Abstract
IS6110 DNA fingerprinting was used to characterize an outbreak of multidrug-resistant tuberculosis in 21 individuals (17 males and 4 females) living in or roaming among four distantly separated areas in the Czech Republic. The restriction fragment length polymorphism (RFLP) analysis separated the collected Mycobacterium tuberculosis strains into group A, including 14 patients with six IS6110 copies, and group B, with 7 patients displaying highly similar RFLP patterns but with two additional IS6110 bands. A switch from pattern A to pattern B was observed in one patient, and the subsequent detection of subclone B in seven more individuals has been explained by the instability of DNA genotypes caused by transposition of IS6110 elements.
The Czech Republic is a territory with a low incidence of tuberculosis, where the total notification rate was 17.9 (4.7 sputum smear positive) cases per 100,000 persons in 1996 (5). The prevalence of primary drug resistance amounted to 2.3% in 1997, the total rate of acquired resistance was 12.5%, and one case of acquired resistance to isoniazid, rifampin, ethambutol, and streptomycin was recorded in the World Health Organization-International Union against Tuberculosis and other Lung Diseases project on antituberculosis drug resistance surveillance in 1997 (4). The occurrence of multidrug-resistant (MDR) tuberculosis, defined by the World Health Organization as unresponsiveness at least to isoniazid and rifampin in vitro, was unknown in this country until recently. Its coming may be associated with the appearance of at-risk population groups newly emerging after 1990 as a consequence of political, social, and ethnic changes in the community.
In view of the conspicuous occurrence of 21 MDR M. tuberculosis strains identified independently in two distantly separated laboratories between 1991 and 1997, we decided to analyze their possible epidemiological links by molecular epidemiology methods. First, three MDR cases were recorded in a penitentiary, where patients A-1, A-2, and A-3 (males aged 45 and 49 years and a female aged 38 years, respectively) were imprisoned between 1991 and 1993, as described in our previous paper (1). The chain of transmission could be traced to a total of 21 tuberculosis patients displaying the following significant characteristics. (i) Males (n = 17) were more numerous than females (n = 4). (ii) The ages of the male patients ranged from 36 to 64 (average, 45.0) years, and those of the females ranged from 38 to 52 years. (iii) A considerable number of the patients belonged to tuberculosis risk groups. Four had been discharged from a penitentiary, four were homeless, one was treated in a psychiatric ward, and one was an immigrant. (iv) The permanent domiciles of the patients were located in four distantly separated regions (the four reported homeless were identified by the local tuberculosis service). (v) All of the patients exhibited resistance to isoniazid and rifampin, all but two showed resistance to streptomycin, and all but four showed resistance to ethambutol. (vi) In six males, clinically significant Mycobacterium kansasii infection preceded superinfection with M. tuberculosis. (vii) Four patients died within 2 to 4 years of the onset of infection.
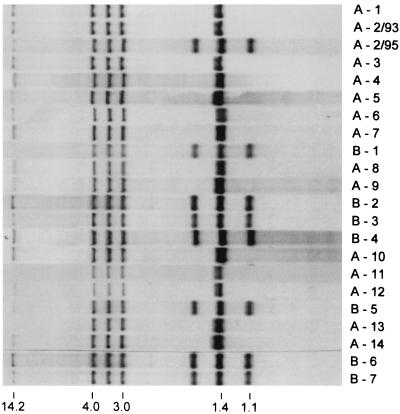
DNA fingerprinting based on detection of IS6110 in PvuII restriction fragments (2) separated the M. tuberculosis isolates into two distinct groups (Fig. 1): subclone A, including 14 isolates characterized by six bands, and subclone B, consisting of seven isolates which displayed basically the same banding pattern as subclone A but with two additional bands. Preliminary results of spoligotyping performed on strains A-1, A-4, B-3, and B-7 showed identical 13-band patterns and corroborated the hypothesis that both subclones A and B are genetically closely related (the investigation was kindly done by C. Martin, University of Zaragoza, Zaragoza, Spain).
FIG. 1.
IS6110 fingerprints of M. tuberculosis isolates of MDR tuberculosis patients.
In five patients, serial M. tuberculosis isolates originating in different time periods were investigated by the restriction fragment length polymorphism (RFLP) technique and four of them showed identical RFLP profiles. However, in patient A-2, two different RFLP patterns were recorded. The first isolate of June 1993 showed a six-band pattern of subclone A, and surprisingly, the isolate of May 1995 displayed the eight-band profile seen in subclone B (Fig. 1 and 2).
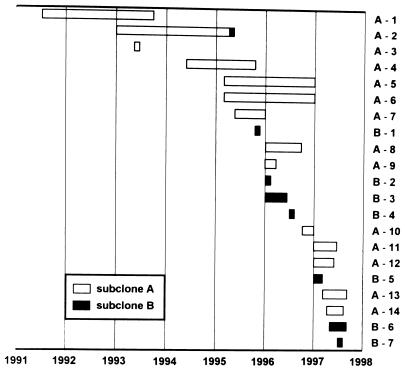
FIG. 2.
Flow chart of the duration of culture positivity in subgroups A and B of MDR tuberculosis patients.
The chronology of M. tuberculosis demonstration in the patients under study showed the first occurrence of pattern B in patient A-2 in May 1995, i.e., with an interval of 2 years from his first culture positivity, and a conspicuous accumulation of the first seven isolates of subclone A between 1991 and 1995. However, none of the isolates of subclone B emerged before November 1995 (Fig. 2). It appears possible that subclone B, first recorded in patient A-2 in May 1995, was then transmitted from person to person among patients bearing the B subclone. Unfortunately, the exact mode of transmission among these patients and their mutual contacts could not be ascertained due to their unwillingness to produce reliable information. Both the appearance of subclone B in our patients and the switch from RFLP pattern A to pattern B in patient A-2 can be explained by the instability of DNA genotypes manifesting itself in transposition of IS6110 elements (3, 6).
Acknowledgments
This work was supported by European Programme for Science, Research and Development grant ERBIC20CT970016 and by the Internal Grant Agency of the Ministry of Health, Czech Republic, grant IGA MZ 3984-2.
We thank Carlos Martin, Department of Microbiology, University of Zaragoza, Zaragoza, Spain, for spoligotyping of selected M. tuberculosis strains.
REFERENCES
- 1.Kremer K, van Soolingen D, Půtová I, Kubín M. Use of IS6110 DNA fingerprinting in tracing man-to-man transmission of Mycobacterium tuberculosis in the Czech Republic. Cent Eur J Public Health. 1996;4:3–6. [PubMed] [Google Scholar]
- 2.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. World Health Organization global tuberculosis programme: antituberculosis drug resistance in the world. WHO/TB/97.229. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 5.World Health Organization. World Health Organization global tuberculosis control. WHO report 1997. WHO/CDS/CPC/TB/99.259. Geneva, Switzerland: World Health Organization; 1999. [Google Scholar]
- 6.Yeh W B, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]