Abstract
Polyadenylation of mRNA precursors is a two-step reaction requiring multiple protein factors. Cleavage stimulation factor (CstF) is a heterotrimer necessary for the first step, endonucleolytic cleavage, and it plays an important role in determining the efficiency of polyadenylation. Although a considerable amount is known about the RNA binding properties of CstF, the protein-protein interactions required for its assembly and function are poorly understood. We therefore first identified regions of the CstF subunits, CstF-77, CstF-64, and CstF-50, required for interaction with each other. Unexpectedly, small regions of two of the subunits participate in multiple interactions. In CstF-77, a proline-rich domain is necessary not only for binding both other subunits but also for self-association, an interaction consistent with genetic studies in Drosophila. In CstF-64, a small region, highly conserved in metazoa, is responsible for interactions with two proteins, CstF-77 and symplekin, a nuclear protein of previously unknown function. Intriguingly, symplekin has significant similarity to a yeast protein, PTA1, that is a component of the yeast polyadenylation machinery. We show that multiple factors, including CstF, cleavage-polyadenylation specificity factor, and symplekin, can be isolated from cells as part of a large complex. These and other data suggest that symplekin may function in assembly of the polyadenylation machinery.
Most steps in gene expression in eukaryotes are catalyzed by massive molecular machines, and polyadenylation of mRNA precursors in the nucleus is no exception. For mammalian cells, numerous protein factors have been identified that must interact with each other and with the pre-mRNA to catalyze the two-step cleavage-poly(A) synthesis reaction (reviewed in references 3, 47, and 53). Two multisubunit complexes, designated cleavage-polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF), cooperate with each other to define the site of polyadenylation (24) by recognizing, respectively, the highly conserved AAUAAA hexanucleotide (10, 25) and a more divergent GU-rich sequence situated downstream of the actual cleavage site (2, 17, 40). Two additional factors, cleavage factors I and II (CFI and CFII), are also essential for the cleavage reaction (44). CFI has been characterized and appears to consist of two subunits that also function in RNA binding and not catalysis (33). Poly(A) polymerase (PAP), a single-subunit enzyme (32, 48), is also required for cleavage of most but not all pre-mRNAs (43). RNA polymerase II, and specifically the carboxy-terminal domain (CTD) of its largest subunit, has also recently been found to be required for the cleavage reaction (8). Despite this large number of factors, the identity of the endonuclease that actually cleaves the pre-mRNA remains unknown. For the second phase of the reaction, poly(A) synthesis, CPSF and PAP, together with a third protein, poly(A) binding protein II (49), are sufficient, although CstF was recently shown to enhance this reaction with a pre-mRNA containing a CstF binding site situated upstream of the AAUAAA signal (23).
CstF is a heterotrimer consisting of subunits of 77, 64, and 50 kDa (42). Mammalian CstF-64 contains an N-terminal ribonucleoprotein-type RNA binding domain (RBD), a long Pro- and Gly-rich region, and a pentapeptide repeat region capable of forming an extended α-helix (reviewed in references 37 and 45). The RBD is responsible for binding the GU-rich element in the polyadenylation signal (40), while the functions of the remainder of the protein are unknown. CstF-64 is essential for cell viability, and changes in the intracellular levels of the protein can affect cell growth and gene expression in B cells (41, 45). CstF-77 holds the complex together (39) and also interacts strongly with a CPSF subunit, CPSF-160 (25). CstF-77 is homologous to the Drosophila protein Suppressor-of-forked [Su(f)] (22, 39). Su(f) is essential for viability, and nonlethal mutations can affect gene expression (27). The protein contains multiple repeats similar to the tetratricopeptide repeat (TPR) motif (31) and a Pro-rich C terminus, suggestive of multiple possible protein-protein interactions. CstF-50 also contains repeats of a potential protein-protein interaction motif, the transducin or WD-40 motif (38). CstF-50 is necessary for CstF activity in vitro (39) and has also been suggested to interact directly with the RNA polymerase II CTD, thus perhaps playing an important role in linking transcription and 3′ processing (20).
The RNA sequences and protein factors that function in polyadenylation appear to be well conserved throughout the metazoa. However, the situation in yeast reveals significant differences as well as similarities. For example, the signal sequences in yeast are degenerate and poorly defined and bear no significant similarity to the corresponding sequences in higher eukaryotes (6). Nonetheless, some of the key proteins appear well conserved (11, 19). For example, all four subunits of human CPSF have yeast counterparts. CstF-77/Su(f) also has a yeast homologue, RNA14, which is associated with RNA 15, a protein that bears strong similarity to CstF-64 in the RBD, although the yeast protein is truncated shortly after this domain. No apparent yeast CstF-50 homologue has been described. Unlike in mammalian cells, it is unclear how these factors interact with RNA, and it has also been difficult to assign the yeast factors, especially the CPSF homologues, to a specific activity (30, 52). This could reflect the existence of a larger complex that fractionates differently dependent on the precise biochemical conditions, and evidence consistent with this is beginning to emerge (30, 54). Furthermore, a combination of biochemistry and genetics has led to the identification of several yeast proteins that appear essential for polyadenylation but which have no known mammalian homologue (1, 13, 29, 30).
In this study, we have investigated a number of the protein-protein interactions involved in the function of CstF, which in turn has provided novel insights into the makeup of the polyadenylation machinery. We first define the regions of each CstF subunit required for interaction with the other subunits. Most notably, a single small region of CstF-64 is shown to be responsible both for interaction with CstF-77 and for a strong and specific association with a previously uncharacterized nuclear protein, symplekin. The sequence of symplekin suggests that it is related to one of the yeast proteins previously implicated in polyadenylation but until now lacking a mammalian counterpart. Finally, we provide the first demonstration that multiple polyadenylation factors, including CstF, CPSF, and symplekin, coexist in a high-molecular-weight complex. On the one hand, these results strengthen the similarities between yeast and mammalian polyadenylation, while on the other, they indicate added complexities in the reaction.
MATERIALS AND METHODS
Synthesis of wild-type and mutant CstF subunit proteins.
CstF subunit cDNAs (37–39) harboring an optimal translation initiation site sequence were cloned in the pGEM-3 vector as described previously (39). To generate C-terminal deletion mutants, plasmid DNAs were digested at appropriate restriction sites. To generate N-terminal and internal deletion mutants, appropriate cDNA fragments were cloned into the pGEM-3 vector as above. Linearized plasmid DNAs were transcribed in vitro, and mRNAs were translated in vitro with reticulocyte lysate (Promega) in the presence of [35S]methionine in 12.5-μl reaction mixtures as described previously (39). The sizes of in vitro-translated proteins were confirmed on sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Far-Western blot analyses.
Human CstF (2 μg) purified by Mono S column chromatography (purity, ∼90%) (42) was loaded into a 1.5-cm-wide well of an SDS–10% polyacrylamide gel. After transfer to nitrocellulose, the membrane was cut into ∼2-mm-wide strips, and one of them was stained with India ink to visualize the CstF subunits. The strips were placed in a multigroove tray (Reservoir Liner; Costar), and the proteins on the strips were denatured, renatured, and probed with 35S-labeled proteins as described previously (15). In some cases, a HeLa cell nuclear protein fraction obtained by (NH4)2SO4 precipitation (20 to 40% saturation) (43) or a CFI- and CFII-containing fraction obtained by Mono Q chromatography (44) was used in place of purified CstF. For far-Western blot analysis using 32P-labeled glutathione S-transferase (GST) fusion proteins, BanI-HindIII and BanI-EcoNI fragments derived from pZ64-18 (37), which encode amino acid residues 108 to 248 (GST1) and 108 to 214 (GST2) of CstF-64, were cloned into the pGEX-2TK vector and GST and GST fusion proteins expressed in Escherichia coli were labeled with [γ-32P]ATP by using the catalytic subunit of protein kinase A as described previously (9). The 32P-labeled proteins (5 × 105 cpm) were used for far-Western blot analysis of the (NH4)2SO4 fraction (20 to 40% saturation) as described above.
Cloning of symplekin cDNAs.
A HeLa cell cDNA expression library in the λEXlox vector (a gift from J. Wu) was screened with a 32P-labeled GST-CstF fusion protein, GST1, as described previously (9) but without denaturation of proteins with guanidine HCl. To obtain cDNAs encoding the entire protein coding regions of symplekin-I and -II, 106 phage derived from the same cDNA library were screened by hybridization with a cDNA fragment and the longest cDNA clones obtained were sequenced in their entirety.
Immunoprecipitations.
To study interactions between symplekin and CstF-64, cDNAs encoding symplekins were cloned in the pGEM-3 vector and the plasmid DNAs were digested with SalI and transcribed as described above. mRNAs were mixed so that approximately equal amounts of proteins were synthesized, and they were translated in vitro as described above. After small aliquots were removed, the rest of the translation mixtures were immunoprecipitated with an anti-CstF-64 monoclonal antibody (MAb) as described previously (39). To study the effects of symplekins on the association between CstF-77 and CstF-64, a mixture of CstF-77 and CstF-64 mRNAs was translated in vitro in the presence of recombinant symplekin-I or -II and then subjected to immunoprecipitation.
Production of symplekin proteins in insect cells.
Symplekin cDNAs were cloned into the pDS56-6His vector (37), and then DNA fragments encoding His-tagged symplekin-I or -II were transferred to the pEV55 vector (21) to generate recombinant baculovirus as described previously (39). To purify recombinant symplekin, Sf9 cells infected with recombinant viruses were harvested and lysed in lysis buffer (50 mM Tris-HCl [pH 7.9], 10% glycerol, 300 mM NaCl, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 1% Nonidet P-40 [NP-40]) for 30 min on ice. After the cell lysates were centrifuged in a microcentrifuge for 3 min in the cold room, the supernatant was loaded onto an Ni-nitrilotriacetic acid agarose column (Qiagen) equilibrated with the same buffer. The column was extensively washed with lysis buffer and washing buffer (the same as lysis buffer but without NP-40). His-tagged proteins were eluted with washing buffer containing 200 mM imidazole-HCl and dialyzed twice against buffer D [20 mM HEPES-NaOH (pH 7.9), 50 mM (NH4)2SO4, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 20% (vol/vol) glycerol].
Immunopurification of a polyadenylation complex.
HeLa cell nuclear proteins obtained by (NH4)2SO4 precipitation (20 to 40% saturation) were first fractionated by Superose 6 column chromatography as described previously (44). For immunopurification under mild conditions (see Fig. 7A), after dialysis against buffer D containing 10% glycerol, 3 ml of the cleavage-specificity factor (CSF)-containing fraction (43) was passed through 100 μl of anti-CstF-64–protein G-Sepharose (PGS) conjugate, packed in a 1-ml pipette tip, three times over 1 h. For immunopurification under stringent conditions, the CSF-containing fraction was loaded on anti-CstF-64–PGS or anti-polyomavirus large T antigen–PGS conjugate after first adjusting the concentrations of (NH4)2SO4 and NP-40 to 150 mM and 0.05%, respectively. After extensive washing with the same buffer, the buffer was removed by centrifugation in clinical tabletop centrifuge. Proteins were eluted by heating at >90°C for 10 min in 200 μl of protein gel-loading buffer, and 20-μl aliquots were loaded onto an SDS-polyacrylamide gel. The proteins were stained with silver or probed with anti-CstF-64 monoclonal antibody (42), anti-CPSF-160 polyclonal antibody (25), anti-CPSF-100 polyclonal antibody, or anti-symplekin polyclonal antibody. To prepare the anti-symplekin antibody, a cDNA fragment encoding the first 506 residues of symplekin was cloned into the pET-3a vector and the expressed protein was purified on an SDS–10% polyacrylamide gel and used to immunize a rabbit (7). Anti-symplekin antibodies were purified using a His-tagged symplekin protein fragment conjugated to cyanogen bromide-activated Sepharose as a ligand.
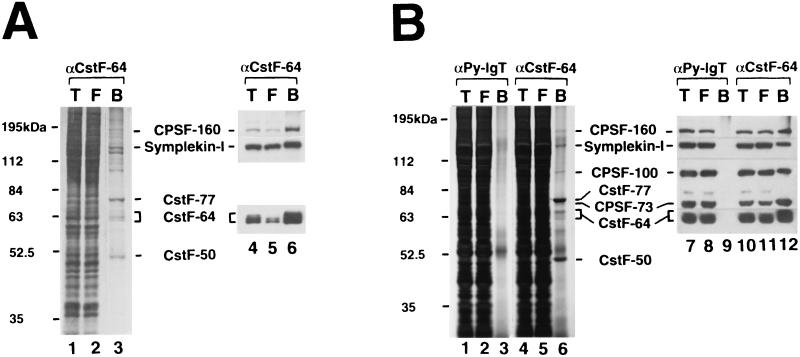
FIG. 7.
Symplekin-I exists in a complex containing both CstF and CPSF. (A) Immunoaffinity purification of a polyadenylation complex under mild conditions. The CSF-containing fraction obtained by Superose 6 column chromatography was subjected to immunoaffinity purification using an anti-CstF-64 MAb in the presence of 50 mM (NH4)2SO4 but no NP-40. Total starting material (T) (lanes 1 and 4) and flowthrough (F) (lanes 2 and 5) and bound (B) (lanes 3 and 6) fractions were loaded on an SDS–10% polyacrylamide gel and stained with silver (lanes 1 to 3) or probed with anti-CstF-64, anti-CPSF-160, or anti-symplekin antibody (lanes 4 to 6). (B) Immunoaffinity purification of a polyadenylation complex under stringent conditions. The CSF-containing fraction was subjected to immunoaffinity purification using an anti-polyomavirus large T (αPy-lgT; lanes 1 to 3 and 7 to 9) or anti-CstF-64 (αCstF-64; lanes 4 to 6 and 10 to 12) MAb in the presence of 150 mM (NH4)2SO4 and 0.05% NP-40. Total starting material (T), (lanes 1, 4, 7, and 10) and flowthrough (F) (lanes 2, 5, 8, and 11) and bound (B) (lanes 3, 6, 9, and 12) fractions were loaded on an SDS–7.5% polyacrylamide gel and stained with silver (lanes 1 to 6) or probed with anti-CstF-64, anti-CPSF-160, anti-CPSF-100/73, or anti-symplekin antibody (lanes 7 to 12).
RESULTS
CstF intersubunit interactions.
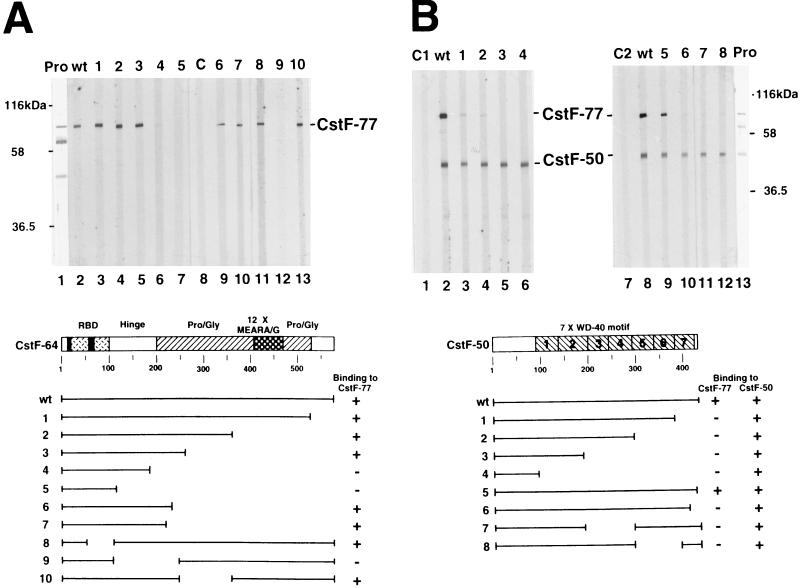
As described in the introduction, CstF plays a key role in determining the efficiency of polyadenylation, and it can be an important target of regulation; each subunit contains repeated motifs that are probably involved in protein-protein interactions. To gain more insights into how CstF functions, we first set out to define the regions of each subunit necessary for interactions with the other subunits. An initial set of experiments was performed by employing the far-Western blotting assay. For this analysis, highly purified CstF was resolved by preparative SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose and the subunits were subjected to a renaturation protocol (see Materials and Methods). The filter was cut vertically into strips, which were then probed with wild-type or mutant derivatives of individual CstF subunits, produced by in vitro translation. Figure 1 shows the results obtained when wild-type and mutant derivatives of CstF-64 (Fig. 1A) and CstF-50 (Fig. 1B) were used in this assay. Wild-type CstF-64 bound strongly to CstF-77, as expected (39), but not to CstF-50 or to itself. C-terminal truncations that removed the repeat structure and the Pro/Gly-rich region (deletions 1 to 3, 6, and 7) were without significant effect on binding. However, a deletion (deletion 4) that impinged on the hinge domain (named simply because it lies between the RBD and Pro- and Gly-rich region) greatly reduced binding, and interaction was eliminated by a further truncation that removed this region (deletion 5). Results obtained with three internal deletions (deletions 8 to 10) were consistent with this and support the idea that residues within the hinge domain, and nowhere else in the protein, are necessary for interaction.
FIG. 1.
Structural domains of CstF-64 and CstF-50 required for association with CstF-77. (A) The hinge domain of CstF-64 is necessary for association with CstF-77. Purified CstF was resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and either stained with India ink (Pro, lane 1) or probed with 35S-labeled wild-type (wt, lane 2) or mutant (deletions 1 to 10, lanes 3 to 7 and 9 to 13) CstF-64. 35S-labeled luciferase (C, lane 8) was used as a negative control. Positions of protein size markers and CstF-77 are indicated on the left and right, respectively. Diagrams of wild-type and mutant CstF-64 proteins are shown at the bottom. (B) The WD-40 motif of CstF-50 is required for association with CstF-77. Purified CstF was transferred onto a nitrocellulose membrane and stained with India ink (Pro, lane 13) or probed with 35S-labeled wild-type (wt, lanes 2 and 8) or mutant (deletions 1 to 8, lanes 3 to 6 and 9 to 12) CstF-50. 35S-labeled brome mosaic virus proteins (C1, lane 1) and luciferase (C2, lane 7) were used as negative controls. Diagrams of wild-type and mutant CstF-50 proteins are shown at the bottom.
The results obtained when CstF-50 and mutant derivatives were used in similar assays were quite different. Wild-type CstF-50 interacted not only as expected with CstF-77 (39) but also with itself. C-terminal truncations removing the final (deletion 1) or last three (deletion 2) WD-40 repeats greatly reduced binding to CstF-77, and two further truncations (deletions 3 and 4) eliminated interaction. Analysis of smaller C-terminal truncations (deletions 5 and 6) indicated that deletion of 6 residues from the C terminus did not affect binding but removal of another 15 residues, which disrupted the final WD-40 repeat, almost eliminated interaction. Two internal deletions, disrupting repeats 3 and 4 (deletion 7) or 5 and 6 (deletion 8), also drastically reduced binding. These findings indicate that sequences within the WD-40 repeats are responsible for interaction with CstF-77. As discussed below, it is possible that any deletion within the repeats disrupts the overall structure of the WD-40 domain, which prevents binding to CstF-77. In contrast, none of the deletions affected self-association, indicating that this interaction must be mediated by the CstF-50 N terminus.
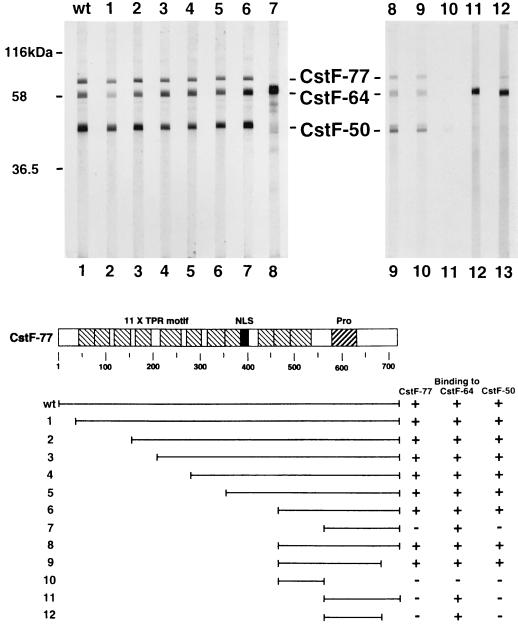
We next examined interactions of CstF-77 with other CstF subunits, and the results of far-Western blotting experiments with purified CstF and in vitro-translated CstF-77 are shown in Fig. 2. As expected, wild-type CstF-77 interacted strongly with both CstF-50 and CstF-64. In addition, as observed with CstF-50, strong self-association was also detected. As discussed below, such an interaction was not predicted from previous biochemical studies but is consistent with genetic interactions observed with Drosophila su(f) (34).
FIG. 2.
The C terminus of CstF-77 is necessary for its association with other CstF subunits. Purified CstF was transferred onto a nitrocellulose membrane and probed with 35S-labeled wild-type (wt, lane 1) or mutant (deletions 1 to 12, lanes 2 to 13) CstF-77 as in Fig. 1. Diagrams of wild-type and mutant CstF-77 proteins are shown at the bottom.
A series of N-terminal deletions of CstF-77 were first analyzed to examine the requirements for the TPR-like, or half a TPR (HAT) (31), motifs (Fig. 2, deletions 1 to 6). Strikingly, deletion of nearly two-thirds of the protein was without detectable effect on any of the three interactions observed with wild-type CstF-77. Deletion of another approximately 100 residues, which removed essentially all sequences N-terminal to the Pro-rich region (deletion 7), had an unusual effect: interaction with CstF-50 and CstF-77 were both greatly reduced or eliminated, but binding to CstF-64 was significantly increased. It is unclear whether this reflects a technical limitation of the assay (i.e., the amount of CstF-77 was limiting) or whether the mutant protein adapted a conformation that enhanced interaction with CstF-64. To define further the region of CstF-77 required for interaction, several C-terminal truncations were constructed in the context of the large but active N-terminal deletions. Removal of about 50 residues did not affect binding (compare deletions 8 and 9 and deletions 11 and 12). However, deletion of the Pro-rich domain, leaving only the small region required for interaction with CstF-50 and CstF-77 (deletion 10), essentially eliminated binding, although reduced interaction with CstF-50 could still be detected. Together, these results indicate that sequences within a ∼100-residue region, consisting primarily of the Pro-rich domain, are sufficient for binding CstF-64. This region is also required for binding to the other two CstF subunits, but sequences just N-terminal to the Pro-rich domain are also necessary for these interactions. The TPR-like repeats are thus available for interactions with other proteins.
CstF-64 interacts strongly with an additional nuclear protein.
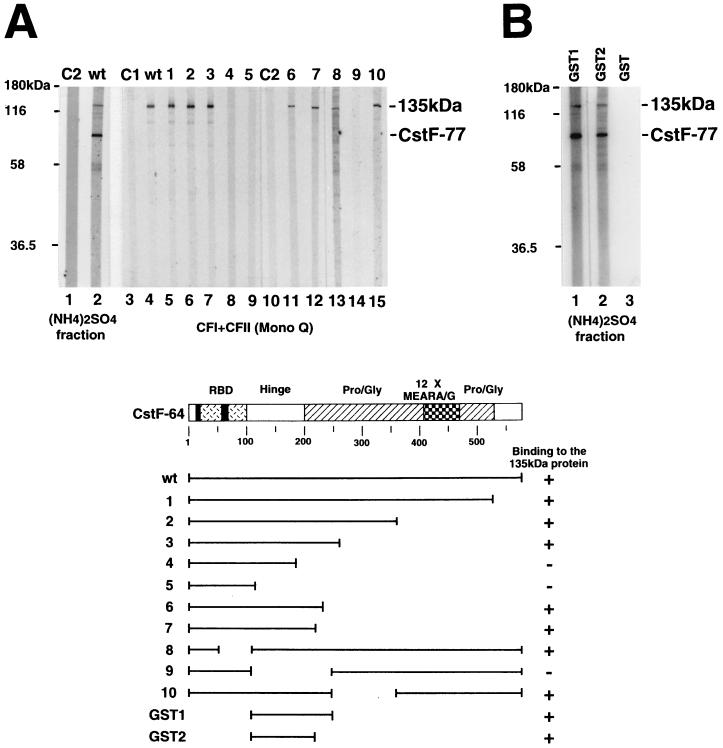
The above experiments provided insights into how the CstF subunits interact with each other but did not address possible interactions with other proteins. For example, we wished to determine whether CstF-64 interacts with other factors and, if so, whether the Pro- and Gly-rich and/or repeat regions might be involved. To this end, we first performed far-Western blotting with in vitro-translated CstF-64 and a crude nuclear fraction of HeLa cells resolved by SDS-PAGE. The results, shown in Fig. 3A, lanes 1 and 2, reveal that CstF-64 interacted detectably in this assay with only two proteins. The strongest of these interactions was with a protein of ∼77 kDa, which almost certainly corresponds to CstF-77, while the second interaction was with a larger protein of ∼135 kDa. The size of this protein indicated that it was not PAP or a subunit of CPSF. However, the estimated native sizes of CFI and CFII (∼130 and 110 kDa, respectively) (44) were consistent with the possibility that the 135-kDa protein corresponded to one of these factors, and we therefore performed far-Western blotting with a fraction enriched in these two activities (Mono Q low salt) (44). Although work performed after this experiment indicates that the 135-kDa protein does not correspond to CFI (33) or CFII (see Discussion), the 135-kDa protein was readily detected in this fraction with wild-type CstF-64 (Fig. 3A, lane 4). In keeping with the known chromatographic behavior of CstF (42, 44), the 77-kDa protein was not detected. We also examined the same set of CstF-64 mutants analyzed above for their ability to interact with the 135-kDa protein. Strikingly, the mutants behaved identically, indicating that the only region of CstF-64 necessary for interaction was again the hinge domain (lanes 5 to 9 and 11 to 15).
FIG. 3.
The hinge domain of CstF-64 is necessary and sufficient for its binding to a 135-kDa protein. (A) The hinge domain of CstF-64 is necessary for binding to a 135-kDa nuclear protein. The (NH4)2SO4 fraction (20 to 40% saturation) derived from HeLa cell nuclear extract (lanes 1 and 2) and a CFI- and CFII-containing fraction obtained by Mono Q chromatography (lanes 3 to 15) were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with 35S-labeled wild-type (wt, lanes 2 and 4) or mutant (deletions 1 to 10, lanes 5 to 9 and 11 to 15) CstF-64. 35S-labeled brome mosaic virus proteins (C1, lane 3) and luciferase (C2, lanes 1 and 10) were used as negative controls. Diagrams of wild-type and mutant CstF-64 proteins are shown at the bottom. (B) The hinge domain of CstF-64 is sufficient for binding the 135-kDa protein. Proteins in the (NH4)2SO4 fraction used in panel A were transferred onto a nitrocellulose membrane and probed with 32P-labeled GST (lane 3) or GST fusion (lanes 1 and 2) proteins.
We next wished to verify that the hinge domain was in fact sufficient for interaction. To do this, we used two purified GST fusion proteins, one containing CstF-64 residues 108 to 248 and the other containing residues 108 to 214. The two proteins, plus GST alone, were 32P labeled with protein kinase A and used in far-Western blots, in this case with the crude nuclear fraction used above (Fig. 3B). Strikingly, both fusion proteins detected two major proteins, again corresponding to CstF-77 and the 135-kDa protein. A weaker band, corresponding to a protein of ∼110 kDa, was also detected, which may reflect an isoform of the 135-kDa protein (see below). Together, these results indicate that the CstF-64 hinge domain contains sequences necessary and sufficient for strong and specific interactions with both CstF-77 and an unknown nuclear protein.
cDNA cloning of CstF-64 binding proteins.
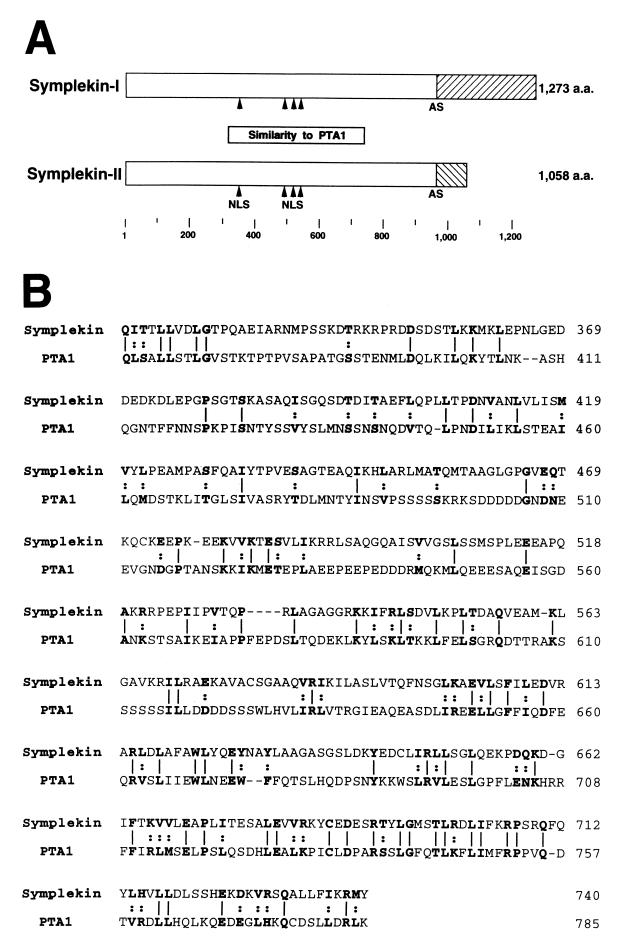
We next set out to identify the 135-kDa CstF-64 binding protein. Given its strong and specific interaction with the GST-hinge domain fusion proteins in the far-Western assays, we decided to screen a HeLa cDNA expression library with the 32P-labeled GST1 fusion protein (see Materials and Methods). Seven positive plaques were identified and purified, and cDNA inserts were isolated and either partly or entirely sequenced. The majority (5) of these encoded CstF-77, providing evidence that the screen was indeed identifying authentic CstF-64-interacting proteins. The other two encoded either of two closely related proteins, which differ only at their C termini and which probably arose from alternative splicing (Fig. 4A). The sequences of the cDNAs were determined and found to encode proteins of 1,273 and 1,058 amino acids, respectively, of which the first 964 residues are identical. The presence of nonsense codons upstream of the putative initiating AUG strongly suggested that the open reading frames are complete.
FIG. 4.
Structure of symplekin and similarity to yeast PTA1. (A) Symplekin-I and symplekin-II have a common amino acid sequence. They are identical up to amino acid residue 964 but diverge thereafter, apparently due to alternative splicing (AS). Putative nuclear localization signals (NLS) are indicated by arrowheads. The region homologous to yeast PTA1 (box) and the sizes of the proteins are also shown. (B) Symplekin has extensive similarity with yeast PTA1. Amino acid (a.a.) sequences of symplekin (top) and PTA1 (bottom) are optimally aligned according to the FASTA program, and identical (lines) and similar (dots) residues are shown in bold type. Similarities are defined as I = L = V = M, Y = F = W, K = R, D = E, S = T, and Q = N. Amino acid residues are numbered on the right.
To determine if the cDNAs encode proteins similar or identical to known proteins, we performed FASTA and BLAST searches of protein databases. The results indicated that the largest protein is apparently identical to a previously described protein, symplekin (12, 46). Symplekin was isolated in a screen for proteins associated with tight junctions. Although characterization with a MAb suggested that the protein may in fact be associated with certain tight junctions, symplekin was convincingly shown to be present in the nuclei of all cell types examined (12). The pattern observed, diffuse granular staining excluding nucleoli, was similar to that detected when CstF was localized by related methods (42). Symplekin was also detected in a yeast two-hybrid screen for proteins interacting with mutant huntingtin protein (5), and we have shown that this interaction can occur in vitro (unpublished data); however, the significance of the interaction is unknown.
Only one other known protein, yeast PTA1, produced a significant match with symplekin, and this reflected a relatively weak similarity that had not been noted previously. However, the similarity is extensive and, for reasons discussed below, very likely to be significant. As shown in Fig. 4B, symplekin is 17% identical and 31% similar (see the legend to Fig. 4) to PTA1 over 427 residues. Similarity is highest at the C terminus of this region, displaying 26% identity and 45% similarity over 140 residues. PTA1 is an essential gene and was isolated initially because cells harboring a conditional pta1 allele were defective in processing intron-containing tRNA precursors (26). However, more recently, PTA1 was purified as a component of the yeast polyadenylation machinery, and extracts prepared from pta1 mutant cells were found to be defective in polyadenylation (30, 54) (see Discussion). These results strongly support the significance of the observed similarity between symplekin and PTA1 and, together with the data presented here, suggest that symplekin plays a role in pre-mRNA polyadenylation.
Symplekin and CstF-64 interact in vitro.
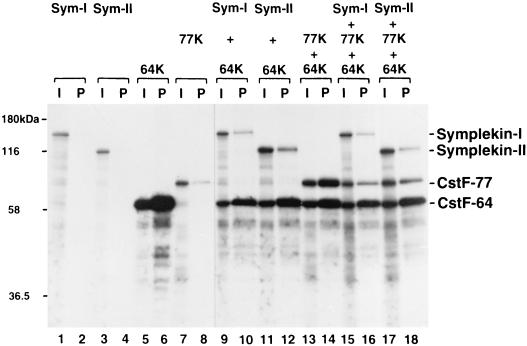
We next wished to verify that symplekin can in fact interact with CstF-64 in vitro. To this end, we first carried out coimmunoprecipitation experiments with in vitro-translated proteins. Specifically, both forms of symplekin (135 and 110 kDa) and CstF-77 were produced by in vitro translation and tested for their ability to be coprecipitated with in vitro-translated CstF-64, using an anti-CstF-64 MAb (42). As shown in Fig. 5, both symplekin isoforms were coprecipitated when mixed with CstF-64 (lanes 9 to 12). The fraction of the input bound was in each case lower than observed with CstF-77 (lanes 13 and 14). However, this difference was not unexpected, given that CstF-64 and CstF-77 are part of the stable CstF heterotrimer whereas symplekin is not. When the three polypeptides were mixed and subjected to immunoprecipitation, all were recovered in the pellet (lanes 15 to 18), indicating that symplekin can bind to CstF-64 even in the presence of CstF-77. Note that this experiment was not designed to determine whether CstF-64 can interact simultaneously with CstF-77 and symplekin. Indeed, a significant reduction in the amount of interacting CstF-77 was observed in the presence of either form of symplekin (compare lanes 13 and 14 with lanes 15 to 18), raising the possibility that these interactions are competitive.
FIG. 5.
Both symplekin-I and symplekin-II interact with CstF-64. Symplekin-I (Sym-I) and symplekin-II (Sym-II), CstF-77 (77K), and CstF-64 (64K) mRNAs were translated in vitro alone or in combination in the presence of [35S]methionine and immunoprecipitated with an anti-CstF-64 MAb. Then 25% of the input in vitro translation mixtures (I, odd-numbered lanes) and 50% of the immunoprecipitated proteins (P, even-numbered lanes) were resolved by SDS-PAGE and visualized by autoradiography.
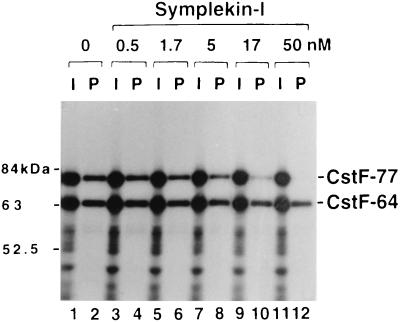
To address whether symplekin and CstF-77 can simultaneously bind to CstF-64, we first asked whether symplekin produced by in vitro translation could be coimmunoprecipitated by the anti-CstF-64 MAb when mixed with purified CstF. The results (not shown) failed to provide evidence for a symplekin-CstF interaction. We therefore decided to address directly whether the interactions of CstF-77 and symplekin with CstF-64 might be competitive. To this end, CstF-77 and CstF-64 were produced by in vitro translation, as in Fig. 5, except that increasing amounts of purified recombinant symplekin-I (the large form) were used (identical results [not shown] were obtained with the small form, symplekin-II), and the proteins were then subjected to immunoprecipitation with the anti-CstF-64 MAb. The results (Fig. 6) show that, as above, CstF-77 is efficiently coprecipitated with CstF-64 in the absence of symplekin, but increasing amounts (up to 50 nM) resulted in inhibition of CstF-77 coimmunoprecipitation. These data suggest that CstF-77 and symplekin can compete for the same or overlapping sites in the CstF-64 hinge domain and raise the possibility that symplekin does not interact with fully assembled CstF.
FIG. 6.
Symplekin-I inhibits association between CstF-77 and CstF-64. CstF-77 and CstF-64 mRNAs were translated in vitro in the absence (lanes 1 and 2) or presence of increasing concentrations of symplekin-I (lanes 3 to 12). Then 10% of the in vitro translation mixtures (I, odd-numbered lanes) and 30% of the immunoprecipitated proteins (P, even-numbered lanes) were resolved by SDS-PAGE.
Multiple polyadenylation factors, including symplekin, coexist in a large complex.
The above results raise interesting questions regarding the role of symplekin in polyadenylation. For example, given the apparent inability of symplekin to interact with intact CstF, perhaps the protein plays only an indirect role in the reaction, such as in transport and/or assembly, and may not be part of the active polyadenylation complex. We therefore wished to determine if the proteins were in fact associated in cell extracts. To this end, we isolated a Superose 6 gel filtration fraction prepared from HeLa nuclear extract (CSF) (43), which is known to contain all the factors necessary for 3′ processing (except PAP). The anti-CstF-64 MAb was then used to immunopurify CstF and associated proteins from the CSF fraction, first under relatively mild conditions [50 mM (NH4)2SO4 and no NP-40 (see Materials and Methods)]. Figure 7A, lanes 1 to 3, presents a silver-stained SDS-polyacrylamide gel displaying the total proteins in the CSF fraction, the flowthrough of the immunoaffinity column, and the bound proteins. It is apparent that the procedure resulted in considerable purification, since only a limited number of polypeptides were detected in the bound fraction. Proteins in addition to the three subunits of CstF were apparent, suggesting that CstF-associated factors were copurified.
To identify some of the CstF-associated proteins and to estimate how efficiently they were associated with CstF, we performed Western blot analysis of the total, flowthrough, and bound fractions with antibodies against several different polypeptides (Fig. 7A, lanes 4 to 6). Not unexpectedly, a large fraction (∼90%) of CstF-64 was detected in the bound fraction, indicating that the immunopurification was efficient as well as selective. Polypeptides the size of CstF-50 and CstF-77 are apparent in the silver-stained gel (lane 3), and we therefore assume the entire CstF complex was selected intact. A significant fraction (∼80%) of CPSF-160, the largest subunit of CPSF, was retained in the bound fraction (lanes 4 to 6). As with CstF, it is likely that the other CPSF subunits were present as well, and polypeptides the size of CPSF-100 and CPSF-73 were among the polypeptides detected by silver staining (see below). Finally, when a polyclonal antibody prepared against the N terminus of symplekin was used, symplekin-I indeed cofractionated with CstF and CPSF during gel filtration (Fig. 7A) and (most importantly) the majority (60 to 70%) of the protein in the CSF fraction could be coimmunopurified with CstF-64 (Fig. 7A, lanes 4 to 6). We have not identified the other polypeptides in the complex, although they may include CFI and/or CFII.
The above experiments provide the first direct evidence that CPSF and CstF can be isolated from cells associated with each other and that symplekin is part of this complex. Given that the existence of a preassembled CPSF-CstF complex has significant implications regarding poly(A) site recognition and that the presence of symplekin in the complex provides strong support for the notion that the protein indeed functions in polyadenylation, we wished to provide additional support for the existence of this complex. To this end, we repeated the immunopurification but used more stringent conditions [150 mM (NH4)2SO4 plus 0.05% NP-40] and a control antibody (anti-polyomavirus large T antigen) and verified the presence of additional CPSF subunits. Compared to the milder purification conditions (Fig. 7A, lanes 1 to 3), even fewer proteins were copurified with CstF (Fig. 7B, lanes 4 to 6), and most of these were undetectable in the bound fraction obtained using the control antibody (Fig. 7B, lanes 1 to 3). Western blot analyses using antibodies directed against CstF-64, CPSF-160, CPSF-100 (which cross-reacts with the related CPSF-73), and symplekin confirm that CPSF and symplekin are indeed present in a stable, high-molecular-weight complex with the CstF (Fig. 7B, lanes 10 to 12). The fraction of each of the polypeptides recovered in the bound fraction was high (although slightly lower than under the milder conditions), and none of these proteins were purified with the control antibody (lanes 7 to 9). These results both provide the first evidence for a mammalian complex containing multiple polyadenylation components and also strongly suggest that symplekin and CstF are in fact associated with each other, directly or indirectly, in vivo.
DISCUSSION
We have described here a series of protein-protein interactions involving subunits of CstF that both provided insights into the structure and function of CstF and also led to the identification of a novel polyadenylation complex-associated factor, symplekin. Our observation that symplekin and the yeast polyadenylation factor PTA1 are related increases the similarities between yeast and mammalian 3′ processing. However, the apparently competitive nature of the symplekin-CstF interaction, along with the ability of two CstF subunits to self-associate, suggests added complexities in the functioning of the polyadenylation complex. Finally, our data provided evidence that at least several of the factors necessary for the first step of the polyadenylation reaction, 3′ cleavage, coexist in a single complex. Below we discuss the significance and implications of these results as they pertain to the mechanism of the polyadenylation reaction.
The nature of the regions in all three subunits required for interactions with each other are intriguing. In CstF-77, all three interactions require the Pro-rich domain near the C terminus of the protein. Pro-rich regions in other proteins are often found to be involved in protein-protein interactions, and it is thus not surprising that this region functions similarly in CstF-77. However, it was unexpected to find a single region required for interaction with all three subunits. It is also notable that two of these interactions (CstF-77 self-association and interaction with CstF-50) show a requirement for sequences just N-terminal to this domain. While CstF-77 must interact simultaneously with Cst-50 and Cst-64 to form the CstF heterotrimer (39), it is conceivable that CstF-77 self-association might be competitive with the CstF-77/CstF-50 interaction (see below), and the identical sequence requirements are consistent with this possibility. The fact that all these interactions are clustered toward the CstF-77 C terminus leaves the remainder of the protein (i.e., the TPR/HAT motifs) available for interactions with other proteins. To date the only other known CstF-77-interacting protein is the largest subunit of CPSF, CPSF-160 (25), and preliminary data suggest that TPR motifs are involved in this interaction (unpublished data).
The WD-40 repeats in CstF-50 are responsible for interaction with CstF-77. All of the deletions we analyzed that affect this region greatly reduced or abolished interaction. This raises the possibility that all or most of the seven WD-40 repeats are involved in interaction with CstF-77. However, another possibility is that these deletions all disrupt a higher-order structure and that only a smaller surface interacts with CstF-77. This latter view is consistent with the crystal structure of the prototypical WD-40 protein, β-transducin, in which the WD-40 repeats form a sevenfold β-propeller made up of seven four-stranded antiparallel β sheets (16, 35, 50). The most highly conserved residues of the repeats form the core of the protein, with variable loops on the surface being available for interaction with other proteins. It is very likely that the CstF-50 WD-40 repeats form a similar structure. In addition to the CstF-77 interaction, the repeats appear to be required for interaction with the CTD of the RNA polymerase II largest subunit (20) and are necessary and sufficient for an interaction with the BRCA1-associated protein BARD1 (14).
The ability of CstF-50 and CstF-77 to self-associate was not predicted from previous biochemical experiments. The molecular weight of native CstF is ∼190,000, and the stoichiometry of the three subunits was estimated to be 1:1:1, suggesting that purified CstF is a monomer with one copy of each subunit (42). However, genetic studies with Drosophila have shown that certain lethal alleles of su(f) can complement one another to produce viable flies (34). The simplest and most consistent explanation of these results is that individual mutant Su(f) polypeptides interact with each other, thereby restoring partial function. The direct protein-protein interaction we have described may form the basis for this genetic interaction. These findings have significant mechanistic implications. One possibility is that CstF dimerizes (or multimerizes) at some point during the polyadenylation reaction. Although there is currently no evidence to support this, there is also nothing to contradict it. Additional studies are necessary to understand the functional significance of CstF-77 (and CstF-50) self-association.
Our results indicate that symplekin is associated with the pre-mRNA polyadenylation machinery. It is unclear whether it is required for polyadenylation, if it plays an auxiliary or stimulatory role, or if it serves some other function. Symplekin is not a component of any of the factors that have been purified to homogeneity (CPSF, CstF, CFI, PAP, and RNA polymerase II), and our unpublished data indicate that the only remaining factor (CFII) can be purified free of symplekin. However, symplekin does cofractionate with CPSF, CFI, and CFII during early purification steps, and the processing efficiency is very low when highly purified (i.e., symplekin-free) preparations of all factors are used in 3′ cleavage assays (unpublished data). Although we have not been able to restore processing by the addition of purified symplekin (unpublished data), these data are consistent with the possibility that symplekin is a required factor. However, whether or not it is essential, our results suggest a possible function. Specifically, we propose that symplekin is an assembly/scaffolding factor. Its ability to interact strongly with free CstF-64 but not assembled CstF is consistent with the idea that it may function in CstF assembly. However, the presence of symplekin in the CPSF-CstF complex, where CstF is presumably fully assembled, suggests that the protein may interact with other polyadenylation factors, such as CPSF, perhaps helping to assemble or stabilize the complex. As discussed below, properties of the putative yeast homologue of symplekin, PTA1, are consistent with this notion.
Symplekin has similarity to the yeast protein PTA1. The similarity is extensive, extending over 400 residues, but the percent identity is relatively low and includes less than half of each protein. However, its significance is very strongly supported by the fact that both proteins were independently implicated in pre-mRNA polyadenylation. In yeast, PTA1 was identified as a component of a multisubunit complex containing the factor PF I and PAP (30). Intriguingly, PF I seems most closely related to mammalian CPSF, since it contains apparent homologues of all four CPSF subunits. PF I, though, is required only for the second, poly(A) synthesis step, not for cleavage. In keeping with this, extracts from cells harboring a conditional allele of PTA1 were reported to be defective in specific poly(A) synthesis but not cleavage (30). Somewhat confusingly, however, the CPSF homologues have also been found as part of an activity designated CF II, which seems to be required for the cleavage but not the poly(A) synthesis step (36, 52). Recently, a separate study found that PTA1 was instead a component of CF II and that mutant strains were defective in cleavage as well as poly(A) synthesis (54). A parsimonious explanation for at least some of these conflicting results is that PF I and CF II are related and form part of a holocomplex in vivo. Individual components might cofractionate differently dependent on the procedures used, and PF I and CF II may actually be derived from a single factor, equivalent to mammalian CPSF. This is consistent with the known role of CPSF in both phases of the reaction, with our demonstration of a multifactor complex in mammals, and with our suggestion that symplekin functions as an assembly/scaffolding factor. Indeed, Zhao et al. (54) also raised the possibility that PTA1 functions as an assembly factor, based on the observation that PTA1 mutant strains contain reduced amounts of CF II subunits associated with other polyadenylation factors. The fact that PTA1 is complexed with a yeast CPSF-like factor also offers an explanation of how symplekin can be part of a CstF-CPSF complex, despite its inability to interact with intact CstF: symplekin may bind a CPSF component in addition to CstF-64.
Another possible function for symplekin/PTA1, not mutually exclusive with a role in assembly, is that it serves in some way to link polyadenylation with other nuclear events. Although there is no evidence addressing this for symplekin, two properties of PTA1 are consistent with this idea. First is the fact that PTA1 was initially discovered because a pta1 mutant strain was defective in pre-tRNA processing, although biochemical experiments failed to show any direct role for PTA1 in this reaction. Second, PTA1 was found to interact genetically with SPT3, which encodes a protein that interacts with the TATA binding protein subunit of transcription factor IID (TFIID) (18). This is particularly intriguing in light of increasing evidence linking transcription and polyadenylation in mammals, especially the interaction between TFIID and CPSF (4).
Our data have provided the first biochemical evidence that CPSF and CstF are physically associated with one another prior to recognition of the poly(A) signal. This is consistent with previous studies suggesting that both factors are components of the RNA polymerase II holoenzyme (20) and that they colocalize in the nucleus (5a, 33a). The existence of the CstF-CPSF association has important implications for initial recognition of the poly(A) site, indicating that the bipartite signal can be identified in a single interaction rather than through sequential recognition of AAUAAA by CPSF and of the GU-rich sequence by CstF. This may be important, for example, in helping to discriminate between cryptic and authentic poly(A) sites and to increase processing efficiency due to preassembly of the processing complex. It is noteworthy that PAP does not appear to be associated with the complex, as expected from the absence of PAP activity in the CSF fraction from which the CstF-CPSF complex was derived (43). This contrasts with yeast studies, which have shown that PAP can be isolated associated with yeast CPSF (i.e., PF I or CF II) (30, 54) whereas the yeast equivalent of a CPSF-CstF complex has not been described. Whether this reflects a physiologically significant difference between yeast and mammals or simply distinct fractionation properties remains to be determined.
On the one hand, our results have added further complexities to the already complex set of factors and interactions responsible for polyadenylation of mRNA precursors. We have identified a novel, unexpected factor, symplekin, and provided evidence for dynamic and unanticipated interactions between CstF subunits. On the other hand, our discovery of a polyadenylation complex and of the similarity between symplekin and PTA1 has further narrowed the gap between yeast and mammalian polyadenylation machineries. Future studies should lead to an unraveling of the details of the polyadenylation reaction and, importantly, to an appreciation of how it is integrated with other cellular processes.
ACKNOWLEDGMENTS
We thank K. G. K. Murthy and C. Prives for antibodies, J. Wu for the cDNA library, J. D. Kohtz for helpful discussions, Z. Lai for technical assistance, and I. Boluk for help in preparing the manuscript.
This work was supported by National Institutes of Health grant GM28983.
REFERENCES
- 1.Amrani N, Minet M, Wyers F, Dufour M-E, Aggerbeck L P, Lacroute F. PCF11 encodes a third protein component of the yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 3.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Dantonel J-C, Murthy K G K, Manley J L, Tora L. CPSF links transcription and mRNA 3′ end formation. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 5.Faber P W, Barnes G T, Srinidhi J, Chen J, Gusella J F, MacDonald M E. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 5a.Gall J G, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Z, Sherman F. 3′-End-forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 7.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 8.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 10.Keller W, Bienroth S, Lang K M, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 12.Keon B H, Schafer S, Kuhn C, Grund C, Franke W W. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M M, Henry M F, Shen E, Zhao J, Gross S, Silver P A, Moore C L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleiman F E, Manley J L. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 15.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 16.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. The 2.0Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 17.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manley J L, Takagaki Y. The end of the message-another link between yeast and mammals. Science. 1996;274:1481–1482. doi: 10.1126/science.274.5292.1481. [DOI] [PubMed] [Google Scholar]
- 20.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 21.Miller D W, Safer P, Miller L K. An insect baculovirus host-vector system for high-level expression of foreign genes. Genet Eng. 1986;8:277–298. [Google Scholar]
- 22.Mitchelson A, Simonelig M, Williams C, O'Hare K. Homology with Saccharomyces cerevisiae RNA 14 suggests that phenotypic suppression in Drosophila melanogaster by suppressor of forked occurs at the level of RNA stability. Genes Dev. 1993;7:241–249. doi: 10.1101/gad.7.2.241. [DOI] [PubMed] [Google Scholar]
- 23.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy K G K, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 25.Murthy K G K, Manley J L. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor J P, Peebles C L. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Hare K. mRNA 3′ ends in focus. Trends Genet. 1995;11:255–257. doi: 10.1016/s0168-9525(00)89067-8. [DOI] [PubMed] [Google Scholar]
- 28.Peterson M L, Perry R P. The regulated production of μm and μs mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the Cμ4-to-M1 splice. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 30.Preker P J, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preker P J, Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- 32.Raabe T, Bollum F J, Manley J L. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 33.Rüegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 33a.Schul W, Groenhaut B, Koberna K, Takagaki Y, Jenny A, Manders E M M, Ravska I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- 34.Simonelig M, Elliott K, Mitchelson A, O'Hare K. Interallelic complementation at the suppressor of forked locus of Drosophila reveals complementation between suppressor of forked proteins mutated in different regions. Genetics. 1996;142:1225–1235. doi: 10.1093/genetics/142.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a Ga protein βγ dimer at 2.1Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 36.Stumpf G, Domdey H. Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science. 1996;274:1517–1520. doi: 10.1126/science.274.5292.1517. [DOI] [PubMed] [Google Scholar]
- 37.Takagaki Y, MacDonald C C, Shenk T, Manley J L. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takagaki Y, Manley J L. A human polyadenylation factor is a G protein β-subunit homologue. J Biol Chem. 1992;267:23471–23474. [PubMed] [Google Scholar]
- 39.Takagaki Y, Manley J L. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 40.Takagaki Y, Manley J L. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takagaki Y, Manley J L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 42.Takagaki Y, Manley J L, MacDonald C C, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 43.Takagaki Y, Ryner L C, Manley J L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 44.Takagaki Y, Ryner L C, Manley J L. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 45.Takagaki Y, Seipelt R L, Peterson M L, Manley J L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 46.Ueki K, Ramaswamy S, Billings S J, Mohrenweiser H W, Louis D N. Chromosomal localization to 19q 13.3, partial genomic structure and 5′ cDNA sequence of the human symplekin gene. Somatic Cell Mol Genet. 1997;23:229–231. doi: 10.1007/BF02721375. [DOI] [PubMed] [Google Scholar]
- 47.Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 48.Wahle E, Martin G, Schiltz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahle E, Lustig A, Jeno P, Maurer P. Mammalian poly(A)-binding protein II. J Biol Chem. 1993;268:2937–2945. [PubMed] [Google Scholar]
- 50.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Gi∝1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Kessler M M, Moore C L. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 53.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J, Kessler M, Helmling S, O'Connor P, Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]