Abstract
Antibiotic-resistant Pseudomonas aeruginosa infections are the primary cause of mortality in people with cystic fibrosis (CF). Yet, it has only recently become appreciated that resistance mutations can also increase P. aeruginosa virulence, even in the absence of antibiotics. Moreover, the mechanisms by which resistance mutations increase virulence are poorly understood. In this study we tested the hypothesis that mutations affecting efflux pumps can directly increase P. aeruginosa virulence. Using genetics, physiological assays, and model infections, we show that efflux pump mutations can increase virulence. Mutations of the mexEF efflux pump system increased swarming, rhamnolipid production, and lethality in a mouse infection model, while mutations in mexR that increased expression of the mexAB-oprM efflux system increased virulence during an acute murine lung infection without affecting swarming or rhamnolipid gene expression. Finally, we show that an efflux pump inhibitor, which represents a proposed novel treatment approach for P. aeruginosa, increased rhamnolipid gene expression in a dose-dependent manner. This finding is important because rhamnolipids are key virulence factors involved in dissemination through epithelial barriers and cause neutrophil necrosis. Together, these data show how current and proposed future anti-Pseudomonal treatments may unintentionally make infections worse by increasing virulence. Therefore, treatments that target efflux should be pursued with caution.
Keywords: Pseudomonas, efflux pumps, virulence, evolution, antibiotic resistance, cystic fibrosis
1. Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that colonizes a wide range of environments [1]. This environmental flexibility is due to its high adaptability to changing conditions and is driven by substantial metabolic versatility, expression of a large array of virulence factors, and extensive adaptive transport and efflux systems [2]. P. aeruginosa is infamous for causing serious nosocomial infections, such as burn wound infections and ventilator-associated pneumonia [3], and it is the predominant pathogen causing chronic infections in cystic fibrosis (CF) [4].
During chronic infections, P. aeruginosa adapts to the CF lung environment by decreasing production of virulence factors and evolves to become antibiotic resistant [5,6,7]. Even more complicated, populations of P. aeruginosa within patients evolve high degrees of diversity and within-patient variability, where contemporary sibling cells that have descended from a common ancestor can differ in antibiotic susceptibilities and virulence factor production [8,9,10,11]. P. aeruginosa virulence mechanisms include attachment to cells (e.g., pili), induction of host damage (e.g., DNase, proteases), competition with the host for nutrients, and the secretion of surfactant molecules such as rhamnolipids which cause neutrophil necrosis and permit dissemination [12,13,14]. Virulence factors usually trigger an exacerbated inflammatory response by the innate immune system, with macrophages and neutrophils as the first line of defense [15]. Thus, it has been proposed that decreasing virulence factor production attenuates the host inflammatory response and favors bacterial colonization of the lung during CF chronic infections.
In addition to the host pressures that affect virulence, bacteria colonizing CF lungs also experience intense antibiotic pressure due to mono and combination therapies including aminoglycosides, quinolones, and β-lactams [16]. This environment strongly favors the selection of antibiotic-resistant strains. Some of the best characterized antibiotic resistance features are efflux pump systems that expel antibiotics out of the cell. For example, we previously showed that nalD and mexR genes, both encoding for transcriptional repressors of the MexAB-OprM efflux pump [17,18,19], were recurrently mutated under aztreonam selection [20]. P. aeruginosa with mutations in nalD and mexR overexpress the MexAB-OprM efflux pump, rendering them multidrug resistant [17,19,21]. Moreover, mutations in these two genes have been found in anywhere from 7–47% of clinical isolates in various studies [20,21,22,23,24]. Importantly, strains with mutations in nalD and mexR evolved under aztreonam selection showed increased virulence in vivo, challenging the longstanding theory that antibiotic-resistance mutations inherently pay fitness costs that attenuate virulence [20]. However, whether these mutations increase virulence in an efflux-dependent manner remains unknown.
In this study, we sought to investigate the mechanisms involved in the hypervirulence of a mutant P. aeruginosa strain, PAO1-AzEvB8, that was experimentally evolved in a previous study during cyclic aztreonam exposure [20]. Because this strain was previously shown to have a mutation in mexR, we hypothesized that its hypervirulence could be attributed to the increased expression of the MexAB-OprM efflux pump. We explore this hypothesis using a combination of genome sequencing, genetics, transcriptional reporter assays, and infection models.
2. Results
2.1. Deletion of mexAB Restores Aztreonam Susceptibility of Evolved P. aeruginosa
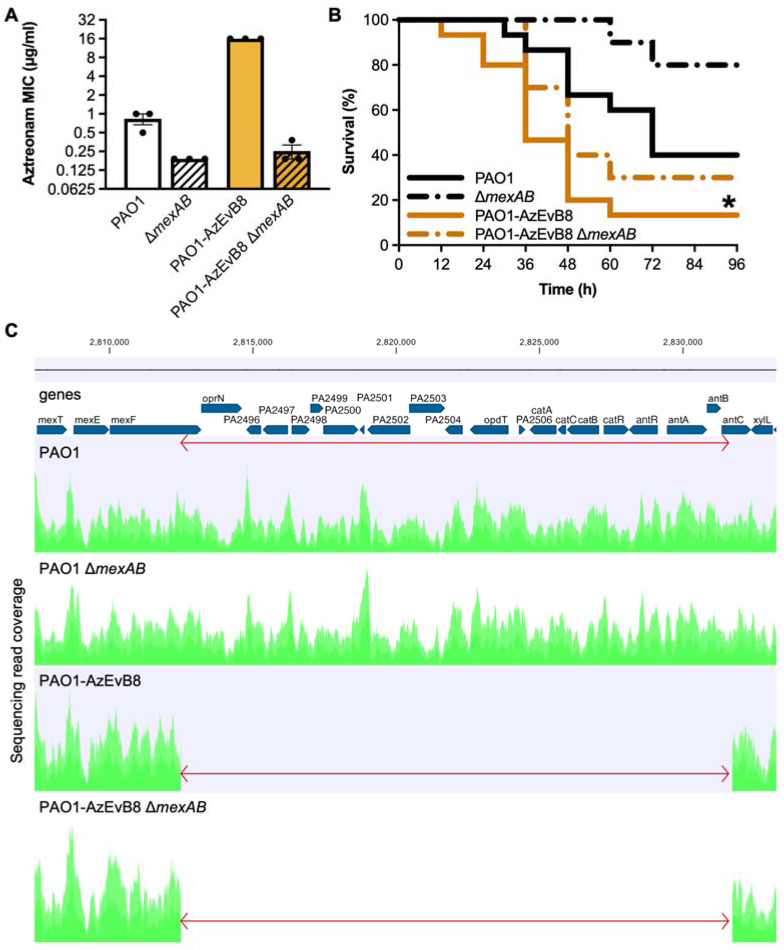
Previously, we used experimental evolution with cyclical exposure to aztreonam to evolve the aztreonam resistant strain PAO1-AzEvB8 [20]. Strain PAO1-AzEvB8 was found to have a nonsense mutation in mexR (E118*) and complementation with a wild-type (WT) mexR gene restored aztreonam susceptibility [20]. Because mexR mutations have been shown to increase expression of the mexAB-oprM efflux pump operon [19,21], which encodes the pump that effluxes aztreonam, we hypothesized that deletion of mexAB in PAO1-AzEvB8 would restore aztreonam susceptibility. We generated clean mexAB deletions in both WT PAO1 and in PAO1-AzEvB8, and the PAO1-AzEvB8 ΔmexAB strain was equally susceptible to aztreonam by a gradient diffusion assay as the PAO1 ΔmexAB strain (Figure 1A); the mode minimum inhibitory concentrations (MIC) for ΔmexAB strains was 0.19 μg/mL, compared to 16 μg/mL for PAO1-AzEvB8 and 1.0 μg/mL for WT PAO1. This showed that mexAB was required for aztreonam resistance of the of the PAO1-AzEvB8 mutant.
Figure 1.
MexAB-OprM is not involved in the PAO1-AzEvB8 mutant in vivo virulence. (A) Deletion of the mexAB genes abrogates aztreonam resistance in strain PAO1-AzEvB8. Aztreonam MICs were determined by gradient diffusion assays (n = 3 replicates/group; MIC: minimum inhibitory concentration). (B) mexAB deletion in PAO1-AzEvB8 mutant does not significantly improve mouse survival following acute lung infection. Log-rank test was used to compare survival curves. * p < 0.05 compared to WT PAO1; n = 10–15 mice/group. (C) Genome diagram showing coverage of sequencing reads aligning to the region spanning mexEF through xylZ. The 19,233 bp deleted region (indicated by the red arrow) in strains PAO1-AzEvB8 and PAO1-AzEvB8 ΔmexAB begins at the 3′ end of mexF and continues through the 5′ region of antC. Genome coverage plots generated from sequencing read alignments to the PAO1 reference genome are indicated in green.
2.2. Deletion of mexAB Does Not Affect Virulence of Evolved P. aeruginosa
In addition to exhibiting increased aztreonam resistance relative to WT PAO1, strain PAO1-AzEvB8 also displayed increased killing of mice in an acute murine infection model. We hypothesized that, similar to the aztreonam resistance, the increased virulence of PAO1-AzEvB8 would be dependent upon mexAB which was overexpressed in this strain. Surprisingly, the PAO1-AzEvB8 ΔmexAB strain was equally virulent to strain PAO1-AzEvB8 (Figure 1B), indicating that the increased virulence was not dependent on mexAB alone. This was even more unexpected since PAO1 ΔmexAB was less virulent than WT PAO1 (Figure 1B). This suggested that another gene or mutation was involved in the enhanced virulence of PAO1-AzEvB8.
2.3. Genome Sequencing Reveals a Previously Undetected 19 kb Deletion in PAO1-AzEvB8
To determine whether another mutation may be present in the strain PAO1-AzEvB8, we performed whole genome sequencing on strains PAO1 (WT parent strain), PAO1 ΔmexAB, PAO1-AzEvB8, and PAO1-AzEvB8 ΔmexAB. Using this approach, we found that relative to WT PAO1, a large ~19 kb deletion affecting 21 genes spanning mexF through antC was present in the PAO1-AzEvB8 and PAO1-AzEvB8 ΔmexAB strains (Figure 1C). This deletion was not detected by the genome sequencing in our original study despite >10-fold sequencing coverage of the genome [20]. The genes affected by this mutation included mexF and oprN which form part of the mexEF-oprN efflux pump operon. Previously, these genes have been linked to the swarming phenotype in P. aeruginosa, with studies indicating that strains overexpressing mexE have decreased swarming [25,26]. This led us to predict that the mexF mutation could lead to increased swarming and rhamnolipid production in the PAO1-AzEvB8 strain.
2.4. Mutation of mexEF Increases Swarming While mexR Mutation Does not
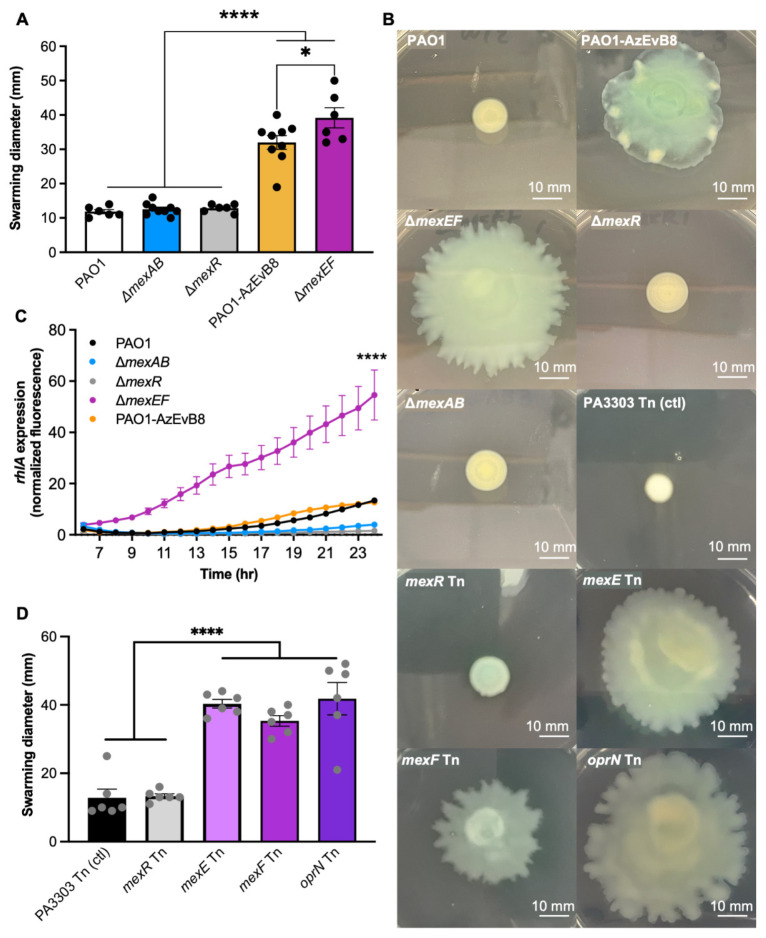
Based on the deletion of a large part of the mexEF-oprN operon in strain PAO1-AzEvB8, we hypothesized that this strain would exhibit increased swarming, as would mutants lacking mexEF. To test this, we generated a strain with the mexEF genes deleted in a WT PAO1 background. Gradient diffusion susceptibility testing showed that the PAO1 ΔmexEF strain was more susceptible to both ciprofloxacin and chloramphenicol than WT PAO1 (mode ciprofloxacin MICs: PAO1 ΔmexEF 0.125 μg/mL vs. WT PAO1 0.5 μg/mL; mode chloramphenicol MICs: PAO1 ΔmexEF 48 μg/mL vs. WT PAO1 >256 μg/mL; n = 3). Swarming assays were performed using the WT PAO1, PAO1-AzEvB8, PAO1 ΔmexEF, and PAO1 ΔmexR strains. As predicted, the PAO1-AzEvB8 strain displayed more swarming than WT, as did PAO1 ΔmexEF (Figure 2A,B). In contrast, deletion of mexR in a WT PAO1 background did not affect swarming (Figure 2A,B). Next, we tested whether mexEF mutations led to increased rhamnolipid production, because rhamnolipids are involved in swarming and cause necrosis of neutrophils [13]. A rhlA-gfp transcriptional reporter was transformed into strains PAO1, PAO1 ΔmexEF, PAO1 ΔmexAB, PAO1 ΔmexR, and PAO1-AzEvB8 and rhlA expression was monitored during the course of growth. Consistent with the swarming phenotypes, PAO1 ΔmexEF had the highest rhlA expression (Figure 2C). However, unexpectedly, PAO1-AzEvB8 had less rhlA expression than PAO1 ΔmexEF, similar rhlA expression to WT PAO1, and slightly more rhlA expression than PAO1 ΔmexAB and PAO1 ΔmexR strains (Figure 2C). Together these data showed that the PAO1-AzEvB8 strain had a hyper-swarming phenotype, a mexEF deletion mutation was sufficient to increase swarming, and a mexR deletion mutation alone does not affect swarming.
Figure 2.
MexEF-OprN mutation increases swarming motility and biosurfactant production. (A) Swarming is increased in the PAO1-AzEvB8 and ΔmexEF strains. Measurement of the swarming motility in different mexAB-oprM and mexEF-oprN mutants after 24 h, n = 6–7 replicates/group. (B) Representative images of swarming motility from panels (A,D). Scale bars indicate 10 mm. (C) Rhamnolipid gene expression is increased in the ΔmexEF strain. Rhamnolipid production measured by quantification of rhlA gene expression using a rhlA-gfp promoter reporter fusion to measure GFP fluorescence over time. Gene expression was calculated as area under the curve for each strain and compared to all other groups, n = 6–7 replicates/group. (D) Swarming is increased in mexE, mexF, and oprN PAO1 transposon mutant strains. Measurement of the swarming motility in different mexR and mexEF-oprN transposon mutants vs. a neutral Tn mutant control (PA3033) after 24 h, n = 5–12 replicates/group. For all panels, * p < 0.05, **** p < 0.0001, one-way ANOVA, followed by a Tukey’s multiple comparisons test.
To further study the effects of mexEF mutations on swarming and rhamnolipid gene expression, we also investigated these phenotypes using transposon (Tn) mutants. Swarming assays showed the PAO1 Tn mutants in mexE, mexF, and oprN all exhibited increased swarming relative to a Tn mutant control (PA3033), which has been shown to be a neutral Tn mutation for numerous phenotypes. As above, a mexR Tn mutant showed the same levels of swarming as the Tn mutant control.
2.5. Deletion of either mexR or mexEF Increases P. aeruginosa Virulence
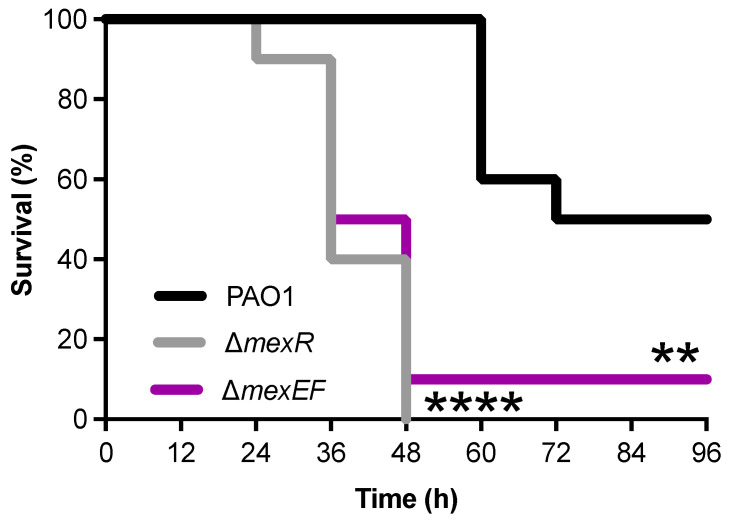
To better understand the increased virulence of the PAO1-AzEvB8 strain, we analyzed the virulence of PAO1 ΔmexR and PAO1 ΔmexEF strains to determine if either the mexR or mexF mutations detected in the PAO1-AzEvB8 strain could have contributed to the increased virulence. Using an acute murine lung infection model, we found that both PAO1 ΔmexR and PAO1 ΔmexEF killed the infected mice significantly faster than the WT PAO1 strain (Figure 3). These findings suggest that either of the two mutations present in the PAO1-AzEvB8 were sufficient to increase the virulence, relative to the WT parent strain.
Figure 3.
MexAB-OprM overexpression and MexEF-OprN deletion exhibit increased in vivo virulence. Both ΔmexR and ΔmexEF mutations in WT PAO1 significantly reduce mouse survival during an acute lung infection compared to WT PAO1 (WT). Survival times were tested across strata with a Bonferroni-adjusted log-rank test: ** p < 0.005 and **** p < 0.0001 compared to WT PAO1, n = 10 mice/group.
2.6. Efflux Pump Inhibition Increases Rhamnolipid Virulence Factor Expression
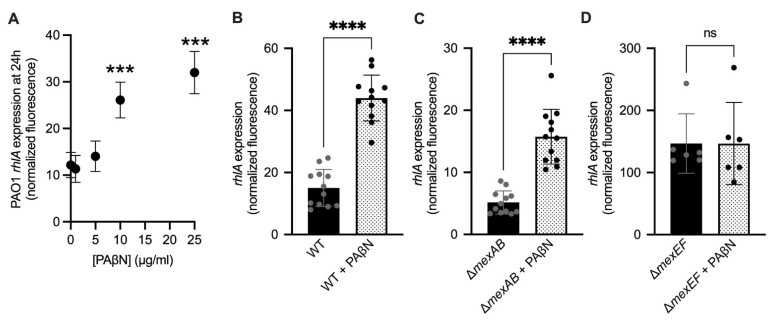
The swarming assays and mouse virulence experiment suggested that loss of the mexEF efflux pump was sufficient to increase swarming, rhamnolipid expression, and virulence in a mouse lung infection model. Therefore, we hypothesized that inhibition of efflux pump activity would lead to increased rhamnolipid gene expression. To test this, we exposed the PAO1 attB::rhlA-gfp reporter strain to increasing concentrations of the PAβN efflux pump inhibitor [27]. As hypothesized, increasing concentrations of PAβN from 0–25 μg/mL led to increased rhlA rhamnolipid gene expression in a dose-dependent manner (Figure 4A). This showed that a broad-spectrum efflux pump inhibitor could increase the expression of an important virulence factor.
Figure 4.
Rhamnolipid gene expression is induced by the PaβN efflux pump inhibitor. (A) rhlA expression in WT PAO1 attB::rhlA-gfp treated with 0–25 μg/mL PAβN as measured by GFP fluorescence from the attB::rhlA-gfp promoter reporter fusion at 24 h (*** p < 0.0005, compared 0, 1, and 5 μg/mL, one-way Brown−Forsythe and Welch ANOVA followed by Dunnett’s T3 multiple comparisons test, n= 6 replicates/group). (B–D) rhlA expression in PAO1 attB::rhlA-gfp (B), PAO1 ΔmexAB attB::rhlA-gfp (C), and PAO1 ΔmexEF attB::rhlA-gfp (D), treated with or without 25 μg/mL PAβN as measured by GFP fluorescence from the attB::rhlA-gfp promoter reporter fusion at 24 h. Gene expression was calculated as GFP fluorescence normalized to cell density (**** p < 0.0001; ns: not significant, p > 0.05, one-way Brown−Forsythe and Welch ANOVA followed by Dunnett’s T3 multiple comparisons test, n = 6–12 replicates/group).
Because PAβN can inhibit both MexEF-OprN and MexAB-OprM, we tested the hypothesis that the PAβN inhibitor was increasing rhamnolipid gene expression in a mexEF-dependent manner. We incubated WT PAO1, PAO1 ΔmexAB, and PAO1 ΔmexEF rhlA-gfp reporter strains with or without 25 μg/mL PAβN for 24 h. As predicted, PAβN increased rhlA gene expression in WT PAO1 and PAO1 ΔmexAB strains relative to untreated controls (Figure 4A–C), but not in the PAO1 ΔmexEF strain (Figure 4D). This shows that the increase of rhlA expression is occurring via inhibition of MexEF-OprN, not MexAB-OprM.
3. Discussion
3.1. Deletion of mexEF and Overexpression of mexAB through mexR Mutation Can Each Increase Virulence of P. aeruginosa
Here we showed that two different mutations affecting efflux pumps that evolved in response to aztreonam selective pressure can increase P. aeruginosa virulence in the absence of antibiotic treatment. These findings are interesting because one mutation, mexR, leads to overexpression of the MexAB-OprM efflux pump, while the other mutation conferred the loss-of-function of a second efflux pump, mexEF-oprN. We also showed that deletion of mexEF caused increased swarming, likely due to increased rhlA gene expression, which was demonstrated using a rhamnolipid gene expression reporter. Finally, we showed that MexEF-OprN efflux pump inhibition can have the unexpected effect of increasing rhamnolipid gene expression. This observation is important because rhamnolipids can cause necrosis of neutrophils and are also involved in the penetration of P. aeruginosa through epithelial barriers [13,14]. Altogether, these data highlight how antibiotic selection can unexpectedly increase virulence through multiple pathways.
3.2. Understanding Why rhlA Was Not Expressed in the PAO1-AzEvB8 Strain That Exhibited Greater Swarming Than WT
One curious result was that the PAO1-AzEvB8 strain did not overexpress rhlA relative to WT PAO1, despite exhibiting increased swarming. Because this strain had a large deletion mutation that included the 3′ end of mexF, we expected it to exhibit elevated rhlA expression similar to the PAO1 ΔmexEF strain. One possible explanation is that with an intact mexE gene, the increase in rhlA relative to WT may be less pronounced than when both mexE and mexF are deleted. This is somewhat apparent in the quantification of the swarming assays where the swarming zone of PAO1 ΔmexEF was slightly larger than PAO1-AzEvB8. Another possible explanation is that the swarming assays were performed using semi-solid 0.5% agar plates, whereas the rhlA expression assays were done in broth cultures. In future work we plan to explore whether differences in rhlA expression are detectable during infection, which is where it is likely most relevant.
3.3. Relationship of These Findings to Previous Research
Overall, these results are consistent with previous studies which found that P. aeruginosa nfxC mutants, which overexpress mexEF-oprN, do not swarm and exhibit decreased rhlA expression relative to WT [26]. Likewise, these data also align with a study by Cosson et al. which found that P. aeruginosa nfxC mutants were avirulent in a rat acute pneumonia infection model, and virulence could be partially restored in the nfxC mutant by deleting mexE [28]. An important distinction is that, to our knowledge, it had not previously been shown that deletion of mexEF in a WT genetic background can increase virulence.
3.4. Differences among Infection Models Help Explain Why Previous Studies Concluded Different Effects of Efflux Pump Inhibition on Virulence
While our data agree with the studies discussed above, the finding that the ΔmexEF strain was more virulent than WT in the acute murine lung infection does appear to conflict other previous research related to efflux pump inhibition. In a study from Hirakata et al., PAβN was shown to reduce the intracellular invasion of Madin−Darby canine kidney epithelial cell monolayers by P. aeruginosa [29], which appears inconsistent with our findings. One simple explanation for this difference is that we were not measuring intracellular invasion in our model. Instead, we measured virulence by survival following acute pneumonia, which we argue is more similar to human lung infections because neutrophils and epithelial barriers are present that can be harmed by the excess rhamnolipid produced in mexEF mutants and bacteria treated with PAβN. In another study, PAβN reduced the swarming and killing of Galleria mellonella larvae [30]. Again, slight differences in the infection model and the swarming assay could help explain differences from the present study. For example, the medium used for the swarming assay is different from the present study [30]. The G. mellonella infection model is also quite distinct from the mouse infection model used here, as it seems to be primarily affected by type III secretion [31] and lipopolysaccharide [32], and the role of rhamnolipids in the G. mellonella model is less clear. Therefore, we believe that differences in experimental design can account for the differences in interpretation of the effects of the PAβN efflux pump inhibitor. Future experiments will be required to determine whether PAβN and similar efflux pump inhibitors enhance or decrease virulence in mammalian infections.
3.5. Relevance of mexEF Mutations in P. aeruginosa Clinical Isolates
While mexR mutations have been identified in countless P. aeruginosa clinical isolates in previous studies [24,33], mutations directly affecting mexEF have not been as commonly described. Previous work has shown that mutations in mexT, the transcriptional activator of mexEF, are very common and these mutants were dubbed nfxC-type mutants for their resistance to norfloxacin [34,35]. Additionally, several studies have also examined mexE gene expression in large sets of clinical isolates, and in some strains mexE expression was reported to be less than WT, or completely undetected [22,36,37]. This suggests that these strains have either lost function of mexT or may potentially have deletions in mexE. This raises the possibility that these clinical isolates would also exhibit increased swarming, increased rhamnolipid production, and be more virulent than other clinical isolates, though this has not yet been investigated. We plan to test these phenotypes in clinical isolates with reduced mexE expression in the future.
3.6. Implications for CF and the Use of Efflux Pump Inhibitor as Potential Therapies
These results have several implications that are particularly relevant to CF treatment, including currently used anti-Pseudomonal therapies, as well as proposed new therapies.
First, these data show how aztreonam, a common antibiotic used to treat P. aeruginosa infections in people with CF, can select for not one, but two different mutations that increase P. aeruginosa virulence. This may help explain why a recent clinical observational study found that people with CF that were infected with aztreonam-resistant P. aeruginosa were more likely to experience pulmonary exacerbations and be hospitalized than people infected with aztreonam-susceptible P. aeruginosa [38]. If the isolates in those patients evolved mutations in either mexR or mexEF-oprN, then this could lead to increased virulence and lung damage in these individuals. Future work investigating the prevalence and association of these mutations with clinical outcomes will help test this theory.
Second, these data show that efflux pump inhibitors might have unexpected negative consequences on infections in people. Efflux pump inhibitors such as PAβN can inhibit efflux and potentiate other antibiotics that would normally be expelled through those pumps [27]. However, the experiments presented here show that inhibiting or deleting one efflux pump, MexEF-OprN, can have the unintended consequence of increasing rhamnolipid gene expression. Thus, inhibitors that affect MexEF-OprN may not be good candidates for use clinically. Instead, it would be more beneficial to identify more specific efflux pump inhibitors. For example, the MexAB-OprM-specific efflux pump inhibitor D13-9001 [39] could be used to block MexAB-OprM without inhibiting MexEF-OprN, and this could make P. aeruginosa more susceptible to other antibiotics and reduce virulence, since mexAB deletion mutants are less virulent than WT P. aeruginosa, as shown here and in other research [40]. Our data highlight the need to explore these types of precise approaches in future research using animal infection models.
3.7. Limitations of the Present Work
It is also important to acknowledge the limitations of this work. First, the strain PAO1-AzEvB8 that was the focus of the present study was not an actual clinical isolate and it is unclear how frequently mutants arise that have both mexR and mexEF mutations. As mentioned above, literature suggests that mexR mutations are very common in CF, and that clinical isolates can display varying degrees of mexE gene expression [20,22,24,33,36,37], so we feel that this study still has relevance to real-world P. aeruginosa infections.
The second primary limitation to this work is that the mouse model analyzed does not perfectly model the chronic infections that affect people with CF. One weakness is that this model does not capture the types of nutrient and biochemical properties present in CF that can affect P. aeruginosa antibiotic tolerance and virulence factor expression [41,42,43,44]. However, one strength to this model is that it does recapitulate the airway epithelial cell barrier and neutrophils present in CF infections that are susceptible to rhamnolipid-mediated toxicity [13,14]. To explore the effects of these mutations on other aspects of CF lung disease, future work could utilize chronic infection models in rodents or the more recently developed CF ferret and pig models [14,45,46,47,48].
One final limitation to this work is that the deletion mutants have not been complemented. Therefore, it is possible that the swarming and virulence phenotypes could be driven by the off-target effects of the generated mutations. We believe this is unlikely because we were able to reproduce identical phenotypes in multiple strains possessing related mutations. For example, the PAO1-AzEvB8 strain has both a mexR null mutation and mexEF deletion mutation, yet we only saw the swarming phenotype increased in the PAO1-AzEvB8 and ΔmexEF strains, not the ΔmexR or ΔmexAB strains. This suggests that the swarming phenotype in the PAO1-AzEvB8 strain is driven by its mexEF mutation. This was also supported by experiments with Tn mutants, where we observed increased swarming in mexE, mexF, and oprN Tn mutants, but not in a mexR Tn mutant. An additional caveat is that the clean deletion mutation in the PAO1 ΔmexEF strain was not generated in-frame with the mexE and mexF coding sequences, whereas the mexR and mexAB deletion constructs were generated in-frame with the original genes. Therefore, it is possible that a small peptide may be transcribed in the ΔmexEF strain that is out-of-frame (+1) relative to the mexEF-oprN coding sequences. This could have two consequences independent of the mexEF deletion. First, the ΔmexEF strain phenotypes could be caused by this novel peptide and not the mexEF deletion which was confirmed by whole genome sequencing. We think this is unlikely because the other phenotypes of this strain, including increased antibiotic susceptibilities to ciprofloxacin and chloramphenicol are consistent with previous studies of mexE mutants [26,35]. The other possibility is that these phenotypes could be caused by the effects of the mexEF deletion that prevent normal expression of oprN. Even if this were true, the phenotypes would still be consistent with the overall loss-of-function of the mexEF-oprN efflux pump, which is consistent with our conclusions. Long-term, we plan to perform complementation studies to tease out whether the swarming, rhamnolipid, and virulence phenotypes are due to loss-of-function of all three genes in the mexEF-oprN operon, or if phenotypes can be caused by mutation by each of the individual genes.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
Bacterial strains and plasmids are listed in Table S1. Mutant strains were derived from WT PAO1, which was obtained from Colin Manoil’s laboratory at the University of Washington [49]. Tn mutants were also obtained from Colin Manoil’s laboratory at the University of Washington and they were grown from freezer stock on Luria−Bertani (LB) agar with 10 μg/mL tetracycline [49]. All other strains were routinely grown at 37 °C on LB agar and in LB broth (cat# 244520 and 244620, Becton & Dickinson Co., Franklin Lakes, NJ, USA) unless otherwise specified.
4.2. Deletion Plasmid Construction
PAO1 strains carrying full deletions of mexAB, mexEF, or mexR genes listed in Table S1 were generated with a suicide plasmid as described previously [50]. For the ΔmexAB and ΔmexR strains, two PCR fragments were generated from chromosomal DNA for each construct using the following primer pairs up- and down-stream of each target for mutation; mexAB genes: mexAB-KO-UP-F and mexAB-KO-UP-R for the upstream mexAB fragment and mexAB-KO-DN-F and mexAB-KO-DN-R for the downstream mexAB fragment; and mexR: mexR-KO-UP-F and mexR-KO-UP-R for the upstream mexR fragment and mexR-KO-DN-F and mexR-KO-DN-R for the downstream mexR fragment. The suicide plasmid pEX18Gm [51] was prepared for assembly by restriction digest or PCR amplification with primers pEX18Gm-F and pEX18Gm-R (Supplementary Material Table S1). The two fragments for each construct were then assembled into the prepared vector pEX18Gm [51] using NEBuilder HiFi DNA Assembly Cloning Kit (cat# E5520, New England BioLabs, Ipswich, MA, USA) for the ΔmexAB mutant and the NEB Gibson Assembly Master Mix (cat# E2611L, New England BioLabs, Ipswich, MA, USA) for the ΔmexR mutant. Suicide plasmids were transformed into E. coli DH5α [52] competent cells (NEB cat# C2987H, New England BioLabs, Ipswich, MA, USA) according to manufacturer’s protocol, selected on LB agar with 10 μg/mL gentamicin (Gm). Plasmids were isolated using the NEB Monarch Plasmid Purification MiniPrep Kit (cat# T1010L, New England BioLabs, Ipswich, MA, USA) and verified by Sanger sequencing. For the ΔmexEF mutant construct, two sets of primers (oRP_21, oRP_22 and oRP_23, oRP_24) were designed to amplify 527 and 495 bp regions upstream and downstream, respectively, of the mexEF genes. These PCR fragments were assembled by splicing by overlap extension PCR and the deletion allele, containing attachment sites (attB1- and attB2-) for Gateway recombination, was integrated into the pDONRPEX18Gm vector via the BP Clonase reaction, as previously described [50]. The reaction mixture was transformed into E. coli DH5α by electroporation, and clones harboring the plasmid with the ΔmexEF allele were selected on LB agar with 10 μg/mL Gm and identified by colony PCR using M13 universal primers. This yielded plasmid pRP12 (pDONRPEX18Gm::ΔmexEF), which was sequence verified using M13 primers.
4.3. Transformation of P. aeruginosa Deletion Mutants
PAO1 deletion mutants were generated from WT P. aeruginosa PAO1 through two-step allelic exchange through either electroporation or mating, as described previously [50]. For electroporation of pEX18Gm::ΔmexAB, 1.5 mL of PAO1 or PAO1-AzEvB8 grown overnight in LB were centrifuged at 14,000× g for 3 m and washed twice with 300 mM sterile sucrose. Cells were then centrifuged at 14,000× g for 3 m and resuspended in 100 μL of 300 mM sterile sucrose. One microgram of pEX18Gm::ΔmexAB was then added to each cell suspension, and the bacterial suspensions were transformed by electroporation at 2.5 kv and incubated on LB with 30 μg/mL Gm at 37 °C overnight. For counter selection, isolated clones were streaked on low-salt LB containing 15% sucrose as published [50] and incubated at room temperature for 48 h. Gene deletions were confirmed by PCR using primers mexAB-KO-Chk-F and mexAB-KO-Chk-R (Table S1) and Sanger sequencing. For the ΔmexR strain, pEX18Gm::ΔmexR was transformed into chemically competent E. coli SM10(λpir) [53] which were prepared using transformation and storage solution (TSS, LB broth with 10% (w/v) polyethylene glycol, 5% (v/v) dimethyl sulfoxide, and 0.5% (w/v) MgSO4·7H2O), as described previously [54] and selection on LB agar with 20 μg/mL Gm. E. coli SM10(λpir) pEX18Gm::ΔmexR was mixed with PAO1, spotted onto an LB plate, and incubated overnight at 30 °C. Matings were collected and plated on VBMM agar with 60 μg/mL Gm to select for P. aeruginosa merodiploids and incubated overnight at 37 °C, as described [50]. Merodiploid colonies were streaked onto LB agar with 15% sucrose and incubated at 25 °C for 4 days. PAO1 ΔmexR strains were confirmed by PCR amplification using primers mexR-KO-UP-F and mexR-KO-DN-R (Table S1). To generate the PAO1 ΔmexEF strain, pDONRPEX18Gm::ΔmexEF was transformed into the donor E. coli S.17.1 (λpir) strain. Biparental mating was used to introduce the suicide plasmid into the P. aeruginosa PAO1 recipient. Merodiploids resistant to sucrose but sensitive to Gm were isolated by counter-selection on no-salt-LB agar with 15% w/v sucrose after two days of incubation at 30 °C. Colony PCR was performed to identify ΔmexEF mutants and PCR products generated from the cloned mutant allele were sent for sequencing with oRP_27 and oRP_28 primers. Using this protocol, a 4397 bp fragment from the 4434 bp mexEF sequence was removed resulting in a frameshift mutation in the mexEF-oprN multidrug efflux operon, yielding strain RP05 (PAO1 ΔmexEF) (Table S1).
4.4. PAO1 Transformation with rhlA Reporter Plasmid
To quantify the expression of genes involved in rhamnolipid production, strains PAO1, PAO1-AvEvB8, PAO1 ΔmexR, PAO1 ΔmexEF, and PAO1 ΔmexAB were transformed with pYL122, a plasmid containing rhlA-gfp promoter fusion in a mini-CTX-lacZ backbone [55]. Following transformation, strains were maintained on LB plates with 100 μg/mL tetracycline. Clones were then tested using PCR with the primers pYL122-Chk-F and pYL122-Chk-R (Table S1).
4.5. DNA Extraction, Purification, and PCR
Plasmid DNA was prepared using Monarch Plasmid Miniprep Kit (cat# T1010, New England BioLabs, Ipswich, MA, USA). Genomic DNA was prepared using DNeasy Blood & Tissue Kit (cat# 69504, Qiagen, Hilden, Germany). When necessary, cDNA was purified using Monarch PCR & DNA Cleanup Kit (cat# T1030, New England BioLabs, Ipswich, MA, USA). PCR was performed using either KAPA HIFI 2X ready mix (cat# KK2602, KAPA Biosystems, Wilmington, MA, USA).
4.6. Swarming Assay
Swarming plates were created using 2.5% LB and 0.5% agar. For the assay, all strains were grown overnight for 16–20 h in LB broth without antibiotics, including Tn mutants. For each strain, 2 μL of the overnight broth was then placed at the center of a swarming plate and left to incubate at 37 °C for 24 h. The maximum swarming diameter was then measured and recorded for each strain.
4.7. Gradient Diffusion Antibiotic Susceptibility Testing
Aztreonam, ciprofloxacin, and chloramphenicol Etest strips were purchased from bioMérieux (cat# 501758, 412310, 412308, Durham, NC, USA) and antimicrobial susceptibility testing was performed with the following modifications to the manufacturer’s instructions. For aztreonam, a sterile swab was soaked in an overnight culture for each strain after growth for 18 h in LB broth and excess fluid was removed by pressing it against the inside wall of the test tube. For ciprofloxacin and chloramphenicol, colonies were picked directly from LB agar plates and diluted in sterile PBS to OD600 0.15 (~1.5 × 108 CFU/mL). Mueller Hinton agar plates were fully streaked 4 times with the swabs. After allowing the plates to dry, an Etest gradient strip was placed in the middle of the plates. Plates were incubated at 37 °C for 16–20 h.
4.8. rhlA GFP Reporter Assay
WT PAO1, PAO1 attB::rhlA-gfp, PAO1-AzEvB8, PAO1-AzEvB8 attB::rhlA-gfp, ΔmexR, ΔmexR attB::rhlA-gfp, ΔmexEF, ΔmexEF attB::rhlA-gfp, ΔmexAB, and ΔmexAB attB::rhlA-gfp strains were grown overnight for 16–20 h in 2 mL LB. The overnight cultures were then diluted to OD600 ~0.005 (~5 × 106 CFU/mL). Two hundred microliters of the working dilutions were added in triplicate to a sterile black (clear bottom) 96-well plate. To prevent evaporation, 50 μL of mineral oil was added to each well. The plate was incubated for 24 h at 37 °C, shaking at 250 rpm, and absorbance at 600 nm and fluorescence with excitation at 488 nm and emission at 510 nm were read every hour for 24 h. For each strain, fluorescence was first normalized to the absorbance value. The background of each nontransformed strain was then subtracted from its pYL122-carrying counterpart. Gene expression was calculated as area under the curve using Prism GraphPad. For the efflux pump inhibitor assay, the rhlA reporter assay was performed as described above with the exception that the efflux pump inhibitor PAβN (Phe-Arg β-naphthylamide dihydrochloride, Sigma-Aldrich, Burlington, MA, USA, cat# P4157-25MG) was added to cultures to reach final concentrations of 0 μg/mL, 5 μg/mL, 10 μg/mL, and 25 μg/mL.
4.9. Murine Lung Infection
Experiments were approved by the Institutional Animal Care and Use Committee at Cedars-Sinai Medical Center under protocol IACUC008115. Strains were grown to mid-exponential phase, washed with sterile PBS, and diluted to 1 × 108 CFU/mL in sterile PBS. Female C57BL/6 (Jackson Laboratories, Bar Harbor, ME, USA) 12-week-old mice (10 mice/group) were anesthetized using isoflurane. A 24-gauge angiocatheter was used to intubate the mice. Acute lung infections were performed by single intratracheal instillations of 5 × 106 CFUs in 50 μL sterile PBS. During inoculation the mice were placed on a surgical board in the supine position. The front paws of each mouse were stretched outwards. A rubber band was placed around the two front incisors of the mouse to help keep their mouths open during the intubation procedure. After inoculation mice were kept in a clean cage under infrared lamp until full recovery. Mice were evaluated thrice per day to assess morbidity, and moribund mice were sacrificed with inhaled CO2. Surviving mice were euthanized 96 h post-infection. Survival curves were analyzed with log-rank tests as described below.
4.10. Genome Sequencing and Analysis
To verify mutations in engineered P. aeruginosa strains, DNA was isolated from strains PAO1, PAO1 ΔmexAB, PAO1-AzEvB8, PAO1-AzEvB8 ΔmexAB, and PAO1 ΔmexEF and subjected to whole genome sequencing. DNA isolation was performed using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). DNA was submitted to the Microbial Genome Sequencing Center at the University of Pittsburgh where libraries were prepared and sequenced using an Illumina NextSeq platform. Sequencing reads were analyzed using Breseq [56], comparing reads for each strain to the P. aeruginosa PAO1 parent reference genome. Figure showing deletion in the P. aeruginosa PAO1-AzEvB8 strain was created using CLC Genomics Workbench Software.
4.11. Statistics
Statistical analyses were performed using Prism GraphPad Software v9 and SAS. For swarming and rhlA reporter assays, one-way ANOVA tests were performed followed by Tukey’s multiple comparison test. For survival analyses, survival times were tested across strata with a log-rank test, with Bonferroni-adjusted p-values where >2 strata were compared.
5. Conclusions
Altogether, this work sheds important light on how antibiotic-resistance mutations can affect efflux pumps and subsequent P. aeruginosa virulence. Long-term, we believe that new strategies for treating efflux pump overexpression or deletion mutants should be pursued. It will be particularly important to study the effects of efflux pump mutations in the context of biofilms and aggregates, both in vitro and in vivo, because this is the predominant bacterial mode of growth during CF infections. Additionally, we plan to explore how mexR mutations lead to increased virulence, since this mechanism still remains mysterious.
Acknowledgments
We would like to thank members of the Jorth Lab, Joe Harrison, and Holly Huse for helpful discussions and feedback on this manuscript. We are grateful to Pradeep K. Singh, Colin Manoil, and Joe J. Harrison for the generous gifts of strains for this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10101164/s1, Table S1: Strains and primers used in this study.
Author Contributions
Conceptualization, M.V. and P.J.; methodology, M.V. and P.J.; formal analysis, M.V., S.P.L., C.B. and P.J.; investigation, M.V., S.P.L., C.B., R.P. and P.J.; resources, R.P. and P.J.; data curation, M.V., S.P.L., C.B. and P.J.; writing—original draft preparation, M.V., S.P.L., R.P. and P.J.; writing—review and editing, M.V., S.P.L., C.B., R.P. and P.J.; supervision, M.V. and P.J.; project administration, M.V. and P.J.; funding acquisition, P.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grant numbers JORTH17F5 and JORTH19P0 from the Cystic Fibrosis Foundation and grant numbers K22AI127473 and R01AI14642 from the NIH/National Institute of Allergy and Infectious Diseases, as well as a sub-award from grant number UL1TR001881 from the NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Animal Care and Use Committee of Cedars-Sinai Medical Center (protocol code IACUC008115 approved on May 23, 2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome sequencing data are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive through BioProject accession number: PRJNA766087, http://www.ncbi.nlm.nih.gov/bioproject/766087.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Silby M.W., Winstanley C., Godfrey S.A., Levy S.B., Jackson R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 2.Moradali M.F., Ghods S., Rehm B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison A.J., Jr., Wenzel R.P. Epidemiology of infections due to Pseudomonas aeruginosa. Rev. Infect. Dis. 1984;6((Suppl. S3)):S627–S642. doi: 10.1093/clinids/6.Supplement_3.S627. [DOI] [PubMed] [Google Scholar]
- 4.Parkins M.D., Somayaji R., Waters V.J. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clin. Microbiol. Rev. 2018;31:e00019-18. doi: 10.1128/CMR.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain M., Ramirez D., Seshadri R., Cullina J.F., Powers C.A., Schulert G.S., Bar-Meir M., Sullivan C.L., McColley S.A., Hauser A.R. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D’Argenio D.A., Miller S.I., Ramsey B.W., Speert D.P., Moskowitz S.M., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huse H.K., Kwon T., Zlosnik J.E., Speert D.P., Marcotte E.M., Whiteley M. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio. 2010;1:e00199-10. doi: 10.1128/mBio.00199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorth P., Staudinger B.J., Wu X., Hisert K.B., Hayden H., Garudathri J., Harding C.L., Radey M.C., Rezayat A., Bautista G., et al. Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winstanley C., O’Brien S., Brockhurst M.A. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darch S.E., McNally A., Harrison F., Corander J., Barr H.L., Paszkiewicz K., Holden S., Fogarty A., Crusz S.A., Diggle S.P. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci. Rep. 2015;5:7649. doi: 10.1038/srep07649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien S., Williams D., Fothergill J.L., Paterson S., Winstanley C., Brockhurst M.A. High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol. 2017;17:30. doi: 10.1186/s12866-017-0941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faure E., Kwong K., Nguyen D. Pseudomonas aeruginosa in Chronic Lung Infections: How to Adapt within the Host? Front. Immunol. 2018;9:2416. doi: 10.3389/fimmu.2018.02416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen P.O., Bjarnsholt T., Phipps R., Rasmussen T.B., Calum H., Christoffersen L., Moser C., Williams P., Pressler T., Givskov M., et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 14.Zulianello L., Canard C., Kohler T., Caille D., Lacroix J.S., Meda P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 2006;74:3134–3147. doi: 10.1128/IAI.01772-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavoie E.G., Wangdi T., Kazmierczak B.I. Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect. 2011;13:1133–1145. doi: 10.1016/j.micinf.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mogayzel P.J., Jr., Naureckas E.T., Robinson K.A., Brady C., Guill M., Lahiri T., Lubsch L., Matsui J., Oermann C.M., Ratjen F., et al. Cystic Fibrosis Foundation pulmonary guideline. pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann. Am. Thorac Soc. 2014;11:1640–1650. doi: 10.1513/AnnalsATS.201404-166OC. [DOI] [PubMed] [Google Scholar]
- 17.Sobel M.L., Hocquet D., Cao L., Plesiat P., Poole K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole K., Tetro K., Zhao Q., Neshat S., Heinrichs D.E., Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 1996;40:2021–2028. doi: 10.1128/AAC.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito K., Yoneyama H., Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 1999;179:67–72. doi: 10.1111/j.1574-6968.1999.tb08709.x. [DOI] [PubMed] [Google Scholar]
- 20.Jorth P., McLean K., Ratjen A., Secor P.R., Bautista G.E., Ravishankar S., Rezayat A., Garudathri J., Harrison J.J., Harwood R.A., et al. Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa. mBio. 2017;8:e00517-17. doi: 10.1128/mBio.00517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suresh M., Nithya N., Jayasree P.R., Vimal K.P., Manish Kumar P.R. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2018;34:83. doi: 10.1007/s11274-018-2465-0. [DOI] [PubMed] [Google Scholar]
- 22.Horna G., Lopez M., Guerra H., Saenz Y., Ruiz J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018;8:16463. doi: 10.1038/s41598-018-34694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llanes C., Hocquet D., Vogne C., Benali-Baitich D., Neuwirth C., Plesiat P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 2004;48:1797–1802. doi: 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLean K., Lee D., Holmes E.A., Penewit K., Waalkes A., Ren M., Lee S.A., Gasper J., Manoil C., Salipante S.J. Genomic Analysis Identifies Novel Pseudomonas aeruginosa Resistance Genes under Selection during Inhaled Aztreonam Therapy In Vivo. Antimicrob. Agents Chemother. 2019;63:e00866-19. doi: 10.1128/AAC.00866-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshri R.D., Zrihen K.S., Shner I., Omer Bendori S., Eldar A. Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 2018;12:2458–2469. doi: 10.1038/s41396-018-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohler T., van Delden C., Curty L.K., Hamzehpour M.M., Pechere J.C. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomovskaya O., Warren M.S., Lee A., Galazzo J., Fronko R., Lee M., Blais J., Cho D., Chamberland S., Renau T., et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosson P., Zulianello L., Join-Lambert O., Faurisson F., Gebbie L., Benghezal M., van Delden C., Curty L.K., Kohler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirakata Y., Kondo A., Hoshino K., Yano H., Arai K., Hirotani A., Kunishima H., Yamamoto N., Hatta M., Kitagawa M., et al. Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents. 2009;34:343–346. doi: 10.1016/j.ijantimicag.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Rampioni G., Pillai C.R., Longo F., Bondi R., Baldelli V., Messina M., Imperi F., Visca P., Leoni L. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci. Rep. 2017;7:11392. doi: 10.1038/s41598-017-11892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyata S., Casey M., Frank D.W., Ausubel F.M., Drenkard E. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2003;71:2404–2413. doi: 10.1128/IAI.71.5.2404-2413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kropinski A.M., Chadwick J.S. The pathogenicity of rough strains of Pseudomonas aeruginosa for Galleria mellonella. Can. J. Microbiol. 1975;21:2084–2088. doi: 10.1139/m75-297. [DOI] [PubMed] [Google Scholar]
- 33.Marvig R.L., Sommer L.M., Molin S., Johansen H.K. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda H., Hosaka M., Hirai K., Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob. Agents Chemother. 1990;34:1757–1761. doi: 10.1128/AAC.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohler T., Michea-Hamzehpour M., Henze U., Gotoh N., Curty L.K., Pechere J.C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 36.Quale J., Bratu S., Gupta J., Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2006;50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomas M., Doumith M., Warner M., Turton J.F., Beceiro A., Bou G., Livermore D.M., Woodford N. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2010;54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keating C.L., Zuckerman J.B., Singh P.K., McKevitt M., Gurtovaya O., Bresnik M., Marshall B.C., Saiman L. Pseudomonas aeruginosa Susceptibility Patterns and Associated Clinical Outcomes in People with Cystic Fibrosis following Approval of Aztreonam Lysine for Inhalation. Antimicrob. Agents Chemother. 2021;65:e02327-20. doi: 10.1128/AAC.02327-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida K., Nakayama K., Ohtsuka M., Kuru N., Yokomizo Y., Sakamoto A., Takemura M., Hoshino K., Kanda H., Nitanai H., et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: Highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg. Med. Chem. 2007;15:7087–7097. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 40.Roux D., Danilchanka O., Guillard T., Cattoir V., Aschard H., Fu Y., Angoulvant F., Messika J., Ricard J.D., Mekalanos J.J., et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci. Transl. Med. 2015;7:297ra114. doi: 10.1126/scitranslmed.aab1621. [DOI] [PubMed] [Google Scholar]
- 41.Cowley E.S., Kopf S.H., LaRiviere A., Ziebis W., Newman D.K. Pediatric Cystic Fibrosis Sputum Can Be Chemically Dynamic, Anoxic, and Extremely Reduced Due to Hydrogen Sulfide Formation. mBio. 2015;6:e00767-15. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer K.L., Brown S.A., Whiteley M. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 2007;189:4449–4455. doi: 10.1128/JB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer K.L., Aye L.M., Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner K.H., Wessel A.K., Palmer G.C., Murray J.L., Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers C.S., Stoltz D.A., Meyerholz D.K., Ostedgaard L.S., Rokhlina T., Taft P.J., Rogan M.P., Pezzulo A.A., Karp P.H., Itani O.A., et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Starke J.R., Edwards M.S., Langston C., Baker C.J. A mouse model of chronic pulmonary infection with Pseudomonas aeruginosa and Pseudomonas cepacia. Pediatr. Res. 1987;22:698–702. doi: 10.1203/00006450-198712000-00017. [DOI] [PubMed] [Google Scholar]
- 47.Van Heeckeren A.M., Schluchter M.D. Murine models of chronic Pseudomonas aeruginosa lung infection. Lab. Anim. 2002;36:291–312. doi: 10.1258/002367702320162405. [DOI] [PubMed] [Google Scholar]
- 48.Van Heeckeren A.M., Schluchter M.D., Drumm M.L., Davis P.B. Role of Cftr genotype in the response to chronic Pseudomonas aeruginosa lung infection in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;287:L944–L952. doi: 10.1152/ajplung.00387.2003. [DOI] [PubMed] [Google Scholar]
- 49.Held K., Ramage E., Jacobs M., Gallagher L., Manoil C. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 2012;194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hmelo L.R., Borlee B.R., Almblad H., Love M.E., Randall T.E., Tseng B.S., Lin C., Irie Y., Storek K.M., Yang J.J., et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoang T.T., Karkhoff-Schweizer R.R., Kutchma A.J., Schweizer H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 52.Taylor R.G., Walker D.C., McInnes R.R.E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993;21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R., Priefer U., Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 54.Chung C.T., Niemela S.L., Miller R.H. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lequette Y., Greenberg E.P. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deatherage D.E., Barrick J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequencing data are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive through BioProject accession number: PRJNA766087, http://www.ncbi.nlm.nih.gov/bioproject/766087.