Abstract
The ribosomal intergenic spacer regions (ISRs) of 19 laboratory strains and 30 clinical samples of Porphyromonas gingivalis were amplified by PCR and sequenced to provide a strain identifier. The ISR is a variable region of DNA located between the conserved 16S and 23S rRNA genes. This makes it an ideal locus for differentiation of strains within a species: primers specific for the conserved flanking genes were used to amplify the ISR, which was then sequenced to identify the strain. We have constructed a P. gingivalis ISR sequence database to facilitate strain identification. ISR sequence analysis provides a strain identifier that can be easily reproduced among laboratories and catalogued for unambiguous comparison.
Porphyromonas gingivalis has been strongly implicated as a periodontal pathogen (13, 16, 34, 36, 42). Many studies have shown phenotypic differences, including differences in virulence, among strains of P. gingivalis (3, 4, 8, 9, 15, 21, 22, 31, 38). Accurate strain identification is a prerequisite for studies investigating the roles of specific strains of P. gingivalis in periodontitis and for studies tracking their transmission and distribution. Previous techniques for the identification of P. gingivalis strains include whole-genome restriction fragment length polymorphism analysis or DNA fingerprinting (10, 39), ribotyping (17, 39), arbitrarily primed (AP)-PCR (29, 39), serotyping (6, 18, 32), and multilocus enzyme electrophoresis (26). While these techniques have made it possible to track strains, none have provided a strain identifier that is easily reproduced among laboratories or that can be catalogued for unambiguous comparison. In addition, many of these techniques require culturing of the organisms prior to analysis. Not only is this time-consuming, but it also reduces sensitivity and may introduce bias.
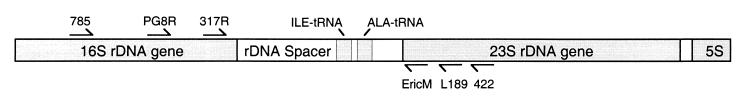
The DNA sequence of the ribosomal small subunit (16S in bacteria and 18S in eukaryotes) has been employed extensively for both identification and phylogenetic resolution of bacteria at the species level (5, 7). This gene contains both conserved regions and areas of variability sufficient to resolve species. Within a species, however, this gene does not provide sufficient variability to resolve strains. In contrast, the ribosomal intergenic spacer region (ISR), a stretch of DNA that lies between the small and large (23S) ribosomal subunit genes (Fig. 1), is variable among strains. Analysis of the ISR has been employed for the resolution of strains within several species (14, 20, 33, 41). The location of the ISR makes it ideal for strain identification: the ribosomal operon can be amplified and sequenced with species-specific primers whose targets are located within the conserved 16S and 23S genes. The 16S gene can be sequenced to verify the species, and the sequence of the ISR can be used to distinguish among strains of a species. Here we demonstrate the utility of direct PCR amplification, without culturing, followed by sequencing of the ISR for strain identification of P. gingivalis. Using this technique, we have constructed a catalogue of ISR sequences for 19 known laboratory strains of P. gingivalis as well as 30 novel sequences obtained from clinical samples. Twenty-seven of these clinical samples were selected based on their failure to match any of the patterns obtained by heteroduplex analysis of the ISR for the 19 laboratory strains (24). The strains sequenced in this study are listed in Table 1.
FIG. 1.
Map of the ribosomal operon including the ISR and primer-binding locations. rDNA, ribosomal DNA.
TABLE 1.
Strains of P. gingivalis included in the ISR sequence database
| Strain | Sourcea |
|---|---|
| Cultured lab strains | |
| 381 | J. Zambon |
| 3492 | D. Mayrand |
| 17-5 | C. Cutler |
| 22KN612 | D. Mayrand |
| 23A4 | D. Mayrand |
| 817H | C. Cutler |
| A7A1 (28) | J. Zambon |
| ATCC 33277 | ATCC |
| ATCC 49417 | ATCC |
| B57 | C. Cutler |
| DCR2011 | C. Cutler |
| ESO/27 | C. Cutler |
| HG1691 | R. Schifferle |
| HG445 | C. Cutler |
| HG564 | T. J. M. van Steenbergen |
| JKG7 | C. Cutler |
| MSM3 | C. Cutler |
| W50 | J. Zambon |
| W83 | M. Duncan |
| Clinical amplification products | |
| 7.4 | Periodontally healthy subject |
| 35 | Unidentified subject |
| 36 | Unidentified subject |
| 37 | Unidentified subject |
| 61.2 | Periodontally healthy subject |
| 62 | Unidentified subject |
| A102 | Periodontitis patient |
| A111 | Periodontitis patient |
| A116 | Periodontitis patient |
| A117 | Periodontitis patient |
| A119 | Periodontitis patient |
| A120 | Periodontitis patient |
| A134 | Periodontitis patient |
| A140 | Periodontitis patient |
| A151 | Periodontitis patient |
| A17 | Periodontitis patient |
| A198 | Periodontitis patient |
| A211 | Periodontitis patient |
| A27 | Periodontitis patient |
| A39 | Periodontitis patient |
| A50 | Periodontitis patient |
| A52 | Periodontitis patient |
| A62 | Periodontitis patient |
| A64 | Periodontitis patient |
| A8 | Periodontitis patient |
| FS8 | Student from the People’s Republic of China |
| FS106 | Student from France |
| FS155 | Student from the People’s Republic of China |
| FS159 | Student from Taiwan |
| FS170 | Student from the People’s Republic of China |
The sources for all clinical amplification products were sampled in Columbus, Ohio. ATCC, American Type Culture Collection.
The ribosomal DNA spacer regions from both cultured laboratory strains and clinical samples were amplified as described previously (23, 28). The sequences and locations of the primers are shown in Table 2 and Fig. 1. Genomic DNA isolated from plaque samples or laboratory strains was used as a template with universal prokaryotic primers 785 and 422. To generate species-specific DNA fragments from mixed clinical samples, a second amplification was performed. Aliquots consisting of 2% of the product from the first amplification served as templates for the second amplification with the P. gingivalis species-specific primer PG8R and the universal prokaryotic primer L189. This generated ISR DNA fragments specific to P. gingivalis.
TABLE 2.
Primers used for ISR amplification and sequencing
| Primer | Specificity | Sequence | Target gene |
|---|---|---|---|
| 785 | Universal | GGATTAGATACCCTGGTAGTC | 16S |
| PG8R | P. gingivalis | TGTAGATGACTGATGGTGAAAACC | 16S |
| 317R | Universal | GGCTGGATCACCTCCTT | 16S |
| EricM | Universal | GCCAAGGCATCCACCG | 23S |
| L189 | Universal | GGTACTTAGATGTTTCAGTTC | 23S |
| 422 | Universal | GGAGTATTTAGCCTT | 23S |
PCR products were purified via the Geneclean protocol (Bio 101, Inc., La Jolla, Calif.) and sequenced with an ABI 310 automated DNA sequencer. Universal prokaryotic primers 317R and EricM were used for sequencing. Both strands were sequenced at least once to ensure accuracy. Direct sequencing of PCR products eliminated the problem of misincorporation that is associated with cloning PCR products. Because of the large number of templates available at the beginning of the amplification, a base change in any one molecule would have resulted in an insignificant fraction of the amplified products representing the misincorporation.
Sequences were assembled in SeqPup (11) and aligned via Clustal X (19, 37) for automated alignment and via SeqApp for final manual alignment. A total of 830 bases were sequenced and aligned for each strain examined. The ISR sequences for strains W50, ATCC 49417, and ATCC 33277 are available from GenBank (see below); complete ISR alignments for the 19 laboratory strains and 30 clinical samples are available in National Biomedical Research Foundation format (15a).
The 19 laboratory strains were resolved into 17 unique groups based on their ISR sequences. Strains W50 and W83 were indistinguishable from one another, as were strains ATCC 49417 and HG445. Also, strains W50 and W83 were unresolved by techniques such as AP-PCR (2, 30), fimbrial restriction fragment length polymorphism analysis (25), genomic DNA fingerprinting (27), and serotyping (6, 40). It is possible that they are either the same strain or two very closely related strains. Strains ATCC 49417 and HG445 were not compared in any of the previous strain-typing studies; therefore, the difficulty of distinguishing between these two isolates by using other methods is unknown. Strains 381 and ATCC 33277, which have been previously unresolvable by techniques such as Southern blotting (1), serotyping (6), genomic DNA fingerprinting (27), and AP-PCR (2) but were separable based on infectivity and metabolic requirements (12), were distinguishable by ISR sequencing. A previous study has also been able to distinguish between these two strains via AP-PCR (35).
Twenty-seven clinical samples were selected for sequencing because they showed ISR heteroduplex patterns distinct from that of any of the 19 laboratory strains (24). As expected, their sequences did not match that of any of the laboratory strains, although of the 830 bases compared, some of the sequences differed from those of the laboratory strains by as little as a single indel (insertion or deletion event). Three samples that matched either strain W50 or 381 by ISR heteroduplex type were sequenced and found to be between 99.28 and 99.76% identical to their heteroduplex type strain. The existence of laboratory strains with perfect ISR sequence homology (e.g., W50 and W83) suggests that although the ISR is variable, it is sufficiently stable within an existing strain to make it a useful marker for strain identification.
Sequence analysis of the P. gingivalis ISR provides a strain identifier that can be easily reproduced among laboratories and catalogued for unambiguous comparison. The ISR sequence alignment is available for downloading and comparison (15a). We will continue to add additional ISR sequences to the catalogue as they become available.
Nucleotide sequence accession numbers.
The ISR sequences for strains W50, ATCC 49417, and ATCC 33277 are available from GenBank (accession no. AF118633, AF118634, and AF118635, respectively).
Acknowledgments
We thank the individuals listed in Table 1 for providing strains.
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Slots J. Clonal analysis of Porphyromonas gingivalis by the arbitrarily primed polymerase chain reaction. Oral Microbiol Immunol. 1994;9:99–103. doi: 10.1111/j.1399-302x.1994.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 3.Chu L, Bramanti T E, Ebersole J L, Holt S C. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect Immun. 1991;59:1932–1940. doi: 10.1128/iai.59.6.1932-1940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebersole J L, Kesavalu L, Schneider S L, Machen R L, Holt S C. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1995;1:115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 5.Field K G, Olsen G J, Lane D J, Giovannoni S J, Ghiselin M T, Raff E C, Pace N R, Raff R A. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 6.Fisher J G, Zambon J J, Genco R J. Identification of serogroup-specific antigens among Bacteroides gingivalis. J Dent Res. 1987;66:222. [Google Scholar]
- 7.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl D A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara T, Nakagawa I, Morishima S, Takahashi I, Hamada S. Inconsistency between the fimbrilin gene and the antigenicity of lipopolysaccharides in selected strains of Porphyromonas gingivalis. FEMS Microbiol Lett. 1994;124:333–341. doi: 10.1111/j.1574-6968.1994.tb07305.x. [DOI] [PubMed] [Google Scholar]
- 9.Genco C A, Cutler C W, Kapczynski D, Maloney K, Arnold R R. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–1263. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genco R J, Loos B G. The use of genomic DNA fingerprinting in studies of the epidemiology of bacteria in periodontitis. J Clin Periodontol. 1991;18:396–405. doi: 10.1111/j.1600-051x.1991.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, D. G. 1 February 1998, posting date. [Online.] SeqPup version 0.8. IUBio Archive, Indiana University, Bloomington, Ind. http://iubio.bio.indiana.edu. [9 June 1999, last date accessed.]
- 12.Grenier D, Mayrand D. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987;25:738–740. doi: 10.1128/jcm.25.4.738-740.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffen A L, Becker M R, Lyons S R, Moeschberger M L, Leys E J. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffen A L, Leys E J, Fuerst P A. Strain identification of Actinobacillus actinomycetemcomitans using the polymerase chain reaction. Oral Microbiol Immunol. 1992;7:240–243. doi: 10.1111/j.1399-302x.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffen A L, Moeschberger M L, Lyons S R, Leys E J. Identification of virulent strains of Porphyromonas gingivalis. J Dent Res. 1997;76:174. [Google Scholar]
- 15a.Griffen, A. L., and E. J. Leys. 9 June 1999, revision date. Sequence data. [Online.] http://www.dent.ohio-state.edu/griffen_leys. [9 June 1999, last date accessed.]
- 16.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L, Jr, Socransky S S. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 17.Hillman J D, Maiden M F, Pfaller S P, Martin L, Duncan M J, Socransky S S. Characterization of hemolytic bacteria in subgingival plaque. J Periodontal Res. 1993;28:173–179. doi: 10.1111/j.1600-0765.1993.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa E, Kumada H, Umemoto T. Serological classification of Bacteroides gingivalis and purification of group specific antigens. Shika Kiso Igakkai Zasshi. 1989;31:647–655. doi: 10.2330/joralbiosci1965.31.647. [DOI] [PubMed] [Google Scholar]
- 19.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 20.Kambhampati S, Rai K S. Temporal variation in the ribosomal DNA nontranscribed spacer of Aedes albopictus (Diptera: Culicidae) Genome. 1991;34:293–297. doi: 10.1139/g91-047. [DOI] [PubMed] [Google Scholar]
- 21.Katz J, Ward D C, Michalek S M. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:309–318. doi: 10.1111/j.1399-302x.1996.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 22.Laine M L, van Winkelhoff A J. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322–325. doi: 10.1111/j.1399-302x.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 23.Leys E J, Griffen A L, Strong S J, Fuerst P A. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J Clin Microbiol. 1994;32:1288–1294. doi: 10.1128/jcm.32.5.1288-1294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. Strain identification and detection of multiple strains of Porphyromonas gingivalis by heteroduplex analysis. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Loos B G, Dyer D W. Restriction fragment length polymorphism analysis of the fimbrillin locus, fimA, of Porphyromonas gingivalis. J Dent Res. 1992;71:1173–1181. doi: 10.1177/00220345920710050901. [DOI] [PubMed] [Google Scholar]
- 26.Loos B G, Dyer D W, Whittam T S, Selander R K. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–212. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loos B G, Mayrand D, Genco R J, Dickinson D P. Genetic heterogeneity of Porphyromonas (Bacteroides) gingivalis by genomic DNA fingerprinting. J Dent Res. 1990;69:1488–1493. doi: 10.1177/00220345900690080801. . (Erratum, 69:1623.) [DOI] [PubMed] [Google Scholar]
- 28.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1996;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menard C, Brousseau R, Mouton C. Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification of Porphyromonas (Bacteroides) gingivalis. FEMS Microbiol Lett. 1992;74:163–168. doi: 10.1111/j.1574-6968.1992.tb05360.x. [DOI] [PubMed] [Google Scholar]
- 30.Ménard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 32.Parent R, Mouton C, Lamonde L, Bouchard D. Human and animal serotypes of Bacteroides gingivalis defined by crossed immunoelectrophoresis. Infect Immun. 1986;51:909–918. doi: 10.1128/iai.51.3.909-918.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng W, Anderson T J, Zhou X, Kennedy M W. Genetic variation in sympatric Ascaris populations from humans and pigs in China. Parasitology. 1998;117:355–361. doi: 10.1017/s0031182098003102. [DOI] [PubMed] [Google Scholar]
- 34.Preus H R, Anerud A, Boysen H, Dunford R G, Zambon J J, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 35.Rumpf, R. W. 1998. Unpublished data.
- 36.Tanner A C, Haffer C, Bratthall G T, Visconti R A, Socransky S S. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Steenbergen T J, Kastelein P, Touw J J, de Graaff J. Virulence of black-pigmented Bacteroides strains from periodontal pockets and other sites in experimentally induced skin lesions in mice. J Periodontal Res. 1982;17:41–49. doi: 10.1111/j.1600-0765.1982.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 39.Van Steenbergen T J, Menard C, Tijhof C J, Mouton C, De Graaff J. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. J Med Microbiol. 1993;39:416–421. doi: 10.1099/00222615-39-6-416. [DOI] [PubMed] [Google Scholar]
- 40.van Winkelhoff A J, Appelmelk B J, Kippuw N, de Graaff J. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol. 1993;8:259–265. doi: 10.1111/j.1399-302x.1993.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 41.Wakefield A E. Genetic heterogeneity in human-derived Pneumocystis carinii. FEMS Immunol Med Microbiol. 1998;22:59–65. doi: 10.1111/j.1574-695X.1998.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 42.Wolff L F, Aeppli D M, Pihlstrom B, Anderson L, Stoltenberg J, Osborn J, Hardie N, Shelburne C, Fischer G. Natural distribution of 5 bacteria associated with periodontal disease. J Clin Periodontol. 1993;20:699–706. doi: 10.1111/j.1600-051x.1993.tb00694.x. [DOI] [PubMed] [Google Scholar]