Abstract
The overuse of antibiotics and the scarcity of new drugs have led to a serious antimicrobial resistance crisis, especially for multi-drug resistant (MDR) Gram-negative bacteria. In the present study, we investigated the antimicrobial activity of a marine antibiotic equisetin in combination with colistin against Gram-negative bacteria and explored the mechanisms of synergistic activity. We tested the synergistic effect of equisetin in combination with colistin on 23 clinical mcr-1 positive isolates and found that 4 µg/mL equisetin combined with 1 µg/mL colistin showed 100% inhibition. Consistently, equisetin restored the sensitivity of 10 species of mcr-1 positive Gram-negative bacteria to colistin. The combination of equisetin and colistin quickly killed 99.9% bacteria in one hour in time-kill assays. We found that colistin promoted intracellular accumulation of equisetin in colistin-resistant E. coli based on LC-MS/MS analysis. Interestingly, equisetin boosted ROS accumulation in E. coli in the presence of colistin. Moreover, we found that equisetin and colistin lost the synergistic effect in two LPS-deficient A. baumannii strains. These findings suggest that colistin destroys the hydrophobic barrier of Gram-negative bacteria, facilitating equisetin to enter the cell and exert its antibacterial effect. Lastly, equisetin restored the activity of colistin in a G. mellonella larvae infection model. Collectively, these results reveal that equisetin can potentiate colistin activity against MDR Gram-negative bacteria including colistin-resistant strains, providing an alternative approach to address Gram-negative pathogens associated with infections in clinics.
Keywords: equisetin, colistin, Gram-negative bacteria, drug combination, antimicrobial resistance
1. Introduction
The alarming emergence and dissemination of antimicrobial resistance constitute a crisis to One Health. It is predicted that antimicrobial resistance will lead to the death of 10 million people/year by 2050 [1]. The overuse and misuse of antibiotics and the scarcity of new drugs result in the migration of multidrug-resistant (MDR) bacteria from hospitals to the community. In 2017, the World Health Organization (WHO) introduced a list of “critical priority” pathogens—including the “ESKAPE” pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species)—responsible for a global health problem [2]. Compared to Gram-positive bacteria, Gram-negative bacteria (Escherichia coli, K. pneumoniae, P. aeruginosa and A. baumannii) are among the primary infectious agents, owing to their dual-membrane envelope, which prevent many antibiotics from accessing targets. Therefore, future development strategies should pay more attention to antibiotics that are active against MDR Gram-negative bacteria. There was almost no novel compounds discovered that were active against Gram-negative bacteria in the past fifty years [3]. Thus, it is imperative to explore novel antibiotics or other approaches against MDR Gram-negative bacteria-associated infections. Otherwise, we will probably experience a pandemic that is similar to the current COVID-19 crisis [4,5].
The discovery of antibiotics against Gram-negative bacteria has been much slower due to the presence of an intrinsic outer membrane (OM) [6]. The OM is an asymmetric lipid bilayer consisting of an outer leaflet of lipopolysaccharide (LPS) and an inner leaflet of phospholipids. The LPS prevents an influx of large or hydrophobic antibiotics, such as vancomycin and erythromycin, restricting their access to intracellular targets [7]. Polymyxins are regarded as the last-resort antibiotics to treat MDR Gram-negative pathogens [8] and play a critical role in the era of antibiotic resistance [9], but the serious nephrotoxicity and neurotoxicity also limit their clinical use. The revival of colistin in recent years is largely due to the lack of novel antibiotics that are effective against the increasingly prevalent MDR Gram-negative infections, particularly the carbapenem-resistant Enterobacteriaceae. However, bacteria harboring an mcr-1 plasmid-borne colistin resistance contribute to the rapid dissemination and ubiquitous distribution of colistin resistance [10,11]. Lacking the discovery of new drugs, the utilization of currently available drugs or the expansion of the antibacterial spectra would contribute to our arsenal against MDR Gram-negative bacteria [12]. For example, the development of advanced drug-delivery systems for antibacterial therapies [13] and the discovery of novel antibiotic adjuvants are becoming promising strategies to revitalize the existing antibiotics [14].
We recently found that a marine fungal symbiont efficiently produces equisetin with broad-spectrum antibacterial activities against MDR Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE) [15]. However, equisetin alone is ineffective against Gram-negative bacteria, probably due to the presence of OM barriers. Interestingly, equisetin as an adjuvant restores colistin sensitivity in colistin-resistant Gram-negative pathogens although the underlying mechanism remains unclear. In this study, we found that low levels of colistin combined with equisetin was effective against the 10 species of colistin-resistant Gram-negative bacteria that were tested, which, in turn, reduced the therapeutic toxicity of colistin. The combination of equisetin and colistin boosts the accumulation of ROS in E. coli. These results indicate that colistin destroys the membrane structure of Gram-negative bacteria, facilitating equisetin to enter the cell and exert the antibacterial effect.
2. Results
2.1. Equisetin Synergies with Colistin against MDR Gram-Negative Bacteria
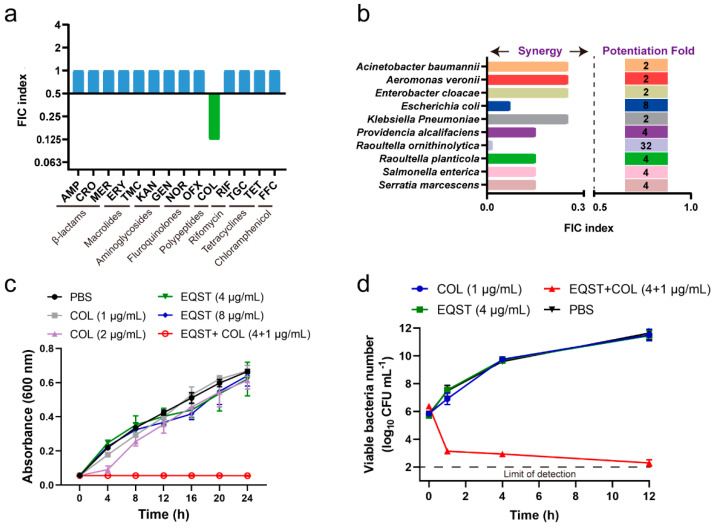
To expand the antibacterial spectrum of equisetin and to improve the efficiency of currently available antibiotics, we systematically assessed the synergistic activity between equisetin and other major classes of antibiotics against E. coli B2 (NDM-5 + MCR-1). We found that equisetin had no synergistic activity with other antibiotics except colistin, as was previously reported [15] (Figure 1a, Table S1). Then, we focused on the synergistic effect of equisetin and colistin against colistin-resistant bacteria. We tested the synergistic antibacterial effect of equisetin and colistin on 23 clinical mcr-1 positive resistant isolates at different concentrations and found that equisetin (4 µg/mL) combined with colistin (1 µg/mL) inhibited 100% of the tested bacteria (Table 1 and Table S1). Consistently, equisetin restored colistin sensitivity in 10 species of mcr-1 positive Gram-negative bacteria in the presence of equisetin (4 µg/mL) plus colistin (1 µg/mL) (Figure 1b, FIC index ≤ 0.5), which is consistent with our previous report [15]. Neither equisetin alone (4 µg/mL), nor sublethal levels of colistin (1 µg/mL, 1/8 MIC), affected the growth of E. coli (Figure 1c) but their combination sharply inhibited the growth of E. coli. Furthermore, the time-killing curve showed that equisetin combined with colistin rapidly killed more than 99% of E. coli within 1 h (Figure 1d).
Figure 1.
Equisetin restores colistin sensitivity against MDR Gram-negative bacteria. (a) The FIC indexes of antibiotics combined with equisetin against E. coli B2. Synergy is defined as an FIC of ≤0.5. AMP: ampicillin, CRO: ceftriaxone; MER: meropenem, ERY: erythromycin, TMC: tilmicosin, KAN: kanamycin, GEN: gentamycin, OFX: ofloxacin, NOR: norfloxacin, COL: colistin, RIF: rifampicin, TGC: tigecycline, TET: tetracyclines, FFC: florfenicol. (b) Synergistic antibacterial activities between equisetin and colistin against 10 species of mcr-1 positive Gram-negative bacteria. FIC index ≤ 0.5, synergistic. (c) Growth curve of E. coli B2 after treatment with different combinations of equisetin and colistin. EQST: equisetin. (d) Time-dependent killing of E. coli B2 treated with equisetin combined with colistin. Data are representative of three independent experiments, mean ± s.d.
Table 1.
The activity of equisetin combined with colistin against 23 mcr-1 positive isolates.
| Equisetin + Colistin (µg/mL) |
Inhibition Ratio |
|---|---|
| 1 + 1 | 73.91% (17/23) |
| 2 + 0.5 | 82.60% (19/23) |
| 2 + 1 | 91.30% (21/23) |
| 4 + 0.5 | 95.65% (22/23) |
| 4 + 1 | 100.00% (23/23) |
2.2. Mechanism of Equisetin Combined with Colistin against Bacteria
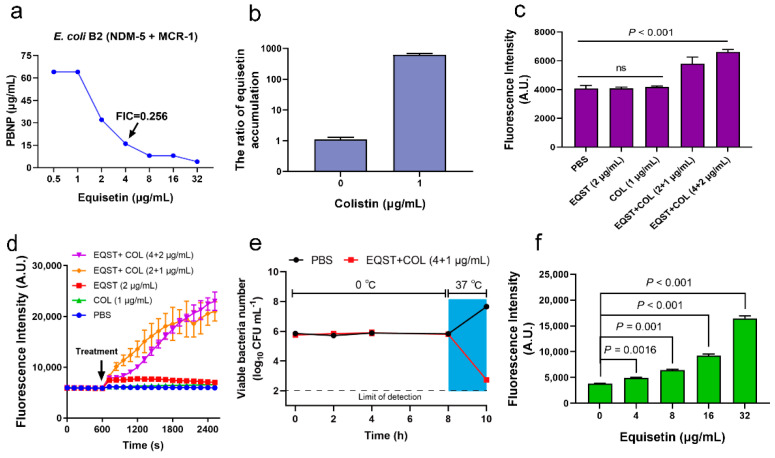
Colistin is thought to require the insertion of the fatty-acyl chain into the OM to kill the bacteria [16]. Interestingly, the synergistic antibacterial activity can also be achieved by PBNP (Figure 2a), which lacks the fatty-acyl moiety found on colistin and is therefore incapable of inducing lysis [17]. Deacylated PMBN can sufficiently interact with and disrupt the OM, which is largely irrespective of the antibacterial mechanism [18]. Additionally, the OM disruption of bacteria and potentiation of antibiotics by PBNP and colistin are unaffected by the mcr-1 expression [17,18]. Accordingly, we speculated that the role of colistin in drug combination is based on the destruction of the membrane. Indeed, colistin can significantly enhance the amount of other antibiotics entering the cell and exerting antibacterial synergy [19,20,21]. We used an optimized LC-MS/MS detection method to determine the intracellular accumulation of equisetin when combined with colistin or equisetin alone and found that colistin can significantly increase the accumulation of equisetin in the cell (Figure 2b). The result may indicate that equisetin enters the cytoplasm to exert its antibacterial function. Next, we used the hydrophobic fluorescent probe 1-N-phenylnaphthylamine (NPN) to evaluate the damaging effects of equisetin combined with colistin or equisetin and colistin alone on the OM of E. coli B2. As shown in Figure 3c, we found that equisetin combined with colistin showed significant damaging effects to the OM and that equisetin or colistin alone cannot damage the OM. In addition to the bacterial OM, polymyxin also plays an important role in the permeability of the bacterial cytoplasmic membrane (CM) [22]. Therefore, we used propidium iodide (PI) mediated fluorescent dyes to test the effect of the drugs on the permeability of the inner membrane. As expected, equisetin plus colistin exposure resulted in a strong PI signal from E. coli B2 cells (Figure 2d), indicative of the damaged CM causing the PI to fluoresce upon contact with DNA in the bacterial cytoplasm. Taken together, equisetin combined with colistin destroy the OM and CM of the bacteria. Additionally, equisetin also inhibited transcription of mcr-1, but not in a dose-dependent manner (Figure S2).
Figure 2.
Mechanism of equisetin in combination with colistin against bacteria. (a) Synergistic antibacterial activities between equisetin and PBNP against E. coli B2. (b) Colistin promotes the accumulation of equisetin in bacterial cells. (c) Increased permeability of the outer membrane of E. coli B2 treated with drug combination, probed with NPN. (d) Increased permeability of the cytoplasmic membrane of E. coli B2 treated with drug combination, probed with PI. (e) The synergistic activity of equisetin and colistin is dependent on bacterial metabolic states. E. coli B2 cells at the exponential phase were incubated in the presence of EQST + COL (4 + 1 µg/mL) in ice-water mixtures (0 °C, 0–8 h) and culture tubes at 37 °C (8–10 h). (f) In the presence of 1 µg/mL colistin, the higher the concentration of equisetin, the more ROS accumulated in E. coli. Data are representative of three independent experiments and the mean of three biological replicates is shown. Error bars represent the s.d. and p values were determined using an unpaired, two-tailed Student’s t-test.
Figure 3.
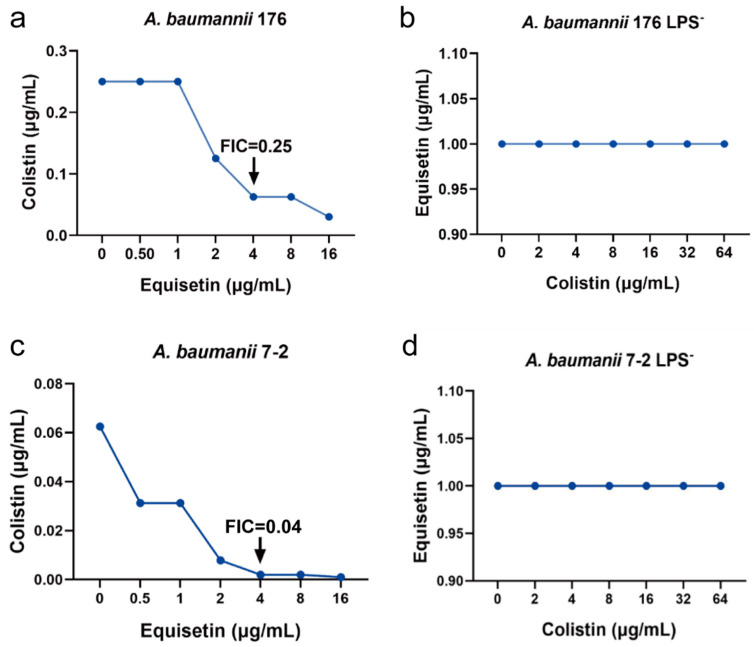
FIC for parent and LPS-deficient A. baumannii strains: FIC for wild type A. baumannii 176 (a) and A. baumannii 176 LPS− (b); wild-type A. baumannii 7-2 (c) and A. baumannii 7-2 LPS− (d).
Since the bactericidal activity was rapid, the mechanistic basis of equisetin was considered to be associated with bacterial propagation. Thus, we evaluated whether the bacterial metabolism affects the antibacterial activity of equisetin combined with colistin and found that the combination rapidly eliminated metabolically active bacteria in 2 h (37 °C) but lost its antibacterial activity against bacteria in metabolically suppressed states (0 °C, Figure 2e). According to the rapid sterilization rate of the drug combination, we speculated that the bacteria accumulated a large number of toxic substances in a short time. Hence, we detected the content of reactive oxygen species (ROS) in bacterial cells after drug treatments. Indeed, in low levels of colistin, the accumulation of ROS in cells gradually increased with the addition of equisetin (Figure 2f).
2.3. FIC for Parent and LPS-Deficient A. baumannii Strains
To further explore the synergistic mechanism of equisetin and colistin, we studied two LPS-deficient A. baumannii strains that are inactivated in the lipid A biosynthesis [23,24]. We first determined the MIC of equisetin and colistin on wild-type and LPS-deficient A. baumannii isolates and found that colistin resistance increased more than 100-fold, whereas equisetin resistance decreased more than 100-fold relative to the wild-type parent A. baumannii (Table 2). Our results demonstrated that LPS deficiency of strains fostered resistance to colistin but was sensitive to equisetin, which is exclusively active against Gram-positive bacteria. Furthermore, equisetin lacked synergy with colistin on LPS-deficient A. baumannii, contrary to the wild type (Figure 3). These findings denote that outer-membrane LPS are the major obstacle to equisetin-dependent killing in A. baumannii.
Table 2.
MICs of parent and LPS-deficient A. baumannii strains.
| A. baumannii Strain | Colistin (µg/mL) | Equisetin (µg/mL) |
|---|---|---|
| 176 (wild type) | 0.25 | >128 |
| 176 (mutant lacking LPS) | >128 | 1 |
| 7-2 (wild type) | 0.125 | >128 |
| 7-2 (mutant lacking LPS) | >128 | 1 |
2.4. In Vivo Efficacy of Equisetin Combined with Colistin
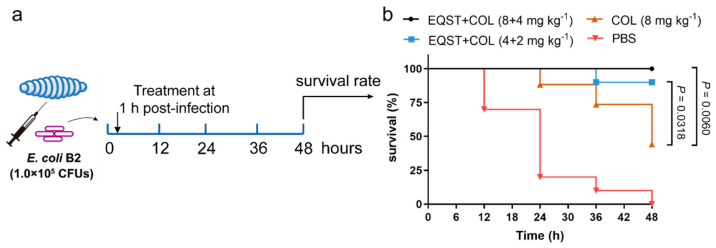
The encouraging antibacterial activity of equisetin combined with colistin in vitro inspired us to further investigate their therapeutic potential in vivo. We constructed a resistant bacteria infection model of E. coli B2 in G. mellonella larvae, as was previously reported [19]. After 1 h post-infection, equisetin combined with colistin was supplied with different dose gradients (Figure 4a). The experimental results showed that equisetin combined with colistin at 8 mg kg−1 + 4 mg kg−1 fully protected G. mellonella from infection, which suggests that equisetin combined with colistin is a promising solution to treat MDR-associated infections in vivo (Figure 4b).
Figure 4.
Efficacy of equisetin combined with colistin in G. mellonella infection model. (a) Scheme of the experimental protocol for the G. mellonella infection model; (b) survival rates of G. mellonella larva infected with E. coli B2 (1.0 × 105 CFUs) in the presence of equisetin combined with colistin at 8 mg kg−1 + 4 mg kg−1 and 4 mg kg−1 + 2 mg kg−1 (n = 10). Colistin at 8 mg kg−1 was used as a positive control. p values were determined using a two-sided, Mann–Whitney U-test.
3. Discussion
We are in a “post-antibiotic era” and there are few clinical antibacterial candidates, especially for Gram-negative pathogens [25]. The OM of Gram-negative bacteria is an effective barrier in the outer leaflet that protects bacteria from toxic environmental insults, including antibiotics. Given the long development duration of antibacterial drugs and the high probability of failure, we must preserve the existing drugs. Combination therapy with existing drugs is a key to combat MDR infections and extend their shelf lives [26]. Potentiators that restore the activity of current antibiotics can expand the antibacterial spectrum as well. For example, the inhibitors of β-lactamases and sulfonamide synergists overcome a number of pathogens [27]. Notably, colistin disrupts the OM of Gram-negative bacteria to permit other compounds access to their cytoplasmic targets [28]. Thus, combinations of colistin with hydrophobic antibiotics [29] or adjuvants [30] are often used in the treatment of MDR Gram-negative infections. Colistin binding to the negatively charged lipid A phosphate groups of an LPS component causes membrane destabilization, leading to the increase of cell envelope permeability, leakage of cellular contents and ultimately cell death [18]. Despite that the medication of phosphate groups decreases the therapeutic utility of colistin, colistin still can paralyze the integrity of colistin-resistant bacteria [31,32]. For instance, colistin in combination with rifampicin [33], macrolides [34], tigecycline [35], carbapenems and glycopeptides [36] achieves promising therapeutic outcomes. Indeed, although hydrophobic antibiotics are conventionally active against Gram-positive bacteria such as rifampicin and macrolides, they have shown favorable prospects against Gram-negative pathogens in the presence of colistin or other Gram-negative OM disrupting compounds [37].
Compared to natural antibiotics of terrestrial origins, marine organisms that survive in high salinity, high pressure, low temperature and oligotrophic environments become appealing for the discovery of new compounds. Marine fungi derived from sponges are outstanding sources of natural products with various biological activities [38]. Equisetin isolated from a marine sponge-associated fungal strain Fusarium can kill diverse MDR Gram-positive bacteria without cross-resistance and detectable resistance [15,39]. In addition to effective antimicrobial effects, equisetin also has activity against HIV-1 integrasel [40] and mitochondria [41]. In combination with colistin, equisetin displays antibacterial activity against Gram-negative pathogens including colistin-resistant bacteria, enhancing colistin activity substantially and reducing its cytotoxicity accordingly.
Interestingly, the rapid bactericidal ability of equisetin combined with colistin against E. coli (Figure 1d) is similar to the equisetin-mediated killing curve of S. aureus [15]. Moreover, the result that LPS-deficient Gram-negative bacteria are sensitive to equisetin indicates that equisetin may target the same component in both Gram-positive and Gram-negative bacteria. Previous studies suggest that the formation of ROS is a common mechanism of bacterial death induced by bactericidal antibiotics [42,43,44]. Accumulated ROS break DNA, peroxidate lipids, carbonylate proteins and so on [45]. Correspondingly, ROS-mediated damages accelerate ROS accumulation, leading to metabolic perturbation and cell death [46]. Equisetin shows an ROS-independent bactericidal mechanism (data unpublished). Thus, we speculate that colistin facilitates the entry of equisetin to the MDR Gram-negative bacteria, while colistin promotes intracellular ROS accumulation that contributes to rapid bacterial killing. Additionally, combination antibiotic therapy is frequently used to treat severe Gram-negative infections in clinics [47]. Equisetin combined with colistin is highly effective in eradicating MDR Gram-negative bacteria in G. mellonella infection models. Altogether, these findings suggest that equisetin exhibits a distinct mode of action against bacteria and possesses promising pharmacokinetic properties.
In summary, our study shows that equisetin acts synergistically with colistin to kill various MDR Gram-negative bacteria (including colistin-resistant strains) and restores the efficacy of colistin in vivo. We find drug combinations alter the permeability of the OM and CM of Gram-negative bacteria, permitting the increased accumulation of intracellular equisetin to exert antibacterial activity. Collectively, we demonstrate that equisetin combined with colistin represents an attractive strategy for the treatment of infections caused by MDR Gram-negative pathogens.
4. Materials and Methods
4.1. Bacterial Isolates
The standard strains in this study were purchased from China General Microbial Collection and Management Center (CGMCC) and the other clinically resistant strains were isolated by our laboratory, including E. coli B2 and 23 mcr-1 positive bacteria [19].
4.2. Drugs
Ampicillin, ceftriaxone, meropenem, erythromycin, tilmicosin, kanamycin, gentamycin, ofloxacin, norfloxacin, rifampicin, tigecycline, tetracyclines, florfenicol and colistin were purchased from the China Institute of Veterinary Drugs Control (IVDC). Equisetin was isolated from the fungus Fusarium equiseti 33-10 [15].
4.3. Antibacterial Test
The minimum inhibitory concentrations (MICs) of equisetin and colistin were determined by the standard broth microdilution method, according to the CLSI 2021 guideline. Briefly, equisetin or colistin were mixed with equal volumes of bacterial suspensions in Mueller–Hinton broth (MHB) containing approximately 1.5 × 106 colony-forming units (CFUs)/mL in a clear UV-sterilized 96-well microtiter plate. After 16–20 h incubation at 37 °C, the MIC values were defined as the lowest concentrations of antibiotics with no visible growth of bacteria.
4.4. FIC Index Determination
FIC index (FICI) was determined using chequerboard assays. Colistin was diluted along the ordinate, while equisetin was diluted along the abscissa by MHB in a 96-well microtiter plate in a final volume of 100 µL. Then, bacterial suspensions were diluted by approximately 1.5 × 106 CFUs/mL in MHB and 100 µL were added to 96-well microtiter plates containing antibiotics. After 16–20 h of incubation at 37 °C, the FICI was calculated according to the following formula:
| FIC index = MICAB/MICA + MICBA/MICB = FICA + FICB |
MICA is the MIC of compound A alone, MICAB is the MIC of compound A in combination with compound B, MICB is the MIC of compound B alone, MICBA is the MIC of compound B in combination with compound A; FICA is the FIC of compound A and FICB is the FIC of compound B. Synergy is defined as an FIC index ≤ 0.5.
4.5. Growth Curves of Bacteria
The overnight cultures of E. coli B2 were standardized to match a 0.5 McFarland turbidity standard and followed by 1:100 dilution in MHB. Different concentrations of equisetin, colistin or the combination of equisetin with colistin were added to a 96-well microplate and mixed with an equal volume of culture diluted 1:100 in MHB with the plate lid covered. The growth curves were recorded under the wavelength of 600 nm with an interval of 4 h at 37 °C, using an Infinite M200 Microplate reader (Tecan) in the long-term model. PBS was used as a negative control in the experiment.
4.6. Time-Kill Assays
E. coli B2 was cultured to the exponential phase at 37 °C with shaking at 200 revolutions per minute (rpm) for 2 h and then diluted in MHB to a desired concentration at 106 to 107 CFU mL−1. Then, bacteria were treated by equisetin, colistin or the combination of equisetin with colistin in culture tubes at 37 °C with shaking at 200 rpm. Subsequently, 10-fold serially diluted suspensions were plated on Tryptose Soya Agar (TSA) plates and incubated at 37 °C overnight for enumeration of bacterial colonies. PBS was used as a negative control in the experiment.
4.7. Antibiotic Accumulation Analysis
The accumulation of equisetin in E. coli B2 was determined based on LC–MS/MS analysis. Mobile phase A: 0.1% formic acid aqueous solution; mobile phase B: 0.1% formic acid methanol solution; chromatographic column: C18 column (2.1 × 100 mm, 1.7 μm); flow rate is 0.3 mL/min; gradient elution ratio is as follows: 0.1–1.0 min, 60% A; 1.0–3.0 min, 95% A; 3.0–5.0 min, 95% A; 6.0–8.0 min, 60% A; injection volume is 2 µL. Multiple reaction monitoring (MRM) in the positive ion mode was used to quantify the content of intracellular equisetin (Table S3, Figure S1).
4.8. Outer Membrane Permeability Assay
The fluorescent probe NPN was used to evaluate the outer membrane integrity of E. coli B2 when treated by different concentrations of equisetin combined with colistin or with drugs alone. Briefly, E. coli B2 grown overnight at 37 °C were washed and resuspended in 5 mmol/L of HEPES (pH 7.2, +5 mmol/L of glucose) to obtain an OD600 of 0.5. NPN was added to a final concentration of 10 µmol/L and the mixture was incubated at 37 °C for 30 min. A total of 190 µL of probe-labelled bacterial cells were added to a 96-well plate and then 10 µL of the drug combination or the drug alone was added. After incubating for 30 min, fluorescence was measured on an Infinite M200 Microplate reader (Tecan) with the excitation wavelength at 350 nm and the emission wavelength at 420 nm.
4.9. Cytoplasmic Membrane Permeability Assay
E. coli B2 grown overnight at 37 °C were washed and resuspended in 0.01 mol/L of PBS (pH 7.4) to obtain an OD600 of 0.5. PI was added to a final concentration of 10 µmol/L and the mixture was incubated at 37 °C for 10 min. Then, after being treated by the drug combination or drug alone, fluorescence was measured with the excitation wavelength at 535 nm and emission wavelength at 615 nm.
4.10. ROS Measurement
The levels of ROS in E. coli B2, when treated by different concentrations of equisetin combined with colistin, were measured with 10 µmol/L of 2′,7′-dichlorofluorescein diacetate (DCFH-DA), following the manufacturer’s instructions (Beyotime, catalogue No. S0033). Briefly, E. coli B2 grown overnight at 37 °C were washed and resuspended in 0.01 mol/L of PBS (pH 7.4) to obtain an OD600 of 0.5. DCFH-DA was added to a final concentration of 10 µmol/L and the mixture was incubated at 37 °C for 30 min. After washing with 0.01 mol/L of PBS three times, 190 µL of probe-labelled bacterial cells were added to a 96-well plate before adding 10 µL of 1 µg/mL of colistin combined with different concentrations of equisetin. After incubating for another 30 min, fluorescence intensity was immediately measured with the excitation wavelength at 488 nm and the emission wavelength at 525 nm using the Infinite M200 Microplate reader (Tecan).
4.11. Reverse Transcription-(RT)PCR Analysis
E. coli B2 was grown overnight at 37 °C and treated with equisetin (0–64 µg/mL). Total RNA was extracted using the EASYspin Plus kit (Aidlab, catalogue no. RN0801) and quantified by the ratio of absorbance (260 nm:280 nm) using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, American). Before the complementary DNA synthesis, the RNA from all bacterial cells was adjusted to an identical concentration of 1 µg and a reverse transcription of extracted RNA was performed using the PrimeScript RT reagent Kit (Takara, catalogue no. RR047A), following the manufacturer’s protocol.
The levels of mcr-1 in treated E. coli groups were determined by RT-PCR relative to the 16S rRNA mRNA levels of the control gene. According to the previously reported primers for 16S rRNA and mcr-1 [35], RT-PCR was performed with SYBR Green qPCR kit (TaKaRa). Thermal cycling was performed by a standard two-step PCR amplification method, 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The ABI quantstudio TM 7 detection system (Applied Biosystem) for RT-PCR detection and a 2−ΔΔCt method was used to determine the fold change of gene expression.
4.12. G. mellonella Infection Model
The synergy between equisetin and colistin was accessed in the G. mellonella infection model. The larvae of G. mellonella (purchased from Tianjin Huiyude Biotech Company) were randomly divided into four groups (n = 10 per group) and infected with 10 µL of E. coli B2 suspension (1.0 × 105 CFUs) at the right posterior gastropoda. After 1 h post-infection, G. mellonella was treated with PBS, colistin (8 mg kg−1) or the combination of equisetin with colistin (8 mg kg−1 + 4 mg kg−1 and 4 mg kg−1 + 2 mg kg−1) at the left posterior gastropoda. Survival rates of G. mellonella were recorded over 48 h. PBS was used as negative control and colistin at 8 mg kg−1 was used as a positive control in the experiment.
5. Conclusions
In conclusion, these results demonstrate that equisetin combined with colistin shows great efficacy for treating MDR Gram-negative bacteria infections in vitro and in vivo, which suggests that drug combination holds promise for overcoming antibacterial resistance. Therefore, given the fact that development of new antibiotics declines rapidly, drug combination can be a promising approach to extend the life of the already existing antibacterial drug arsenal.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10101263/s1, Table S1: The FICI of antibiotics combined with equisetin against E. coli B2, Table S2. Resistant isolates used in this study. Table S3: MRM parameters for the determination of equisetin by LC-MS/MS, Figure S1: Precursor ion mass spectrum of equisetin standard solutions (1 μg/mL). Figure S2: Equisetin inhibits transcription of mcr-1 determined by reverse transcription (RT)-PCR.
Author Contributions
Conceptualization, W.L. and K.Z.; methodology, Q.Z.; software, Q.Z. and S.C.; validation, Q.Z., S.C. and X.L.; formal analysis, Q.Z.; investigation, Q.Z. and S.C; resources, S.C. and W.L.; data curation, Q.Z.; writing—original draft preparation, Q.Z.; writing—review and editing, Q.Z. and S.C.; visualization, Q.Z.; supervision, K.Z.; project administration, K.Z.; funding acquisition, K.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31922083 and 21861142006), Chinese Universities Scientific Fund (2021TC058) and the Fund of Beijing Dairy Industry Innovation Team.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.O’Neill J. “Tackling Drug-Resistant Infections Globally: Final Report and Recommendations” (Wellcome Trust, 2016) 2014. [(accessed on 1 May 2016)]. Available online: https://wellcomecollection.org/works/thvwsuba.
- 2.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lan. Inf. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. The Science of Antibiotic Discovery. Cell. 2020;181:29–45. doi: 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 4.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhouma M., Tessier M., Aenishaenslin C., Sanders P., Carabin H. Should the Increased Awareness of the One Health Approach Brought by the COVID-19 Pandemic Be Used to Further Tackle the Challenge of Antimicrobial Resistance? Antibiotics. 2021;10:464. doi: 10.3390/antibiotics10040464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart E.M., Mitchell A.M., Konovalova A., Grabowicz M., Sheng J., Han X., Rodriguez-Rivera F.P., Schwaid A.G., Malinverni J.C., Balibar C.J., et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. USA. 2019;116:21748–21757. doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnamoorthy G., Leus I.V., Weeks J.W., Wolloscheck D., Rybenkov V.V., Zgurskaya H.I. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio. 2017;8:e01172-17. doi: 10.1128/mBio.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung E.S., Ko K.S. Eradication of persister cells of Acinetobacter baumannii through combination of colistin and amikacin antibiotics. J. Antimicrob. Chemother. 2019;74:1277–1283. doi: 10.1093/jac/dkz034. [DOI] [PubMed] [Google Scholar]
- 9.El-Sayed Ahmed M.A.E., Zhong L.L., Shen C., Yang Y., Doi Y., Tian G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019) Emerg. Microbes Infect. 2020;9:868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lan. Inf. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y., Zhou H., Xu J., Wang Y., Zhang Q., Walsh T.R., Shao B., Wu C., Hu Y., Yang L., et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 2018;3:1054–1062. doi: 10.1038/s41564-018-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Li R., Xiao X., Wang Z. Antibiotic adjuvants: An alternative approach to overcome multi-drug resistant Gram-negative bacteria. Crit. Rev. Microbiol. 2019;45:301–314. doi: 10.1080/1040841X.2019.1599813. [DOI] [PubMed] [Google Scholar]
- 13.Hou X., Zhang X., Zhao W., Zeng C., Deng B., McComb D.W., Du S., Zhang C., Li W., Dong Y. Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 2020;15:41–46. doi: 10.1038/s41565-019-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyers M., Wright G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21 st century. Nat. Rev. Microbiol. 2019;17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Liu D., Zhang Q., Guo P., Ding S., Shen J., Zhu K., Lin W. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS. Infect. Dis. 2021;7:884–893. doi: 10.1021/acsinfecdis.0c00913. [DOI] [PubMed] [Google Scholar]
- 16.Yu Z., Qin W., Lin J., Fang S., Qiu J. Antibacterial mechanisms of polymyxin and bacterial resistance. BioMed Res. Int. 2015;2015:679109. doi: 10.1155/2015/679109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stokes J.M., MacNair C.R., Ilyas B., French S., Cote J.-P., Bouwman C., Farha M.A., Sieron A.O., Whitfield C., Coombes B.K., et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017;2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacNair C.R., Stokes J.M., Carfrae L.A., Fiebig-Comyn A.A., Coombes B.K., Mulvey M.R., Brown E.D. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018;9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song M., Liu Y., Huang X., Ding S., Wang Y., Shen J., Zhu K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020;5:1040–1050. doi: 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
- 20.Dong X., Chen F., Zhang Y., Liu H., Liu Y., Ma L. In vitro activities of rifampin, colistin, sulbactam and tigecycline tested alone and in combination against extensively drug-resistant Acinetobacter baumannii. J. Antibiot. 2014;67:677–680. doi: 10.1038/ja.2014.99. [DOI] [PubMed] [Google Scholar]
- 21.Thummeepak R., Kitti T., Kunthalert D., Sitthisak S. Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage OABP-01 endolysin (LysABP-01) in combination with colistin. Front. Microbiol. 2016;7:1402. doi: 10.3389/fmicb.2016.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabnis A., Hagart K.L., Klockner A., Becce M., Evans L.E., Furniss R.C.D., Mavridou D.A., Murphy R., Stevens M.M., Davies J.C., et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife. 2021;10:e65836. doi: 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boll J.M., Crofts A.A., Peters K., Cattoir V., Vollmer W., Davies B.W., Trent M.S. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA. 2016;113:E6228–E6237. doi: 10.1073/pnas.1611594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song M., Liu Y., Li T., Liu X., Hao Z., Ding S., Panichayupakaranant P., Zhu K., Shen J. Plant natural flavonoids sgainst multidrug resistant pathogens. Adv. Sci. 2021;8:e2100749. doi: 10.1002/advs.202100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theuretzbacher U., Outterson K., Engel A., Karlen A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2019;18:275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright G.D. Antibiotic Adjuvants: Rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Worthington R.J., Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chai W.C., Whittall J.J., Song D., Polyak S.W., Ogunniyi A.D., Wang Y., Bi F., Ma S., Semple S.J., Venter H. Antimicrobial action and reversal of resistance in MRSA by difluorobenzamide derivatives targeted at FtsZ. Antibiotics. 2020;9:873. doi: 10.3390/antibiotics9120873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrosillo N., Ioannidou E., Falagas M.E. Colistin monotherapy vs. combination therapy: Evidence from microbiological, animal and clinical studies. Clin. Microbiol. Infect. 2008;14:816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 30.Annunziato G., Spadini C., Franko N., Storici P., Demitri N., Pieroni M., Flisi S., Rosati L., Iannarelli M., Marchetti M., et al. Investigational studies on a hit compound cyclopropane-carboxylic acid derivative targeting O-Acetylserine sulfhydrylase as a colistin adjuvant. ACS Infect. Dis. 2021;7:281–292. doi: 10.1021/acsinfecdis.0c00378. [DOI] [PubMed] [Google Scholar]
- 31.Korycka-Machala M., Ziolkowski A., Rumijowska-Galewicz A., Lisowska K., Sedlaczek L. Polycations increase the permeability of Mycobacterium vaccae cell envelopes to hydrophobic compounds. Microbiology. 2001;147:2769–2781. doi: 10.1099/00221287-147-10-2769. [DOI] [PubMed] [Google Scholar]
- 32.Klinger-Strobel M., Stein C., Forstner C., Makarewicz O., Pletz M.W. Effects of colistin on biofilm matrices of Escherichia coli and Staphylococcus aureus. Int. J. Antimicrob. Agents. 2017;49:472–479. doi: 10.1016/j.ijantimicag.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y., Liu B., Wang T., Cai Y., Liang B., Wang R., Liu Y., Wang J. In Vitro activities of combinations of rifampin with other antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015;59:1466–1471. doi: 10.1128/AAC.04089-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timurkaynak F., Can F., Azap O.K., Demirbilek M., Arslan H., Karaman S.O. In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int. J. Antimicrob. Agents. 2006;27:224–228. doi: 10.1016/j.ijantimicag.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y.M., Kong L.C., Liu J., Ma H.X. Synergistic effect of eugenol with Colistin against clinical isolated Colistin-resistant Escherichia coli strains. Antimicrob. Resist. Infect. Control. 2018;7:17. doi: 10.1186/s13756-018-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni W., Shao X., Di X., Cui J., Wang R., Liu Y. In vitro synergy of polymyxins with other antibiotics for Acinetobacter baumannii: A systematic review and meta-analysis. Int. J. Antimicrob. Agents. 2015;45:8–18. doi: 10.1016/j.ijantimicag.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Zhao S., Adamiak J.W., Bonifay V., Mehla J., Zgurskaya H.I., Tan D.S. Defining new chemical space for drug penetration into Gram-negative bacteria. Nat. Chem. Biol. 2020;16:1293–1302. doi: 10.1038/s41589-020-00674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bovio E., Garzoli L., Poli A., Luganini A., Villa P., Musumeci R., McCormack G.P., Cocuzza C.E., Gribaudo G., Mehiri M., et al. Marine Fungi from the Sponge Grantia compressa: Biodiversity, Chemodiversity, and Biotechnological Potential. Mar. Drugs. 2019;17:220. doi: 10.3390/md17040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo M., Ming Y., Wang L., Li Y., Li B., Chen J., Shi S. Local delivery of deep marine fungus-derived equisetin from polyvinylpyrrolidone (PVP) nanofibers for anti-MRSA activity. Chem. Eng. J. 2018;350:157–163. doi: 10.1016/j.cej.2018.05.159. [DOI] [Google Scholar]
- 40.Singh S.B., Zink D.L., Goetz M., Dombrowski A. ChemInform Abstract: Equisetin (I) and a Novel Opposite Stereochemical Homolog Phomasetin (II), Two Fungal Metabolites as Inhibitors of HIV-1 Integrase. Tetrahedron Lett. 1998;39:2243–2246. doi: 10.1016/S0040-4039(98)00269-X. [DOI] [Google Scholar]
- 41.Ratnaweera P.B., de Silva E.D., Williams D.E., Andersen R.J. Antimicrobial activities of endophytic fungi obtained from the arid zone invasive plant Opuntia dillenii and the isolation of equisetin, from endophytic Fusarium sp. BMC Complement. Altern. Med. 2015;15:220. doi: 10.1186/s12906-015-0722-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 43.Belenky P., Ye J.D., Porter C.B., Cohen N.R., Lobritz M.A., Ferrante T., Jain S., Korry B.J., Schwarz E.G., Walker G.C., et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dwyer D.J., Belenky P.A., Yang J.H., MacDonald I.C., Martell J.D., Takahashi N., Chan C.T., Lobritz M.A., Braff D., Schwarz E.G., et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dwyer D.J., Collins J.J., Walker G.C. Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 2015;55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 46.Hong Y., Zeng J., Wang X., Drlica K., Zhao X. Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. USA. 2019;116:10064–10071. doi: 10.1073/pnas.1901730116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tangden T. Combination antibiotic therapy for multidrug-resistant Gram-negative bacteria. Ups. J. Med. Sci. 2014;119:149–153. doi: 10.3109/03009734.2014.899279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.