Abstract
Candida glabrata is one of the most prevalent causative pathogens of invasive candidiasis, and multidrug-resistant strains are emerging. We identified two clinical isolates of C. glabrata, BMU10720 and BMU10722 sequentially isolated from one patient with multidrug-resistance to posaconazole (POS), caspofungin (CAS), micafungin (MCF), and anidulafungin (ANF). Overexpression of ERG11 in BMU10720 and CDR1 in BMU10722 were detected at basal level. When exposed to POS, CDR1 was significantly up-regulated in both isolates compared with susceptible reference strain, while ERG11 was up-regulated considerably only in BMU10720. PDR1 sequencing revealed that both isolates harbored P76S, P143T, and D243N substitutions, while ERG11 was intact. Cdr1 inhibitor FK520 reversed POS-resistance by down-regulating ERG11 expression. FKS sequencing revealed that both isolates harbored S663P substitution in FKS2, and four single nucleotide polymorphisms (SNPs) existed in FKS2 genes between BMU10720 and BMU10722, while FKS1 was intact. Both FKS1 and FKS2 were up-regulated by CAS in BMU10720 and BMU10722. FK520 down-regulated FKS2 expression induced by CAS through inhibiting calcineurin, resulting in synergic effect with echinocandins as well as Congo Red and Calcofluor White, two cell wall-perturbing agents. In conclusion, the multidrug-resistance of C. glabrata isolates in our study was conferred by different mechanisms. CDR1 and ERG11 overexpression in one isolate and only CDR1 overexpression in the other isolate may mediate POS-resistance. S663P mutation in FKS2 and up-regulation of FKS2 may contribute to echinocandin-resistance in both isolates.
Keywords: Candida glabrata, multidrug-resistance, echinocandins, triazoles, posaconazole, FK520
1. Introduction
Invasive candidiasis (IC) is a life-threatening disease with substantial morbidity and mortality, especially among immunosuppressed hosts [1]. Although Candida albicans remains the most prevalent Candida species causing IC, the past decades have witnessed an epidemiological shift to non-albicans spp., among which Candida glabrata is one of the most common pathogens and exhibits an increasing trend [1,2,3]. C. glabrata has become the most prevalent non-albicans Candida spp. for candidemia in the United States and the third most common in China [4,5]. Triazoles, such as fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), and posaconazole (POS), acting by binding and inhibiting the 14-α-demethylase, a key enzyme of the ergosterol biosynthetic pathway encoded by ERG11, remain the most widely used antifungals. C. glabrata is of particular concern due to its high rate of reduced susceptibility to triazoles, with nearly 20% isolates exhibiting intrinsic resistance and susceptible isolates acquiring resistance rapidly [6]. To address this issue, echinocandins, such as caspofungin (CAS), micafungin (MCF), and anidulafungin (ANF), targeting β-1,3-glucan synthase (encoded by FKS1 in most Candida spp. and FKS1/FKS2 in C. glabrata), a key enzyme in the biosynthesis of a major structural component of the fungal cell wall β-1,3-glucan, are recommended as first-line agents for the treatment of invasive C. glabrata infections [7]. Troublingly, echinocandin-resistance is emerging, and on the rise, and becoming a growing threat to successful clinical management [8]. Echinocandin-resistance among C. glabrata isolates ranges from 3–5% in population-based studies, and some centers even report as high as 10–15% [9], but it is less than 1% in China [10,11]. Alarmingly, C. glabrata often presents as multidrug-resistance, with nearly one-third of echinocandin-resistant isolates also being non-susceptible to triazoles, leaving extremely few options to treat patients infected with multidrug-resistant isolates [12].
Triazole-resistance in C. glabrata is mainly mediated by functional mutations in the transcription regulator PDR1, which is responsible for the overexpression of ATP binding cassette (ABC) family multidrug transporters, such as CDR1, CDR2, and SNQ2 [13,14]. Mutations in the ERG11 [15] and overexpression of ERG11 were also reported in sporadic resistant clinical isolates [16,17], and one case of EGR11 overexpression was resulted from a chromosomal duplication [18]. Mechanisms of echinocandin-resistance appear to be more straightforward, mainly associating with mutations in hotspot (HS) regions of FKS1 and FKS2 genes encoding a catalytic subunit of β-1,3-glucan synthase [12]. Drug adaptation is also a key intermediate leading to echinocandin-resistance in C. glabrata, among which, and of particular significance, is the cell wall integrity (CWI) pathway regulating glucan synthesis through upregulation of the FKS genes and activation of PKC [12].
However, multidrug-resistance in C. glabrata was rarely reported in China, and the mechanisms underlying multidrug-resistance in this pathogen have been poorly explored and remain obscure. Here, we report two isolates of C. glabrata recovered from one patient at a three-day interval, which are both resistant to all the three echinocandins and non-wide-type (NWT) to POS as well. In addition, the mechanisms responsible for the multidrug-resistance phenotype were also elucidated.
2. Materials and Methods
2.1. Strains and Molecular Identification
A 53-year-old male was admitted to Renmin Hospital of Wuhan University with a complaint of low back pain accompanied by fever and chills. Abdominal computed tomography (CT) revealed renal abscess. Renal fungal abscess was speculated and empiric antifungal therapy with MCF 150 mg daily was initiated. A week later, the symptom persisted, and bedside B-ultrasound revealed deteriorated renal abscess. Ultrasound-guided percutaneous nephrostomy and drainage was performed and the drainage purulent urine culture was positive for Candida spp., named BMU10720. Antifungal therapy with MCF 150 mg daily was continued for another three days, but the symptom did not relive and drainage purulent urine from nephrostomy tube was cultured positive for another isolate of Candida spp., named BMU10722. Due to the poor response, MCF was replaced by VRC 200 mg daily. Unfortunately, the condition continued to deteriorate and nine days later he died because of sepsis and multi-organ failure.
These two isolates of Candida spp. BMU10720 and BMU10722 were sent to the Research Center for Medical Mycology, Peking University First Hospital, Beijing, China, for species identification and further study. Genomic DNA of BMU10720 and BMU10722 was extracted, the internal transcribed spacers (ITS) gene and large-subunit (28S) ribosomal rRNA gene D1/D2 domain sequencing was undertaken [19]. The sequences were analyzed against the CBS database (https://wi.knaw.nl/page/Pairwise_alignment, accessed on 21 May 2021), and both isolates were identified as C. glabrata.
2.2. Antifungal Susceptibility Testing
In vitro antifungal susceptibility to POS, FLC, ITC, VRC, CAS, MCF, ANF, and amphotericin B (AMB) was performed according to the Clinical and Laboratory Standards Institute (CLSI) M27-A4 document [20]. Two reference strains, Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258, were included as quality-control. The susceptible reference strain C. glabrata ATCC2001 was included for comparison purposes. MICs were determined after 24 h of incubation. The MIC was estimated as the lowest concentration of the antifungal, inducing at least a 50% reduction (for all antifungals except AMB) or a 100% reduction (for AMB) in fungal growth from that of the control. The MIC interpretive criteria included C. glabrata-specific CLSI breakpoints for FLC, CAS, MCF, and ANF [21], as well as epidemiologic cutoff values (ECVs) for VRC, ITC, POS, and AMB [22].
Antifungal susceptibility testing by E-test was performed according to the manufacturer’s instructions. Briefly, inoculum suspensions at a final concentration of 1 × 106 CFU/mL were inoculated on the entire surface of each 90-mm plate containing 25 mL of RPMI 1640 medium with a sterile cotton swab. The E-test strips (Autobio, Zhengzhou, China) were placed on the center of the plate and incubated at 35 °C. The MICs were determined from the inhibition ellipse that intersected the scale on the strip after 24 h.
Antifungal susceptibility testing by disk diffusion was performed by using disks prepared in-house of POS (10 μg), FLC (80 μg), ITC (80 μg), VRC (10 μg), CAS (10 μg), MCF (10 μg), ANF (10 μg), with or without Cdr1 inhibitor FK520 (20 μg), referring to the previously described method [23].
2.3. Amplification and Sequence Analysis of Resistance-Related Genes
In order to detect mutations in resistance-related genes in these two multidrug-resistant C. glabrata isolates, the open reading frame (ORF) of the PDR1, ERG11, FKS1, and FKS2 genes were amplified with the primers listed in Table 1. The amplified products were sent to the BGI Company (Beijing, China) for sequencing. The sequences were analyzed against those of reference strain (GenBank reference sequence No. PDR1 FJ550269, ERG11 XM_445876, FKS1 XM_446406, and FKS2 XM_448401) and single nucleotide polymorphisms (SNPs) were detected using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 15 June 2021).
Table 1.
Primers and fluorescent probes used in this study.
| Primers | Sequences (5′-3′) | Purposes |
|---|---|---|
| ERG11-F | ATGTCCACTGAAAACACTTC | ERG11 amplification and sequencing |
| ERG11-R | CTAGTACTTTTGTTCTGGATGTC | |
| PDR1-F | GGTAAAGTCATTCTTTAGCTACG | PDR1 amplification and sequencing |
| PDR1-R | TACAGGCTATGCACACTGTCT | |
| FKS1-F | ATGTCTTACAATAATAACGGAC | FKS1 amplification and sequencing |
| FKS1-W1 | TTCTCCGATTTCAGCAGTTAC | FKS1 sequencing |
| FKS1-W2 | ACTCCAATCGAAAGAGTTCGT | |
| FKS1-W3 | AGTTTCATCCAACTTCTAGCT | |
| FKS1-W4 | TCAACACTGTCTTTTCCGTTG | |
| FKS1-W5 | GATCAAGATCCTGAGAAGGAA | |
| FKS1-W6 | TCGATGCTAACCAAGACAACT | |
| FKS1-W7 | TGCTTTGATTTTCTACAGAGG | |
| FKS1-W8 | CCTGGTTTCCATTTGAATAAC | |
| FKS1-W9 | CTTCTTGGATTACAGAGACTA | |
| FKS1-R | TTATTTGATTGTAGACCAGGTC | FKS1 amplification and sequencing |
| FKS2-F | ATGTCTTACGATCAAGGTGG | FKS2 amplification and sequencing |
| FKS2-W1 | CAAGGTCAAATGCCACAACAA | FKS2 sequencing |
| FKS2-W2 | ACAAAAAAGCAATGGAAGAGG | |
| FKS2-W3 | TCTCCTACTTTCTACACTCAC | |
| FKS2-W4 | GATTGCTACAGATTTCATTTTG | |
| FKS2-W5 | TGTTAAGGATACCAAGATTCTG | |
| FKS2-W6 | TTGATGCTAACCAAGACAACTA | |
| FKS2-W7 | CTGGTTTCCATTTGAATAACTT | |
| FKS2-W8 | AGATGGTTATCAAGAGGTAACA | |
| FKS2-W9 | TTGGACTCAACCAATGAGAG | |
| FKS2-R | TTATTTTATAGTGGACCAGGTCTT | FKS2 amplification and sequencing |
| RND5.8a | CTTGGTTCTCGCATCGATGA | real-time PCR for RND5.8 |
| RND5.8b | GGCGCAATGTGCGTTCA | |
| RND5.8pr | 6FAM-ACGCAGCGAAATGCGATACGTAATGTG-TAMRA | |
| CDR1a | TAGCACATCAACTACACGAACGT | real-time PCR for CDR1 |
| CDR1b | AGAGTGAACATTAAGGATGCCATG | |
| CDR1pr | 6FAM-TGCTGCTGCTTCTGCCACCTGGTT-TAMRA | |
| CDR2a | GTGCTTTATGAAGGCTACCAGATT | real-time PCR for CDR2 |
| CDR2b | TCTTAGGACAGAAGTAACCCATCT | |
| CDR2pr | 6FAM-TACCTTTGCGTGCTGGGCGTCACC-TAMRA | |
| SNQ2a | ACCATGTGTTCTGAATCAATCAAT | real-time PCR for SNQ2 |
| SNQ2b | TCGACATCATTACAATACCAGAAA | |
| SNQ2pr | 6FAM-AACTAATCGCCGCAGGTTGTGACA-TAMRA | |
| ERG11a | ATTGGTGTCTTGATGGGTGGTC | real-time PCR for ERG11 |
| ERG11b | TCTTCTTGGACATCTGGTCTTTCA | |
| ERG11pr | 6FAM-ACTTCCGCTGCTACCTCCGCTTGG-TAMRA | |
| FKS1a | TACCAACCAGAAGACCAACAGAATGG | real-time PCR for FKS1 |
| FKS1b | TCACCACCGCTGATGTTTGGGT | |
| FKS1pr | 6FAM-TGGTCAAGCCATGTACGGTGACG-TAMRA | |
| FKS2a | CAATTGGCAGAACACCGATCCCAA | real-time PCR for FKS2 |
| FKS2b | AGTTGGGTTGTCCGTACTCATCGT | |
| FKS2pr | 6FAM-CCAGAACAACAACAAGGTGGTGAAGGT-TAMRA |
2.4. RNA Extraction and Quantitative Real-Time Reverse-Transcription (RT)-PCR
Suspensions of C. glabrata isolates BMU10720, BMU10722, and ATCC2001 cells (OD600, 0.1) freshly prepared in YPD medium were grown at 30 °C to reach an OD600 of 0.5. Subsequently, each isolate was divided into 6 groups: drug-free, FK520 (added into YPD medium at a final concentration of 100 mg/L), POS (2 mg/L for BMU10720 and BMU10722, 0.12 mg/L for ATCC2001), POS+FK520 (above mentioned concentration), CAS (4 mg/L for BMU10720 and BMU10722, 0.12 mg/L for ATCC2001), CAS+FK520 (above mentioned concentration); then the cultures were incubated at 30 °C for 3 h. After treatment, the cells were centrifuged at 14,000 g for 3 min and washed twice with sterile water. Total RNA was extracted using the RNeasy Mini kit (QIAGEN Science, Germanton, MD, USA) following the manufacturer’s instructions. The RNA was then treated with RNase-free DNase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s recommendations. According to the manufacturer’s instructions provided for the ReverAid first-strand cDNA synthesis kit (Thermo Science, Waltham, MA, USA), first-strand cDNA was amplified from 1 μg total RNA by the use of oligo (dT) primers. Real-time PCR was run on an Applied Biosystems ViiA7 real-time PCR system. Each reaction mixture volume (20 μL) contained 1 × TaqPath ProAmp master mix (Applied Biosystems, Foster City, CA, USA), 500 nM each primer, 250 nM probe, 2 μL cDNA, and nuclease-free water. The cycling conditions were as follows: 1 cycle of 95 °C for 10 min, 5 cycles of 92 °C for 15 s and 58 °C for 1 min, and 40 cycles of 92 °C for 15 s and 60 °C for 1 min. The relative quantification in gene expression was determined using the 2−ΔΔCt method with the expression level of the RDN5.8 gene for normalization [24]. The primers and fluorescent probes used were listed in Table 1.
2.5. Testing Susceptibility to FK520 with Other Agents
Spot assay was used to determine susceptibilities of different agents plus FK520 in C. glabrata strains. The indicated strains were diluted to 106 CFU/mL, and 1:10 serially diluted conidial suspensions (5 μL) were inoculated onto YPD plates (plus 100 mg/L FK520 or not), with 2 mg/L CAS, 1 mg/L MCF, 1 mg/L ANF, 50 mg/L Calcofluor White (CFW), and 50 mg/L Congo Red (CR). These plates were incubated at 35 °C for 24 h before observation.
2.6. Statistical Analysis
The experimental data were assessed by two-way ANOVA using GraphPad Prism 8. p-values < 0.05 were considered statistically significant. Experiments were performed at least three independent biological replicates.
2.7. Data Availability
The sequences of the PDR1, ERG11, FKS1, and FKS2 genes of BMU10720 and BMU10722 have been submitted to the GenBank database and assigned accession numbers MW899496-MW899503.
3. Results
3.1. C. glabrata Isolates BMU10720 and BMU10722 Are Multidrug-Resistant to POS and Echinocandins
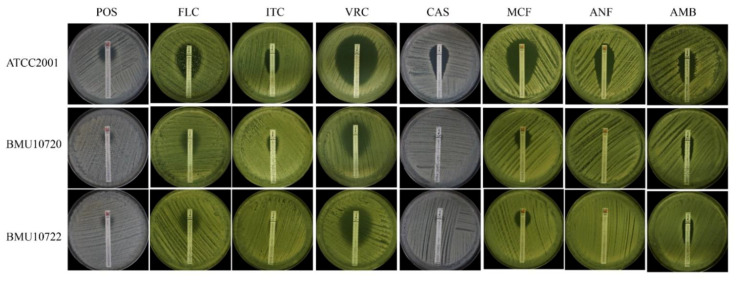
The MICs of POS against BMU10720 and BMU10722 were 4 mg/L (Table 2), which were defined as NWT to POS (ECV is 1 mg/L) [22]. Although the MICs of FLC, ITC, VRC against BMU10720 and BMU10722 (4, 2, 0.12 mg/L, respectively) were higher than those of ATCC2001, they were still defined as susceptible-dose dependent (SDD) to FLC and WT to ITC and VRC (ECVs are 4 and 0.25 mg/L, respectively) [22]. In addition, the MICs of CAS, MCF, ANF against BMU10720 and BMU10722 were 32, 8, and 4 mg/L, respectively. According to the proposed interpretative breakpoints [21], BMU10720 and BMU10722 were defined as resistant to CAS, MCF, and ANF. E-test assays showed similar results (Figure 1).
Table 2.
Minimal inhibitory concentrations (MICs) (mg/L) of antifungal agents alone or combined with FK520.
| Isolates | POS 1 | FLC | ITC | VRC | CAS | MCF | ANF | AMB | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | |
| ATCC2001 | 0.25 | 0.03 | 2 | 1 | 0.25 | 0.03 | 0.06 | 0.03 | 0.25 | 0.03 | 0.06 | 0.03 | 0.06 | 0.03 | 1 |
| BMU10720 | 4 | 0.12 | 4 | 2 | 2 | 0.06 | 0.12 | 0.06 | 32 | 8 | 8 | 4 | 4 | 2 | 1 |
| BMU10722 | 4 | 0.12 | 4 | 2 | 2 | 0.06 | 0.12 | 0.06 | 32 | 8 | 8 | 4 | 4 | 2 | 1 |
1 POS, posaconazole; FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; CAS, caspofungin; MCF, micafungin; ANF, anidulafungin; AMB, amphotericin B; (−): in the absence of FK520; (+): in the presence of 100 mg/L FK520.
Figure 1.
Antifungal susceptibility of ATCC2001, BMU10720, and BMU10722 performed by E-test. POS, posaconazole; FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; CAS, caspofungin; MCF, micafungin; ANF, anidulafungin; AMB, amphotericin B. The inhibition zones of POS, FLC, ITC, VRC, CAS, MCF, ANF in BMU10720 and BMU10722 were smaller than those of ATCC2001, which were in accordance with the result of CLSI-M27 microdilution method. The inhibition zones of AMB in BMU10720 and BMU10722 were also smaller than that of ATCC2001, but they exhibited the same AMB MICs tested by CLSI-M27 microdilution method.
3.2. Four SNPs Exist in FKS2 Genes between C. glabrata Isolates BMU10720 and BMU10722
There were four SNPs in FKS2 genes between BMU10720 and BMU10722, while the sequences of FKS1, PDR1, and ERG11 genes were identical among them. In addition, both two isolates were identified as ST3 according to MLST analysis and also had the same genotype defined by microsatellite genotyping analysis (data not shown), indicating being closely originated with identical antifungal susceptibility profile despite being with four SNPs in FKS2 genes.
3.3. Contribution of Overexpression of ERG11 and CDR1 in POS-Resistance
3.3.1. Both ERG11 and CDR1 Overexpressed in BMU10720 while Only CDR1 Overexpressed in BMU10722
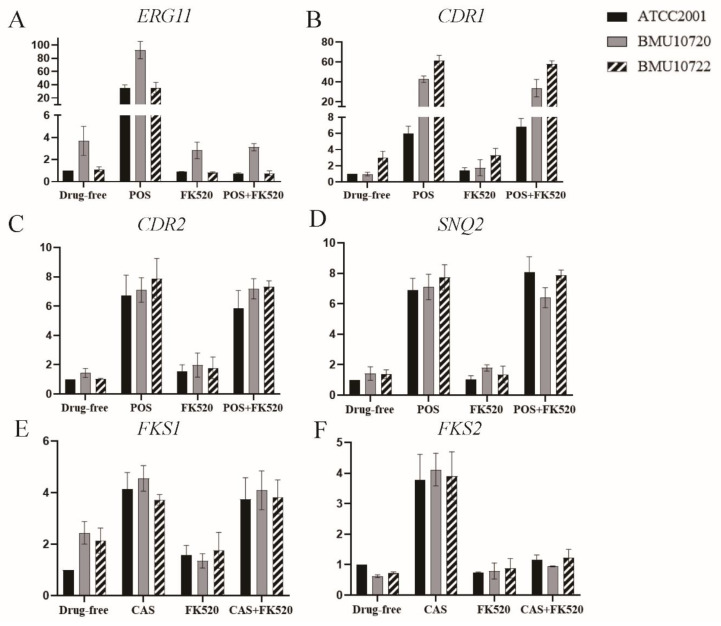
Compared to C. glabrata ATCC2001, the expression level of ERG11 was 4.35-fold higher in BMU10720 but was comparable in BMU10722 (Figure 2A), and the expression level of CDR1 was 3.01-fold higher in BMU10722 but was comparable in BMU10720, when in the absence of POS (Figure 2B). When in the presence of POS, the expression level of ERG11 and CDR1 in BMU10720 was significantly higher than that in ATCC2001, while the expression level of CDR2 and SNQ2 in BMU10720 was comparable to ATCC2001. Only CDR1 was significantly up-regulated in BMU10722 when exposed to POS than in ATCC2001, while expression of ERG11, CDR2, and SNQ2 was comparable to ATCC2001 (Figure 2A–D).
Figure 2.
The expression level of resistance-related genes in ATCC2001, BMU10720, and BMU10722. For each target gene, the relative amount of expression was compared to that of RDN5.8 as an internal control. The treatment of each group in detail was in the text. Data presented as means ± SE (error bars) from biological triplicates with technical triplicates. Expression data were assessed by two-way ANOVA. POS, posaconazole; CAS, caspofungin. (A) At basal level, the expression level of ERG11 in BMU10720 was 4.35-fold higher than that in C. glabrata ATCC2001 (p < 0.01), while that of BMU10722 was comparable to ATCC2001 (1.12-fold vs. 1-fold, p > 0.99). When exposed to POS alone, the expression level of ERG11 in BMU10720 was significantly higher than ATCC2001 (92.3-fold vs. 35.3-fold, p < 0.0001), while that of BMU10722 was comparable to ATCC2001 (35.1-fold vs. 35.3-fold, p > 0.99). FK520 alone did not change the expression level of ERG11. The combination of FK520 and POS sharply down-regulated ERG11 compared to POS alone in ATCC2001, BMU10720, and BMU10722 (0.71-fold vs. 35.3-fold, p < 0.0001, 3.1-fold vs. 92.3-fold, p < 0.0001, and 0.75-fold vs. 35.1-fold, p < 0.0001, respectively). (B) At basal level, the expression level of CDR1 in BMU10722 was 3.01-fold higher than that in C. glabrata ATCC2001 (p < 0.01), while that of BMU10720 was comparable to ATCC2001 (0.97-fold vs. 1-fold, p > 0.99). When exposed to POS alone, the expression level of CDR1 in BMU10720 and BMU10722 was significantly higher than ATCC2001 (42.64-fold vs. 5.98-fold, p < 0.0001 and 61.31-fold vs. 5.98-fold, p < 0.0001, respectively). FK520 did not change the expression level of CDR1 either in the absence or presence of POS in three isolates. (C) The expression level of CDR2 was comparable in BMU10720 and BMU10722 compared to ATCC2001 at basal level (1.44-fold vs. 1-fold, p > 0.99 and 1.04-fold vs. 1-fold, p > 0.99, respectively). POS up-regulated CDR2 in three isolates to a comparable level (7.11-fold vs. 6.73-fold, p > 0.99 and 7.88-fold vs. 6.73-fold, p = 0.85, respectively). FK520 did not change the expression level of CDR2 either in the absence or presence of POS in three isolates. (D) The expression level of SNQ2 was comparable in BMU10720 and BMU10722 compared to ATCC2001 at basal level (1.42-fold vs. 1-fold, p > 0.99 and 1.37-fold vs. 1-fold, p > 0.99, respectively). POS up-regulated SNQ2 in three isolates to a comparable level (7.73-fold vs. 6.90-fold, p > 0.99 and 6.13-fold vs. 6.90-fold, p = 0.85, respectively). FK520 did not change the expression level of SNQ2 either in the absence or presence of POS in three isolates. (E) The expression level of FKS1 was comparable in BMU10720 and BMU10722 compared to ATCC2001 at basal level (2.44-fold vs. 1-fold, p = 0.11 and 2.14-fold vs. 1-fold, p = 0.35, respectively). CAS up-regulated FKS1 in three isolates to a comparable level (4.56-fold vs. 4.13-fold, p > 0.99 and 3.72-fold vs. 4.13-fold, p > 0.99, respectively). FK520 did not change the expression level of FKS1 either in the absence or presence CAS in three isolates. (F) The expression level of FKS2 was comparable in BMU10720 and BMU10722 compared to ATCC2001 at basal level (0.62-fold vs. 1-fold, p = 0.99 and 0.73-fold vs. 1-fold, p > 0.99, respectively). CAS up-regulated FKS2 in three isolates to a comparable level (4.12-fold vs. 3.78-fold, p > 0.99 and 3.91-fold vs. 3.78-fold, p > 0.99, respectively). FK520 alone did not change the expression level of FKS2. The combination of FK520 and CAS sharply down-regulated FKS2 compared to CAS alone in ATCC2001, BMU10720, and BMU10722 (1.61-fold vs. 3.78-fold, p < 0.0001, 0.94-fold vs. 4.12-fold, p < 0.0001, and 1.23-fold vs. 3.91-fold, p < 0.0001, respectively).
Since disruption of the transcription factors genes UPC2A and RPN4 have been identified resulting in down-regulation of ERG11 by disturbing binding to the promoter of this gene in C. glabrata [25,26], the genes of these two transcriptional factors were sequenced and were found being intact in both BMU10720 and BMU10722 (data not shown). In addition, because functional alterations, except the substitutions P76S, P143T, and D243N that being considered to be MLST genotype-specific (high prevalence of ST3) [27], resulting from the mutations in the transcription regulator gene PDR1 contribute to the overexpression of multidrug transporters genes (CDR1, CDR2, SNQ2) [28], PDR1 were sequenced and substitutions P76S, P143T, and D243N were detected in both BMU10720 and BMU10722. These demonstrated that UPC2A and RPN4 were not responsible for overexpression in BMU10720 and PDR1 was not responsible for CDR1 overexpression in BMU10720 and BMU10722.
3.3.2. ERG11 Expression Level Can Be Down-Regulated by Cdr1 Inhibitor FK520 So as to Reverse POS-Resistance in BMU10720 and BMU10722
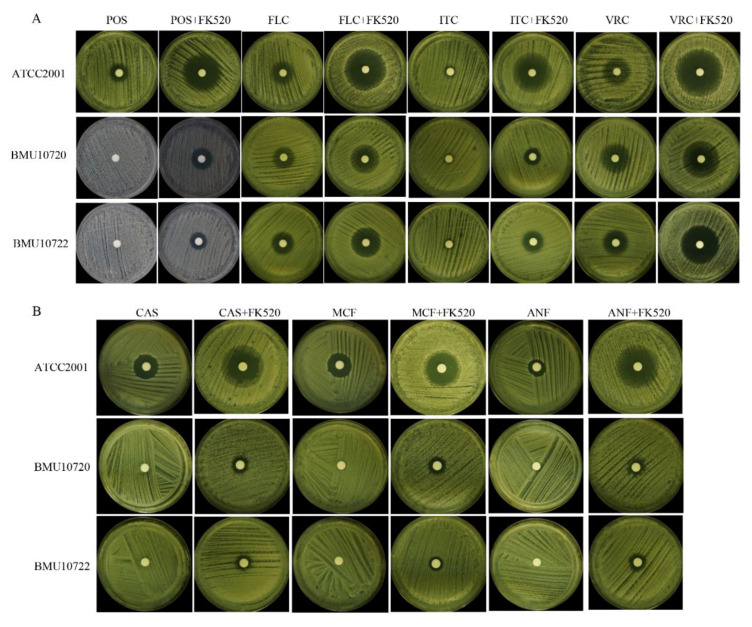
Both BMU10720 and BMU10722 were resistant to POS with MICs of 4 mg/L. When being combined with Cdr1 inhibitor FK520, these two isolates showed increased susceptibility to POS with MIC of 0.12 mg/L. Besides, this synergistic effect of FK520 was also observed when being combined with FLC, ITC, and VRC (Table 1, Figure 3A). Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), another efflux pump inhibitor specifically inhibiting major facilitator superfamily (MFS) transporter superfamily, was also used for comparison. And CCCP did not alter susceptibility to POS and other triazoles in C. glabrata isolates tested (data not shown), indicating no correlation of MFS transporter with triazole-resistance in C. glabrata.
Figure 3.
Synergic activity of FK520 with triazoles, echinocandins, and cell wall-perturbing agents. POS, posaconazole; FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; CAS, caspofungin; MCF, micafungin; ANF, anidulafungin. (A) Disk diffusion of FK520 with triazoles: POS (10 μg), FLC (80 μg), ITC (80 μg), VRC (10 μg), without or with FK520 (20 μg). The inhibition zones of POS, FLC, ITC, and VRC combined with FK520 in ATCC2001, BMU10720, and BMU10722 were significantly larger than those of triazoles used alone. (B) Disk diffusion of FK520 with echinocandins: CAS (10 μg), MCF (10 μg), ANF (10 μg), without or with FK520 (20 μg). The inhibition zones of CAS, MCF, and ANF combined with FK520 in ATCC2001, BMU10720, and BMU10722 were significantly larger than those of echinocandins used alone.
Since FK520 also reversed POS-resistance in BMU10720 whose ERG11 was overexpressed, and its structural analogue FK506 reported to enhance the susceptibility of FLC against resistant C. glabrata by suppressing the expression of ERG11 and SNQ2 [29], the expression level of ERG11 was significantly up-regulated by exposure to POS, while no significant alteration occurred when being exposed to FK520 alone, and was sharply down-regulated to a comparable level when being exposed to the combination of POS and FK520 in C. glabrata ATCC2001 as well as in BMU10720 and BMU10722 (Figure 2A). The transporter genes CDR1, CDR2, and SNQ2 were also up-regulated by exposure to POS, while the expression level of CDR1 but not CDR2 and SNQ2 was slightly up-regulated when being exposed to FK520 alone (no statistical significance) in these three isolates (Figure 2B–D). When being exposed to the combination of POS and FK520, no significant alteration of the expression level in CDR1, CDR2, and SNQ2 was observed (Figure 2B–D).
Taken together, ERG11 and CDR1 overexpression in BMU10720 and CDR1 overexpression in BMU10722 may mediate POS-resistance, which could be reversed by FK520 via down-regulating EGR11 the expression level and Cdr1 inhibition.
3.4. Contribution of Mutation and Up-Regulation of FKS1 and FKS2 in Echinocandin-Resistance
3.4.1. BMU10720 and BMU10722 Harbor S663P Substitution in FKS2 as Well as Up-Regulation of FKS1 and FKS2 Induced by CAS
The T1987C mutation, resulting in the S663P amino acid substitution in the Hotspot 1 (HS1) region of FKS2, which has proven to confer echinocandin-resistance in C. glabrata [30], was detected in BMU10720 and BMU10722, while FKS1 was intact in these two isolates.
Since upregulation of the FKS genes, which regulates glucan synthesis, is also a key intermediate leading to echinocandin-resistance in C. glabrata [12], the expression level of FKS1 and FKS2 in the absence and presence of CAS was measured. The result showed that the basal expression level of FKS1 was higher in BMU10720 and BMU10722 compared to that in ATCC2001 (no statistical significance) (Figure 2E), and that of FKS2 was comparable in these three isolates (Figure 2F). Being exposed to CAS, all the three isolates showed that FKS1 and FKS2 were up-regulated to a comparable level (Figure 2E,F).
3.4.2. FKS2 Expression Level Can Be Down-Regulated by FK520 So as to Partially Reverse Echinocandin-Resistance in BMU10720 and BMU10722
Despite no significant impact on the expression level of FKS1 and FKS2 in these three isolates, FK520 exerted a synergistic effect with echinocandins (Table 1, Figure 3B). In addition, the expression level of FKS2, instead of FKS1, was sharply down-regulated in these three isolates when being exposed to the combination of FK520 with CAS, compared to being exposed to CAS alone (Figure 2E,F). Since FK520 and its structural analogue FK506 both inhibit calcineurin activity by binding FKBP12 and forming complexes [31] and FK506 has demonstrated the potential of down-regulating FKS2 expression via inhibiting calcineurin so as to partially reverse FKS2-mediated echinocandin-resistance [32], we assumed that FK520 also down-regulated FKS2 expression via calcineurin inhibition. This inhibitory effect of calcineurin by FK520 was further confirmed by the increased susceptibility to CFW and CR when being combined with FK520, as tested by spot assay (data not shown), since calcineurin is a key factor mediating cell wall stress-response in C. glabrata.
Our data are consistent with the hypothesis that echinocandin-resistance in BMU10720 and BMU10722 is mediated by S663P substitution in FKS2, and up-regulation of FKS2 induced by CAS, is also a key intermediate, while the role of FKS1 up-regulation needs to be further verified.
4. Discussion
In this study, we reported two isolates of C. glabrata BMU10720 and BMU10722 from the nephrostomy drainage purulent urine of the renal fungal abscess patient after MCF treatment for seven days and ten days, respectively. They exhibited the same antifungal susceptibility profile defined as multidrug-resistant between POS and echinocandins. Both MCF and VRC, which are not present in urine, failed to treat the patient infected with multidrug-resistant C. glabrata. The difference with four SNPs in FKS2 genes between these two isolates indicates that they are two distinct isolates, despite their same source and being closely originated, revealed by MLST and microsatellite genotyping analysis.
Unlike other Candida spp., mutations and overexpression of ERG11 seem to play a less critical role in triazole-resistance in C. glabrata [6]. Currently, only anecdotal cases of overexpression of ERG11 in C. glabrata have been reported [17,18], and one was later found due to duplication of the entire chromosome containing the ERG11 gene [18]. Although overexpression of ERG11 plays a subtle role in triazole-resistance in C. glabrata, another study showed that ERG11 up-regulation was easily inducible in susceptible C. albicans growing in subinhibitory concentrations of FLC [33]. In serial cultured C. glabrata isolates from a patient, the expression of ERG11 was up-regulated and then decreased, with the same MIC to FLC [17]. In our study, ERG11 was intact in both isolates while being overexpressed in BMU10720 except BMU10722. Besides, FK520 restrained ERG11 overexpression induced by POS, so as to play a role in reversing POS-resistance in BMU10720 and BMU10722, further demonstrating the contribution of ERG11 overexpression to POS-resistance. It has been proven that disruption of UPC2A and RPN4, the key regulators of ERG11 expression in C. glabrata, had great impact on the regulation of ERG11 [25,26]. However, no functional mutations were detected in UPC2A and RPN4 of these two isolates in our study, indicating other mechanisms regulating ERG11 expression that need to be further elucidated.
Several studies have demonstrated that overexpression of ABC transporter (mainly CDR1, CDR2, SNQ2) is a pivotal triazole-resistance mechanism in C. glabrata clinical isolates [6,34], but the MFS transporters FLR1 and QDR2 are not found to confer triazole-resistance [29]. Our study revealed CDR1 overexpression in BMU10722 at the basal level while in both BMU10720 and BMU10722 under the exposure of POS. Cdr1 inhibitor FK520 restored their susceptibility to POS, further indicating the significant contribution of Cdr1 to POS-resistance. However, the CDR1 expression was slightly up-regulated by FK520, maybe due to FK520 directly interacting with Cdr1 protein [35,36], leading to a compensatory increase in the expression level. We also used another efflux pump inhibitor CCCP for comparison, which was proven to inhibit Mdr1 belonging to MFS transporter superfamily but not Cdr1 and Cdr2 belonging to ABC transporter superfamily in C. albicans [37]. As expected, CCCP did not alter susceptibility to POS and other triazoles in C. glabrata isolates tested, confirming that MFS transporter superfamily does not confer triazole-resistance in C. glabrata. It has been reported that PDR1 is comprised of four domains, namely, DNA-binding domain (DBD), inhibitory domain (ID), middle-homology domain (MHD), and activator domain (AD) [14]. Mutations in PDR1 leading to overexpression of ABC transporter genes mainly located in ID, MDH, and AD regions [14]. In our study, P76S, P143T, and D243N in PDR1, located outside of these domains, was detected in both BMU10720 and BMU10722, indicating PDR1 mutation did not contribute to POS-resistance in the two isolates. Additionally, factors other than PDR1 regulating CDR1 overexpression in C. glabrata need to be further investigated.
Intriguingly, although these two C. glabrata isolates were recovered from the same site of a patient at a three-day interval after MCF treatment but before VRC treatment and exhibit POS-resistance without functional mutation in ERG11 and PDR1, the different expression patterns of ERG11 and CDR1 further indicate that they are two distinct isolates. Since the two isolates were recovered without triazole exposure, they probably harbor intrinsic POS-resistance, in line with the fact that some C. glabrata isolates have intrinsic resistance to triazoles [38]. On the other hand, studies also demonstrated that prior echinocandin therapy was a risk factor of both echinocandin-resistance and triazole-resistance [39], indicating that the reduced susceptibility to POS may be induced by MCF therapy in the patient. Furthermore, considering BMU10720 and BMU10722 are resistant to POS but susceptible to other triazoles, we hypothesize that the selective binding of each efflux pump with different triazoles, harboring different chemical structures, leads to various susceptibility profiles.
Unlike other Candida spp., C. glabrata exhibits a relatively high rate of echinocandin-resistance, which is mainly mediated by mutations in FKS1 and FKS2 [8]. In this study, the T1987C mutation resulting in the S663P substitution in the HS1 region of FKS2, which has been proven to be sufficient causing echinocandin-resistance in C. glabrata [30] and reported to be one of the most common substitutions associated with high echinocandin-MICs and therapy failure [40,41], was detected in both BMU10720 and BMU10722. In addition, the difference with four SNPs was also detected in FKS2 between these two isolates, again confirming they are two distinct isolates. The FKS2 mutation may be induced by MCF treatment, in concordance with a previous study that a significantly higher FKS mutant rate was found after MCF exposure in vivo and in vitro, among which S663P was the most frequent mutation [42]. Although FKS mutation is the most prevalent mechanism, drug adaptation is also a key intermediate leading to echinocandin-resistance [12]. Adaptive cellular responses make cells survive in the presence of the drug, which affords time to escape the effects of the drug by forming FKS mutations [8]. We further observed that CAS induced FKS1 and FKS2 up-regulation in echinocandin-susceptible and -resistant isolates, and up-regulation of FKS2 but not FKS1 was suppressed by FK520. It has been reported that inhibition of glucan synthase by echinocandins activated calcineurin-dependent stress-responses [43], and cell wall integrity (CWI) pathway activation leads to an increase in FKS2 expression, dependent upon calcineurin [44]. Calcineurin inhibitor FK506 suppresses FKS2 expression resulting in reversal of FKS2-mediated resistance in C. glabrata [22,45]. As an analogue of FK506, we speculated that FK520 could down-regulate FKS2 expression also by inhibiting calcineurin activity, resulting in disturbing cell wall stress-response and sensitizing the isolates to echinocandins as well as other cell wall-perturbing agents such as CFW and CR.
Although to the best of our knowledge, this is the first study trying to elucidate the mechanisms underlying multidrug-resistance of C. glabrata in China, there are some limitations in this study. First, CDR1 and ERG11 overexpression is detected in one isolate and only CDR1 overexpression in the other isolate was detected but cannot be explained by the mutation of the well-known transcriptional factor PDR1. Other cellular mechanisms may regulate the expression of ERG11 and CDR1 in C. glabrata, which require further investigations. Second, S663P mutation in FKS2, which has been verified sufficiently to confer echinocandin-resistance previously and induced up-regulation of FKS1 and FKS2 were detected in the two isolates, but the exact role of up-regulation of FKS needs verification.
5. Conclusions
Two sequential clinical isolates of C. glabrata with multidrug-resistance to POS and echinocandins were isolated from nephrostomy drainage purulent urine of the patient with renal abscess who failed MCF and VRC therapy. CDR1 and ERG11 overexpression in one isolate and only CDR1 overexpression in the other isolate may be associated with POS-resistance. S663P mutation in FKS2 and up-regulation of FKS2 may contribute to echinocandin-resistance in both isolates.
Author Contributions
Conceptualization, Q.W. and W.L.; Methodology, Q.W. and Z.W.; Software, Q.W. and E.Y.; Formal Analysis, Q.W. and W.L.; Investigation, Q.W.; Resources, Y.L., X.C., B.Z., R.L., Z.W. and W.L.; Writing–Original Draft Preparation, Q.W.; Writing–Review & Editing, T.L., X.Y., W.L.; Visualization, Q.W. and W.L.; Supervision, Y.L., B.Z., R.L., E.Y. and W.L.; Project Administration, R.L. and W.L.; Funding Acquisition, W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China grants (grant numbers 81861148028, 81971912), and Natural Science Foundation of Guangxi Province of China project Guangxi Innovation Research Team for Fungal Infectious Diseases Prevention and Treatment (grant number 2020GXNSFGA238001).
Data Availability Statement
The original data contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pappas P.G., Lionakis M.S., Arendrup M.C., Ostrosky-Zeichner L., Kullberg B.J. Invasive Candidiasis. Nat. Rev. Dis. Primers. 2018;4:18026. doi: 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- 2.Alobaid K., Khan Z. Epidemiologic Characteristics of Adult Candidemic Patients in a Secondary Hospital in Kuwait: A Retrospective Study. J. Mycol. Méd. 2019;29:35–38. doi: 10.1016/j.mycmed.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Kakeya H., Shibata W., Yamada K., Kaneko Y. National Trends in the Japanese Distribution of Major Candida Species Causing Candidemia During 2003–2017: A Report by the Epidemiological Investigation Committee for Human Mycoses in Japan. Open Forum. Infect. Dis. 2019;6:S142. doi: 10.1093/ofid/ofz360.329. [DOI] [Google Scholar]
- 4.Perlin D.S., Rautemaa-Richardson R., Alastruey-Izquierdo A. The Global Problem of Antifungal Resistance: Prevalence, Mechanisms, and Management. Lancet Infect. Dis. 2017;17:e383–e392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 5.Hou X., Xiao M., Wang H., Yu S.Y., Zhang G., Zhao Y., Xu Y.C. Profiling of PDR1 and MSH2 in Candida glabrata Bloodstream Isolates from a Multicenter Study in China. Antimicrob. Agents Chemother. 2018;62:e00153-18. doi: 10.1128/AAC.00153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whaley S.G., Rogers P.D. Azole Resistance in Candida glabrata. Curr. Infect. Dis. Rep. 2016;18:41. doi: 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 7.Pappas P.G., Kauffman C.A., Andes D.R., Clancy C.J., Marr K.A., Ostrosky-Zeichner L., Reboli A.C., Schuster M.G., Vazquez J.A., Walsh T.J., et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016;62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlin D.S. Mechanisms of Echinocandin Antifungal Drug Resistance. Ann. N. Y. Acad. Sci. 2015;1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmakiotis D., Tarrand J.J., Kontoyiannis D.P. Drug-Resistant Candida glabrata Infection in Cancer Patients. Emerg. Infect. Dis. 2014;20:1833–1840. doi: 10.3201/eid2011.140685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou X., Xiao M., Chen S.C., Kong F., Wang H., Chu Y.Z., Kang M., Sun Z.Y., Hu Z.D., Li R.Y., et al. Molecular Epidemiology and Antifungal Susceptibility of Candida glabrata in China (August 2009 to July 2014): A Multi-Center Study. Front. Microbiol. 2017;8:880. doi: 10.3389/fmicb.2017.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y., Chen X., Yan Y., Wan Z., Liu W., Li R. Prevalence and Antifungal Susceptibility of Pathogenic Yeasts in China: A 10-Year Retrospective Study in a Teaching Hospital. Front. Microbiol. 2020;11:1401. doi: 10.3389/fmicb.2020.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healey K.R., Perlin D.S. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J. Fungi. 2018;4:105. doi: 10.3390/jof4030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonicova L., Moye-Rowley W.S. Functional Information from Clinically-derived Drug Fesistant forms of the Candida glabrata Pdr1 Transcription Factor. PLoS Genet. 2020;16:e1009005. doi: 10.1371/journal.pgen.1009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filler S.G., Ferrari S., Ischer F., Calabrese D., Posteraro B., Sanguinetti M., Fadda G., Rohde B., Bauser C., Bader O., et al. Gain of Function Mutations in CgPDR1 of Candida glabrata Not Only Mediate Antifungal Resistance but Also Enhance Virulence. PLoS Pathog. 2009;5:e1000268. doi: 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull C.M., Parker J.E., Bader O., Weig M., Gross U., Warrilow A.G., Kelly D.E., Kelly S.L. Facultative Sterol Uptake in an Ergosterol-Deficient Clinical Isolate of Candida glabrata Harboring a Missense Mutation in ERG11 and Exhibiting Cross-Resistance to Azoles and Amphotericin B. Antimicrob. Agents Chemother. 2012;56:4223–4232. doi: 10.1128/AAC.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanden Bossche H., Marichal P., Odds F.C., Le Jeune L., Coene M.C. Characterization of an Azole-Resistant Candida glabrata Isolate. Antimicrob. Agents Chemother. 1992;36:2602–2610. doi: 10.1128/AAC.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redding S.W., Kirkpatrick W.R., Saville S., Coco B.J., White W., Fothergill A., Rinaldi M., Eng T., Patterson T.F., Lopez-Ribot J. Multiple Patterns of Resistance to Fluconazole in Candida glabrata Isolates from a Patient with Oropharyngeal Candidiasis Receiving Head and Neck Radiation. J. Clin. Microbiol. 2003;41:619. doi: 10.1128/JCM.41.2.619-622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marichal P., Vanden Bossche H., Odds F.C., Nobels G., Warnock D.W., Timmerman V., Van Broeckhoven C., Fay S., Mose-Larsen P. Molecular Biological Characterization of an Azole-Resistant Candida glabrata Isolate. Antimicrob. Agents Chemother. 1997;41:2229–2237. doi: 10.1128/AAC.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neppelenbroek K.H., Seo R.S., Urban V.M., Silva S., Dovigo L.N., Jorge J.H., Campanha N.H. Identification of Candida Species in the Clinical Laboratory: A Review of Conventional, Commercial, and Molecular Techniques. Oral. Dis. 2014;20:329–344. doi: 10.1111/odi.12123. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard-Fourth Edition. CLSI Document M27-A4. Wayne, PA, USA. 2017. [(accessed on 28 May 2021)]. Available online: https://clsi.org/standards/products/microbiology/documents/m27/
- 21.Clinical and Laboratory Standards Institute Performance Standards for Antifungal Susceptibility Testing of Yeasts, 1st ed. CLSI Document M60. Wayne, PA, USA. 2017. [(accessed on 28 May 2021)]. Available online: https://clsi.org/standards/products/microbiology/documents/m60/
- 22.Clinical and Laboratory Standards Institute Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 2nd ed. CLSI Document M59. Wayne, PA, USA. 2018. [(accessed on 28 May 2021)]. Available online: https://clsi.org/standards/products/microbiology/documents/m59/
- 23.Qiao J., Kontoyiannis D.P., Wan Z., Li R., Liu W. Antifungal Activity of Statins against Aspergillus Species. Med. Mycol. 2007;45:589–593. doi: 10.1080/13693780701397673. [DOI] [PubMed] [Google Scholar]
- 24.Li Q.Q., Skinner J., Bennett J.E. Evaluation of Reference Genes for Real-Time Quantitative PCR Studies in Candida glabrata Following Azole Treatment. BMC Mol. Biol. 2012;13:22. doi: 10.1186/1471-2199-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pais P., California R., Galocha M., Viana R., Ola M., Cavalheiro M., Takahashi-Nakaguchi A., Chibana H., Butler G., Teixeira M.C. Candida glabrata Transcription Factor Rpn4 Mediates Fluconazole Resistance through Regulation of Ergosterol Biosynthesis and Plasma Membrane Permeability. Antimicrob. Agents Chemother. 2020;64:e00554-20. doi: 10.1128/AAC.00554-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whaley S.G., Caudle K.E., Vermitsky J.P., Chadwick S.G., Toner G., Barker K.S., Gygax S.E., Rogers P.D. UPC2A is Required for High-Level Azole Antifungal Resistance in Candida glabrata. Antimicrob. Agents Chemother. 2014;58:4543–4554. doi: 10.1128/AAC.02217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Won E.J., Choi M.J., Kim M.N., Yong D., Lee W.G., Uh Y., Kim T.S., Byeon S.A., Lee S.Y., Kim S.H., et al. Fluconazole-Resistant Candida glabrata Bloodstream Isolates, South Korea, 2008–2018. Emerg. Infect. Dis. 2021;27:779–788. doi: 10.3201/eid2703.203482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whaley S.G., Zhang Q., Caudle K.E., Rogers P.D. Relative Contribution of the ABC Transporters Cdr1, Pdh1, and Snq2 to Azole Resistance in Candida glabrata. Antimicrob. Agents Chemother. 2018;62:e01070-18. doi: 10.1128/AAC.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Chen Z., Zhang C., Gao Y., Zhang X., Sun S. Resistance reversal induced by a combination of fluconazole and tacrolimus (FK506) in Candida glabrata. J. Med. Microbiol. 2015;64:44–52. doi: 10.1099/jmm.0.081760-0. [DOI] [PubMed] [Google Scholar]
- 30.Singh-Babak S.D., Babak T., Diezmann S., Hill J.A., Xie J.L., Chen Y.L., Poutanen S.M., Rennie R.P., Heitman J., Cowen L.E. Global Analysis of the Evolution and Mechanism of Echinocandin Resistance in Candida glabrata. PLoS Pathog. 2012;8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erin N., Lehman R.A., Boyer P.J., Billingsley M.L. In Vitro Hypoxia and Excitotoxicity in Human Brain Induce Calcineurin-Bcl-2 Interactions. Neuroscience. 2003;117:557–565. doi: 10.1016/S0306-4522(02)00934-X. [DOI] [PubMed] [Google Scholar]
- 32.Katiyar S.K., Alastruey-Izquierdo A., Healey K.R., Johnson M.E., Perlin D.S., Edlind T.D. Fks1 and Fks2 are Functionally Redundant but Differentially Regulated in Candida glabrata: Implications for Echinocandin Resistance. Antimicro.b Agents Chemother. 2012;56:6304–6309. doi: 10.1128/AAC.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribeiro M.A., Paula C.R. Up-regulation of ERG11 gene among fluconazole-resistant Candida albicans generated in vitro: Is there any clinical implication? Diagn. Microbiol. Infect. Dis. 2007;57:71–75. doi: 10.1016/j.diagmicrobio.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Abbes S., Mary C., Sellami H., Michel-Nguyen A., Ayadi A., Ranque S. Interactions between Copy Number and Expression Level of Genes Involved in Fluconazole Resistance in Candida glabrata. Front. Cell. Infect. Microbiol. 2013;3:74. doi: 10.3389/fcimb.2013.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla S., Ambudkar S.V., Prasad R. Substitution of threonine-1351 in the Multidrug Transporter Cdr1p of Candida albicans Results in Hypersusceptibility to Antifungal Agents and Threonine-1351 is Essential for Synergic Effects of Calcineurin Inhibitor FK520. J. Antimicrob. Chemother. 2004;54:38–45. doi: 10.1093/jac/dkh308. [DOI] [PubMed] [Google Scholar]
- 36.Nim S., Rawal M.K., Prasad R. FK520 Interacts with the Discrete Intrahelical Amino Acids of Multidrug Transporter Cdr1 Protein and Acts as Antagonist to Selectively Chemosensitize Azole-Resistant Clinical Isolates of Candida albicans. FEMS Yeast Res. 2014;14:624–632. doi: 10.1111/1567-1364.12149. [DOI] [PubMed] [Google Scholar]
- 37.Basso L.R., Jr., Gast C.E., Mao Y., Wong B. Fluconazole Transport into Candida albicans Secretory Vesicles by the Membrane Proteins Cdr1p, Cdr2p, and Mdr1p. Eukaryot. Cell. 2010;9:960–970. doi: 10.1128/EC.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan Y., Chew S.Y., Than L.T.L. Candida glabrata: Pathogenicity and Resistance Mechanisms for Adaptation and Survival. J. Fungi. 2021;7:667. doi: 10.3390/jof7080667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander B.D., Johnson M.D., Pfeiffer C.D., Jiménez-Ortigosa C., Catania J., Booker R., Castanheira M., Messer S.A., Perlin D.S., Pfaller M.A. Increasing Echinocandin Resistance in Candida glabrata: Clinical Failure Correlates with Presence of FKS Mutations and Elevated Minimum Inhibitory Concentrations. Clin. Infect. Dis. 2013;56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castanheira M., Woosley L.N., Messer S.A., Diekema D.J., Jones R.N., Pfaller M.A. Frequency of Fks Mutations Among Candida glabrata Isolates from a 10-year Global Collection of Bloodstream Infection Isolates. Antimicrob. Agents Chemother. 2014;58:577–580. doi: 10.1128/AAC.01674-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pham C.D., Iqbal N., Bolden C.B., Kuykendall R.J., Harrison L.H., Farley M.M., Schaffner W., Beldavs Z.G., Chiller T.M., Park B.J., et al. Role of FKS Mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 2014;58:4690–4696. doi: 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivero-Menendez O., Navarro-Rodriguez P., Bernal-Martinez L., Martin-Cano G., Lopez-Perez L., Sanchez-Romero I., Perez-Ayala A., Capilla J., Zaragoza O., Alastruey-Izquierdo A. Clinical and Laboratory Development of Echinocandin Resistance in Candida glabrata: Molecular Characterization. Front. Microbiol. 2019;10:1585. doi: 10.3389/fmicb.2019.01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlin D.S. Current Perspectives on Echinocandin Class Drugs. Future Microbiol. 2011;6:441–457. doi: 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Healey K.R., Paderu P., Hou X., Jimenez Ortigosa C., Bagley N., Patel B., Zhao Y., Perlin D.S. Differential Regulation of Echinocandin Targets Fks1 and Fks2 in Candida glabrata by the Post-Transcriptional Regulator Ssd1. J. Fungi. 2020;6:143. doi: 10.3390/jof6030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou X., Healey K.R., Shor E., Kordalewska M., Ortigosa C.J., Paderu P., Xiao M., Wang H., Zhao Y., Lin L.Y., et al. Novel FKS1 and FKS2 modifications in a high-level echinocandin resistant clinical isolate of Candida glabrata. Emerg. Microbes Infect. 2019;8:1619–1625. doi: 10.1080/22221751.2019.1684209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original data contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.