Abstract
A multiplex PCR was designed to detect the eae gene and simultaneously identify specific alleles in pathogenic Escherichia coli. The method was tested on 87 strains representing the diarrheagenic E. coli clones. The results show that the PCR assay accurately detects eae and resolves alleles encoding the α, β, and γ intimin variants.
Two groups of pathogenic Escherichia coli have evolved similar mechanisms of adhering to the intestinal epithelium that result in a characteristic attaching-and-effacing (A/E) histopathology (7). Both enteropathogenic E. coli (EPEC), a major cause of infantile diarrhea in the developing world, and enterohemorrhagic E. coli (EHEC), the agent responsible for foodborne epidemics of hemorrhagic colitis in North America, Europe, and Japan (3–5), can produce A/E lesions which contribute to the severity of diarrheal disease. Production of A/E lesions is associated with the expression of intimin, an outer membrane protein encoded by a gene (eae) that is part of the LEE (locus of enterocyte effacement) pathogenicity island (2, 7).
Evolutionary analysis has shown that E. coli strains with the virulence properties and serotypes of EPEC and EHEC are subdivided into four distinct groups of clones (EPEC 1, EPEC 2, EHEC 1, and EHEC 2) (8–10). The clonal lineages differ in the site where LEE is inserted in the genome (11), and they carry distinct intimin alleles (1, 6). Three variants of intimin—Int-α, Int-β, and Int-γ—are characteristic of EPEC 1, EPEC 2, and EHEC 1 respectively. A fourth intimin (Int-δ), found in EPEC strains of serotype O86:H34, has greater homology to the intimin homologue of Citrobacter rodentium than to Int-α of EPEC strain E2348/69 (1). Most members of EHEC 2 (e.g., O26:H11) express Int-β, with the exception of a closely related group of bacteria of serotypes O111:H8, O111:H11, and O111:H− whose intimin allele has yet to be determined.
The objective of the present study was to devise a multiplex PCR for rapid detection of eae and identification of the specific intimin alleles in E. coli strains. To accomplish this, we designed oligonucleotide primers for multiplex PCR based on the multiple sequence alignment of eae alleles by McGraw et al. (6). Primers eae P1 (5′-CTGAACGGCGATTACGCGAA-3′) and eae P2 (5′-CCAGACGATACGATCCAG-3′) were constructed in the N-terminal conserved region of the gene at positions 544 and 1461, respectively. PCR with eae P1 and eae P2 generated a 917-bp fragment, indicating the presence of the eae gene (Fig. 1). Primers designed to determine the specific eae allele were constructed in the same orientation as eae P2 on the noncoding strand in the part of the gene specifying the variable C-terminal region of the protein (Fig. 1). Ecoeaeα (5′-CTGGAGTTGTCGATGTT-3′) was located at position 2192, generating a 1,648-bp fragment indicative of the eae allele specifying Int-α. Ecoeaeβ (5′-GTAATTGTGGCACTCC-3′), positioned at bp 2470, generated a 1,926-bp fragment indicative of the allele specifying Int-β. Ecoeaeγ (5′-GCCTCTGACATTGTTAC-3′), positioned at bp 2314, produced a 1,770-bp fragment indicative of the allele specifying Int-γ. All primers were synthesized by a Beckman 1000 oligonucleotide synthesizer (Beckman, Fullerton, Calif.).
FIG. 1.
Primer locations and fragment sizes for multiplex PCR with five primers: eae P1, eae P2, Ecoeaeα, Ecoeaeβ, and Ecoeaeγ. PCR results are given for three standards (DEC 12a [EPEC 2], DEC 4f [EHEC 1], and E2348/69 [EPEC 1]) representing the three intimin alleles (Int-β, Int-γ, and Int-α, respectively).
To test the allele-specific PCR assay, we examined 87 strains of the diarrheagenic E. coli (DEC) collection, which have been characterized by electrophoretic type based on multilocus enzyme electrophoresis of 20 housekeeping genes (Table 1). The DEC strains represent 15 common clones associated with diarrheal disease and were examined previously for the presence of eae and several other virulence factors (10). The eae genes of DEC strains 3a, 3f, 5d, 11a, and 12a have also been sequenced (6).
TABLE 1.
Characteristics of 86 DEC strains examined by multiplex PCR to detect Int-α, Int-β, and Int-γ alleles
| DEC strain no. | Original strain no. | Serotype | Isolation dataa
|
PCR fragment size (bp)
|
eae allele | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Yr | Locale | Host | 917 | 1,648 | 1,770 | 1,926 | ||||
| 1a | 572-56 | O55:H6 | 1956 | USA | Human | + | + | α | ||
| 1b | C54-58 | O55:H6 | 1958 | Suriname | Human | + | + | α | ||
| 1c | F196-51 | O55:H6 | 1951 | Germany | Human | + | + | α | ||
| 1d | F563-55 | O55:H6 | 1955 | Egypt | Human | + | + | α | ||
| 1e | AC-C21 | O55:H6 | 1986 | Mexico | Human | + | + | α | ||
| 2a | 3787-62 | O55:H6 | 1962 | Congo | Human | + | + | α | ||
| 2b | 5513-56 | O55:H− | 1956 | USA | Human | + | + | α | ||
| 2c | 607-54 | O55:H6 | 1954 | USA | Human | + | + | α | ||
| 2d | F60-51 | O55:H6 | 1951 | France | Human | + | + | α | ||
| 2e | 2087-77 | O55:H6 | 1977 | USA | Human | + | + | α | ||
| 3a | 3299-85 | O157:H7 | 1985 | USA | Human | + | + | γ | ||
| 3b | 46240 | O157:H7 | 1990 | USA | Human | + | + | γ | ||
| 3c | 3104-88 | O157:H7 | 1988 | USA | Human | + | + | γ | ||
| 3d | 3009-88 | O157:H7 | 1988 | USA | Human | + | + | γ | ||
| 3e | 3077-88 | O157:H7 | 1988 | Canada | Human | + | + | γ | ||
| 3f | 493/89 | O157:H− | 1989 | Germany | Human | + | + | γ | ||
| 4a | C1520-77 | O157:H7 | 1977 | Argentina | Calf | + | + | γ | ||
| 4b | C999-87 | O157:H7 | 1987 | Denmark | Human | + | + | γ | ||
| 4c | C374-83 | O157:H7 | 1983 | Egypt | Buffalo | + | + | γ | ||
| 4d | C681-87 | O157:H7 | 1987 | Japan | Calf | + | + | γ | ||
| 4e | C7-88 | O157:H7 | 1988 | Denmark | Human | + | + | γ | ||
| 4f | EDL-933 | O157:H7 | 1982 | USA | Meat | + | + | γ | ||
| 5a | 5624-50 | O55:H7 | 1950 | USA | Human | + | + | γ | ||
| 5b | 660-79 | O55:H7 | 1979 | USA | Human | + | + | γ | ||
| 5c | 5380-66 | O55:H7 | 1966 | USA | Human | + | + | γ | ||
| 5d | C586-65 | O55:H7 | 1965 | Sri Lanka | Human | + | + | γ | ||
| 5e | C997-63 | O55:H7 | 1963 | Iran | Human | + | + | γ | ||
| 6a | 5338-66 | O111:H21 | 1966 | USA | Human | |||||
| 6b | C142-54 | O111:H12 | 1954 | Germany | Human | |||||
| 6c | 2277-67 | O111:H12 | 1967 | Guatemala | Human | |||||
| 6d | F436-51 | O111:H12 | 1951 | Italy | Human | |||||
| 6e | 184-83 | O111:H12 | 1983 | Brazil | Human | |||||
| 7a | 750001 | O157:H43 | 1975 | USA | Pig | |||||
| 7b | 902034 | O149:H− | 1990 | USA | Pig | |||||
| 7c | 820691 | O157:H43 | 1982 | USA | Pig | |||||
| 7d | 831015 | O157:H43 | 1983 | USA | Pig | |||||
| 7e | 861575 | O157:H− | 1986 | USA | Pig | |||||
| 8a | 2198077 | O111:H− | 1977 | USA | Human | + | ? | |||
| 8b | 3030A-86 | O111:H8 | 1986 | USA | Human | + | ? | |||
| 8c | 8610049 | O111:H− | 1986 | USA | Calf | + | ? | |||
| 8d | C130-53 | O111:H11 | 1953 | Cuba | Human | + | ? | |||
| 8e | C194-65 | O111:H8 | 1965 | Denmark | Human | + | ? | |||
| 9a | 3323-61 | O26:H11 | 1961 | USA | Human | + | + | β | ||
| 9b | 2262-79 | O26:H− | 1979 | USA | Human | + | + | β | ||
| 9c | C240-52 | O26:H− | 1952 | Switzerland | Human | + | + | β | ||
| 9d | C814-67 | O26:H11 | 1967 | Denmark | Human | + | + | β | ||
| 9e | 45 | O26:H11 | 1986 | Mexico | Human | + | + | β | ||
| 10a | H30 | O26:H11 | ND | Canada | Human | + | + | β | ||
| 10b | 3047-86 | O26:H11 | 1986 | Australia | Human | + | + | β | ||
| 10c | 1557-77 | O26:H11 | 1977 | USA | Human | + | + | β | ||
| 10d | C12-52 | O26:H11 | 1952 | France | Human | + | + | β | ||
| 10e | 900105 | O26:H11 | 1990 | USA | Calf | + | + | β | ||
| 10f | RDEC-1 | O15:H− | 1970s | USA | Rabbit | + | + | β | ||
| 10g | C309-64 | O128:H8 | 1964 | ND | Human | + | + | β | ||
| 10h | C186-61 | O119:H11 | 1961 | ND | Human | + | + | β | ||
| 10i | 87-1713 | O145:H6 | 1987 | Canada | Human | + | + | β | ||
| 10j | 88817 | O70:H11 | 1988 | Canada | Human | + | + | β | ||
| 11a | 2254-75 | O128:H2 | 1975 | USA | Human | + | + | β | ||
| 11b | 3733-71 | O128:H2 | 1971 | USA | Human | + | + | β | ||
| 11c | A9619-c2 | O45:H2 | 1983 | USA | Human | + | + | β | ||
| 11d | E335021 | O128:H2 | 1989 | UK | Human | + | + | β | ||
| 11e | WM-63 | O128:H2 | ND | Brazil | Human | + | + | β | ||
| 12a | F1-50 | O111:H2 | 1950 | UK | Human | + | + | β | ||
| 12b | 2966-56 | O111:H2 | 1956 | USA | Human | + | + | β | ||
| 12c | 3942-67 | O111:H− | 1967 | Panama | Human | + | + | β | ||
| 12d | 9101-83 | O111:H2 | 1983 | Peru | Human | + | + | β | ||
| 12e | 3291-86 | O111:H− | 1986 | Kenya | Human | + | + | β | ||
| 13a | 3350-73 | O128:H7 | 1973 | USA | Human | |||||
| 13b | 5024-71 | O128:H7 | 1971 | USA | Human | |||||
| 13c | C500-74 | O128:H7 | 1974 | Tanzania | Human | |||||
| 13d | C1083-79B | O128:H7 | 1979 | Rwanda | Human | |||||
| 13e | 2384-81 | O128:H47 | 1981 | USA | Human | |||||
| 14a | C916-70 | O128:H21 | 1970 | Peru | Human | |||||
| 14b | C691-71 | O128:H21 | 1971 | India | Human | |||||
| 14c | 9088-83 | O128:H21 | 1983 | Peru | Human | |||||
| 14d | 1791-79 | O128:H− | 1979 | USA | Human | |||||
| 14e | C639-77 | O128:H21 | 1977 | Bangladesh | Human | |||||
| 15a | 5430-66 | O111:H21 | 1966 | USA | Human | |||||
| 15b | 448-71 | O111:H21 | 1971 | USA | Human | |||||
| 15c | 2660-77 | O111:H21 | 1977 | USA | Human | |||||
| 15d | 2708-78 | O111:H21 | 1978 | USA | Human | |||||
| 15e | 2394-80 | O111:H21 | 1980 | USA | Human | |||||
USA, United States; ND, not determined; UK, United Kingdom.
In preparation for PCR, each E. coli strain was grown overnight at 37°C in 10 ml of nutrient broth (Difco, Detroit, Mich.) in a shaking water bath. Chromosomal DNA was isolated according to the instructions in the Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.). Aliquots (1 μl) of DNA samples were each amplified in a 50-μl reaction mixture that contained 5.0 μl of PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 1% Triton, 0.05% gelatin), 2.5 μl of primer eae P1 at 200 ng/μl, 1.0 μl each of primers eae P2, Ecoeaeα, Ecoeaeβ, and Ecoeaeγ at 200 ng/μl, 1.25 mM deoxynucleoside triphosphate mixture, 5 units of displayTAQ (Display Systems Biotech), and distilled H2O to volume. Amplification in a Perkin-Elmer 480 DNA thermal cycler utilized an initial denaturing step at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 53°C for 2 min, and 72°C for 3 min. Positive and negative controls were included with each set of strains tested. PCR products were visualized on ethidium bromide-stained gels by transillumination with UV light.
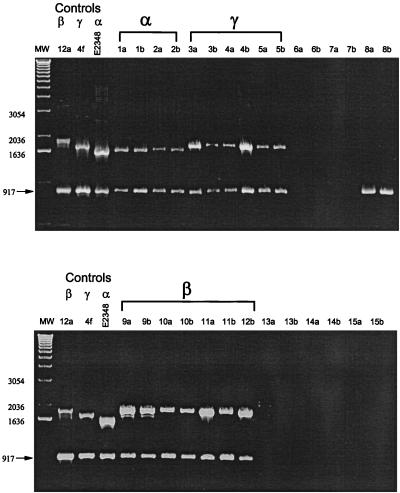
We tested the multiplex PCR assay with three positive controls for which the complete eae sequence is known (E2348/69, DEC 4f, and DEC 12a) and observed PCR fragments of the predicted sizes (Fig. 1). We then tested the 87 DEC strains by multiplex PCR and found that 57 strains produced the 917-bp fragment indicative of the presence of the eae sequence (Table 1). The allele-specific fragments showed that Int-α occurs in DEC 1 and 2 strains (EPEC 1 group), Int-β occurs both in DEC 11 and 12 (EPEC 2) and in DEC 9 and 10 (EHEC 2) strains, and Int-γ occurs both in DEC 3 and 4 (EHEC 1) and in DEC 5 (atypical EPEC of serotype O55:H7) strains. PCR results for representative DEC strains are shown in Fig. 2. Interestingly, DEC 8 strains have an eae gene, but it is sufficiently different in sequence that it is not amplified by the allele-specific primers (Fig. 2). The genetic basis of this difference remains to be determined.
FIG. 2.
Detection of the eae gene and specific intimin alleles in strains of the DEC collection. The first two strains of each of the 15 electrophoretic types of the DEC collection (Table 1) are shown. The presence of eae is indicated by a 917-bp fragment. Identification of specific intimin alleles is indicated by fragments of characteristic sizes (α, 1,648 bp; β, 1,926 bp; γ, 1,770 bp). Three controls (Fig. 1) are presented on the left side of each gel for comparison.
The results demonstrate that the multiplex PCR can accurately detect the presence of the eae gene and simultaneously identify specific eae alleles. Because the eae alleles encoding Int-α, Int-β, and Int-γ are lineage specific, this multiplex PCR method provides a rapid way to classify suspected pathogens into the major clonal groups of EPEC and EHEC.
Acknowledgments
This study was supported by Public Health Service grant AI 42391 from the National Institutes of Health.
REFERENCES
- 1.Adu-Bobie J, Frankel G, Bain C, Goncalves A G, Trabulsi L R, Douce G, Knutton S, Dougan G. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elliot S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y, Lai L-C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 3.Feng P. Escherichia coli serotype O157:H7: novel vehicles of infection and emergence of phenotypic variants. Emerg Infect Dis. 1995;1:47–52. doi: 10.3201/eid0102.950202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 5.Izumiya H, Terajima J, Wada A, Inagaki Y, Itoh K I, Tamura K, Watanabe H. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:1675–1680. doi: 10.1128/jcm.35.7.1675-1680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGraw E A, Li J, Selander R K, Whittam T S. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol Evol Biol. 1999;16:12–22. doi: 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 7.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whittam T S. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. In: Kaper J B, O’Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM Press; 1998. pp. 195–209. [Google Scholar]
- 9.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol. 1996;27(Suppl. 1):7–16. [Google Scholar]
- 10.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]