Abstract
Salmonella continues to be a major food and public health burden worldwide that can threaten human health via eating contaminated meats, particularly those originating from chicken. In this study, the antimicrobial resistance profiles, epidemiological characteristics of resistance genes, and pulsed field gel electrophoresis (PFGE-XbaI) typing of 120 non-Pullorum/Gallinarum Salmonella isolates recovered from chicken embryos in Henan province were determined. The antimicrobial resistant phenotypes and evaluation of the extended-spectrum beta-lactamases (ESBLs) producing strains of Salmonella were investigated by the Kirby–Bauer test and the double-disk synergy test. Additionally, 37 antimicrobial resistance genes encoding resistance to five different categories, including aminoglycosides, cephalosporins, sulphonamides, tetracyclines, and β-lactams, were examined by conventional PCR. However, genotyping analysis was conducted by macro-restriction using enzyme XbaI followed by the separation of the restricted DNA fragments by PFGE. The results of this study showed that the studied Salmonella strains were highly resistant to ampicillin (66.67%) and sulfisoxazole (66.67%), while they were all susceptible to meropenem, imipenem, colistin, and chloramphenicol. Additionally, 67.5% (81/120) of the studied strains were multidrug resistant, and 21.67% (26/120) were phenotypically confirmed as ESBLs positive. The statistical analysis showed that resistance depends on the serovars, and ESBLs positive strains showed more multi-resistance than ESBLs negative strains (p < 0.05). The genotypic antimicrobial resistance showed the detection of 14 among the 37 tested genes, and the concordance between genotypic and phenotypic antimicrobial resistance ranged from 0% to 100% depending on the serovars. However, the PFGE-XbaI typing results showed that the examined Salmonella strains were divided into 22 individual subtypes and were grouped in nine clusters, with similarity values ranging from 64.7% to 100%. From this study, we can conclude that the antimicrobial resistance of Salmonella serovars isolated from chicken embryos in Henan province was alarming, with rigorous multidrug resistance, which requires the urgent mitigation of the use of antimicrobial drugs in chicken hatcheries. Additionally, our results showed evidence of the presence of different PFGE patterns among the studied Salmonella serovars, suggesting the presence of different sources of contamination.
Keywords: Salmonella, antimicrobial resistance, antimicrobial resistance genes, PFGE, ESBLs, chicken embryos
1. Introduction
The World Health Organization (WHO) estimates that 600 million people fall ill and that 420,000 people die each year after consuming contaminated food, while USD 110 billion is lost each year in productivity and medical expenses resulting from unsafe food in low- and middle-income countries [1]. Foodborne diseases hamper socioeconomic development via stressing health care systems and harm national economies, tourism, and trade [1]. In this regard, Salmonella is classified at the top of the list of major foodborne pathogens, affecting millions of people every year, and is of great importance to food and public health worldwide. In China, 70% to 80% of bacterial food poisonings were linked to Salmonella [2]. In fact, this bacterium can directly threaten human health after eating contaminated foods, especially poultry meat, which appears to be one of the major sources of human infection [3,4,5]. Recently, several studies have evaluated the prevalence of Salmonella in poultry products [4,5,6,7,8].
The overuse of antimicrobial drugs in agriculture for a long time, especially in animal husbandry, has led to the increase of the antimicrobial resistance of pathogenic bacteria. Under the selection pressure of antibiotic use, bacteria develop antimicrobial genetic determinants that are responsible for bacterial resistance [9,10]. Then, these antimicrobial genetic determinants are horizontally transmitted to other critical pathogenic bacteria via different mechanisms [9,11]. In recent years, the antimicrobial resistance of bacteria was considered among the most major threats to public health worldwide [10,12,13,14]. Regarding Salmonella isolates, recent studies have demonstrated their resistance to many critical antimicrobial drugs, including polymyxin, β-lactams, and fluoroquinolones [15,16,17,18,19,20,21,22]. Indeed, these antimicrobial resistant strains can eventually be transmitted to humans through the food chain, causing severe infectious diseases [13,23,24,25,26,27].
The molecular typing of foodborne pathogens is of great importance in epidemiological investigations, allowing the genetic discrimination of isolates and determining the causal agent when an outbreak occurs. Pulsed-field gel electrophoresis (PFGE) is considered among the major standard typing methods for foodborne pathogens and has good repeatability and high discrimination and has been widely applied in typing Salmonella isolates recovered from different samples [28,29,30]. However, we noted a lack in the genotypic discrimination of Salmonella isolates from chicken embryos in breeder hatcheries.
China is considered a major chicken consuming country, and Henan is the major breeding province. Additionally, chicken and its products are considered important vehicles for the transmission of antimicrobial resistant Salmonella. Therefore, this study aims to investigate the antimicrobial resistance of Salmonella serovars from the chicken embryos of breeder hatcheries in different regions of Henan province, China, and to trace the genetic relationship of Salmonella in different regions and, in particular, among different farms.
2. Results
2.1. Antimicrobial Resistance and Multidrug Resistance Patterns
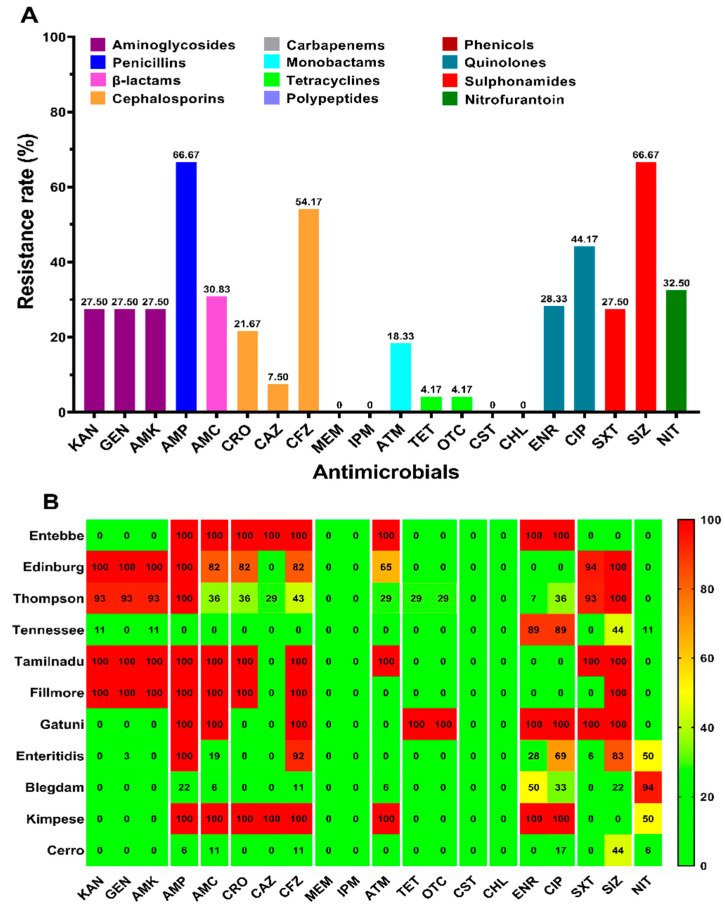
The antimicrobial susceptibility test was conducted for 20 antimicrobial agents of 12 different categories towards 120 Salmonella enterica isolates, and the results are shown in Figure 1A. Our results showed that the studied isolates present high resistance to penicillins (ampicillin, 66.67%) and sulphonamides (sulfisoxazole, 66.67%) and quite a high resistance to cephalosporins (cefazolin, 54.17%) and quinolones (ciprofloxacin, 44.17%), while they were all sensitive to carbapenems (meropenem, imipenem), polypeptides (colistin), and phenicols (chloramphenicol). However, the distribution of antimicrobial resistance among the eleven serovars, including Entebbe, Edinburg, Thompson, Tennessee, Tamilnadu, Fillmore, Gatuni, Enteritidis, Blegdam, Kimpese, and Cerro, revealed that the differences in the resistance among the serovars was statistically significant (p < 0.05) (Figure 1B).
Figure 1.
Prevalence of antimicrobial resistance among Salmonella isolates. The names of the antimicrobials are abbreviated as kanamycin (KAN), gentamicin (GEN), amikacin (AMK), ampicillin (AMP), amoxicillin-clavulanic acid (AMC), ceftriaxone (CRO), ceftazidime (CAZ), cefazolin (CFZ), meropenem (MEM), imipenem (IPM), aztreonam (ATM), tetracycline (TET), oxytetracycline (OTC), colistin (CST), chloramphenicol (CHL), enrofloxacin (ENR), ciprofloxacin (CIP), trimethoprim-sulfamethoxazole (SXT), sulfisoxazole (SIZ), and nitrofurantoin (NIT). (A) The prevalence of antimicrobial resistance among the 120 Salmonella isolates against 20 antimicrobial agents of 12 categories. The same category of antimicrobial agents is represented by the same color; (B) the distribution of the average antimicrobial resistance (in percent) of various serovars towards 20 antimicrobial agents of 12categories. The color of individual cells varies with the percentage of antimicrobial resistance shown in the cells.
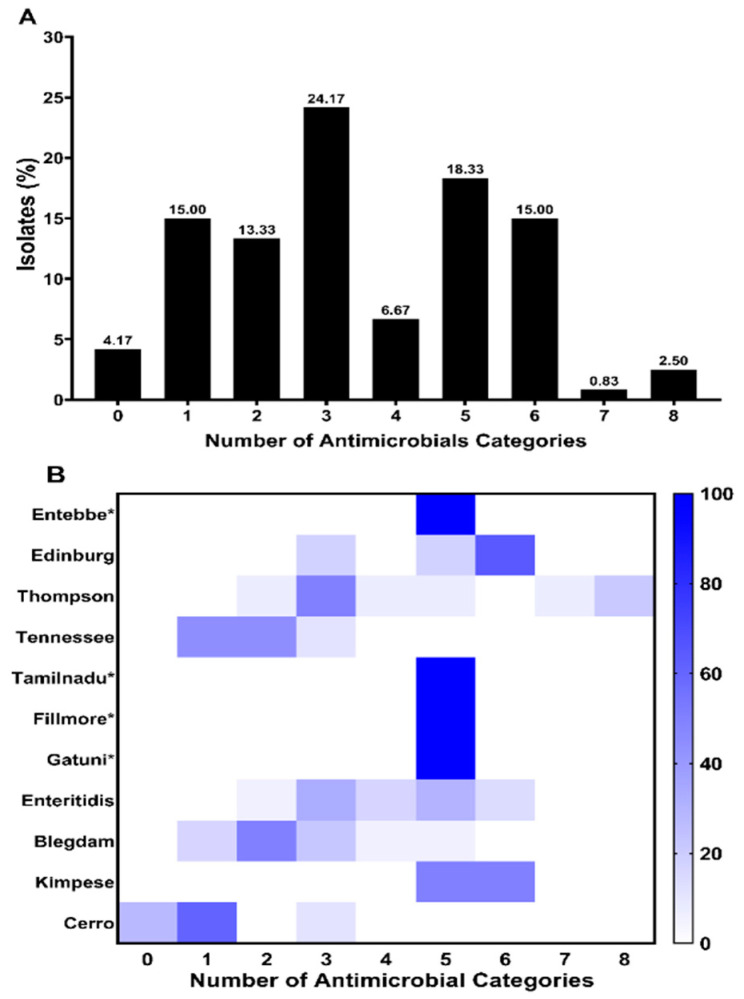
Multidrug resistance (MDR) is defined as the resistance of isolates to three or more than three antimicrobial classes. Our results showed that 81/120 (67.5%) strains were multidrug resistant (Figure 2A). Additionally, the distribution of MDR among serovars was presented in Figure 2B, where the high MDR rate was observed in S. Edinburg (100%, n = 17), S. Kimpese (100%, n = 4), S. Enteritidis (94.44%, n = 34), S. Thompson (92.86%, n = 13), and S. Blegdam (33.33%, n = 6). It is noted that only one strain was identified in the serovars Entebbe, Tamilnadu, Fillmore, and Gatuni, which all presented resistance to five antimicrobial classes (Figure 2B).
Figure 2.
Distribution of multidrug resistance (MDR) strains. (A) Prevalence of antimicrobial-resistant strains according to the number of antimicrobial classes; (B) serovar distribution of MDR prevalence. The X-axis represents the number of antimicrobial categories. The color of individual cells varies with the percentage of antimicrobial resistance shown in the cells. * serovar containing only one strain.
2.2. Phenotypic ESBL Screening
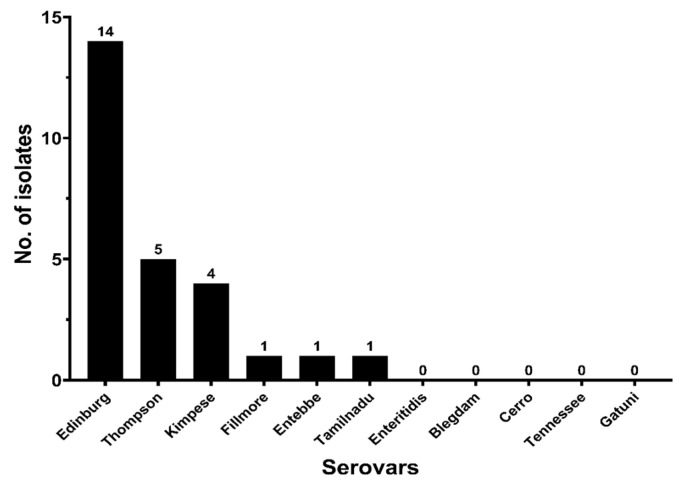
The double-disk synergy test method was used to phenotypically screen and confirm the ESBLs-producing strains. The results showed that 26 of 120 (21.67%) strains were ESBLs positive (ESBLs+). The distribution of the ESBLs-producing strains among the studied serovars is shown in Figure 3, which reveals that the difference in the ESBLs produced among the serovars was statistically significant (p < 0.05). This finding is only valid for the serovars presenting more than four isolates.
Figure 3.
The distribution of ESBLs isolates among the studied serovars.
2.3. Phenotypic-Genotypic Concordance of Drug Resistance
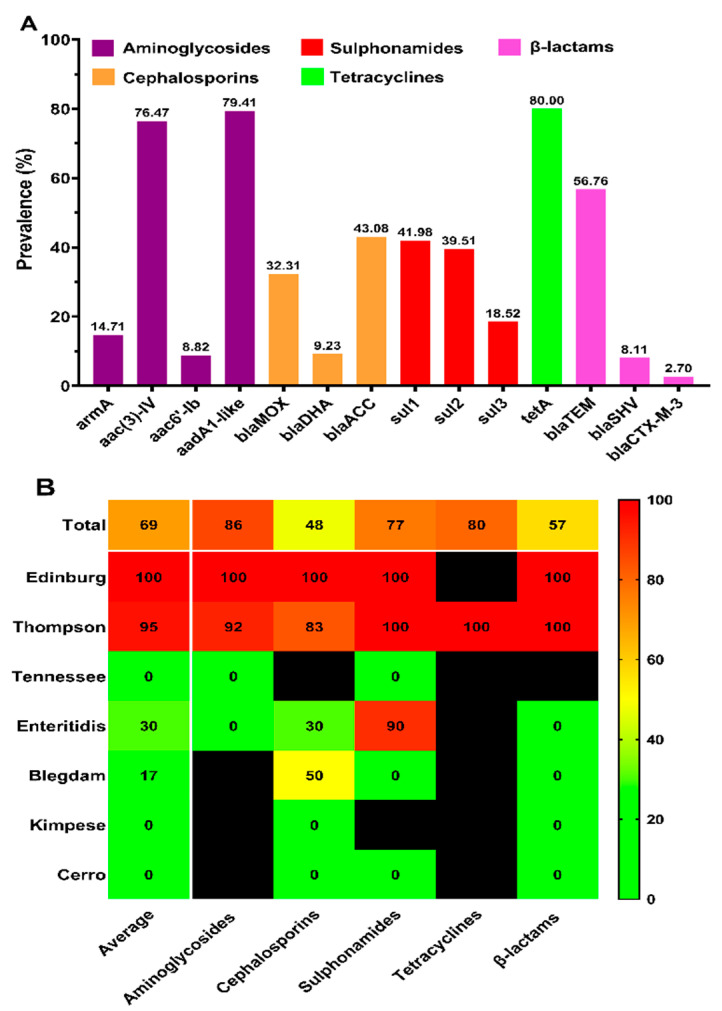
The detection of antimicrobial resistance genes was conducted by conventional PCR, and the results are presented in Figure 4A. Our results show the detection of 14 antimicrobial-resistant genes among the 37 tested antimicrobial-resistant genes. These genes are distributed as follows: Among the ten tested genes encoding resistance to aminoglycosides, only 4 genes (armA, aac(3)-IV, aac6′-Ib and aadA1-like) were detected in 35 aminoglycosides resistant strains, and among the 6 tested genes encoding resistance to cephalosporins, only 3 genes (blaMOX, blaDHA and blaACC) were detected in the 65 cephalosporins resistant strains, while among the 12 tested β-lactamase-encoding genes, only 3 genes (blaTEM, blaSHV and blaCTX-M-3) were detected in the 37 strains resistant to amoxicillin/clavulanic acid (β-lactams combinations). Regarding resistance to sulphonamides, all of the tested genes (sul1, sul2 and sul3) were detected in the 81 sulphonamides resistant strains. However, only one (tetA) of the six genes encoding resistance to tetracyclines was detected in the five tetracyclines resistant strains.
Figure 4.
Phenotype-genotype concordance of antimicrobial resistance. (A) Prevalence of genotypic drug-resistance among Salmonella isolates possessing corresponding phenotypic drug-resistance. Resistant genes belonging to the same class of resistant phenotypes are shown in the same color; (B) serovar distribution of concordance between genotypic and phenotypic drug resistance. Each column represents a resistance category, except the first column, which averages the concordance between the genotype and phenotype of the five categories. Due to the small sample size of S. Entebbe, S. Tamilnadu, S. Fillmore, and S. Gatuni, each serovar had only one strain, so we only analyzed the remaining seven serovars with relatively large sample sizes. The color of the individual cells varies with the percentage of phenotype–genotype concordance shown in the cells. Cells represented by black mean that the corresponding serovar was phenotypically susceptible and was not tested for the presence of the corresponding resistance genes; thus, the concordance percentage was not calculated.
The drug resistance phenotype–genotype concordance patterns related to different serovars are shown in Figure 4B. Overall, the concordance of the resistance genes and the resistance phenotypes was 69.34%. The detection of the drug-resistant genes corresponding to each drug-resistant phenotype was different, and the phenotype–genotype concordance was uneven in different serovars. In serovars with a relatively large number of phenotypic drug-resistant strains, the corresponding resistance genes were detected approximately in all of the phenotypic resistance strains; for example, the phenotype–genotype drug resistance concordance of the S. Edinburg strains was 100%. However, only one phenotypic drug-resistant strain of S. Thompson failed to detect the drug resistance gene with a high concordance (95.13%), while the detection rate of drug resistance genes in S. Kimpese, S. Tennessee, and S. Cerro was 0. The remaining serovars, including S. Fillmore, S. Entebbe, S. Gatuni, and S. Tamilnadu, which only presented one isolate, were not included in the phenotype–genotype concordance analysis.
2.4. PFGE Patterns
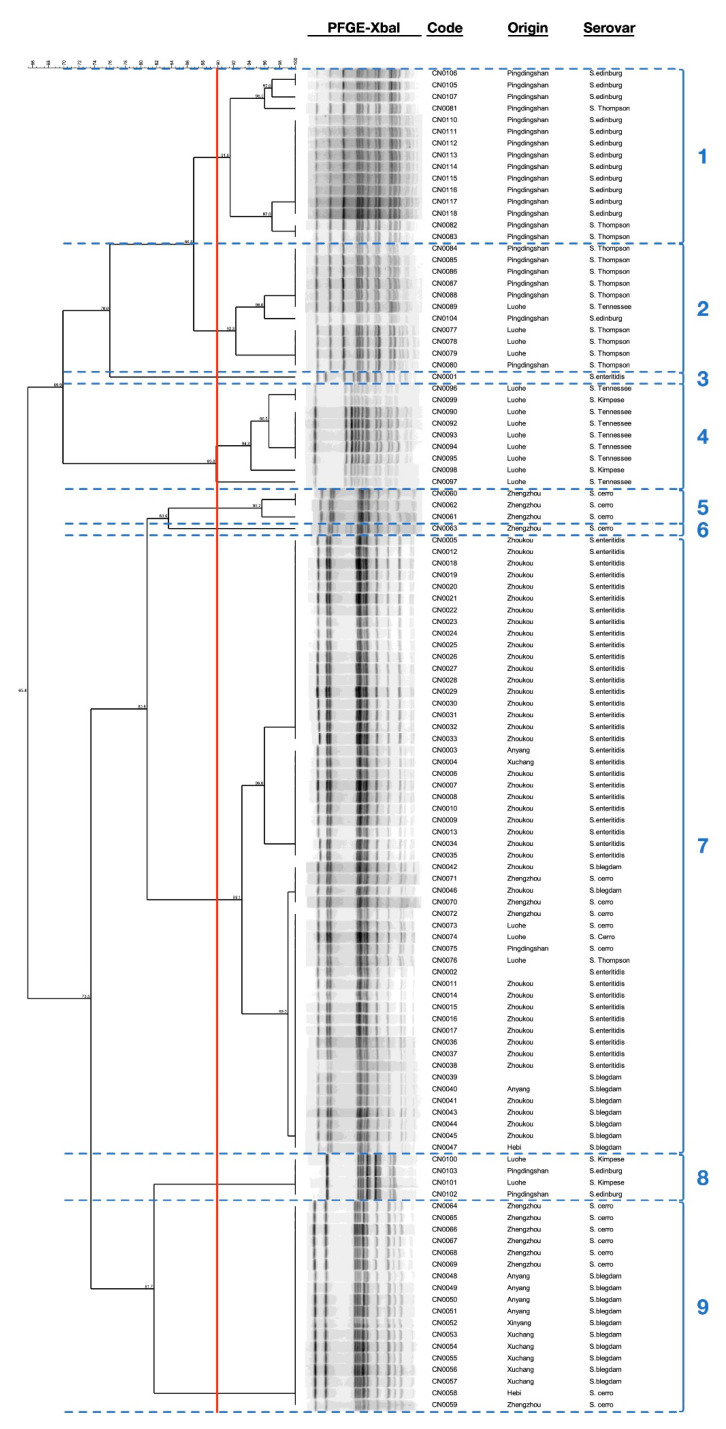
PFGE XbaI analysis was performed for 118 strains, including 115 of 120 strains, and 3 strains preserved in laboratory (one of S. Blegdam and two of S. Enteritidis) to expand analysis. The result shows that these strains were divided into 22 different subtypes with similarity values ranging from 64.7% to 100% and were grouped into 9 clusters, and each subtype contained 1 to 21 strains (Figure 5). Among the S. Enteritidis isolates analyzed, it was possible to identify four different subtypes, in which one isolate was allocated separately as cluster no. 3 and the other isolates were grouped in cluster no. 7, with similarity values varying between 93.1% to 100%, whereas among the analyzed S. Blegdam isolates, three different subtypes were identified, and the isolates were grouped in two different clusters, including no. 7 and 9, with similarity values varying between 73.1% and 100%. Additionally, our results showed that the analyzed S. Cerro isolates had high diversity, which were genotypically divided into six different subtypes and that were grouped in four clusters, including no. 5, 6, 7, and 9, with similarity values ranging from 73.5% to 100%. Similarly, we reported high diversity among the S. Edinburg isolates with the identification of five different subtypes, in which two isolates were grouped in cluster no. 8, with similarity values of 100%, and the other isolates were grouped in clusters no. 1 and 2, with similarity values ranging between 86.8% and 100%. Moreover, the analyzed S. Thompson isolates were also divided into five different subtypes, in which one isolate was placed in cluster no. 7, and the other isolates were grouped in clusters no. 1 and 2, with similarity values varying between 86.8% and 100%. However, among the analyzed S. Tennessee isolates, we identified four different subtypes, in which one isolate was placed in cluster no. 4, and the other isolates were grouped together in cluster no. 2, with similarity values ranging between 90% and 100%. Finally, PFGE analysis identified three different subtypes among the analyzed S. Kimpese isolates, in which two isolates were grouped in cluster no. 4, with similarity values ranging between 94.3% and 100%, and the two other isolates were grouped in cluster no. 8, with a similarity value of 100%.
Figure 5.
Dendrogram of PFGE profiles of 118 Salmonella strains. The analyzed strains were isolated from 28 hatcheries in 9 cities of Henan province, including 115 of 120 isolates, and three strains, including one S. Blegdam and two S. Enteritidis, were added for expanding analysis.
3. Discussion
Salmonella infection remains a major health concern that is common in poultry and is transmitted to humans through the food chain [31,32,33]. It seriously endangers the health of humans and poultry as well as the development of the poultry industry and the economy [34]. Therefore, it is urgent to strengthen non-therapeutic medication management in the poultry industry with guidance and restrictions on prophylactic drug use to reduce the use of antimicrobials in the breeding process so as to alleviate the emergence of drug-resistant and multidrug-resistant strains, thereby reducing the risk of drug-resistant strain transmission to humans through the food chain [16,31,35,36].
All of the 120 strains tested in this study were susceptible to colistin, meropenem, imipenem, and chloramphenicol. It is interesting to note that the use of these antimicrobials is banned by veterinary medicine. While the resistance to nitrofurantoin, which was listed as a banned veterinary drug, was 32.50%. Additionally, the results of this study showed that the antimicrobial resistant strains were mainly obtained from two hatcheries in Zhoukou region, which indirectly indicates the necessity of improving the drug regimen in this area. Moreover, the highest resistance was observed against ampicillin and sulfisoxazole (66.67% for each one), which was consistent with the results reported in other Chinese regions, including Shanghai (AMP: 50.7%; SIZ: 49.32%) [30], Sichuan (AMP: 87.8%) [37], Shandong (AMP: 97.7%) [38], and Guangdong (AMP: 31.8%; SIZ: 70.2%) [39], indicating that the Salmonella recovered from chickens had developed serious resistance to ampicillin and sulphonamides; therefore, the use of these drugs should be suspended. However, our results showed moderate resistance to amoxicillin-clavulanic acid (37/120; 30.83%), which was consistent with the results obtained in retail raw poultry meat in China after merging intermediate with resistant results (28%) and higher than those reported in broiler chickens along the slaughtering process in China (8.5%) [37]. On the other hand, we found that the resistance rate to first-generation cephalosporins (CFZ: 54.17%) was much higher than that of third-generation cephalosporins (CRO: 21.67%; CAZ: 7.50%), which was in line with related reports [40]. Importantly, our study showed a high rate of multidrug-resistant strains (67.5%), which was higher than that found in slaughterhouses in Henan (39.8%) [41], and in poultry, swine, and cattle farms in central China, including Henan (34.72%) [42], but was in line with many other studies [8,43,44,45]. The antimicrobial resistance of the studied Salmonella strains varies according to serovars, in which the serovars Edinburg and Thompson present the highest resistance. Additionally, this resistance may depend also on the region of isolation and drug regimen.
ESBLs are enzymes that are mainly produced by bacteria belonging to the Enterobacteriaceae family under the selective pressure of the use of broad-spectrum β-lactam antimicrobials, especially third-generation cephalosporins. ESBLs have a wide range of enzyme activities and can hydrolyze penicillin, first- or second-generation cephalosporins, and broad-spectrum cephalosporins containing oxime groups, including third-generation cephalosporins such as ceftazidime, cefotaxime, ceftriaxone, and cefixime as well as fourth-generation cephalosporin-like cefepime, and can also hydrolyze monobactams such as aztreonam [46]. Recent studies around the world indicate that the prevalence of ESBLs-producing strains is increasing each year [5,47,48,49]. In our study, the prevalence of ESBLs-producing strains was 21.67%, which was lower than that found in retail chicken in Henan province in 2017 (50.0%) [5]. However, the distribution of ESBLs producing strains according to serovars was as follows: Edinburg (82.35%; 14/17), Thompson (35.71%; 5/14), Blegdam (0%; 0/18), Cerro (0%; 0/18), Tennessee (0%; 0/9), and Enteritidis (0%; 0/36), which was consistent with other reports [5]. It is noted that the serovars presented a number of isolates that were lower than or equal to five were not taken for this comparison and discussion. Additionally, our results showed the resistance of ESBLs-producing strains to other non-β-lactam antimicrobial categories, with high resistance not only to cephalosporins and penicillins but also to aminoglycosides and sulphonamides, which was consistent with other studies demonstrating that ESBLs+ strains are often multidrug resistant [48]. Importantly, the antimicrobial resistance of ESBLs+ bacteria is significantly more serious than that of ESBLs- bacteria. Therefore, it is necessary to combine the antimicrobial susceptibility test with the ESBLs detection test to choose effective antimicrobial agents for prevention and control when dealing with salmonellosis in the clinic. At the same time, taking some measures such as the adoption of antimicrobial synergists or enzyme inhibitors, synergy, and medication rotation can effectively reduce the development of bacterial resistance.
The detection of antimicrobial resistance genes showed that among the targeted genes encoding resistance to aminoglycosides, the prevalence of aadA1-like (79.41%) and aac(3)-IV (76.47%) was the highest, which was higher than that found previously for aac(3)-IV (53.2%) in Salmonella recovered from broilers in Sichuan province [37]. However, among the ESBLs+ strains, the dominant resistance gene was blaTEM (80.77%), which was in agreement with the results obtained previously in Salmonella recovered from retail raw chicken carcasses [5]. Additionally, the detection of genes encoding resistance to tetracyclines showed the presence of only tetA, with a prevalence rate of 80%; this result is in accordance with that of previous studies showing the dominance of tetA (100%) and tetB (67.7%) genes among the tetracycline-resistant Salmonella isolates [30]. However, all of the targeted genes encoding resistance to sulphonamides were detected with different proportions (sul1: 41.98%, sul2: 39.51%, and sul3: 18.52%), these results were lower than those obtained from the Salmonella isolates recovered from slaughterhouses in Sichuan and from retail chicken in Shanghai [30,37]. According to the statistical analysis, the phenotypic and genotypic antimicrobial resistance of Salmonella strains varied greatly in each serovar. In this study, the results of phenotypic resistance of S. Edinburg and S. Thompson, especially those presenting high-multidrug resistance, were consistent with genotypic profiles (100% and 95.13%, respectively), while there is no concordance (0%) between the phenotypic and genotypic profiles of S. Tennessee, Kimpese, and S. Cerro. Therefore, this indirectly indicates that the detection of antimicrobial resistance genes is related to multidrug resistance and, to a certain extent, with serovars. However, due to the small number of strains in certain serovars and the narrow sampling range, it is necessary to expand the sampling range to verify this conclusion.
PFGE typing was widely used in molecular epidemiological studies of a variety of pathogens worldwide, which can analyze bacterial chromosomal DNA directly with high discrimination and repeatability. Although the whole genome sequencing (WGS) is actually considered the gold standard method used to fingerprint the foodborne pathogens, PFGE is still considered among the major typing methods for the screening and discrimination of bacterial isolates, especially in the case of limited access to WGS. In fact, PFGE has been used by PulseNet for many years to track the source of diseases or food safety emergencies caused by bacterial infections, which has played an important role in the management of many public health incidents. In this study, a total of 118 Salmonella strains were analyzed and discriminated by PFGE-XbaI restriction. According to the obtained PFGE profiles, 22 different subtypes (PFGE profiles) were identified, which were grouped into 9 different cluster by taking a cut-off value of 90%. Additionally, the combination of the PFGE profiles and the serological results showed that different serovars shared the same PFGE profile (subtype) and that the isolates of each serovar presented more than one subtype divided into different clusters, which demonstrates that PFGE is unable to discriminate Salmonella serovars. These results are in agreement with those reported in previous studies [50,51,52]. Moreover, these findings suggest the presence of different sources of contamination, which are in line with previous studies [51,53]. In addition, our results showed that the differences in the PFGE patterns cannot be linked to the difference in antimicrobial-resistant genotypes among the same serovar. In fact, the targeted resistance genes are often carried by mobile genetic elements, including plasmids, integrons, and transposons, which cannot be discriminated by PFGE; moreover, in the case of antimicrobial resistance resulting from punctual mutations on bacterial DNA, the mutations are not at the restriction site of the XbaI enzyme and thus cannot be detected or discriminated by PFGE. This conclusion should be confirmed by further research.
4. Materials and Methods
4.1. Strains
This study was conducted with 120 strains selected from 504 Salmonella strains recovered during a previous study from 2139 chicken embryos from 28 hatcheries for breeding chickens in 9 cities of Henan province between August 2014 to April 2015 [8]. This study was focused on analyzing the non-Pullorum/non-Gallinarum Salmonella isolates (n = 120). However, the isolation, identification, and serotyping of Salmonella strains were performed as previously reported [8]. The studied Salmonella strains (n = 120) were divided into eleven serovars, including S. Enteritidis (n = 36; 30%), S. Blegdam (n = 18; 15%), S. Cerro (n = 18, 15%), S. Edinburg (n = 17; 14.17%), S. Thompson (n = 14; 11.67%), S. Tennessee (n = 9; 7.5%), S. Kimpese (n = 4; 3.33%), and one strain each (0.83%) for S. Entebbe, S. Tamilnadu, S. Fillmore, and S. Gatuni. Additionally, three other isolates belonging to S. Enteritidis (n = 2) and S. Blegdam (n = 1), which were isolated, identified, and stored by the Infectious disease Laboratory of Henan University of Animal Husbandry and Economics were also added to the PFGE analysis. Escherichia coli ATCC 25922 was donated by the Jiangsu Key Laboratory of Zoonoses, Yangzhou University. Salmonella Braenderup H9812 was provided by Henan CDC and was used as a standard strain for PFGE investigation.
4.2. Antimicrobial Susceptibility Test
The Kirby–Bauer (K-B) disc diffusion method was used to conduct the antimicrobial susceptibility tests, while the Clinical Laboratory Standards Institute (CLSI) [54] and veterinary recommendations of the Antibiogram Committee of the French Society of Microbiology (CA-SFM) [55] were used for the results interpretation of the studied isolates against 20 antimicrobial agents, representing 12 different classes: aminoglycosides (kanamycin: KAN, 30 μg; gentamicin: GEN, 10 μg; amikacin: AMK, 30 μg); Penicillins (ampicillin: AMP, 10 μg); β-lactams combination (amoxicillin/clavulanic acid: AMC, 20/10 μg); cephalosporins (ceftriaxone: CRO, 30 μg; ceftazidime: CAZ, 30 μg; cefazolin: CFZ, 30 μg); carbapenems (meropenem: MEM, 10 μg; imipenem: IPM, 10 μg); monobactams (aztreonam: ATM 30 μg); tetracyclines (tetracycline: TET 30 μg, oxytetracycline: OTC 30 μg); polypeptides (colistin: CST 50 μg); phenicols (chloramphenicol: CHL 30 μg); quinolones (enrofloxacin: ENR, 5 μg; ciprofloxacin: CIP, 5 μg); sulphonamides (trimethoprim/sulfamethoxazole: SXT, 1.25/23.75 μg; sulfisoxazole: SIZ, 250 μg); and nitrofurans (nitrofurantoin: NIT, 300 μg). However, the results obtained by the disc diffusion method against colistin were confirmed by means of the broth dilution method, according to the recommendations of CA-SFM (Supplementary Figure S1) [55]. To facilitate analysis, isolates showing intermediate results were classified as resistant strains. Moreover, the phenotypic evaluation of the extended-spectrum β-lactamases- (ESBLs) producing strains was performed by double-disk synergy test according to the recommendation of the Clinical and Laboratory Standards Institute [54].
4.3. Detection of Antimicrobial Resistance Genes
In this study, the genomic DNA was extracted using FastPure Bacteria DNA Isolation Mini Kit (Vazyme Biotech Co.,Ltd) according to the user manual. The amplification of 37 genes encoding resistance to different antimicrobial categories was conducted by 2×Easy KOD PCR SuperMix (Zhejiang Easy-Do Biotech Co., Ltd) subsequently. Among them, twelve genes encoding ESBLs (blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-2, blaCTX-M-3, blaCTX-M-8, blaCTX-M-9, blaCTX-M-25, blaOXA-1, blaOXA-2, blaOXA-10, and blaPSE), with ten encoding resistance to aminoglycosides (armA, rmtA, rmtB, rmtC, rmtD, rmtE, npmA, aac(3)-IV, aac6′-Ib, and aadA1-like), six encoding resistance to cephalosporins (blaMOX, blaCIT, blaDHA, blaACC, blaEBC, and blaFOX), three encoding resistance to sulphonamides (sul1, sul2, and sul3), and six encoding resistance to tetracyclines (tetA, tetG, tetX, tetB, tetC, and tetR). Primer design and PCR amplification conditions were summarized in Table S1. A part of the amplified products was randomly selected and sequenced by Sangon Biotech after PCR amplification (Sangon Biotech (Shanghai) Co., Ltd). Nucleotide sequence comparisons were performed using the BLAST software (NCBI, Bethesda, MD, USA) available from the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 18 September 2021), DNAStar software (DNASTAR, Inc., Madison, WI, USA) as well as MEGA 5.1 software (Pennsylvania State University, State College, PA, USA).
4.4. Pulsed-Field GEL Electrophoresis Analysis
Pulsed-field gel electrophoresis (PFGE) was conducted according to the standardized protocol for Salmonella PFGE from PulseNet USA, a surveillance network of public health laboratories across the United States and other countries. Specifically, bacterial suspensions of Salmonella isolates were prepared from overnight cultures, fixed in agarose plugs, and lysed to liberate the DNA in agarose plugs. Then, DNA was digested with 50U XbaI restriction enzyme in a 37 °C water bath for at least 2 h. Afterward, the restricted fragments were separated in 1% agarose using Chef Mapper pulse-field gel electrophoresis, according to the following conditions: the buffer solution was 0.5× TBE or Tris/Borate/EDTA, the electrophoresis time was 18 h, the electrophoresis temperature was 14 °C, and the pulse time was 2.16 s to 63.8 s. After migration, the gel was stained with ethidium bromide and visualized under ultraviolet (UV) light, and an image was taken with a digital camera and was stored for further analysis.
The obtained PFGE profiles were analyzed by BioNumerics 7.6 software and the Dice coefficient, and the dendrogram was generated by the unweighted pair-group method with arithmetic means (UPGMA) based on the 1.5% position optimization and tolerance values. The band of 100% similarity was regarded as the same PFGE type.
4.5. Statistical Analysis
Two-way ordinary ANOVA was used to test the significant difference of antimicrobial resistance and ESBLs being produced among the serovars. p values less than 0.05 were considered statistically significant. GraphPad Prism 7 software (San Diego, CA, USA) was used for data analysis and the generation of the figures.
5. Conclusions
This study investigated the antimicrobial resistance profiles, epidemiological characteristics of resistance genes, and PFGE-XbaI typing of Salmonella isolates from chicken embryos in Henan province, China. Our findings showed that the studied Salmonella isolates presented high resistance to antimicrobial agents with diversified multidrug-resistant patterns, which requires more attention to the dissemination of multidrug-resistant strains, especially those producing ESBLs. Moreover, we demonstrated the coherence between genotypic and phenotypic resistance of the studied strains, while the PFGE patterns analysis provided 22 different subtypes grouped into 9 clusters with high diversity in each serovar, suggesting multiple sources of contamination. Therefore, we recommend the implementation of systematic and judicious medication management in the breeding industry to mitigate the development of drug-resistant and multidrug-resistant strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10101156/s1, Figure S1: The distribution of minimum inhibitory concentration (MIC) values among the examined Salmonella isolates (n = 120) against Colistin (CST). The dashed line indicates the cutoff level of the MIC, in which the lowest values correspond to susceptibility and the highest values correspond to the resistance, Table S1: Primers used for the detection of antimicrobial resistance genes.
Author Contributions
Y.X. and X.Z. contributed equally to this work. Conceptualization, M.Y.; methodology, Y.X. and X.Z.; software, Y.X. and X.Z.; validation, Z.J., Y.Q. and A.E.-D.; formal analysis, X.Z.; investigation, Y.X.; resources, Y.X.; data curation, Z.J.; writing—original draft preparation, Y.X. and X.Z.; writing—review and editing, Z.J., Y.Q. and A.E.-D.; visualization, X.Z.; supervision, M.Y.; project administration, M.Y.; funding acquisition, M.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Program on Key Research Project of China (grant # 2019YFE0103900) as well as the European Union’s Horizon 2020 Research and Innovation Programme (grant # 861917–SAFFI), the Zhejiang Provincial Natural Science Foundation of China (grant # LR19C180001), and the Zhejiang Provincial Key R&D Program of China (grant # 2020C02032 & 2021C02008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (WHO) Food Safety. [(accessed on 24 May 2021)]. Available online: https://www.who.int/en/news-room/fact-sheets/detail/food-safety.
- 2.Cai Y., Tao J., Jiao Y., Fei X., Zhou L., Wang Y., Zheng H., Pan Z., Jiao X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016;222:56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Pan H., Zhou X., Chai W., Paudyal N., Li S., Zhou X., Zhou K., Wu Q., Wu B., Li G., et al. Diversified sources for human infections by Salmonella enterica serovar Newport. Transbound. Emerg. Dis. 2019;66:1044–1048. doi: 10.1111/tbed.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X., Huang J., Zhang Y., Liu S., Chen L., Xiao C., Zeng H., Wei X., Gu Q., Li Y., et al. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 2020;713:136385. doi: 10.1016/j.scitotenv.2019.136385. [DOI] [PubMed] [Google Scholar]
- 5.Qiao J., Zhang Q., Alali W.Q., Wang J., Meng L., Xiao Y., Yang H., Chen S., Cui S., Yang B. Characterization of extended-spectrum β-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 2017;248:72–81. doi: 10.1016/j.ijfoodmicro.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Ed-Dra A., Filali F.R., Karraouan B., El Allaoui A., Aboulkacem A., Bouchrif B. Prevalence, molecular and antimicrobial resistance of Salmonella isolated from sausages in Meknes, Morocco. Microb. Pathog. 2017;105:340–345. doi: 10.1016/j.micpath.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 7.Jibril A.H., Okeke I.N., Dalsgaard A., Menéndez V.G., Olsen J.E. Genomic analysis of antimicrobial resistance and resistance plasmids in Salmonella serovars from poultry in Nigeria. Antibiotics. 2021;10:99. doi: 10.3390/antibiotics10020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y., Zhou X., Jiang Z., Qi Y., Ed-Dra A., Yue M. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell. Infect. Microbiol. 2020;10:497. doi: 10.3389/fcimb.2020.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christaki E., Marcou M., Tofarides A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- 10.Varela M.F., Stephen J., Lekshmi M., Ojha M., Wenzel N., Sanford L.M., Hernandez A.J., Parvathi A., Kumar S.H. Bacterial resistance to antimicrobial agents. Antibiotics. 2021;10:593. doi: 10.3390/antibiotics10050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alekshun M.N., Levy S.B. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization (WHO) Global Priority List of Antibiotic Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. [(accessed on 23 July 2021)]. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 13.Elbediwi M., Pan H., Biswas S., Li Y., Yue M. Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerg. Microbes Infect. 2020;9:535–538. doi: 10.1080/22221751.2020.1733439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Z., Paudyal N., Xu Y., Deng T., Li F., Pan H., Peng X., He Q., Yue M. Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front. Microbiol. 2019;10:1513. doi: 10.3389/fmicb.2019.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burnett E., Ishida M., de Janon S., Naushad S., Duceppe M.O., Gao R., Jardim A., Chen J.C., Tagg K.A., Ogunremi D., et al. Whole-genome sequencing reveals the presence of the blactx-m-65 gene in extended-spectrum β-lactamase-producing and multi-drug-resistant clones of Salmonella serovar Infantis isolated from broiler chicken environments in the Galapagos islands. Antibiotics. 2021;10:267. doi: 10.3390/antibiotics10030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbediwi M., Li Y., Paudyal N., Pan H., Li X., Xie S., Rajkovic A., Feng Y., Fang W., Rankin S.C., et al. Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980–2018) Microorganisms. 2019;7:461. doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbediwi M., Pan H., Jiang Z., Biswas S., Li Y., Yue M. Genomic characterization of mcr-1-carrying Salmonella enterica serovar 4, [5], 12:i:- ST 34 clone isolated from pigs in China. Front. Bioeng. Biotechnol. 2020;8:663. doi: 10.3389/fbioe.2020.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbediwi M., Pan H., Zhou X., Rankin S.C., Schifferli D.M., Yue M. Detection of mcr-9 -harbouring ESBL-producing Salmonella Newport isolated from an outbreak in a large-animal teaching hospital in the USA. J. Antimicrob. Chemother. 2020;76:1107–1109. doi: 10.1093/jac/dkaa544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jovčić B., Novović K., Filipić B., Velhner M., Todorović D., Matović K., Rašić Z., Nikolić S., Kiškarolj F., Kojić M. Genomic characteristics of colistin-resistant Salmonella enterica subsp. enterica serovar Infantis from poultry farms in the Republic of Serbia. Antibiotics. 2020;9:886. doi: 10.3390/antibiotics9120886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karraouan B., Ziyate N., Ed-dra A., Amajoud N., Boutaib R., Akil A., El Allaoui A., El Ossmani H., Zerouali K., Elmdaghri N., et al. Salmonella Kentucky: Antimicrobial resistance and molecular analysis of clinical, animal and environment isolates, Morocco. J. Infect. Dev. Ctries. 2017;11:368–370. doi: 10.3855/jidc.8171. [DOI] [PubMed] [Google Scholar]
- 21.Paudyal N., Yue M. Antimicrobial resistance in the “dark matter”. Clin. Infect. Dis. 2019;69:379–380. doi: 10.1093/cid/ciz007. [DOI] [PubMed] [Google Scholar]
- 22.Yue M. Bacterial persistent infection at the interface between host and microbiota. Clin. Infect. Dis. 2016;62:1325–1326. doi: 10.1093/cid/ciw136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paudyal N., Pan H., Wu B., Zhou X., Zhou X., Chai W., Wu Q., Li S., Li F., Gu G., et al. Persistent asymptomatic human infections by Salmonella enterica serovar Newport in China. mSphere. 2020;5:e00163-20. doi: 10.1128/mSphere.00163-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Biswas S., Gu G., Elbediwi M., Li Y., Yue M. Characterization of multidrug resistance patterns of emerging Salmonella enterica serovar Rissen along the food chain in China. Antibiotics. 2020;9:660. doi: 10.3390/antibiotics9100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu Y., Nambiar R.B., Xu X., Weng S., Pan H., Zheng K., Yue M. Global genomic characterization of Salmonella enterica Serovar Telelkebir. Front. Microbiol. 2021;12:2007. doi: 10.3389/fmicb.2021.704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B., Ed-Dra A., Pan H., Dong C., Jia C., Yue M. Genomic investigation of Salmonella isolates recovered from pigs slaughtering process in Hangzhou, China. Front. Microbiol. 2021;12:704636. doi: 10.3389/fmicb.2021.704636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., Chen Y., Pan H., Pang Z., Li F., Peng X., Ed-Dra A., Li Y., Yue M. Genomic characterization of Salmonella Uzaramo for human invasive infection. Microb. Genom. 2020;6:mgen000401. doi: 10.1099/mgen.0.000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ed-Dra A., Karraouan B., El Allaoui A., Khayatti M., El Ossmani H., Rhazi Filali F., ElMdaghri N., Bouchrif B. Antimicrobial resistance and genetic diversity of Salmonella Infantis isolated from foods and human samples in Morocco. J. Glob. Antimicrob. Resist. 2018;14:297–301. doi: 10.1016/j.jgar.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Ribot E.M., Hise K.B. Future challenges for tracking foodborne diseases: PulseNet, a 20-year-old US surveillance system for foodborne diseases, is expanding both globally and technologically. EMBO Rep. 2016;17:1499–1505. doi: 10.15252/embr.201643128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Xu L., Xu X., Zhu Y., Suo Y., Shi C., Shi X. Antimicrobial resistance and molecular characterization of Salmonella enterica serovar Enteritidis from retail chicken products in Shanghai, China. Foodborne Pathog. Dis. 2018;15:346–352. doi: 10.1089/fpd.2017.2387. [DOI] [PubMed] [Google Scholar]
- 31.Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., de Silva N.R., Gargouri N., et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12:119–123. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Biswas S., Paudyal N., Pan H., Li X., Fang W., Yue M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019;10:985. doi: 10.3389/fmicb.2019.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakaria Z., Hassan L., Ahmad N., Husin S.A., Ali R.M., Sharif Z., Sohaimi N.M., Garba B. Discerning the antimicrobial resistance, virulence, and phylogenetic relatedness of Salmonella isolates across the human, poultry, and food materials sources in Malaysia. Front. Microbiol. 2021;12:2513. doi: 10.3389/fmicb.2021.652642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbediwi M., Tang Y., Shi D., Ramadan H., Xu Y., Xu S., Li Y., Yue M. Genomic investigation of antimicrobial-resistant Salmonella enterica isolates from dead chick embryos in China. Front. Microbiol. 2021;12:684400. doi: 10.3389/fmicb.2021.684400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Jiang J., Ed-Dra A., Li X., Peng X., Xia L., Guo Q., Yao G., Yue M. Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res. Int. 2021;142:110198. doi: 10.1016/j.foodres.2021.110198. [DOI] [PubMed] [Google Scholar]
- 36.Paudyal N., Pan H., Elbediwi M., Zhou X., Peng X., Li X., Fang W., Yue M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 2019;19:226. doi: 10.1186/s12866-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y., Lai H., Zou L., Yin S., Wang C., Han X., Xia X., Hu K., He L., Zhou K., et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 2017;259:43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y., Wu C.M., Wu G.J., Zhao H.Y., He T., Cao X.Y., Dai L., Xia L.N., Qin S.S., Shen J.Z. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 2011;8:45–53. doi: 10.1089/fpd.2010.0605. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L., Fu Y., Xiong Z., Ma Y., Wei Y., Qu X., Zhang H., Zhang J., Liao M. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 2018;9:2104. doi: 10.3389/fmicb.2018.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W.W., Bai L., Zhang X.L., Xu X.J., Tang Z., Bi Z.W., Guo Y.C. Prevalence and antimicrobial susceptibility of Salmonella isolated from broiler whole production process in four provinces of China. Chin. J. Prev. Med. 2018;52:352–357. doi: 10.4103/0366-6999.223842. [DOI] [PubMed] [Google Scholar]
- 41.Bai L., Lan R., Zhang X., Cui S., Xu J., Guo Y., Li F., Zhang D. Prevalence of Salmonella isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan, China. PLoS ONE. 2015;10:144532. doi: 10.1371/journal.pone.0144532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang X., Hao H., Dai M., Wang Y., Ahmad I., Liu Z., Zonghui Y. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front. Microbiol. 2015;6:602. doi: 10.3389/fmicb.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdeen E., Elmonir W., Suelam I.I.A., Mousa W.S. Antibiogram and genetic diversity of Salmonella enterica with zoonotic potential isolated from morbid native chickens and pigeons in Egypt. J. Appl. Microbiol. 2018;124:1265–1273. doi: 10.1111/jam.13697. [DOI] [PubMed] [Google Scholar]
- 44.Chuah L.O., Shamila Syuhada A.K., Mohamad Suhaimi I., Farah Hanim T., Rusul G. Genetic relatedness, antimicrobial resistance and biofilm formation of Salmonella isolated from naturally contaminated poultry and their processing environment in northern Malaysia. Food Res. Int. 2018;105:743–751. doi: 10.1016/j.foodres.2017.11.066. [DOI] [PubMed] [Google Scholar]
- 45.Li R., Lai J., Wang Y., Liu S., Li Y., Liu K., Shen J., Wu C. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 2013;163:14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 46.Karaiskos I., Giamarellou H. Carbapenem-sparing strategies for ESBL producers: When and how. Antibiotics. 2020;9:61. doi: 10.3390/antibiotics9020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doi Y., Iovleva A., Bonomo R.A. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 2017;24:S44–S51. doi: 10.1093/jtm/taw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuang D., Zhang J., Xu X., Shi W., Yang X., Su X., Shi X., Meng J. Increase in ceftriaxone resistance and widespread extended-spectrum β-lactamases genes among Salmonella enterica from human and nonhuman sources. Foodborne Pathog. Dis. 2018;15:770–775. doi: 10.1089/fpd.2018.2468. [DOI] [PubMed] [Google Scholar]
- 49.Liu J.H., Wei S.Y., Ma J.Y., Zeng Z.L., Lü D.H., Yang G.X., Chen Z.L. Detection and characterisation of CTX-M and CMY-2 β-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents. 2007;29:576–581. doi: 10.1016/j.ijantimicag.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Murgia M., Bouchrif B., Timinouni M., Al-Qahtani A., Al-Ahdal M.N., Cappuccinelli P., Rubino S., Paglietti B. Antibiotic resistance determinants and genetic analysis of Salmonella enterica isolated from food in Morocco. Int. J. Food Microbiol. 2015;215:31–39. doi: 10.1016/j.ijfoodmicro.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Dos Santos Bersot L., Cavicchioli V.Q., Viana C., Burin R.C.K., Camargo A.C., de Almeida Nogueira Pinto J.P., Nero L.A., Destro M.T. Prevalence, antimicrobial resistance, and diversity of Salmonella along the pig production chain in Southern Brazil. Pathogens. 2019;8:204. doi: 10.3390/pathogens8040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gad A.H., Abo-Shama U.H., Harclerode K.K., Fakhr M.K. Prevalence, serotyping, molecular typing, and antimicrobial resistance of Salmonella isolated from conventional and organic retail ground poultry. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.02653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodríguez-Hernández R., Bernal J.F., Cifuentes J.F., Fandiño L.C., Herrera-Sánchez M.P., Rondón-Barragán I., Garcia N.V. Prevalence and molecular characterization of Salmonella isolated from broiler farms at the Tolima region—Colombia. Animals. 2021;11:970. doi: 10.3390/ani11040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI; Wayne, PA, USA: 2020. CLSI Supplement M100. [Google Scholar]
- 55.Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM). Recommandations Vétérinaires. 2020. [(accessed on 9 August 2021)]. Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2020/09/CASFM_VET2020.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Material.