 | 10 | 10 | 100 % | 38.24 ± 0.39 | 1.0 % |
| 100 | 19 | 19 | 100 % | 39.44 ± 0.75 | 1.9 % |
| 50 | 10 | 10 | 100 % | 40.54 ± 0.31 | 0.8 % |
| 25 | 10 | 9 | 90 % | 40.69 ± 0.28 | 0.7 % |
| 10 | 9 | 8 | 89 % | 40.74 ± 0.25 | 0.6 % |
| 5 | 9 | 3 | 33 % | 40.79 ± 0.35 | 0.9 % |
cps = copies; n = number of samples; Ct = cycle threshold; SD = standard deviation; CV = coefficient of variation.
3.2. Cross-reactivity with other respiratory pathogens
Potential interference of other respiratory pathogens with RT-SARS and AlinSARS was evaluated by retesting 20 patient samples with RT-SARS and another 20 samples with AlinSARS. All samples were reported negative by RT-SARS and AlinSARS (Table S1 in the supplementary material), confirming the high specificity of both assays.
3.3. Clinical performance of RT-SARS, AlinSARS, and Alin4Plex
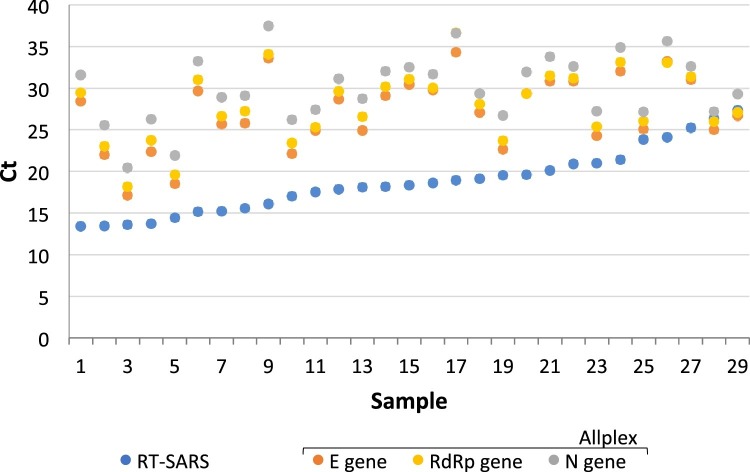
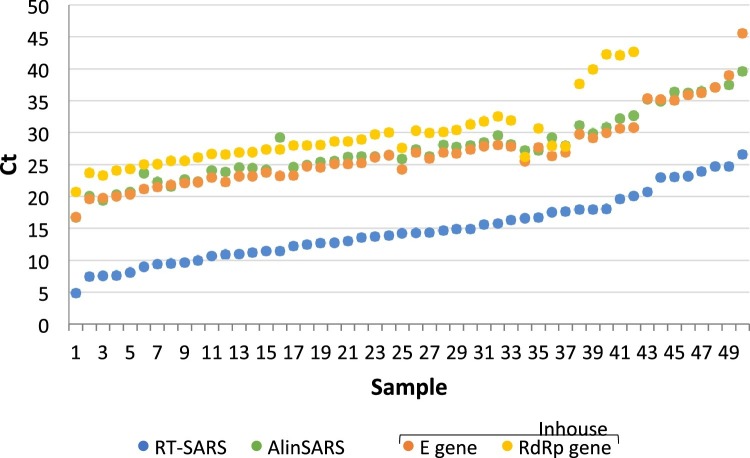
Of 197 residual swab specimens, 29 SARS-CoV-2 positive and 29 SARS-CoV-2 negative specimens by Allplex were retested with RT-SARS resulting in 100 % positive (PPA; 95 % CI 88–100) and 100 % negative percent agreement (NPA; 95 % CI 88–100). Individual Ct values for the 29 positive specimens obtained by RT-SARS and Allplex are shown in Fig. 1 . The lower Ct values observed for RT-SARS were related to the first 10 unread cycles on RT-SARS. Another subset of 50 SARS-CoV-2 positive and 50 SARS-CoV-2 negative specimens by Inhouse was retested with both RT-SARS and AlinSARS. Fig. 2 shows the individual Ct values for the 50 positive specimens as reported by RT-SARS, AlinSARS, and Inhouse, respectively. The percent agreement between the candidate (RT-SARS and AlinSARS) and reference methods (Inhouse) were calculated as follows: 100 % NPA (95 % CI 93–100) in each combination, 100 % PPA (95 % CI 93–100) between Inhouse E gene target, RT-SARS, and AlinSARS. In contrast, 8 samples were negative in the Inhouse RdRp gene (84 % PPA; 95 % CI 71–93), suggesting a lower sensitivity of this target region. As all positive Inhouse results had been confirmed with Allplex before inclusion, the PPA among Allplex, RT-SARS and AlinSARS was also 100 % (95 % CI 93–100) in each combination.
Fig. 1.
Comparison of RT-SARS and Allplex cycle thresholds in positive SARS-CoV-2 specimens.
Twenty-nine positive samples by Allplex were retested with RT-SARS showing concordantly positive results (100 % PPA). The mean Ct difference (mean of Allplex target regions minus RT-SARS) was 9.6 Ct which is in the range of the 10 unread cycles of RT-SARS. The positive samples in the figure are presented in ascending order of Ct values by RT-SARS. Additionally, 29 SARS-CoV-2 negative specimens by Allplex retested with RT-SARS were found negative (100 % NPA).
Fig. 2.
Comparison of RT-SARS, AlinSARS and Inhouse cycle thresholds in positive SARS-CoV-2 specimens.
Fifty samples positive for SARS-CoV-2 by Inhouse were retested with RT-SARS and AlinSARS showing concordantly positive results with the Inhouse E gene target (100 % PPA). Eight samples were negative in the RdRp gene target of Inhouse (84 % PPA). However, samples with one or two positive targets by Inhouse had been considered positive after confirmation by Allplex. The observed mean Ct differences between the assays were 1.2 Ct (mean of Inhouse target regions minus AlinSARS), 13.9 Ct (mean of Inhouse target regions minus RT-SARS), and 12.7 Ct (AlinSARS minus RT-SARS), the latter two mean Ct differences reflecting the 10 unread cycles of RT-SARS. The positive samples in the figure are presented in ascending order of Ct values by RT-SARS. In addition, 50 SARS-CoV-2 negative specimens by Inhouse retested with RT-SARS and AlinSARS were found negative, respectively (100 % NPA).
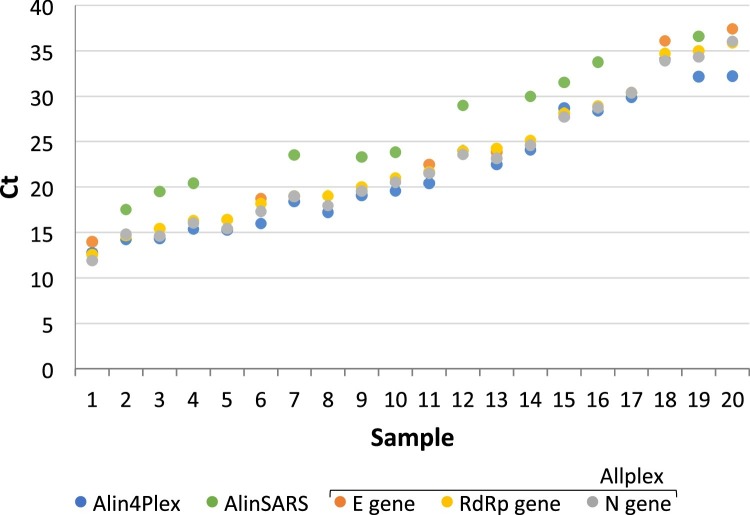
A third set of 39 specimens positive for either SARS-CoV-2 (n = 20), FluA (n = 11), FluB (n = 5), or RSV (n = 3) was correctly identified as positive or negative for SARS-CoV-2 upon retesting with Alin4Plex and AlinSARS, respectively. This resulted in 100 % PPA (95 % CI 83–100) and 100 % NPA (95 % CI 82–100; confidence intervals of small sample sizes (n≤20) might be slightly overestimated). The individual Ct values obtained by Allplex, Alin4Plex, and AlinSARS are shown in Fig. 3 . Similarly, high concordance was observed when retesting SARS-CoV-2 negative samples containing FluA, FluB or RSV with Alin4Plex. The assay accurately identified 11/11 positive FluA (Ct values 23.9–36.6), 4/5 positive FluB (Ct values 26.4–30.5), and 3/3 positive RSV specimens (Ct values 22.5–33.1). Allplex identified FluB with a high Ct value (40.9) in one specimen which was negative by Alin4Plex, while Alin4Plex identified one additional FluA co-infection (Ct 38.2) in a positive FluB specimen that was not detected by Allplex (data not shown).
Fig. 3.
Comparison of cycle thresholds in clinical specimens positive for SARS-CoV-2 across Allplex, Alin4Plex, and AlinSARS.
Positive SARS-CoV-2 samples by Allplex were retested with Alin4Plex (n = 20) and AlinSARS (n = 11) and showed concordant positive results (100 % PPA). The mean Ct differences between the assays were 1.0 Ct (mean of Allplex target regions minus Alin4Plex), 4.6 Ct (AlinSARS minus Alin4Plex), and 4.0 Ct (AlinSARS minus mean of Allplex target regions), respectively. The positive samples in the figure are presented in ascending order of Allplex Ct values. Additionally, a set of 19 SARS-CoV-2 negative specimens by Allplex was retested by Alin4Plex confirming all negative results for SARS-CoV-2 (100 % NPA).
3.4. Detectability of SARS-CoV-2 VOCs with AlinSARS
In February/March 2021, the prevalence of VOCs in our laboratory rapidly increased from 10 % to 75 %. While variant Beta only accounted for ≤1 %, the vast majority (up to 74 %) consisted of the variant Alpha. As expected, all 24 samples (23 Alpha and one Beta specimens) with Ct values ranging from 16 to 39 by Allplex were also detected by AlinSARS, exhibiting comparable Ct values (data not shown).
3.5. Testing throat washes with AlinSARS
Testing 10 negative throat wash samples spiked with high target concentrations of SARS-CoV-2 (∼6 log cps/mL) resulted in a mean Ct value by AlinSARS of 22.0 ± 0.6 compared to a Ct value of 21.21 in the corresponding spiked saline specimen. Six out of ten spiked throat wash samples with low target concentrations around the LOD, were detected positive by AlinSARS with a mean Ct value of 38.9 ± 1.6 compared to a Ct value of 38.2 in saline.
4. Discussion
In our study we demonstrated accurate detection of SARS-CoV-2 RNA with high sensitivity and specificity by the automated RT-SARS, AlinSARS, and Alin4Plex assays in clinical samples.
At 50 cps/mL, 100 % detection rates were observed with RT-SARS and AlinSARS, exceeding the sensitivity claimed by the manufacturer. A high analytical sensitivity is essential to identify and contain outbreaks during the SARS-CoV-2 pandemic (Arnaout et al., 2021) and is fundamental in cases of low viral load due to poor swabbing technique of a potentially infectious patient (Sieker et al., 2021). The observed high sensitivity of RT-SARS and AlinSARS agrees well with previous studies evaluating their LOD and comparing the assays with multiple commercial SARS-CoV-2 assays (Sieker et al., 2021; Mostafa et al., 2020; Fung et al., 2020; Hirschhorn, 2021; Kohmer et al., 2021; Degli-Angeli, 2020; Perchetti et al., 2021). The high precision and reproducibility observed with both tests confirm previous findings (Mostafa et al., 2020; Hirschhorn, 2021; Perchetti et al., 2021; Smith et al., 2020).
RT-SARS and AlinSARS showed no cross-reactivity with common respiratory pathogens of the upper respiratory tract. Similarly, in a previous evaluation, RT-SARS did not cross-react with various respiratory viruses, including other human coronaviruses (Degli-Angeli, 2020).
When assessing the clinical performance of RT-SARS, AlinSARS, and Alin4Plex by retrospective testing of SARS-CoV-2 swab specimens previously tested with Allplex and Inhouse, we found 100 % PPA and 100 % NPA in all comparisons between RT-SARS, AlinSARS, Alin4Plex, Allplex, and the E gene of Inhouse. Only with the RdRp target region of Inhouse, a lower PPA of 84 % compared to all other assays was observed. High PPAs and NPAs were similarly described comparing RT-SARS with a CDC-based in-house assay and the ThermoFisher TaqPath RT-PCR COVID-19 EUA assay (Degli-Angeli, 2020; Hirschhorn, 2021). Furthermore, excellent agreement was obtained between RT-SARS and AlinSARS (Hirschhorn, 2021). Finally, in our study, high concordance was observed between Alin4Plex and Allplex for the detection of FluA, FluB, and RSV. The two identified discordant specimens had very late Ct values suggesting low viral loads around the LOD which likely serves as a potential explanation for the discrepancy.
Previously, mutations in the S gene encoding the spike protein have been shown to affect the performance of some diagnostic PCR assays targeting the S gene (European Centre for Disease Prevention and Control, 2021d). However, RT-SARS, AlinSARS, and Alin4Plex target two highly conserved sequences in the RdRp and N genes. Consequently, AlinSARS reliably identified 24 specimens containing the Alpha or Beta variants.
Throat washes are easy to obtain and can be self-sampled by the individual, e.g. in screening settings. In a laboratory evaluation of this off-label specimen type using AlinSARS, similar Ct values were obtained as in 0.9 % NaCl solution. For our laboratory, testing throat wash specimens on Alinity m is of major importance to rapidly resolve throat wash pools after being reported positive by our inhouse method.
High level automation and short turnaround time (TAT) of molecular analyzers are of critical importance to manage high demands for SARS-CoV-2 testing. When we evaluated the TATs of 8000 samples tested within 2 months with RT-SARS and AlinSARS, we observed a mean total TAT of 18.0 h for RT-SARS and 4.9 h for AlinSARS (data not shown). A reduction of the total TAT for RT-SARS would have required an additional shift which was not necessary with Alinity m. The total TAT comprised the time from sample receipt in the laboratory to result reporting by the instrument. Random access analyzers provide shorter TAT by eliminating the need for sorting and batching of samples, providing continuous loading capability and ready-to-use reagents (Obermeier et al., 2020). The Alin4Plex assay is a multiplex PCR assay allowing differentiation of several respiratory pathogens in a single test, thus enabling laboratories to process more tests per week and saving precious testing materials. Information on the incidence of influenza A and B, SARS-CoV-2, and RSV infections obtained from one single test could support public health authorities to control the spread of diseases and monitor program success, especially when these viruses are circulating simultaneously.
This study has several limitations. Our LOD assessments were conducted prior to release of the First WHO International Standard for SARS-CoV-2 RNA and at that time, alternative reference materials consisted of RNA transcripts only. We acknowledge that the use of the Abbott SARS-CoV-2 positive controls to determine the LOD may limit a comparison of LODs among different SARS-CoV-2 assays. Moreover, the concentrations in the two dilution panels differed since after evaluation of RT-SARS, the second panel for AlinSARS was prepared using a tighter concentration range around the expected LOD. Information on the anatomic origin of swab specimens was not available, preventing differentiation between nasopharyngeal and oropharyngeal swabs. However, this was of minor importance as the focus of our study was comparing different test methods. Parallel testing of clinical samples across all platforms was not possible due to the sequential availability of the Abbott assays and sample volume limitations. The use of diluted clinical specimens was not considered to avoid potential matrix effects. However, testing different sets of clinical samples did not impact the interpretation of the results as high concordance was observed across all assays. Although only a small number of samples positive with AlinSARS was confirmed as SARS-CoV-2 VOCs, this has been substantiated, as at present, nearly all individuals in our population tested SARS-CoV-2 positive with AlinSARS or Alin4Plex are infected with VOCs including the Delta variant (B.1.617.2, India). Finally, no seasonal coronaviruses were included in the cross-reactivity panel, and only a small number of samples positive for FluA, FluB and RSV was tested with the Alin4Plex due to their low prevalence at the time of the study. Thus, additional studies of larger sample size are warranted.
5. Conclusions
The analytical and clinical performance of the Abbott RealTime SARS-CoV-2, the Alinity m SARS-CoV-2, and the Alinity m Resp-4-Plex assays qualify these tests as valuable automated PCR methods. The ability to rapidly test for SARS-CoV-2 on the random access Alinity m analyzer, either as single plex or as multiplex SARS-CoV-2/FluA/FluB/RSV assay, will expand options to further increase testing capacities with the objective to slow down the spread of viral respiratory infections.
CRediT authorship contribution statement
Robert Ehret: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Data curation, Visualization, Writing – original draft, Writing – review & editing. Stefan Breuer: Methodology, Validation, Investigation, Formal analysis, Data curation. Jens Dhein: Conceptualization, Methodology, Resources, Formal analysis, Writing – original draft, Writing – review & editing. Birgit Reinhardt: Conceptualization, Methodology, Formal analysis, Data curation, Visualization, Writing – original draft, Writing – review & editing. Martin Obermeier: Conceptualization, Methodology, Resources, Formal analysis, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: RE and SB have no competing interests. MO has received speaker honorariums from Roche, Abbott, Hologic, Cepheid and Siemens, received travel grants from Roche, Abbott, Hologic and Vela Diagnostics, and acted as advisor for Cepheid, Siemens and Abbott. JD and BR are employees of Abbott GmbH.
Acknowledgements
Abbott RealTime SARS-CoV-2 and Alinity m SARS-CoV-2 testing kits were provided by Abbott Molecular Inc.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114338.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Abbott Molecular Inc . 2020. Package Insert Abbott RealTime SARS-CoV-2, 51-608442/R2. [Google Scholar]
- Abbott Molecular Inc . 2020. Package Insert Alinity m SARS-CoV-2 AMP Kit, 53-608193/R3. [Google Scholar]
- Abbott Molecular Inc . 2020. Package Insert Alinity m Resp-4-Plex AMP Kit, 53-608209/R1. [Google Scholar]
- Arnaout R., et al. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1382. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli-Angeli E., et al. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J. Clin. Virol. 2020;129:104474. doi: 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Diagnostic Testing and Screening for SARS-CoV-2. [Online]. Available: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing. [Accessed 14 Apr 2021] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Objectives for COVID-19 Testing in School Settings – First Update. [Online]. Available: https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-objectives-school-testing.pdf. [Accessed 14 Apr 2021] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Operational Considerations for Influenza Surveillance in the WHO European Region during COVID-19: Interim Guidance. [Online]. Available: https://www.ecdc.europa.eu/sites/default/files/documents/Joint-influenza-interim-guidance.pdf. [Accessed 14 Apr 2021] [Google Scholar]
- European Centre for Disease Prevention and Control . 2021. Risk Assessment on COVID-19. [Online]. Available: https://www.ecdc.europa.eu/en/current-risk-assessment-novel-coronavirus-situation. [Accessed 14 April 2021] [Google Scholar]
- European Centre for Disease Prevention and Control . 2021. Infection Prevention and Control and Preparedness for COVID-19 in Healthcare Settings; Sixth Update. [Online]. Available: https://www.ecdc.europa.eu/sites/default/files/documents/Infection-prevention-and-control-in-healthcare-settings-COVID-19_6th_update_9_Feb_2021.pdf. [Accessed 14 Apr 2021] [Google Scholar]
- European Centre for Disease Prevention and Control . 2021. ECDC Rapid Assessment of Laboratory Practices and Needs Related to COVID-19. Technical Report. [Online]. Available: https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-rapid-assessment-laboratory-practices-needs.pdf. [Accessed 14 Apr 2021] [Google Scholar]
- European Centre for Disease Prevention and Control, World Health Organization, Regional Office for Europe . 2021. Methods for the Detection and Identification of SARS-CoV-2 Variants. [Online]. Available: https://www.ecdc.europa.eu/sites/default/files/documents/Methods-for-the-detection-and-identification-of-SARS-CoV-2-variants-WHO-ECDC.pdf. [Accessed 15 Apr 2021] [Google Scholar]
- Fung B., et al. Direct comparison of SARS-CoV-2 analytical limits of detection across seven molecular assays. J. Clin. Microbiol. 2020;58:e01535–20. doi: 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J.W., et al. Verification and validation of SARS-CoV-2 assay performance on the Abbott m2000 and Alinity m systems. J. Clin. Microbiol. 2021;59:e03119–20. doi: 10.1128/JCM.03119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N., et al. Comparative analysis of point-of-care, high-throughput and laboratory-developed SARS-CoV-2 nucleic acid amplification tests (NATs) J. Virol. Methods. 2021;291:114102. doi: 10.1016/j.jviromet.2021.114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H.H., et al. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J. Clin. Virol. 2020;130:104578. doi: 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeier M., et al. Improved molecular laboratory productivity by consolidation of testing on the new random-access analyzer Alinity m. J Lab Med. 2020;44:319–328. [Google Scholar]
- Perchetti G.A., et al. Performance characteristics of the Abbott Alinity m SARS-CoV-2 assay. J. Clin. Virol. 2021;140:104869. doi: 10.1016/j.jcv.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegene Inc . 2020. Package Insert Allplex SARS-CoV-2 Assay. V1.14. [Google Scholar]
- Sieker J.T., et al. Analytic sensitivity of 3 nucleic acid detection assays in diagnosis of SARS-CoV-2 infection. J. Appl. Lab. Med. 2021;6:421–428. doi: 10.1093/jalm/jfaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.P., et al. Large-scale, in-house production of viral transport media to support SARS-CoV-2 PCR testing in a multihospital health care network during the COVID-19 pandemic. J. Clin. Microbiol. 2020;58:e00913–20. doi: 10.1128/JCM.00913-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIB MolBiol Syntheselabor GmbH . 2020. Instructions for Use: LightMix® SarbecoV E-gene Plus EAV Control. MDx 40-0776-96. [Google Scholar]
- Watson C., et al. 2020. A National Plan to Enable Comprehensive COVID-19 Case Finding and Contact Tracing in the US. [Online]. Available: https://www.centerforhealthsecurity.org/our-work/pubs_archive/pubs-pdfs/2020/200410-national-plan-to-contact-tracing.pdf. [Accessed 14 Apr 2021] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.