Abstract
An unusual Helicobacter sp. was isolated from the blood of a human immunodeficiency virus (HIV)-infected patient. This organism had spiral morphology, with single amphitrichous flagella, and was negative for hippurate hydrolysis, production of urease, and reduction of nitrate. 16S rRNA gene sequence analysis verified that the isolate was a species of Helicobacter, most closely related to an undescribed Helicobacter-like isolate from Vancouver, British Columbia, Canada, and to Helicobacter westmeadii, a recently described species from Australia. Both organisms had also been isolated from the blood of HIV-infected patients. These blood isolates, along with Helicobacter cinaedi, form a cluster of closely related Helicobacter spp. that may represent an emerging group of pathogens in immunocompromised patients.
Several species of Helicobacter have been isolated from the blood of immunocompromised patients. Helicobacter cinaedi and Helicobacter fennelliae are known to cause bacteremia in human immunodeficiency virus (HIV)-infected patients. More recently, Helicobacter westmeadii (10) and Helicobacter sp. strain Mainz (3) from blood cultures of patients with AIDS were described, and there was one report of Helicobacter sp. strain Mainz in a knee effusion of an AIDS patient (5). Another Helicobacter-like organism, Flexispira rappini, has been isolated from blood but from an otherwise healthy child with pneumonia (9). Campylobacter spp. have also been reported to be present in the blood of immunocompromised patients, especially AIDS patients (6, 7). It is possible that infections with Helicobacter spp. have occasionally been erroneously attributed to Campylobacter spp. due to their similar microscopic morphologies. We describe a Helicobacter isolate (VAH), grown from the blood of an AIDS patient, that may represent a subgroup within the Helicobacter genus. This isolate is most closely related to H. westmeadii and another unnamed Helicobacter sp. (British Columbia Isolate [BCH]) isolated from the blood of an AIDS patient.
Case report.
The patient was a 37-year-old man with advanced AIDS, schizophrenia, seizures, and end-stage renal disease secondary to HIV nephropathy. In December 1997, four blood cultures and one stool specimen grew a gram-negative curved rod preliminarily identified as Campylobacter sp. (not Campylobacter jejuni and not Campylobacter fetus). The patient initially refused hospitalization and treatment but by the end of December was placed on parenteral gentamicin. The organism was not isolated from subsequent blood or stool cultures. In January 1998, the patient was admitted to the Veterans Affairs Medical Center with fever, epigastric pain, vomiting, and worsening diarrhea. Ciprofloxacin and gentamicin therapy was instituted, but fever and diarrhea persisted. Five days after admission, the patient suffered a seizure and was transferred to the intensive care unit, where ciprofloxacin therapy was changed to ceftriaxone. Transthoracic echocardiography showed multiple echodense lesions on the mitral valve which were consistent with endocarditis. The patient stabilized over the next week but then became hypothermic and hypotensive and was significantly anemic (hematocrit, 17.8%). He had a grand mal seizure followed by cardiopulmonary arrest and could not be resuscitated. No autopsy was obtained. Prior to the patient’s death, a blood culture isolate (VAH) had been sent to the Microbiology Service at the Clinical Center of the National Institutes of Health (NIH) for further identification.
Culture and morphologic characteristics.
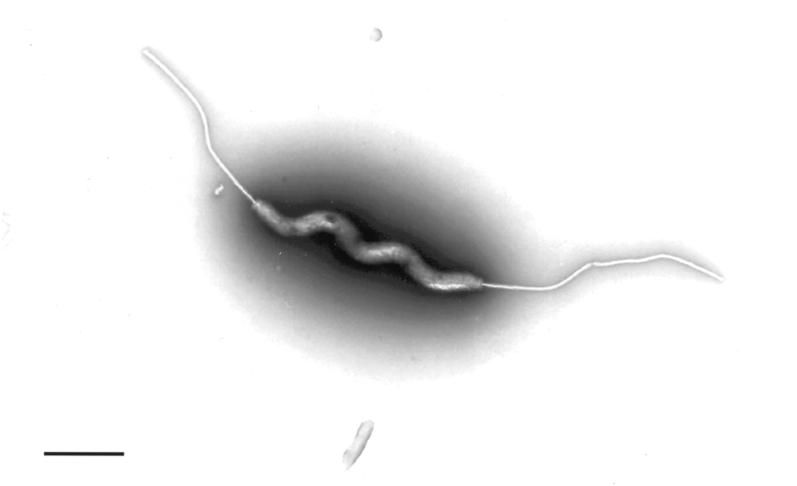
The organism was first grown from the patient’s blood in aerobic Bactec bottles and subsequently grew from a stool culture and additional blood cultures. The stool and blood isolates were the same by colony, Gram stain, and growth characteristics. VAH was a faintly staining gram-negative, curved to spiral rod that was best seen by acridine orange staining. Active motility was observed by dark-field examination of a wet preparation. Flagella were demonstrated by the Ryu flagellum stain (Remel Laboratories, Lenexa, Kans.) and by transmission electron microscopy performed by the Laboratory of Cell and Molecular Structure (Science Applications International Corp., Frederick, Md.). Ryu stain showed that most of the cells were amphitrichous with a single flagellum at each end. Electron microscopy (Fig. 1) showed flagella but no periplasmic fibers. The organism was 3.8 to 4.0 μm long and 0.17 μm wide.
FIG. 1.
Negatively stained transmission electron micrograph of VA isolate. Spiral form and single bipolar flagella are evident. Bar = 0.90 mm.
Because of its growth on Campylobacter selective medium (cefoperazone-vancomycin agar), the organism was initially thought to be a Campylobacter sp., although not C. jejuni since it was negative for hydrolysis of hippurate. It grew on cefoperazone-vancomycin agar and chocolate, horse and sheep blood, and brucella agars but required 5 to 7 days of incubation under microaerobic conditions at 37°C. It grew best with a Campy-Gen generator (Oxoid, Basingstoke, England) and less well with Campy-Pak Plus (Becton Dickinson, Cockeysville, Md.). There was poor growth with bags filled with Campy Gas (Columbia Diagnostics, Springfield, Va.) and no growth under standard aerobic conditions, aerobically with 6% CO2, and anaerobically.
Biochemical characteristics.
Although the organism was urease negative, it was biochemically more consistent with Helicobacter than with Campylobacter based on its failure to hydrolyze hippurate, to reduce nitrate, or to grow at 42°C. A comparison of VAH’s biochemical characteristics with those of other Helicobacter spp. that have been isolated from humans is presented in Table 1. Except for lack of sensitivity to cephalothin, the biochemical pattern of VAH was closest to that of H. fennelliae, a species known to cause bacteremia and diarrhea in AIDS patients.
TABLE 1.
Biochemical and morphological characteristics of Helicobacter spp. from human specimensa
| Characteristic | H. pylori | F. rappini | H. cinaedi | H. westmeadii | H. fennelliae | VA isolate |
|---|---|---|---|---|---|---|
| Catalase | + | vc | + | + | + | + |
| Oxidase | + | + | + | + | + | + |
| Urease | + | + | − | − | − | − |
| Nitrate reduction | + | − | + | + | − | − |
| Alkaline phosphatase | + | vd | ve | + | + | + |
| Indoxyl acetate hydrolysis | − | NAb | − | − | + | + |
| Growth at temp (°C): | ||||||
| 25 | − | − | − | − | − | − |
| 37 | + | + | + | + | + | + |
| 42 | − | + | − | − | − | − |
| Susceptibilityf | ||||||
| Nalidixic acid | R | R | S | S | S | S |
| Cephalothin | S | R | I | R | S | R |
| Periplasmic fibers | − | + | − | − | − | − |
| No. of flagella | 4 to 8 | 10 to 20 | 1 to 2 | 1 | 2 | 2 |
| Distribution | Bipolar | Bipolar | Bipolar | Polar | Bipolar | Bipolar |
| Shape | Curved | Straight/curved | Curved | Straight/spiral | Curved | Spiral |
Gas chromatography of cellular fatty acids.
Automated fatty acid methyl ester analysis by gas-liquid chromatography was performed by Microcheck, Inc. (Northfield, Vt.). The similarity index determined by comparison of VAH’s fatty acid profile with those of reference strains in Microcheck’s database showed an “excellent match” with H. cinaedi. The Euclidean distance relatedness value between VAH and H. cinaedi was 8.8, suggestive of a possible species difference. H. westmeadii was not available for simultaneous testing; however, VAH showed a significant peak (18:1 w7c) not described for H. westmeadii (10).
Antibiotic susceptibility testing.
In vitro antibiotic susceptibilities were determined by the E-test method (AB Biodisk, Piscataway, N.J.) on chocolate agar incubated in a microaerobic atmosphere (Campy-Gen). A heavy inoculum was used to ensure sufficient growth for the plates to be read after 4 days of incubation. Control organisms were tested with the same media and conditions to validate that appropriate control MICs were obtained. Since the testing was by a nonstandardized procedure, these studies were for investigational use only. The organism was resistant to ciprofloxacin (>32 μg/ml), erythromycin (>256 μg/ml), trimethoprim-sulfamethoxazole (>2/38 μg/ml), and azithromycin (>256 μg/ml). The MICs determined for other agents were as follows: clindamycin, 1.5 μg/ml; tetracycline, 0.064 μg/ml; clarithromycin, 1.0 μg/ml; ceftriaxone, 1.5 μg/ml; and gentamicin, 0.25 μg/ml. Treatment of the patient with the last two antibiotics likely contributed to his subsequent negative blood cultures.
16S rRNA gene sequencing.
We were unable to identify the species of VAH based on ultrastructural and biochemical characteristics, and so 16S rRNA sequence analysis was performed. PCR of a 1,372-bp region of the 16S rRNA gene was performed in three segments with primers tailed with M13 sequencing primers, as shown in Table 2. M13 tailing permitted high-quality direct sequencing of PCR products. Primers were selected based on an alignment of 21 Helicobacter spp. sequences from GenBank (National Center for Biotechnology Information, Bethesda, Md.). PCR master mix contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM (each) deoxynucleotides (dATP, dCTP, dGTP, and dTTP), 4 pmol of each primer, 3 mM MgCl2, 2.5 U of Taq DNA polymerase (all reagents from Perkin-Elmer, Branchburg, N.J.), and 500 ng of DNA template in a final volume of 50 ml. Thermocycler conditions were as follows: 95°C for 4 min; 10 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min; 25 cycles of 95°C for 15 s and 72°C for 1 min; and 7 min at 72°C. Unincorporated primers and deoxynucleotides were removed by centrifugal filtration with Millipore Ultrafree-MC 10,000 NMWL filter units (Millipore, Bedford, Mass.). Cycle sequencing reactions were performed with M13 forward and reverse primers (Invitrogen, Carlsbad, Calif.) and the ABI Prism BIGDYE terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, Calif.). Electrophoresis was performed on the ABI PRISM 377 DNA sequencer (PE Applied Biosystems).
TABLE 2.
Primer sequences for PCR amplification and sequencing of the 16S rRNA gene of Helicobacter spp.
| Set | Primer | Sequence | Sequencing primer | Size (bp) | Region on H. pylori U00679 |
|---|---|---|---|---|---|
| 1 | MR-1 | 5′-CAG-GAA-ACA-GCT-ATG-ACA-GAG-TTT-GAT-CCT-GGC-T-3′ | M13 reverse | 455 | 156 to 631 |
| MF-1 | 5′-GTT-TTC-CCA-GTC-ACG-ACG-CTG-GCA-CGG-AGT-TAG-C-3′ | M13 (−40) forward | |||
| 2 | MR-2 | 5′-CAG-GAA-ACA-GCT-ATG-ACC-GCG-TGG-AGG-ATG-AAG-C-3′ | M13 reverse | 573 | 535 to 1107 |
| MF-2 | 5′-GTT-TTC-CCA-GTC-ACG-ACT-GTC-AAG-CCT-AGG-TAA-GG-3′ | M13 (−40) forward | |||
| 3 | MR-3 | 5′-CAG-GAA-ACA-GCT-ATG-ACA-GGA-ATA-GAC-GGG-GAC-C-3′ | M13 reverse | 502 | 1026 to 1530 |
| MF-3 | 5′-GTA-AAA-CGA-CGG-CCA-GAT-GGT-GTG-ACG-GGC-GG-3′ | M13 (−20) forward |
Sequences were analyzed with GeneWorks 2.4 nucleic acid and protein sequence analysis software (IntelliGenetics, Inc., Mountain View, Calif.). VAH was most closely related (99.3% identity) to BCH, an unnamed Helicobacter sp. (GenBank accession no. AF023862, BC), obtained from a blood culture of an AIDS patient in Vancouver, Canada (6a). The most closely related named species was H. westmeadii (99.2% identity), a species isolated from the blood of AIDS patients in Australia (10). The 16S rRNA gene sequence of VA showed lower identity, 98.7 and 96%, with H. cinaedi and H. fennelliae, respectively. A phylogenetic tree was prepared (GeneWorks) with the corresponding 1,372-bp region of 16S rRNA sequences from Helicobacter spp. available from GenBank (Fig. 2).
FIG. 2.
Phylogenetic tree delineating the relationship between the 16S rRNA gene sequence for the VA isolate and sequences available in GenBank. Scale represents percent homology. Asterisks indicate species isolated from human blood.
Discussion.
The nomenclature of Helicobacter has become more complex as the number of recognized Helicobacter spp. increases. These species are closely related by 16S rRNA gene sequence analysis, making specific identification by sequence alone difficult. DNA-DNA hybridization, which can be used to differentiate species with high 16S rRNA homology (4), is needed to definitively identify VAH. H. westmeadii was not available from the American Type Culture Collection, and we were unsuccessful in multiple attempts to have it sent from Australia to the NIH. Therefore, our organism description is limited to our findings of the 16S rRNA sequence relationships in addition to phenotypic characteristics, including morphologic, biochemical, and cellular fatty acid properties.
Biochemically, VAH was most similar to H. fennelliae, while by 16S rRNA sequence analysis, it was more closely related to BCH and H. westmeadii (Fig. 2). H. westmeadii is curved to spiral with single polar flagella like VAH, although flagellar stains of VAH showed most cells to be amphitrichous. A major biochemical difference between VAH and H. westmeadii is the inability of VAH to hydrolyze hippurate, reportedly a key feature of H. westmeadii. Additional differences include those shown in Table 1, as well as the finding of significant amounts of a cellular fatty acid peak not reported for H. westmeadii. The phenotypic and biochemical characteristics of the BC strain were not available for comparison. Although GC analysis suggested that the VA isolate is possibly a strain of H. cinaedi, the lack of nitrate reduction and more importantly a lower rRNA sequence homology than that found with H. westmeadii (99.2 versus 98.7%) call this identification into question.
Analysis of 16S rRNA sequence relationships, as shown in the phylogenetic tree (Fig. 2), shows at least two large groups of Helicobacter spp. The first group contains Helicobacter pylori and several Helicobacter spp. that are associated with gastritis in animals, and except for Helicobacter canis, they exhibit strong urease activity. The second large group consists of the enteric and blood-associated Helicobacter spp. including those isolated from blood of human patients, such as H. fennelliae, H. cinaedi (1), F. rappini (8, 9), Helicobacter sp. strain Mainz (3), and H. westmeadii (10). There is a small, more tightly related cluster (>98% identity) which includes H. cinaedi, H. westmeadii, F. rappini, BCH, and VAH. This particular cluster represents a subgroup of Helicobacter causing sepsis primarily in immunocompromised patients, such as those with AIDS.
H. westmeadii, BC, and VAH were obtained from geographically distant locations (Australia, western Canada, and eastern United States, respectively). It is possible that this group of blood-borne Helicobacter spp. is more common and widespread than currently reported since they are difficult to detect in blood cultures and can be misidentified as Campylobacter sp. due to their curved morphology and positive oxidase and catalase reactions. The source(s) and incidence, as well as the spectrum of infections with these organisms, are largely unknown and need further elucidation.
Acknowledgments
We thank Caroline Fukuda and the microbiology staff of the Clinical Pathology Department of the Clinical Center at the NIH for maintaining the cultures and performing the biochemical tests. We also thank Kunio Nagashima of Laboratory of Cell and Molecular Structure (Science Applications International Corp. Frederick, Frederick, Md.) for the electron microscopy and Eric Konieczynski of the National Cancer Institute for running the sequencing gels.
REFERENCES
- 1.Burman W J, Cohn D L, Reves R R, Wilson M L. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin Infect Dis. 1995;20:564–570. doi: 10.1093/clinids/20.3.564. [DOI] [PubMed] [Google Scholar]
- 2.Eaton K A, Dewhirst F E, Radin M J, Fox J G, Paster B J, Krakowka S, Morgan D R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 3.Fleisch F, Burnens A, Weber R, Zbinden R. Helicobacter species strain Mainz isolated from cultures of blood and from two patients with AIDS. Clin Infect Dis. 1998;26:526–527. doi: 10.1086/517110. [DOI] [PubMed] [Google Scholar]
- 4.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 5.Husmann M, Gries C, Jehnichen P, Woelfel T, Gerken G, Ludwig W, Bhakdi S. Helicobacter sp. strain Mainz isolated from an AIDS patient with septic arthritis: case report and nonradioactive analysis of 16S rRNA sequence. J Clin Microbiol. 1994;32:3037–3039. doi: 10.1128/jcm.32.12.3037-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molina J M, Casin I, Hausfater P, Giretti E, Welker Y, Decazes J M, Garrait V, LaGrange P, Modai A. Campylobacter infections in HIV-infected patients—clinical and bacteriological features. AIDS. 1995;9:881–885. doi: 10.1097/00002030-199508000-00008. [DOI] [PubMed] [Google Scholar]
- 6a.Morshed, M. G., et al. 1997. Unpublished data.
- 7.Pigraou C, Bartolome R, Almirante B, Planes A M, Gavalda J, Pahissa A. Bacteremia due to Campylobacter species: clinical findings and antimicrobial susceptibility patterns. Clin Infect Dis. 1997;25:1414–1420. doi: 10.1086/516127. [DOI] [PubMed] [Google Scholar]
- 8.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tee W, Leder K, Karroum E, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivett-Moore N L, Rawlinson W D, Yuen M, Gilbert G L. Helicobacter westmeadii sp. nov., a new species isolated from blood cultures of two AIDS patients. J Clin Microbiol. 1997;35:1144–1150. doi: 10.1128/jcm.35.5.1144-1150.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]